Abstract
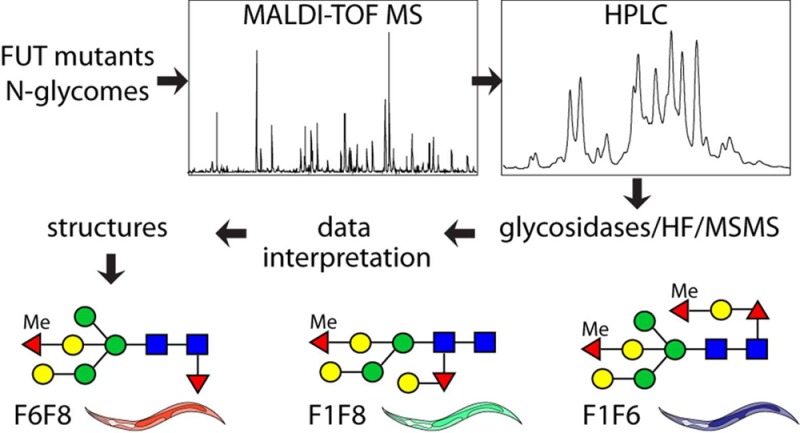
Recent studies have shown a remarkable degree of plasticity in the N-glycome of the model nematode Caenorhabditis elegans; ablation of glycosylation-relevant genes can result in radically altered N-glycan profiles despite only minor biological phenotypic effects. Up to four fucose residues and five different linkages of fucose are known on the N-glycans of C. elegans. Due to the complexity in the wild type, we established three mutant strains defective in two core fucosyltransferases each (fut-1;fut-6, fut-1;fut-8, and fut-6;fut-8). Enzymatically released N-glycans were subject to HPLC and MALDI-TOF MS/MS, in combination with various treatments, to verify structural details. The N-glycome of the fut-1;fut-6 mutant was the most complex of the three double-mutant strains due to the extension of the core α1,6-fucose as well as the presence of fucose on the bisecting galactose. In contrast, maximally two fucoses were found on N-glycans of the fut-1;fut-8 and fut-6;fut-8 strains. The different locations and capping of fucose meant that up to 13 isomeric structures, many highly galactosylated, were determined for some single masses. These data not only show the high variability of the N-glycomic capacity of a “simple” nematode but also exemplify the need for multiple approaches to reveal individual glycan structures within complex invertebrate glycomes.
Keywords: bisect, fucosyltransferase mutant, glycomics, nematode
Introduction
It is probably still a widely held opinion that “lower” organisms only have “simple” glycomes and that glycomic profiling is sufficient for analyzing their protein-linked oligosaccharides; however, data obtained over the years indicate a wide range of variability and complexity of the N-linked glycans from protozoan and invertebrate species.1 This applies not only to many parasitic species or intermediate hosts for pathogens but also to model organisms such as Dictyostelium, Drosophila, and Caenorhabditis.2,3 For instance, the most complicated N-glycans in Dictyostelium contain core α1,3-fucose, inter- and bisecting GlcNAc, sulfate, and methylphosphate residues;4 in addition to the pauci- and oligomannosidic N-glycans typical of invertebrates, Drosophila expresses a range of sialylated, glucuronylated, and sulfated structures,5,6 while in Caenorhabditis structures with up to four fucose residues (in part capped with hexose residues) as well as methyl and phosphorylcholine modifications have been described.7−11 Although extended nonreducing modifications, typical of vertebrates, are rare in insects and nematodes, up to three antennae can be present on N-glycans in such species.
The ability to manipulate glycosylation pathways in these model species by isolating or generating mutants enables the investigation not only of glycan function but also of the plasticity of the biosynthetic potential. The latter effect is probably most obvious with Caenorhabditis, for which the largest range of N-glycans has been detected for any invertebrate model,11 also when considering the structures present in glycosylation mutants as exemplified by a triple knockout of all N-acetylglucosaminyltransferase I isoforms,12 knockouts of a putative S-adenosylmethionine transporter,13 of GDP-fucose metabolism,14 and of a conserved oligomeric Golgi (COG) complex subunit15 as well as single and multiple deletion strains of Golgi glycosidases16−18 and fucosyltransferases.19,20 Many of these mutations had a major impact on the N-glycome of C. elegans but are normally associated with no obvious laboratory phenotype other than often altered sensitivity to toxic lectins or to pathogenic bacteria.21,22
The complex web of glycosyltransferase specificities and “GO-NOGO” biosynthetic signals generated in the absence of certain glycan-modifying enzymes in the Golgi result in a large and, in part, unpredictable range of final N-glycan structures. Despite some progress, many of the biosynthetic routes generating this glycan variability are still unclear, as is the exact nature of some of the linkages. In our own laboratory, we have focused on deciphering the fucosylation pattern of C. elegans N-glycans. In total, five different positions for fucosylation have been found on C. elegans N-glycans (with maximally four per structure): core α1,3- and core α1,6-fucose on the reducing (proximal) N-acetylglucosamine, α1,3-fucose on the second (distal) core GlcNAc, α1,2-fucose capping of the core Galβ1,4Fucα1,6 epitope, and α1,2-fucose on the bisecting β1,4-linked galactose substituting the core β-mannose residue.10,18,20 Whereas maximally three, rather than four, fucose residues were observed for N-glycans from the Golgi mannosidase II (aman-2) and fucosyltransferase (fut-1 and fut-6) mutants,17,19 more radical reductions in the degree of N-glycan fucosylation were seen for the Golgi double hexosaminidase (hex-2;hex-3) and triple fucosyltransferase (fut-1;fut-6;fut-8) deletion strains.18,20 The reduced N-glycome complexity of the latter two strains enabled us to define unusual modifications such as Galα1,2Fuc on the distal core GlcNAc and “bisecting” Fucα1,2Gal on the core β-mannose. Nevertheless, because of the published data there are at least two α1,2-fucosyltransferases, two β1,4-galactosyltransferases, and two α-galactosyltransferases as well as multiple methyltransferases to be discovered that are responsible for the formation of the highly fucosylated core structures.
Indeed the definition of individual N-glycan structures is complicated in C. elegans by the multiplicity of locations for fucosylation as well as capping of residues by galactose, methyl, or phosphorylcholine moieties. This results in a large number of isomeric structures that must be adequately separated and distinguished as part of any glycomic workflow; thereby, sequential differential release with PNGase F, followed by PNGase A (only the latter cleaving core α1,3-fucosylated N-glycans23) as well as RP-HPLC and 2D-HPLC in combination with mass spectrometry and chemical or enzymatic treatments is required.
While the N-glycans of the triple fucosyltransferase mutant lacked all core fucose residues and possessed maximally one fucose residue on the bisecting galactose,20 initial mass spectrometric screening of the PNGase F-released N-glycans of three double fucosyltransferase mutants (fut-1;fut-6, fut-1;fut-8, and fut-6;fut-8) indicated that significant amounts of difucosylated oligosaccharides were still apparent in the first two.24 In the present study, based on the lessons learnt from the analysis of the triple fucosyltransferase mutant20 as well as clues from other studies,8,10 we have performed an in-depth N-glycomic analysis of the three double fucosyltransferase mutants that vary in terms of which fucosylation events can still take place while still being less complex than the wild type. Our analyses reveal an unprecedented isomeric complexity in the glycomic potential of the nematode and so will be valuable in a final definition of the structures of the N-linked oligosaccharides of the wild-type as well as revealing aspects of the biosynthetic pathways in this model organism.
Experimental Procedures
Preparation of C. elegans Double Mutants
Wild-type C. elegans (N2) and single mutants fut-1 (ok892, II), fut-6 (ok475, II) and fut-8 (ok2558, V) were obtained from the Caenorhabditis Genetics Centre (CGC), University of Minnesota, USA. All C. elegans strains were cultured under standard conditions at 20 °C.25 Analogously to the procedure for the fut-1;fut-6 double mutant,26 also fut-1;fut-8 and fut-6;fut-8 strains were prepared. PCR primers were designed to target the mutant alleles. Multiplex PCR screening was employed to simplify this process when handling a large number of DNA samples for screening. Primers used are 5′-CTAAATTGGCATCCACAACCT-3′, 5′-GCCATTTATTAACAGTTCTCAT-3′ and 5′-CCGGAGTAATTAGACCTGC-3′ for probing fut-1; 5′-GAATGCCACCATGCAACAT-3′, 5′-GAATTACCCATGATACTAGAT-3′ and 5′-GCCCCAAATATCAATCTGC-3′ for probing fut-6; and 5′-TCAGTCTTCGCCAATCAT-3′, 5′-TAAAAGGAGTGTCCATTG-3′ and 5′-AATTACCGCATTTGCTAC-3′ for probing fut-8. Expected and observed amplicon sizes are shown in Supplementary Figure S-1. Genomic DNAs from wild-type worm and single mutants were combined to prepare artificial heterozygous genomes as PCR reference sample; for example, a mixture of N2 and fut-1 genomic DNA was used to aid the examination of the fut-1;fut-6 genotype. The 100 bp DNA GeneRuler ladder (Thermo Fisher Scientific) was used to estimate the sizes of the DNA amplicons.
N-Glycan Preparation and MALDI TOF MS Analysis
C. elegans was grown in liquid culture with E. coli OP50 in standard S complete medium, and mixed stages were harvested after cultivation at room temperature (20 °C) for 4–6 days and purified by sucrose density centrifugation. After proteolysis with pepsin, N-glycans were released from worm peptides using peptide:N-glycosidase F (Roche) as previously described,27 with a subsequent digestion of remaining glycopeptides using peptide:N-glycosidase A (Roche). The N-glycomes of the mutants were profiled by MALDI-TOF MS (Autoflex Speed, Bruker Daltonics, Germany) in positive ion mode using FlexControl 3.4 software. Free glycans were labeled with 2-aminopyridine prior to fractionation by reversed-phase HPLC (RP-HPLC), also in combination with either normal phase or hydrophilic interaction columns (see later). All HPLC peaks were collected and examined by MALDI-TOF MS using 6-aza-2-thiothymine (ATT) as matrix; MS/MS to confirm the composition of all proposed structures was performed by laser-induced dissociation (precursor ion selector was generally set to ±0.6%). The detector voltage was generally set at 1977 V for MS and 2133 V for MS/MS; 1000–3000 shots from different regions of the sample spots were summed. Spectra were processed with the manufacturer’s software (Bruker Flexanalysis 3.3.80) using the SNAP algorithm with a signal/noise threshold of 6 for MS (unsmoothed) and 3 for MS/MS (four-times smoothed). In total ∼3500 MS and MS/MS spectra were manually interpreted on the basis of the mass, fragmentation pattern, and results of chemical and enzymatic treatments; isomeric structures present in different RP-HPLC fractions were defined on the basis of comparisons of the aforementioned parameters. At least five MS/MS fragment ions were used to aid definition of each of the structures.
HPLC Purification of N-Glycans
Separation of PA-labeled glycans was carried out on a Shimadzu HPLC system equipped with a fluorescence detector (RF 10 AXL; 320/400 nm). In the case of RP-HPLC, a Hypersil ODS column (C18; Agilent) was used with 100 mM ammonium acetate, pH 4.0 (buffer A) and 30% (v/v) methanol (buffer B); a gradient of increasing buffer B (1% per minute) was programmed. The column was calibrated daily in terms of glucose units (g.u.) with a pyridylaminated partial dextran hydrolysate. For 2D-HPLC, either normal-phase HPLC (Tosoh TSKgel Amide-80) with an inverse gradient of acetonitrile in 10 mM ammonium formate, pH 7, or combined hydrophobic-interaction anionic-exchange HPLC (HIAX, Dionex IonPac AS11) with an inverse gradient of acetonitrile in 800 mM ammonium acetate, pH 3, was applied as previously described.4,27
Structural Elucidation Using Exoglycosidases and Chemical Treatment
In general, a 1 μL aliquot of a HPLC fraction was mixed with 0.2 μL of exoglycosidase and 0.8 μL of 100 mM ammonium acetate solution, pH 5.0 (except pH 6.5 in the case of the microbial α1,2-fucosidase); after an overnight incubation at 37 °C, 0.5 μL aliquot of the mixture was analyzed by MALDI-TOF MS. Exoglycosidases employed were: α-galactosidase from green coffee beans (Sigma, 11 mU), recombinant β-galactosidase from Aspergillus niger [144 μU28], recombinant FDL β1,2-N-acetyl-glucosaminidase [0.2 μU; specific for the nonreducing terminal GlcNAc on the α1,3-arm29], jack bean α-mannosidase (Sigma-Aldrich, 6.25 mU), and recombinant Xanthomonas manihotis α1,2/3- and α1,6-specific mannosidases [New England Biolabs, 6–8 U30]. Also, digestions were attempted with α-l-fucosidases from bovine kidney (Sigma-Aldrich, 10 mU), Xanthomonas (α1,2-specific; New England Biolabs, 4 mU), Corynebacterium (α1,2-specific; Takara, 4 μU), or microbial (α1,2-specific E-FUCM; kind gift of Megazyme). For the removal of α1,2/3-linked fucose or methylfucose, glycan samples were dried in a Speed-Vac and then incubated with 3 μL of 48% (w/v) hydrofluoric acid (HF) on ice for 24 h. The HF was allowed to evaporate overnight. Chemically or enzymatically treated glycans were reanalyzed by MALDI-TOF MS and MS/MS without further purification.
Analysis by LC–MS
A PA-labeled N-glycan fraction (5.5 g.u.) was analyzed by LC–MS/MS using a 10 cm × 150 μm I.D. column, prepared in-house, containing 5 μm porous graphitized carbon (PGC) particles (Thermo Scientific, Waltham, MA). Glycans were eluted using a linear gradient from 0 to 40% acetonitrile in 10 mM ammonium bicarbonate over 40 min at a flow rate of 10 μL/min. The eluted N-glycans were detected using an LTQ ion trap mass spectrometer (Thermo Scientific) in negative-ion mode with an electrospray voltage of 3.5 kV, capillary voltage of −33.0 V, and capillary temperature of 300 °C. Air was used as a sheath gas and mass ranges were defined dependent on the specific structure to be analyzed. Specific ions were selected for MSn fragmentation using CID with the collision energy set to 30%. The data were processed using the Xcalibur software (version 2.0.7, Thermo Scientific) and the glycan was identified from the MS/MS spectra by manual annotation.
Results
Preparation and Initial Glycomic Analysis of Double Fucosyltransferase Mutants
Due to the seeming infinite variety of fucosylated N-glycans in Caenorhabditis elegans and the availability of mutants with deletions in genes encoding the three core fucosyltransferases (fut-1, fut-6, and fut-8; i.e., proximal and distal core α1,3- and proximal core α1,6-fucosyltransferases) for which activities toward N-glycans have been defined, double mutants (fut-1;fut-6, fut-1;fut-8, and fut-6;fut-8) were prepared and verified by PCR (Supplementary Figure S-1) to investigate the putatively simplified N-glycomes. Indeed, initial mass spectrometric screening suggested that there were maximally two fucose residues on the N-glycans of these three strains.24 Nevertheless, we anticipated that “off-line” LC–MALDI-TOF MS would reveal a higher degree of variability in fucosylation as compared with the fut-1;fut-6;fut-8 triple knockout that features maximally one fucose residue on its N-glycans.
The deeper investigation of the structural variability of the N-glycans was based on MALDI-TOF MS of HPLC-separated fractions. Initially the glycans were released by PNGase F, followed by PNGase A; as expected, significant amounts of PNGase A-released material were found only in the case of the fut-6;fut-8 mutant, which retains the ability to core α1,3-fucosylate, a modification abolished in the fut-1;fut-6 and fut-1;fut-8 strains. (Indeed the RP-HPLC chromatograms and MS spectra of the PNGase A-released glycans of the latter two strains are identical but of low intensity as compared with those after PNGase F; data not shown.) Thus, only four N-glycan pools were analyzed in depth: the PNGase F-released glycans of all three mutants and the PNGase A digest (after PNGase F) of the fut-6;fut-8 mutant. RP-HPLC of the pyridylaminated N-glycans was performed in all cases and showed quite different elution profiles. All HPLC fractions were subject to MS/MS backed up with enzymatic treatments or incubation with hydrofluoric acid, a reagent known to remove various fucosidic and phosphodiester modifications; as necessary, due to the complexity, “2D-HPLC” with a second dimension on a HIAX column was performed in the case of fut-1;fut-8 or NP-HPLC, followed by RP-HPLC for the fut-1;fut-6 strain.
Analysis of the fut-6;fut-8 Mutant
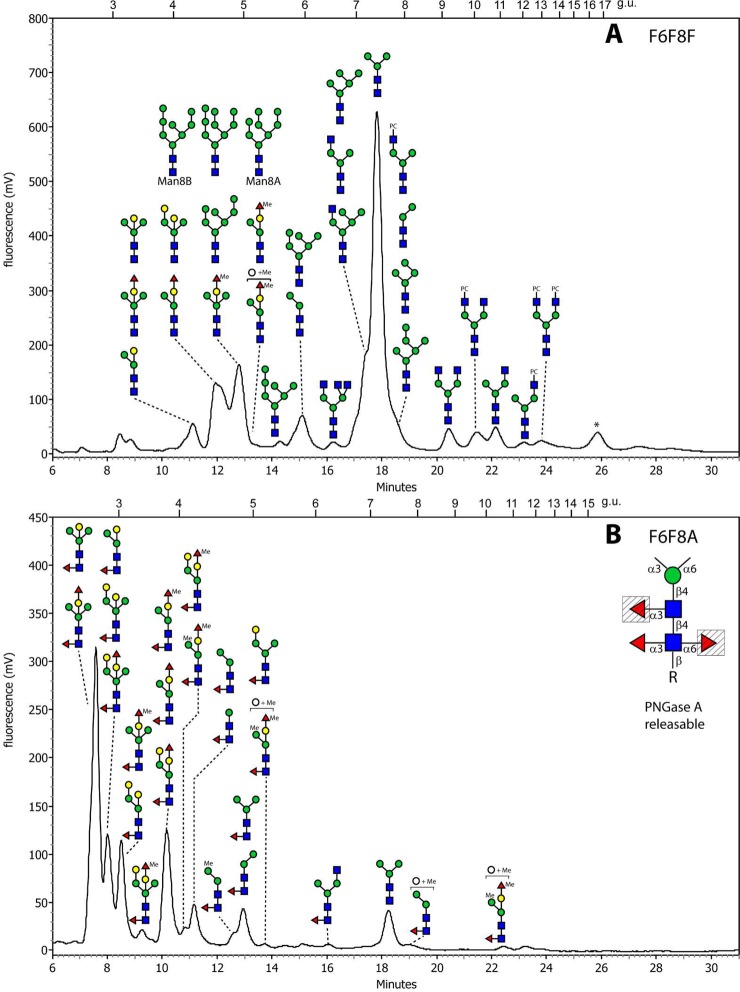
The profile of the PNGase F-released glycans for the fut-6;fut-8 mutant was very similar to that of the recently published triple mutant with a major peak of Man3GlcNAc2–PA (7.2 g.u.) and a range of other glycans, including ones containing maximally one fucose. Due to the early RP-HPLC elution position (4.2 to 5.2 g.u.; Figure 1A) and the complete absence of core-derived m/z 446 GlcNAc1Fuc1-PA fragment ions (Figure 2B), the fucose on these glycans was concluded to be solely on the bisecting galactose. As these glycans are akin to those of the triple mutant,20 they are not described here in greater detail; relevant MS/MS and enzyme digestion data are summarized in Supplementary Table S-1.
Figure 1.
RP-HPLC of pyridylamino-labeled N-glycans of the fut-6;fut-8 double deletion strain. N-glycans released by PNGase F ((A) F6F8F) and PNGase A ((B) F6F8A; PNGase A digest following PNGase F) were pyridylaminated and individual RP-HPLC fractions subject to MALDI-TOF MS; characterized N-glycan structures are annotated on the chromatograms using the nomenclature of the Consortium for Functional Glycomics (circles, hexose; squares, N-acetylhexosamine; triangles, fucose; Me, methyl; PC, phosphorylcholine); the most abundant in a single fraction is shown uppermost. The asterisk indicates a fraction with non-N-glycan contaminants and the inset in panel B shows which core fucose residues are absent from this strain due to the deletion of the relevant fucosyltransferases (shaded triangles). Oligomannosidic and phosphorylcholine-modified glycans are also annotated but are not further discussed in this study.
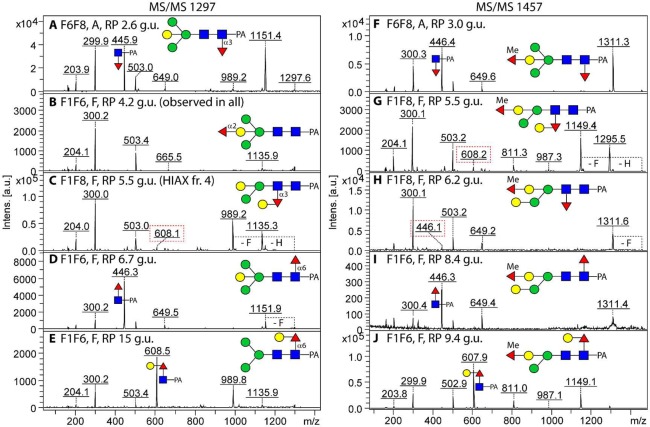
Figure 2.
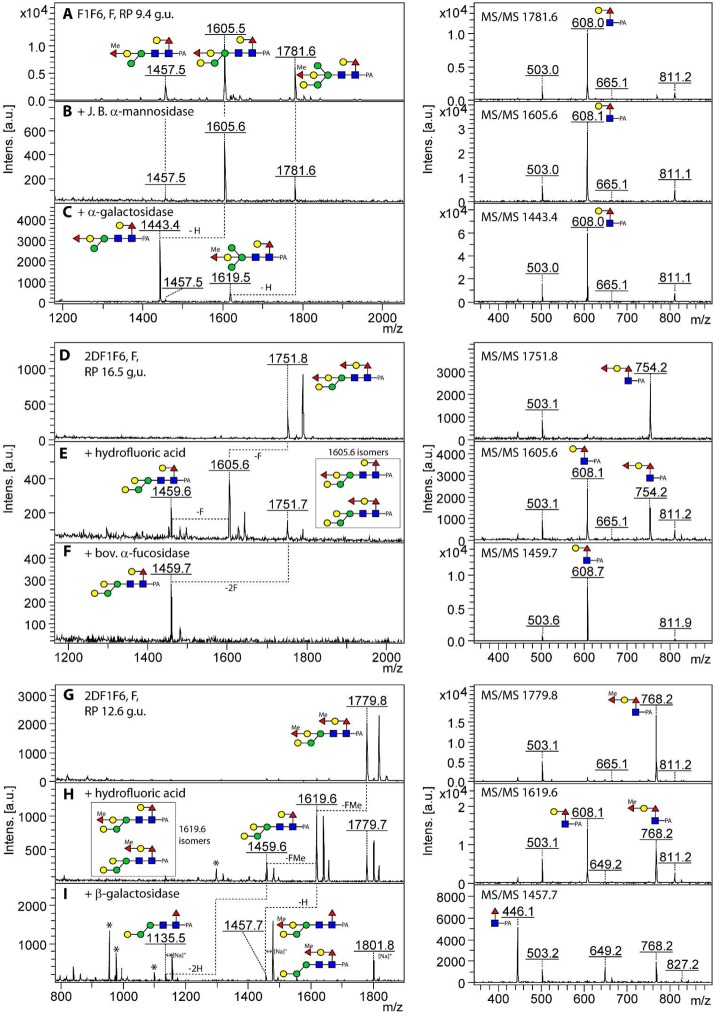
MALDI-TOF MS/MS analysis of isomeric forms of Hex4HexNAc2Fuc1 and Hex4HexNAc2Fuc2Me1 from the fut-1;fut-6, fut-1;fut-8, and fut-6;fut-8 deletion strains. Five selected isomeric forms each of m/z 1297 and m/z 1457 glycans [M + H]+ from PNGase F or A digests of the three double fucosyltransferase mutants (F1F6, F1F8, and F6F8) occurring in RP-HPLC fractions of different glucose units (g.u.) were subject to MALDI-TOF MS/MS in the positive ion mode. Key fragments and the full structures are shown according to the nomenclature of the Consortium for Functional Glycomics (Me, methyl; PA, pyridylamino); the fragments surrounded by dashed squares in panels C, G, and H are putative rearrangement products correlating with the presence of distal (but not antennal) fucose modifications. The 4.2 g.u. form of Hex4HexNAc2Fuc1 (panel B), previously analyzed from the fut-1;fut-6;fut-8 triple mutant, is found in all three double mutants. Loss of fucose or hexose from the parent ions is indicated by −F or −H.
In contrast, the PNGase A pool of this mutant was dominated by glycans of even earlier elution (2–4 g.u.; Figure 1B) for which typical m/z 446 core fucose fragment ions were observed; however, the ratio of the intensity of the m/z 300 and 446 (Fuc0–1GlcNAc-PA) fragment ions of approximately 1:1 correlates with core α1,3-fucosylation, as opposed to the intensity ratio of about 1:9 observed in case of the presence of core α1,6-fucose (Figure 2A,F and Supplementary Figure S-2B as compared with Figure 2D,I; see also refs (31) and (32)). In addition to the reduced RP-HPLC retention, their sensitivity toward hydrofluoric acid and their occurrence solely in the PNGase A pool led to the conclusion that these glycans were core α1,3-fucosylated.
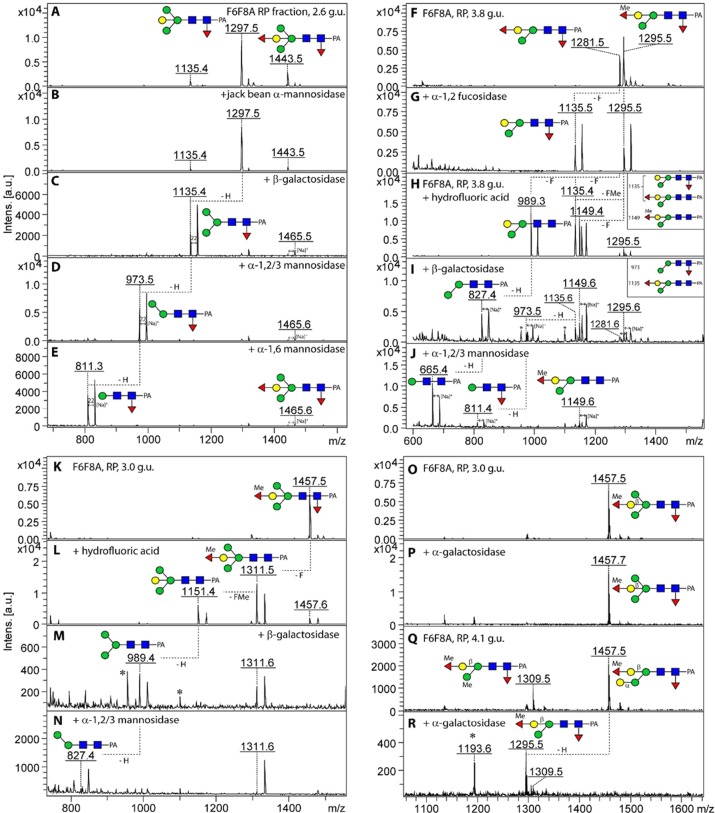
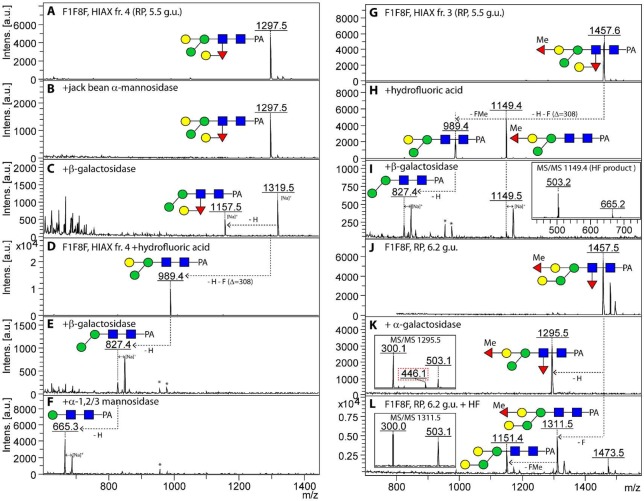
It is noteworthy that, other than the nonfucosylated pauci- and oligomannosidic structures, even some of the simplest glycans (e.g., if considering all mutants, ten forms of Hex3HexNAc2Fuc1-PA; m/z 1135) showed a multiplicity of isomeric structures (as judged by their retention time in terms of glucose units), which could be distinguished by enzymatic and chemical treatment and MS/MS fragmentation pattern. For example, in addition to the PNGase F-releasable m/z 1135 isomer present also in the triple mutant (4.8 g.u.), two PNGase A-releasable forms were found in the fut-6;fut-8 strain, which could not be distinguished on the basis of MS/MS. The earliest eluting form (2.8 g.u.) could be digested down to Man1GlcNAc2Fuc1-PA by sequential treatment with β-galactosidase and α1,2/3-mannosidase (Supplementary Figure S-3A–C), whereas the same product for the 4.9 g.u. glycan was attained by use of two specific mannosidases (Supplementary Figure S-3D–F). Thus, these two glycans were concluded to be core α1,3-fucosylated forms of a bisected Hex3HexNAc2 (Gal1Man2GlcNAc2) and a standard Man3GlcNAc2 respectively, whereby the latter has the same retention properties as a glycan from Pristionchus analyzed on the same column.32 The predominant glycan within the PNGase A pool (Hex4HexNAc2Fuc1; m/z 1297) was analyzed by a similar procedure; this structure was α-mannosidase-resistant prior to removal of the bisecting galactose and is concluded to be an α1,3-fucosylated form of the Hex4HexNAc2 glycan previously found in the triple mutant (Figure 3A–E).
Figure 3.
Digests of PNGase A-released glycans from the fut-6;fut-8 mutant. (A–E) The 2.6 g.u. RP-HPLC fraction, which contains primarily two jack bean α-mannosidase-resistant N-glycan structures ((B) Hex4HexNAc2Fuc1–2-PA, m/z 1297 (for MS/MS see Figure 2A) and 1443), was incubated sequentially with Aspergillus β-galactosidase (C), α1,2/3-mannosidase (D), and α1,6-mannosidase (E), resulting in a final product of m/z 811; digestion of the m/z 1443 glycan is blocked by the α1,2-fucose residue. The trace m/z 1135 structure (A) is more abundant in the neighboring fraction (2.8 g.u.; see Supplementary Figure S-3A–C). (F–J) The 3.8 g.u. fraction was treated with either microbial α1,2-fucosidase (Megazyme), which removed the unmethylated α1,2-fucose substitution of the bisecting galactose (G) or sequentially with hydrofluoric acid (H), Aspergillus β-galactosidase (I), and α1,2/3-mannosidase (J) resulting in a major product of m/z 665. (K–N) The early eluting (3.0 g.u.) isomer of m/z 1457 was first treated with hydrofluoric acid yielding products of m/z 1151 and 1311 (L); while subsequent Aspergillus β-galactosidase and α1,2/3-mannosidase treatments serially removed bisecting galactose and lower arm mannose from the m/z 1151 glycan (M,N), the m/z 1311 glycan, resulting from incomplete release of the methylfucose, is resistant to these enzymes. (O–R) The later eluting (4.1 g.u.) form of m/z 1457 has a contrasting α-galactosidase sensitivity, despite similarity of the MS/MS spectra (for that of the untreated 3.0 g.u. form, see Figure 2 F). Asterisks indicate nonglycan contaminants in the enzyme preparations.
There were various α1,3-fucosylated glycans (Hex3–4HexNAc2Fuc2Me0–1; m/z 1281, 1295, and 1457) concluded to be also capped with a fucose or methylfucose on the bisecting galactose, as they were both β-galactosidase- and α-mannosidase-resistant; this block could be released by prior removal with either α1,2-fucosidase (in the case of the nonmethylated version) or hydrofluoric acid (Figure 3F–J,K–N); the latter treatment was ∼50% efficient for fucose or methylfucose removal from the bisecting galactose, whereas the core α1,3-fucose is nearly completely released. The underlying bisecting β1,4-galactose and the α1,3-mannose residues could then be removed in series to yield final digestion products of m/z 665 (Figure 3J) or m/z 827 (Figure 3N). A further class of N-glycans in the fut-6;fut-8 mutant carried also α-galactose as exemplified by the Hex4HexNAc2Fuc2Me1 glycan (m/z 1457) eluting at 4.1 g.u., isomeric with the one at 3.0 g.u.; the sensitivity to α-galactosidase (Figure 3O–R) is reminiscent of the α- and β-galactose-modified m/z 1313 glycan in the pmk-1 strain.20 Methylation was concluded to be not only on the fucose capping the bisecting galactose but also on hexose, as judged by the presence of m/z 177 and 339 fragment ions (Hex1–2Me1; see Supplementary Figure S-2B and Supplementary Table S-1) for some glycans of low abundance such as m/z 841, 1149, 1309, 1325, 1471, and 1485.
Analysis of the fut-1;fut-8 Mutant
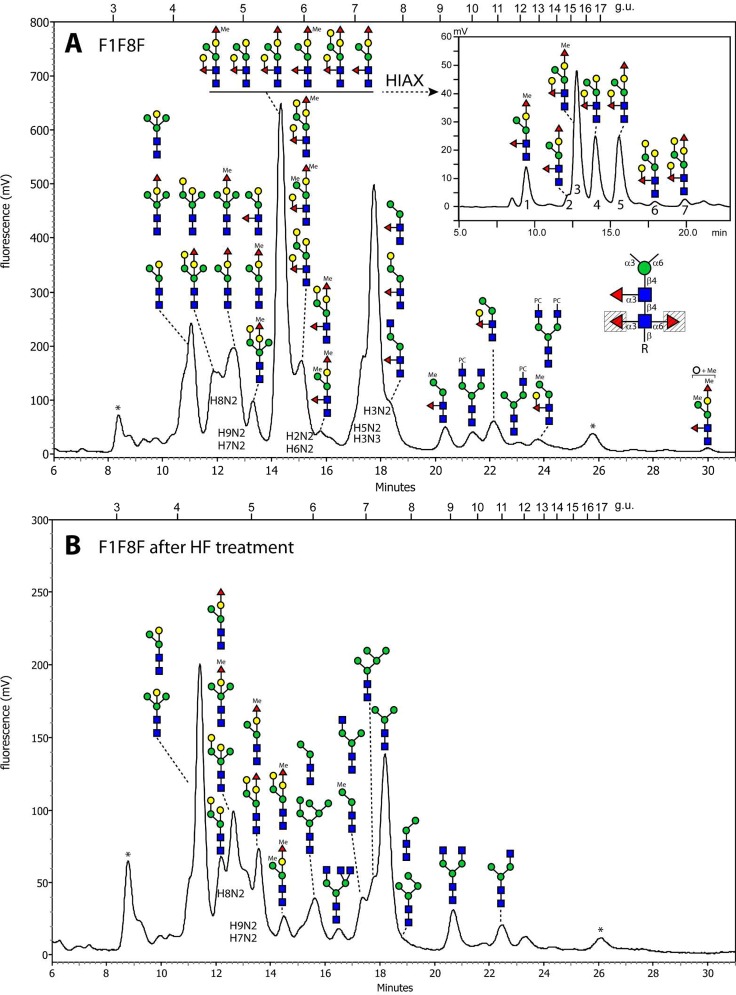
RP-HPLC of the fut-1;fut-8 mutant glycome resulted in a similar number of fractions as for fut-6;fut-8, but the major 5.5 g.u. fraction occurred solely in this mutant and was rather complex in terms of the variety of glycans present (Figure 4A and Supplementary Figure S-4A); thus, to further resolve these, a second chromatographic dimension on the basis of size (HIAX) was performed on this fraction. Alternatively, aliquots of the entire fut-1;fut-8 glycome or of the 5.5. g.u. fraction alone were subject to hydrofluoric acid prior to RP-HPLC; in general, there was a shift to earlier retention times accompanied by loss of fucose or methyl fucose alone, of fucose and hexose together, or of phosphorylcholine (Figure 4B and Supplementary Figure S-4). Compatible with the deletion of genes encoding both enzymes which fucosylate the proximal GlcNAc, glycans with strong m/z 446 and 608 (Gal0–1Fuc1GlcNAc1–PA) MS/MS fragment ions were absent. Nevertheless, extremely low intensities of such ions were observed and can be proposed as minimal rearrangements of fucose or fucose and hexose from the distal GlcNAc (Figure 2C,G,H and Supplementary Figures S-2C,D and S-5E), which are absent once the distal modification is removed by hydrofluoric acid; this phenomenon does not occur upon MS/MS of glycans with a sole fucose on the bisecting galactose. While rearrangements of fucose residues have been reported in other studies,33 the potential here for residual glycans fucosylated on the reducing terminus is negligible due to different HPLC retention time for these glycans and deletion of the relevant genes.
Figure 4.
RP-HPLC chromatograms of PNGase F released N-glycans of the fut-1;fut-8 double deletion strain before and after hydrofluoric acid treatment. The untreated fraction eluting at 5.5 g.u. (A) was collected and further separated using a HIAX column, which resulted in seven glycan containing fractions (labeled 1–7; see inset). The most abundant glycan in a single fraction is shown uppermost, other than for the 5.5 g.u. fraction, in which the order of abundance is from left to right. The inset in panel A highlights which core fucose residues are absent from this strain (shaded triangles). The effect of hydrofluoric acid treatment is shown in panel B. For reasons of space, the elution positions of the pauci- and oligomannosidic glycans are annotated with HXN2. Asterisks indicate fractions containing no N-glycans.
The delayed retention caused by the distal fucosylation, as compared with proximal core α1,3-fucosylation in the fut-6;fut-8 mutant, is evident and is similar to that seen in studies on other nematodes.31,32 The Hex4HexNAc2Fuc1 glycan (m/z 1297; HIAX fraction 4 from the 5.5 g.u. RP-HPLC peak), which was initially resistant to jack bean mannosidase and only partially β-galactosidase-sensitive, could be completely sequenced down to Hex1HexNAc2–PA by serial digestion with hydrofluoric acid, β-galactosidase and α1,2/3-mannosidase (Figure 5A–F). Thus, while the bisecting galactose prevents α-mannosidase digestion, the distal modification appears to have a negative impact on the accessibility of the bisecting residue as compared with the full β-galactosidase sensitivity of the 1297 isomer from the fut-6;fut-8 mutant eluting at 2.6 g.u. (see Figure 3A–E). As for the fut-6;fut-8 mutant, the bisecting galactose could also be modified by fucose or methylfucose, as exemplified by glycans of m/z 1443 and 1457. Upon hydrofluoric acid treatment, the de-2-fucosylated portions were then β-galactosidase-sensitive (Supplementary Figure S-5 and Figure 5G–I).
Figure 5.
Enzymatic and chemical treatments of Hex4HexNAc2Fuc1–2Me0–1 glycans from the fut-1;fut-8 strain. The 2D-HPLC fraction (HIAX fraction 4 of the 5.5 g.u. RP-HPLC fraction; Figure 4A, MS/MS spectrum of m/z 1297 is shown in Figure 2C) was treated sequentially with (B,C) jack bean α-mannosidase and then Aspergillus β-galactosidase or with (D–F) hydrofluoric acid, followed by Aspergillus β-galactosidase and finally α1,2/3-mannosidase. For the two variants of m/z 1457 ((G) HIAX fraction 3 of the 5.5 g.u. RP-HPLC fraction and (J) the 6.2 g.u. RP-HPLC fraction), a differential effect of hydrofluoric acid was observed (pairs of products of m/z 989/1149 or 1151/1311, which are due to losses of 146, 160, or 308 Da (H,L)); the effects of subsequent treatment of the former with Aspergillus β-galactosidase (I) or digestion of the latter solely with α-galactosidase (K) also indicated their structural difference. The absence of the putative initial rearrangement fragments of low intensities at m/z 608 or 446 for these glycans (for MS/MS see Figure 2G,H) only after hydrofluoric acid treatment (insets in panels I and L) is consistent with a correlation between the presence of these rearrangement ions and distal core fucosylation. Note that the properties of the distal GalFuc modification contrast in terms of β-galactosidase and hydrofluoric acid sensitivity with the reducing-terminal Galβ1,4Fucα1,6 motif. (See data on glycans from the fut-1;fut-6 strain.) Components in the β-galactosidase preparation resulted in a shift to sodiated adducts. Asterisks indicate nonglycan contaminants in the enzyme preparations.
In the case of m/z 1135, 1297, 1443, and 1457, two isomers with different fragmentation and putative rearrangement fragments of low intensities at either m/z 608 or 446 (see Supplementary Table S-1) were observed. For those structures with traces of m/z 608, serial losses of hexose and fucose (resulting in fragments of m/z 1135/989, 1295/1149, 1281/1135, and 1309/1163) were observed for m/z 1297, 1457, 1443, and 1471, respectively (Figure 2C,G and Supplementary Figures S-5E and S-2C); where low intensity m/z 446 fragment ions occurred, only loss of fucose from the parent ion was visible, as shown by the fragments of m/z 1311 and 1339 in the case of the m/z 1457 and 1485 glycans (Figure 2H and Supplementary Figure S-2D), which, instead of carrying a hexose on the distal fucose, were modified by α-galactose on the lower arm mannose. Furthermore, these glycans lost either 308 (HexFuc) or 146 (Fuc) upon HF treatment from the distal GlcNAc, as shown for the two m/z 1457 methylated glycans (Figure 5G–L), and lacked an “upper arm” α1,6-mannose in accordance with the specificity of FUT-6.24 Whereas methyl groups were found on mannose residues (e.g., on m/z 841, 1149, 1309, and 1471) in addition to the antennal fucose, the distal (galactosyl)fucose was never methylated.
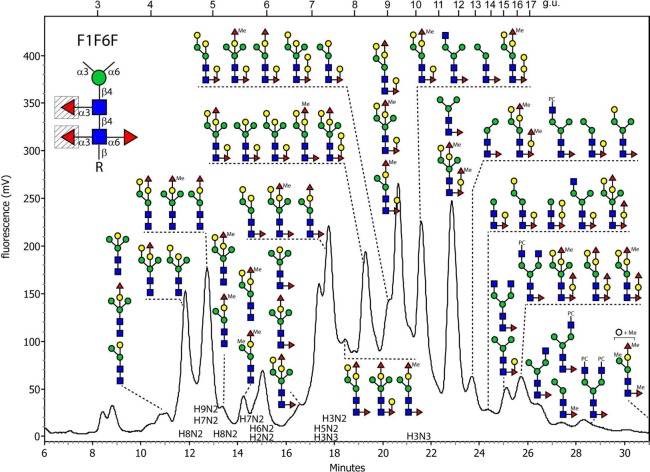
Analysis of the fut-1;fut-6 Mutant
Of the three double mutants, the chromatogram for the glycome of the fut-1;fut-6 mutant was the most complex with some 23 RP-HPLC glycan-containing fractions (Figure 6); therefore, 2D-HPLC (NP-HPLC followed by RP-HPLC) was also performed to yield fractions with a reduced number of glycans (Supplementary Figure S-6). As the core α1,6-fucosylation capacity was intact, a large percentage of the glycans had a relatively late retention time (later than 6.5 g.u.), in keeping with previous data on the RP-HPLC properties of core α1,6-fucosylated glycans.32,34 In comparison with the rather discrete regions of the fut-6;fut-8 and fut-1;fut-8 chromatograms containing difucosylated glycans, the elution positions for such oligosaccharides in the fut-1;fut-6 glycome ranged from 6.5 to >20 g.u. In addition, late-eluting trifucosylated glycans (>11 g.u.) were also detected in this mutant, which possessed m/z 754 or 768 MS/MS core fragment ions (Hex1HexNAc1Fuc2Me0–1-PA; Supplementary Figure S-2G,H,I,N,R,S), while the earlier difucosylated glycans (6.5–11 g.u.) only had key core fragments of m/z 446 or 608 (Hex0–1HexNAc1Fuc1-PA; Figure 2D,E,I,J and Supplementary Figure S-2F,M,O). This suggested that, in addition to fucose directly substituting the proximal core GlcNAc residues in α1,6 linkage, there are two different positions (one core, on the Galβ1,4Fucα1,6, and one peripheral, on the bisecting galactose) for an “extra” fucose residue in this strain. Furthermore, some intermediate-eluting glycans (8–13 g.u.) possessed m/z 770 fragments (Hex2HexNAc1Fuc1-PA; Supplementary Figure 2J,P,Q,T) as previously defined in the hex-2;hex-3 mutant strain as representing two β1,4 galactoses serially linked to the α1,6 core fucose.18 A final core variant is represented by glycans with m/z 460 fragments (HexNAc1Fuc1Me1-PA; Supplementary Figure S-2A,E); this represents methylation of the core α1,6-fucose, which contrasts with methylated core α1,3-fucose found in Pristionchus.32 Additionally, early eluting N-glycans lacking core fucosylation were also observed in this strain as in the triple and the two other mutants; these are concluded to be bisected with and without fucose on the bisecting galactose.
Figure 6.
RP-HPLC chromatogram of PNGase F released N-glycans of the fut-1;fut-6 double knockout. PA-labeled N-glycans were fractionated by an RP-HPLC column; fractions were collected and subject to MALDI MS, MS/MS, and enzymatic and chemical treatments. Characterized structures from both RP-HPLC fractions and 2D-HPLC fractions (see Supplementary Figure S-6) are summarized and annotated on the RP-HPLC chromatogram. The most abundant glycans in a single fraction are shown either uppermost or on the left. The inset indicates which core fucose residues are absent from this strain (shaded triangles). For reasons of space, the elution positions of the pauci- and oligomannosidic glycans are indicated by HXN2.
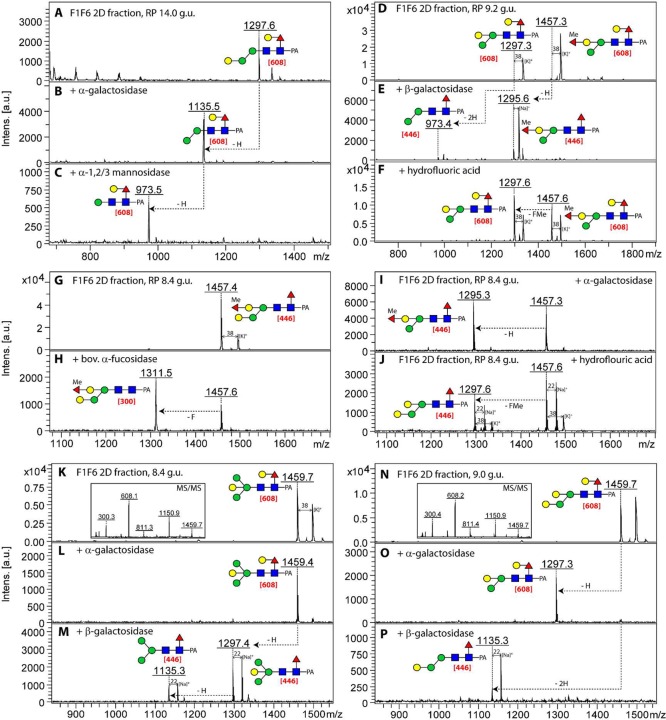
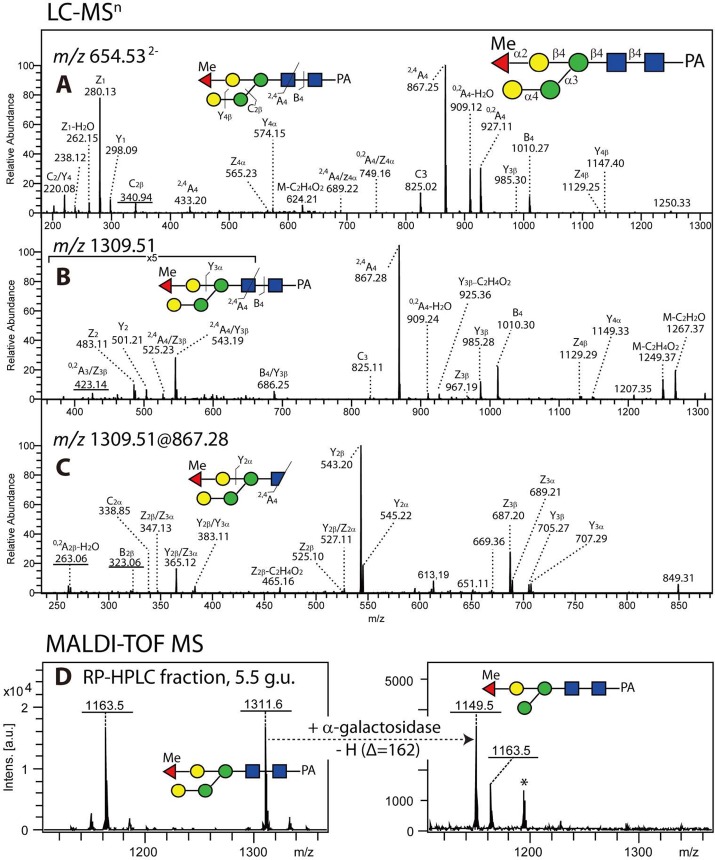
One of the simplest monofucosylated compositions in this strain (Hex4HexNAc2Fuc1; m/z 1297) is represented by eight isomeric structures (see Supplementary Table S-1). Three of these possessed similar MS/MS spectra with an m/z 608 fragment compatible with a proximal GalFuc epitope (9.2, 14, and 15 g.u.). The latter (see Figure 2E) is a simple galactosylated version of the major fucosylated paucimannosidic MMF6 structure as also analyzed from Pristionchus.32 The 14 g.u. glycan was, however, shown to carry a peripheral α-galactose but lacked the α1,6-mannose residue, as after removal of the α-galactose residue the glycan was α1,2/3-mannosidase-sensitive (Figure 7A–C). An earlier-eluting isomeric structure (9.2 g.u.) was, on the contrary, losing two hexose residues upon β-galactosidase treatment (Figure 7E); the MS/MS data suggested that this is due to the loss of a GalFuc epitope and a bisecting galactose. In the same fraction, an m/z 1457 glycan lost only one galactose residue upon β-galactosidase treatment but was also sensitive to hydrofluoric acid; this indicated that the difference between the coeluting m/z 1297 and 1457 glycans was a methylfucose residue attached to the bisecting galactose of the latter (Figure 7D–F). There was also a second m/z 1457 isobaric structure (8.4 g.u.) that lacked the m/z 608 fragment (see Figure 2I) but was sensitive to bovine α-fucosidase (conversion of the m/z 446 core fragment to one at m/z 300), α-galactosidase, and hydrofluoric acid (Figure 7G–J). Thus, this isomer was concluded to be a core α1,6-fucosylated glycan with an α-galactose and a methylfucosylated bisecting β-galactose. Further examples of isomeric glycans varying in having either a bisecting β-galactose on a Man3GlcNAc2 structure or an antennal α-galactose are two glycans of m/z 1459 (compare different galactosidase sensitivities in Figure 7K–M with N–P). The linkage of the galactose to mannose observed in various glycans was investigated by LC–MSn of a non-core-fucosylated α-galactosidase-sensitive m/z 1311 structure: An α1,4-linkage of galactose to the α1,3-linked mannose can be proposed based on the presence of fragment ions at m/z 263 (0,2A2-H2O; for the relevant LC–MS3 data, see Figure 8).
Figure 7.
Sequential treatments of N-glycans from the fut-1;fut-6 strain. 2D-HPLC (normal phase followed by reversed phase) fractions containing isomeric N-glycans (Hex4–5HexNAc2Fuc1–2Me0–1-PA) were sequentially treated with α-galactosidase and α1,2/3-mannosidase (A–C) or independently treated with Aspergillus β-galactosidase and HF (D–F), bovine α-fucosidase, α-galactosidase, or HF (G–J), or with α-galactosidase or Aspergillus β-galactosidase (K–P). All glycans were fragmented and key core fragments are highlighted in red. The relevant MS/MS spectra of the untreated m/z 1457 glycans (panels D and G) are shown, respectively, in Figure 2J,I, whereas the seemingly identical MS/MS spectra of the m/z 1459 glycans are provided as insets (K and N). Note that the reducing terminal Galβ1,4Fucα1,6 motif is β-galactosidase-sensitive but hydrofluoric-acid-resistant; this contrasts with the properties of the distal GalFuc motif found on glycans of the fut-1;fut-8 mutant (see Figure 5; m/z 1297 and 1457).
Figure 8.
LC–MSn spectra of an N-glycan with an α-galactosylated mannose (Gal-Man). (A,B) MS/MS fragmentation spectra of Hex4HexNAc2Fuc1Me1-PA (5.5 g.u.) in, respectively, the doubly and single charged state: m/z 654.53 [M-2H]2– and m/z 1309.51 [M-H]−. Dominant ions are typical cross-ring and glycosidic cleavages for N-glycans (2,4A4, 0,2A4, 0,2A4-H2O, B4, and C3 ions). The fragment ions at m/z 341 (C2β ions in A) and m/z 423 (0,2A3/Z3β ions in B) suggest two hexoses in series. The absence of fragmentation ions at m/z 323 (Hex2-H2O) or 467 (Hex3-H2O) suggests a lack of α1,6-Man in this structure as found in the previous study.20 Thus, the terminal Hex is proposed to be linked to an α1,3Man, while the Y4α ions indicate that the terminal methylated Fuc is linked to bisecting β1,4Gal. (C) In the case of the MS3 spectra of m/z 867.28 (2,4A4 ions) derived from the m/z 1309.51 precursor, the presence of fragmentation ions at m/z 263 (0,2A2-H2O) suggests that the terminal hexose is C4-linked to the α-mannose. (D) The RP-HPLC fraction containing Hex4HexNAc2Fuc1Me1-PA (5.5 g.u.) was treated with green coffee beans α-galactosidase, resulting in the loss of the putatively α1,4Gal residue (mass shift from m/z 1311.6 to m/z 1149.5).
Still more structurally complex than the various Hex4–5HexNAc2Fuc1 and Hex4HexNAc2Fuc2Me1 isomers was the m/z 1619 glycan (10.2 g.u.), which contains all of the various elements (proximal GalFuc, α-galactose, and methylfucosylated bisecting β-galactose). Although analysis of this glycan (one of the major fucosylated ones in this strain) was complicated by the presence of five other minor components, it was inferred that it could be digested down to m/z 811 (Hex1HexNAc2Fuc1-PA; Supplementary Figure S-7) by serial digestion with hydrofluoric acid, α-mannosidase, β-galactosidase, α-galactosidase, and α1,2/3-mannosidase. The loss of β-galactose on the core α1,6-fucose residue was apparent due to the loss of the m/z 608 MS/MS fragment upon β-galactosidase digestion and the appearance of one at m/z 446 (see MS/MS of m/z 1135, right-hand panels of Supplementary Figure S-7).
Related glycans, either lacking a methyl group (m/z 1605), containing an extra mannose (m/z 1781), lacking a methyl group and possessing a third fucose (m/z 1751), or having a methylfucose on the proximal GalFuc (m/z 1779), were also subject to various treatments to confirm their structures. Thereby, the α-mannosidase resistance and α-galactosidase sensitivity of the m/z 1605 and 1781 glycans was associated with no change in the presence of the m/z 608 fragment (Figure 9A–C). On the other hand, the m/z 754 fragment of the m/z 1751 glycan was lost upon bovine fucosidase treatment, indicating that one of two fucose residues removed was linked to the proximal core GalFuc epitope; partial hydrolysis of either of the terminal α1,2-fucose residues with hydrofluoric acid resulted in two isomeric intermediate products, one of which still retained the m/z 754 fragment (Figure 9D–F). In the case of the Hex5HexNAc2Fuc3Me2 glycan (m/z 1779), methylation of two of the fucose residues meant that only a partial removal with hydrofluoric acid resulted in mass shifts to m/z 1459 and 1619; a subsequent treatment with β-galactosidase resulted in the appearance of a product of m/z 1135 in which both the core elongation and bisecting Fucα1,2Gal motifs had been removed (Figure 9G–I). Thus, the m/z 1751 and 1779 glycans are representative of those with the highest degree of fucosylation in the three double fucosyltransferase mutants with methylation of two of the three fucose residues being possible; even larger, but only difucosylated glycans (m/z 1929 and 1943; see Supplementary Table S-1), contained putatively four galactose residues. In this strain, but not in the other two, phosphorylcholine-modified N-glycans were found that were also core fucosylated (see also the Supplementary Table S-1).
Figure 9.
Structural analysis of core modifications of N-glycans isolated from the fut-1;fut-6 strain. N-glycans (Hex4–6HexNAc2Fuc2–3Me0–2-PA) separated by either RP-HPLC or 2D-HPLC were subject to various treatments. A 9.4 g.u. RP fraction containing three major glycans (m/z 1457, 1605, and 1781) was incubated with either jack bean α-mannosidase or α-galactosidase (A–C). Two 2D-HPLC fractions containing trifucosylated structures (m/z 1751 and 1779) were treated either with hydrofluoric acid and bovine α-fucosidase (D–F) or sequentially with hydrofluoric acid and Aspergillus β-galactosidase (G–I). Key core MS/MS fragments of relevant N-glycans are shown in the right panels.
Discussion
Glycomics of Mutant Nematodes
Employing off-line LC–MALDI-TOF MS in combination with MS/MS and various treatments of glycans, we reveal a high degree of isomeric variation in the glycomes of three double-core fucosyltransferase mutant C. elegans strains. The greatest spread of glycan structures, in terms of number, of mass, and of RP-HPLC glucose units, is in the fut-1;fut-6 mutant (Supplementary Figure S-8). Indeed, the deletion of these two α1,3-fucosyltransferase genes seemingly pushes the glycome toward α1,2-fucosylation of the bisecting galactose and core Galβ1,4Fuc motifs. On the other hand, the PNGase F-released pool from the fut-6;fut-8 mutant resembles, perhaps not surprisingly, that of the fut-1;fut-6;fut-8 triple mutant, considering the lack of core α1,3-fucosylated glycans in both cases.
The maximum complexity of the fucosylated N-glycans isolated from the double-knockout strains is represented by glycans of m/z 1619 (Hex5HexNAc2Fuc2Me1) in the case of both the fut-1;fut-8 and fut-6;fut-8 strains and m/z 1927, 1941, and 1943 (Hex6–7HexNAc2Fuc2–3Me1–2) in the fut-1;fut-6 strain (Figure 10A). Only the latter mutant, therefore, contains glycans with up to three fucose residues, as opposed to four in the wild-type N2 strain.19 The maximum number of hexose residues, other than in oligomannosidic glycans, is also higher when the degree of fucosylation increases, which is related to galactose capping of fucose in addition to galactose capping of mannose. The same type of β1 4-digalactosylation of the core α1,6-fucose as found in fut-1;fut-6 was also observed in the hex-2;hex-3 strain in which proximal core α1,3-fucosylation is also absent due to nonreducing terminal GlcNAc on the α1,3-arm being a “NOGO” signal for the relevant FUT-1 enzyme.18 In addition to the Galα1,2Fuc modification of the distal core GlcNAc previously proposed on the basis of GC–MS and enzymatic digestion18 and the bisecting β1,4-galactose defined also by NMR,20 a further form of galactosylation can also now, as judged by negative ion mode LC–MS3 data, be proposed to be a Galα1,4Man motif.
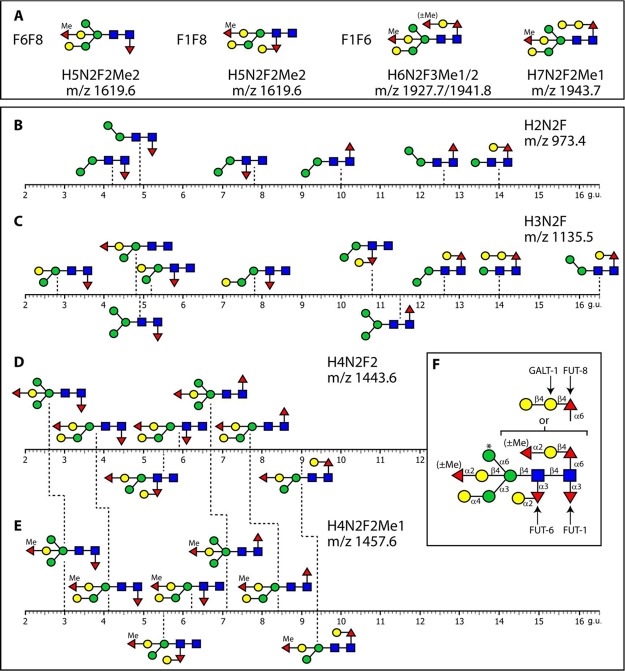
Figure 10.
Elution properties of isobaric glycans on RP-HPLC. The most complicated fucosylated N-glycan structures found in each double mutant are illustrated in panel A. Characterized isobaric glycan structures are listed according to their elution properties on the RP-HPLC (C18) column in terms of glucose units (B–E). Stepped dashed lines between panels D and E indicate the shifts caused by methylation of the α1,2-fucose on the bisecting galactose residue. (F) Summary of the linkages of C. elegans N-glycans with an indication of the glycosyltransferases (FUT-1, -6, -8 and GALT-1) with proven biosynthetic roles. The asterisked α1,6-mannose is absent from glycans modified by FUT-6, whereas the α1,4-linked galactose on the α1,3-mannose is defined in this study for the first time; for the two α1,2-fucose, the methyl groups and the other galactose residues, the relevant enzymes are unknown.
The experimental workflow and data interpretation presented here are indeed the result of what we have learned from glycomic analyses on the aforementioned triple mutant,20 which lacks all complex core modifications and so is biased toward bisecting modifications, and on a double hexosaminidase deletion strain,18 which lacks core α1,3-fucosylation and bisecting galactose but displays increased modifications of the distal core GlcNAc and core α1,6-fucose. Some of these motifs have been found in other nematodes: specifically, the galactosylated core α1,6-fucose and galactosylated distal α1,3-fucose have both been observed in Ascaris, Haemonchus, Pristionchus, and Oesophagostomum.18,31,32 Methylation of core α1,3-fucose, rather than of the α1,6-fucose as in this study, is a low-abundance feature of Pristionchus.32 The bisecting galactose and α1,2-fucose or β1,4-galactose capping of Galβ1,4Fuc is yet to be found in any nematode other than C. elegans. On the other hand, phosphorylcholine is a rather common modification of nematode glycans and is, as in Pristionchus and Trichuris,32,35 often associated with core α1,6-fucose; this applies to the mutants studied here in which Hex3HexNAc3–4PC1–2 structures were detected in all three strains with fucosylated forms thereof found only in the fut-1;fut-6 mutant (see Supplementary Table S-1). A minor phosphorylcholine-modified glycan even carried a core Galβ1,4Fuc modification (Supplementary Figure S-2K).
Elution and Digestion Characteristics Resulting from Nematode N-Glycan Motifs
The use of RP-HPLC as part of the glycomic strategy is valuable in separating isomeric and isobaric structures. The differential effect of core α1,3- and α1,6-fucosylation is well known to cause, respectively, early and late elution.34 Also, phosphorylcholine modifications affect the elution of glycans in a column-specific manner.32 In the present study, we can extend the range of observations regarding retention on a standard C18 chromatography material. Further effects are the linkages of the mannose residues, presence of α-galactose or bisecting β-galactose, distal α1,3-fucosylation, methylation, and capping of the core α1,6-fucose. Thus, considering the range of isomeric structures found in one or more of the three double-mutant strains, we can determine the relative elution properties of, for example, six forms of Hex2HexNAc2Fuc1-PA, ten forms of Hex3HexNAc2Fuc1-PA, and seven forms each of Hex4HexNAc2Fuc2-PA and Hex4HexNAc2Fuc2Me1-PA (Figure 10B–E). Comparing the 973 isomers, (i) the forms with the α1,3-mannose elute earlier than those with the α1,6-mannose, (ii) core α1,3-fucosylation results in early elution, (iii) distal core α1,3-fucosylation results in an intermediate retention, (iv) proximal α1,6-fucose shifts to high retention, and (v) capping of the proximal α1,6-fucose with one galactose causes a large increase in retention (Figure 10B). Analogous effects are observed in the case of the 1135 isomers, for which additionally earlier retention results from the presence of bisecting galactosylation; later retention is caused by α1,4-galactosylation of either the α1,3-mannose or by β-galactosylation of α1,6-fucose, whereas galactosylation of distal α1,3-fucose results in minimal effects (Figure 10C). Double fucosylation (i.e., one core fucose and one antennal fucose) results in generally earlier elution, which is counteracted by methylation of fucose on the bisecting galactose (Figure 10D,E). Interestingly, the presence of an m/z 768, rather than an m/z 754, fragment ion is associated with decreased retention time of glycans with the former as compared with the latter (Supplementary Figure S-8), despite methylation generally resulting in delayed elution.
One can assume that the overall conformation of the glycan affects its interaction with reversed-phase resins, but also steric effects impact the susceptibility to glycosidases. (For possible conformations of selected glycans, see Supplementary Figure S-9.) For instance, the reduced β-galactosidase sensitivity of the bisecting galactose in the case of distally modified glycans (Supplementary Figure S-9B,C) can thereby be explained by the close proximity of the fucose on the distal GlcNAc to the bisecting galactose, whereas modifications of the proximal GlcNAc (E) are more distant and have no inhibitory effect. Interestingly, bovine α-fucosidase will remove all terminal unmethylated fucose residues, other than the proximal core α1,3-fucose, from C. elegans N-glycans. Only if the glycan has a trimannosyl core is the fucose attached to the bisecting galactose resistant to this enzyme (G), whereas glycans without the α1,6-mannose (but having an α-galactose on the α1,3-mannose and even an extended proximal core modification) are still sensitive (H,I), which may be due to the crowded conformation of the former. The ability by α1,2-fucosidase to remove either α1,2-fucose residues (i.e., on the bisecting galactose or the proximal GalFuc motifs) depends on the source of the enzyme with the microbial one being even more effective than chemical treatment (Figure 3G,H).
Biosynthetic and Biological Repercussions of Altered Fucosylation in Nematodes
The glycan structures we find are products of maturation pathways in the Golgi apparatus of the worm, and we can thereby begin to make inferences about “GO” and “NOGO” biosynthetic signals, which differ from those known from mammalian cells;36 for instance, trimming down to trimannosyl core allows α-galactosylation of mannose and the absence of the α1,6-mannose allows distal core fucosylation to take place. However, the biosynthetic basis for only four of the core modifications is known (Figure 10F) and the glycomic repercussions of deletions of the currently identified relevant fut-1, fut-6, fut-8, and galt-1 genes are clear from this and previous studies. In the triple knockout four N-glycan fucosylation events were directly or indirectly abolished, enabling us to verify the bisecting modification;20 although knocking out of the core fucosyltransferases does not affect the bisecting modification directly, it increased the chances of seeing such glycans due to a higher amount of the fewer individual structures. Of course, the absence of various enzymes in the Golgi apparatus can obviously lead to “unnatural” glycoforms that are absent or minimally present in the wild-type, either due to direct substrate-dependent effects or theoretically also to indirect ones due to alteration of the targeting of proteins in the Golgi, if absent glycosyltransferases would be normally associated with others in complexes.
The abolition of some fucosylation events affects the presence of various motifs recognized by nematotoxic lectins. Previously, a deletion in the fut-1 gene was observed to induce resistance to CCL2, a lectin from the ink cap mushroom; interestingly, the fut-1;fut-6 double mutant was completely resistant to the lectin.26 On the other hand, various single fut mutants as well as the fut-1;fut-8 double mutant are still as fully sensitive against the Aleuria aurantia lectin as the wild type (M. Künzler, personal communication); however, as this lectin binds a range of fucosylated glycoconjugates,37 redundancy in terms of its targets is possible. Additionally, deletion of core fucosyltransferases affects galactosylation (e.g., the Galβ1,4Fucα1,6 motif) and so the fut-8 single mutant is also resistant to the mushroom galectin CGL2,21 which is one of four lectins also toxic to Haemonchus contortus.38 With the assignment of the actual glycan structures in various fucosylation mutants, in comparison with parasitic worms, we can begin to understand why certain patterns of resistance and sensitivity toward lectins are observed and so aid the development of potential new therapies against helminth parasites, which share with C. elegans the same range of modifications of the N-glycan core. Furthermore, the present data are a basis for a final definition of the estimated more than 100 N-glycan structures in the wild-type strain of the model nematode.
Acknowledgments
This work was funded in part by the Austrian Fonds zur Förderung der wissenschaftlichen Forschung (FWF; grants P21946 to K.P. and P23922 to I.B.H.W.). We also thank Megazyme for the kind gift of the microbial α1,2-fucosidase, Dr. Martin Dragosits for preparing the recombinant β-galactosidase and FDL hexosaminidase, and Dr. Niclas Karlsson for access to the LTQ mass spectrometer.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.5b00746.
Supplementary Figures S-1–S-9 show mutant genotyping (1), further MS/MS data (2), further enzymatic digests (3,5,7), effect of hydrofluoric acid treatment (4), 2D-HPLC data (6), correlations of glycan size with retention time (8), and possible 3D structural models (9). Supplementary Table S-1 summarizes the data for each proposed structure. (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Schiller B.; Hykollari A.; Yan S.; Paschinger K.; Wilson I. B. H. Complicated N-linked glycans in simple organisms. Biol. Chem. 2012, 393, 661–673 10.1515/hsz-2012-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachter H. Paucimannose N-glycans in Caenorhabditis elegans and Drosophila melanogaster. Carbohydr. Res. 2009, 344, 1391–6 10.1016/j.carres.2009.04.028. [DOI] [PubMed] [Google Scholar]
- Feasley C. L.; Hykollari A.; Paschinger K.; Wilson I. B. H.; West C. M.; Eichinger L.; Rivero F. N-glycomic and N-glycoproteomic Studies in the Social Amoebae. In Dictyostelium discoideum Protocols. Methods Mol. Biol. 2013, 983, 205–229 10.1007/978-1-62703-302-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hykollari A.; Balog C. I.; Rendić D.; Braulke T.; Wilson I. B. H.; Paschinger K. Mass spectrometric analysis of neutral and anionic N-glycans from a Dictyostelium discoideum model for human congenital disorder of glycosylation CDG IL. J. Proteome Res. 2013, 12, 1173–87 10.1021/pr300806b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K.; Perlman M.; Lim J. M.; Cantu R.; Wells L.; Tiemeyer M. Dynamic developmental elaboration of N-linked glycan complexity in the Drosophila melanogaster embryo. J. Biol. Chem. 2007, 282, 9127–9142 10.1074/jbc.M606711200. [DOI] [PubMed] [Google Scholar]
- Kurz S.; Aoki K.; Jin C.; Karlsson N. G.; Tiemeyer M.; Wilson I. B.; Paschinger K. Targetted release and fractionation reveal glucuronylated and sulphated N- and O-glycans in larvae of dipteran insects. J. Proteomics 2015, 126, 172–188 10.1016/j.jprot.2015.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsuka S.; Adachi J.; Kawaguchi M.; Nakakita S.; Hase S.; Ichikawa A.; Ikura K. Structure Analysis of N-Linked Glycans in Caenorhabditis elegans. J. Biochem. 2002, 131, 807–813 10.1093/oxfordjournals.jbchem.a003169. [DOI] [PubMed] [Google Scholar]
- Haslam S. M.; Gems D.; Morris H. R.; Dell A. The glycomes of Caenorhabditis elegans and other model organisms. Biochem. Soc. Symp. 2002, 69, 117–134 10.1042/bss0690117. [DOI] [PubMed] [Google Scholar]
- Haslam S. M.; Dell A. Hallmarks of Caenorhabditis elegans N-glycosylation: complexity and controversy. Biochimie 2003, 85, 25–32 10.1016/S0300-9084(03)00041-5. [DOI] [PubMed] [Google Scholar]
- Hanneman A. J.; Rosa J. C.; Ashline D.; Reinhold V. Isomer and glycomer complexities of core GlcNAcs in Caenorhabditis elegans. Glycobiology 2006, 16, 874–890 10.1093/glycob/cwl011. [DOI] [PubMed] [Google Scholar]
- Paschinger K.; Gutternigg M.; Rendić D.; Wilson I. B. H. The N-glycosylation pattern of Caenorhabditis elegans. Carbohydr. Res. 2008, 343, 2041–2049 10.1016/j.carres.2007.12.018. [DOI] [PubMed] [Google Scholar]
- Zhu S.; Hanneman A.; Reinhold V.; Spence A.; Schachter H. Caenorhabditis elegans triple null mutant lacking UDP-N-acetyl-D-glucosamine:α-3-D-mannoside β1,2 N-acetylglucosaminyltransferase I. Biochem. J. 2004, 382, 995–1001 10.1042/BJ20040793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlager T.; Butschi A.; Grassi P.; Sutov G.; Gauss R.; Hauck D.; Schmieder S. S.; Knobel M.; Titz A.; Dell A.; Haslam S. M.; Hengartner M. O.; Aebi M.; Kunzler M. Methylated glycans as conserved targets of animal and fungal innate defense. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, E2787–96 10.1073/pnas.1401176111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrows B. D.; Haslam S. M.; Bischof L. J.; Morris H. R.; Dell A.; Aroian R. V. Resistance to Bacillus thuringiensis toxin in Caenorhabditis elegans from loss of fucose. J. Biol. Chem. 2007, 282, 3302–11 10.1074/jbc.M606621200. [DOI] [PubMed] [Google Scholar]
- Struwe W. B.; Reinhold V. N. The Conserved Oligomeric Golgi (COG) Complex is Required for Fucosylation of N-Glycans in C. elegans. Glycobiology 2012, 22, 863–875 10.1093/glycob/cws053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutternigg M.; Kretschmer-Lubich D.; Paschinger K.; Rendić D.; Hader J.; Geier P.; Ranftl R.; Jantsch V.; Lochnit G.; Wilson I. B. H. Biosynthesis of truncated N-linked oligosaccharides results from non-orthologous hexosaminidase-mediated mechanisms in nematodes, plants and insects. J. Biol. Chem. 2007, 282, 27825–27840 10.1074/jbc.M704235200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschinger K.; Hackl M.; Gutternigg M.; Kretschmer-Lubich D.; Stemmer U.; Jantsch V.; Lochnit G.; Wilson I. B. H. A deletion in the Golgi α-mannosidase II gene of Caenorhabditis elegans results in unexpected non-wild type N-glycan structures. J. Biol. Chem. 2006, 281, 28265–28277 10.1074/jbc.M602878200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S.; Bleuler-Martinez S.; Plaza D. F.; Künzler M.; Aebi M.; Joachim A.; Razzazi-Fazeli E.; Jantsch V.; Geyer R.; Wilson I. B. H.; Paschinger K. Galactosylated fucose epitopes in nematodes: increased expression in a Caenorhabditis mutant associated with altered lectin sensitivity and occurrence in parasitic species. J. Biol. Chem. 2012, 287, 28276–28290 10.1074/jbc.M112.353128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschinger K.; Rendić D.; Lochnit G.; Jantsch V.; Wilson I. B. H. Molecular basis of anti-horseradish peroxidase staining in Caenorhabditis elegans. J. Biol. Chem. 2004, 279, 49588–49598 10.1074/jbc.M408978200. [DOI] [PubMed] [Google Scholar]
- Yan S.; Brecker L.; Jin C.; Titz A.; Dragosits M.; Karlsson N.; Jantsch V.; Wilson I. B. H.; Paschinger K. Bisecting galactose as a feature of N-glycans of wild-type and mutant Caenorhabditis elegans. Mol. Cell. Proteomics 2015, 14, 2111–25 10.1074/mcp.M115.049817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butschi A.; Titz A.; Wälti M.; Olieric V.; Paschinger K.; Nöbauer K.; Guo X.; Seeberger P. H.; Wilson I. B. H.; Aebi M.; Hengartner M.; Künzler M. Caenorhabditis elegans N-glycan core β-galactoside confers sensitivity towards nematotoxic fungal galectin CGL2. PLoS Pathog. 2010, 6, e1000717. 10.1371/journal.ppat.1000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H.; Tan J.; Schachter H. N-glycans are involved in the response of Caenorhabditis elegans to bacterial pathogens. Methods Enzymol. 2006, 417, 359–89 10.1016/S0076-6879(06)17022-6. [DOI] [PubMed] [Google Scholar]
- Altmann F.; Schweiszer S.; Weber C. Kinetic comparison of peptide: N-glycosidases F and A reveals several differences in substrate specificity. Glycoconjugate J. 1995, 12, 84–93 10.1007/BF00731873. [DOI] [PubMed] [Google Scholar]
- Yan S.; Serna S.; Reichardt N. C.; Paschinger K.; Wilson I. B. H. Array-assisted Characterization of a Fucosyltransferase Required for the Biosynthesis of Complex Core Modifications of Nematode N-Glycans. J. Biol. Chem. 2013, 288, 21015–28 10.1074/jbc.M113.479147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics 1974, 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert M.; Bleuler-Martinez S.; Butschi A.; Walti M. A.; Egloff P.; Stutz K.; Yan S.; Wilson I. B. H.; Hengartner M. O.; Aebi M.; Allain F. H.; Kunzler M. Plasticity of the beta-Trefoil Protein Fold in the Recognition and Control of Invertebrate Predators and Parasites by a Fungal Defence System. PLoS Pathog. 2012, 8, e1002706. 10.1371/journal.ppat.1002706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschinger K.; Hykollari A.; Razzazi-Fazeli E.; Greenwell P.; Leitsch D.; Walochnik J.; Wilson I. B. H. The N-glycans of Trichomonas vaginalis contain variable core and antennal modifications. Glycobiology 2012, 22, 300–313 10.1093/glycob/cwr149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragosits M.; Pflugl S.; Kurz S.; Razzazi-Fazeli E.; Wilson I. B. H.; Rendić D. Recombinant Aspergillus β-galactosidases as a robust glycomic and biotechnological tool. Appl. Microbiol. Biotechnol. 2014, 98, 3553–67 10.1007/s00253-013-5192-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragosits M.; Yan S.; Razzazi-Fazeli E.; Wilson I. B. H.; Rendić D. Enzymatic properties and subtle differences in the substrate specificity of phylogenetically distinct invertebrate N-glycan processing hexosaminidases. Glycobiology 2015, 25, 448–464 10.1093/glycob/cwu132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong-Madden S. T.; Landry D. Purification and characterization of novel glycosidases from the bacterial genus Xanthomonas. Glycobiology 1995, 5, 19–28 10.1093/glycob/5.1.19. [DOI] [PubMed] [Google Scholar]
- Paschinger K.; Wilson I. B. H. Two types of galactosylated fucose motifs are present on N-glycans of Haemonchus contortus. Glycobiology 2015, 25, 585–590 10.1093/glycob/cwv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S.; Wilson I. B. H.; Paschinger K. Comparison of RP-HPLC modes to analyse the N-glycome of the free-living nematode Pristionchus pacificus. Electrophoresis 2015, 36, 1314–29 10.1002/elps.201400528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuhrer M.; Koeleman C. A.; Hokke C. H.; Deelder A. M. Mass spectrometry of proton adducts of fucosylated N-glycans: fucose transfer between antennae gives rise to misleading fragments. Rapid Commun. Mass Spectrom. 2006, 20, 1747–54 10.1002/rcm.2509. [DOI] [PubMed] [Google Scholar]
- Tomiya N.; Awaya J.; Kurono M.; Endo S.; Arata Y.; Takahashi N. Analyses of N-linked oligosaccharides using a two-dimensional mapping technique. Anal. Biochem. 1988, 171, 73–90 10.1016/0003-2697(88)90126-1. [DOI] [PubMed] [Google Scholar]
- Wilson I. B. H.; Paschinger K. Sweet secrets of a therapeutic worm: Mass spectrometric N-glycomic analysis of Trichuris suis. Anal. Bioanal. Chem. 2015, 10.1007/s00216-015-9154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachter H. Biosynthetic controls that determine the branching and microheterogeneity of protein-bound oligosaccharides. Biochem. Cell Biol. 1986, 64, 163–181 10.1139/o86-026. [DOI] [PubMed] [Google Scholar]
- Iskratsch T.; Braun A.; Paschinger K.; Wilson I. B. H. Specificity analysis of lectins and antibodies using remodeled glycoproteins. Anal. Biochem. 2009, 386, 133–46 10.1016/j.ab.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Heim C.; Hertzberg H.; Butschi A.; Bleuler-Martinez S.; Aebi M.; Deplazes P.; Kunzler M.; Stefanic S. Inhibition of Haemonchus contortus larval development by fungal lectins. Parasites Vectors 2015, 8, 425. 10.1186/s13071-015-1032-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.