Abstract
Legume crops present important agronomical and environmental advantages mainly due to their capacity to reduce atmospheric N2 to ammonium via symbiotic nitrogen fixation (SNF). This process is very sensitive to abiotic stresses such as drought, but the mechanism underlying this response is not fully understood. The goal of the current work is to compare the drought response of two legumes with high economic impact and research importance, Medicago truncatula and Glycine max, by characterizing their root nodule proteomes. Our results show that, although M. truncatula exhibits lower water potential values under drought conditions compared to G. max, SNF declined analogously in the two legumes. Both of their nodule proteomes are very similar, and comparable down-regulation responses in the diverse protein functional groups were identified (mainly proteins related to the metabolism of carbon, nitrogen, and sulfur). We suggest lipoxygenases and protein turnover as newly recognized players in SNF regulation. Partial drought conditions applied to a split-root system resulted in the local down-regulation of the entire proteome of drought-stressed nodules in both legumes. The high degree of similarity between both legume proteomes suggests that the vast amount of research conducted on M. truncatula could be applied to economically important legume crops, such as soybean.
Keywords: Medicago truncatula, soybean, nodule proteome, drought, split-root system
Introduction
Grain and forage legumes are grown on around 15% of the Earth’s arable surface.1 The grain legume Glycine max (soybean) is an important source of protein for humans and animals, as well as vegetable oil.1 Soybean’s economic importance and the large number of researchers who work on it have contributed to the development of molecular, genetic, and genomic tools for the species.2 Similarly, in recent decades, Medicago truncatula, a forage legume, has emerged as a useful model for molecular studies.3,4 The availability of the complete genome sequences of M. truncatula(5) and the soybean plant6 has facilitated the genomic and proteomic studies of these two species. Having the genome available is key for protein identification by mass spectrometry and, consequently, for proteomic research. Diverse proteome studies of different plant organs, cell cultures, and organelles have been conducted on M. truncatula and G. max under different abiotic stress conditions (for a review, see refs (7) and (8), respectively). The plant fraction of M. truncatula nodules subjected to drought has been characterized, with a nodule proteome database for this species being established.9 Additionally, a brief study on soybean nodule proteins under drought stress has also been documented.10 Apart from this, the drought response of legume plants has hardly been investigated at the proteome level.
Legumes have the ability to carry out symbiotic nitrogen fixation (SNF) with nitrogen-fixing soil bacteria known as rhizobia. Legumes can be classified into amide or ureide exporters according to the compounds used for transporting the fixed N. In general, amide-exporting legumes, such as M. truncatula, contain indeterminate-type nodules and originated in temperate regions. They transport amides, mainly in the form of asparagine and glutamine. Ureide-exporting legumes, such as soybeans, are mostly tropical legumes with determinate-type nodules and transport mainly allantoin and allantoic acid. Despite the agronomical and environmental advantages of legume crops, their production is limited by environmental constraints, drought being one of the most harmful stresses.11 The regulation of SNF under drought conditions involves various factors, mainly internal oxygen availability, N-feedback regulation, and carbon limitation. Despite research in the field, the molecular-level interactions between the cited factors and the SNF regulation mechanism(s) are not fully understood. Recently, the local regulation of SNF in diverse legumes, including M. truncatula and soybean plants, has been demonstrated under drought stress.10,12,13 These studies dismiss the generally accepted role of amides and ureides as being the molecules involved in inhibiting SNF under drought conditions. However, it remains unclear whether there are local changes in the nodule proteome, as in the SNF process, or whether systemic signals are involved. Moreover, it has been reported that SNF is a more drought-sensitive process in ureide-exporting nodules, such as those of soybean plants, than in the amide exporters such as Medicago.14 Nevertheless, reports concerning the possibly distinct responses to drought stress of ureide- versus amide-exporting legumes are rare.
In the present study, the drought response of the nodule proteome of two model legumes, M. truncatula and G. max, is compared, leading to the identification of shared stress responses as well as unique proteome features of the two legumes. By using a split-root system (SRS), whereby watered and drought-treated nodules shared the same aerial part, one can observe the local effect of drought stress in the legume nodule proteome resulting in a greater understanding of the molecular mechanisms regulating SNF. Additionally, this is the first study demonstrating that comparative proteomics across related species can improve the functional proteome characterization. The work demonstrates a strategy for determining how molecular data may be transferable between legume species, such as from model legumes to crop legume species, which is an important step forward for the legume research community.
Materials and Methods
Plant Growth Conditions, Split-Root System, and Drought-Stress Treatment
Nodulated M. truncatula Gaertn. cv. Jemalong A17 and G. max (L.) Merr. plants were grown under controlled environmental conditions (14 h photoperiod; 400 (μmol m–2 s–1) light intensity; 22 °C and 16 °C day and night temperature; 60 to 70% relative humidity) for 12 and 6 weeks, respectively, in double pots (600 mL each) to produce SRS. The seedlings were inoculated with N2-fixing strains: Ensifer meliloti 2011 for M. truncatula; and Bradyrhizobium japonicum strain UPM752 for soybean plants, watered with N-free nutrient solutions.15,16
The plants were randomly separated into three sets: controls (C) received a daily supply of nutrient solution to field capacity in both sides of the SRS, whereas drought (D) was achieved by withholding water and nutrients. In partial drought plants (PD), one half of the root system was kept at field capacity (PDC), while the other half was kept unwatered (PDD) for 7 days. After this period, four types of nodule samples (C, PDC, PDD, and D) were harvested, immediately frozen in liquid N, and stored at −80 °C for the proteomic analysis.
Physiological Characterization
Transpiration was gravimetrically determined on a daily basis. Leaf water potential (Ψleaf) was measured using a pressure chamber (Soil Moisture Equipment, Santa Barbara, CA). The water potential of detached nodules (Ψnodule) was measured in C52 sample chambers coupled to a Wescor HR-33T Dew Point Microvoltmeter (Wescor, Logan, UT). SNF was measured as apparent nitrogenase activity (ANA) using an electrochemical H2 sensor (Qubit System).
Proteomic Analysis, Composite Protein FASTA File Generation, and Combined Mapman Mapping File
Frozen nodules (0.1 g of fresh weight) were homogenized in a mortar and pestle with 2 mL of extraction buffer (50 mM Hepes pH 7.5, 1 mM EDTA, 1 mM KCl, 2 mM MgCl2, 2.5% (w/v) PVP, and 1 mM PMSF). The homogenates were centrifuged at 20000g at 4 °C for 30 min. Supernatants were collected, and soluble proteins were precipitated overnight at −20 °C after adding 5 volumes of precooled acetone. Pellets recovered by centrifugation at 10000g at 4 °C for 10 min were air-dried and resuspended in 300 μL of solubilization buffer (8 M urea buffer, 100 mM NH4HCO3, pH 8–8.5, 5 mM DTT).
The samples were diluted (1:4) in buffer (25 mM NH4HCO3, pH 8–8.5, 10% (v/v) acetonitrile and 5 mM CaCl2) and aliquots containing 100 μg of protein were digested overnight at 37 °C under rotation using Poroszyme-immobilized trypsin beads (1:20, v/v, Applied Biosystems, Life Technologies). After centrifugation for bead removal, the obtained peptide mixtures were desalted using SPEC C18 columns according to the manufacturer’s instructions (Varian, Agilent Technologies). Finally, the desalted digest solutions were dried, and the pellets were stored at −80 °C until use.
Prior to the mass spectrometric measurement, the protein digest pellets were dissolved in 0.1% (v/v) formic acid. The protein digests (5 μg) were analyzed via shotgun nano-LC-ultra (Eksigent system, Axel Semrau GmbH) using a monolithic reversed-phase column (Chromolith 150 × 0.1 mm; Merck, Darmstadt) directly coupled to an LTQ-Orbitrap XL mass spectrometer (Thermo Scientific, Rockford, IL) operated with Xcalibur (2.0.7 SP1) as described elsewhere.17 The peptides were eluted with a 100 min gradient from 5% to 60% acetonitrile. An Orbitrap-FT analyzer was used for precursor MS1, and an LTQ-XL was used for MS2 mass analyses, respectively. CID was performed at a normalized collision energy of 35. Dynamic exclusion settings were as described in ref (18). The mass window for precursor selection was set to 10 ppm with a resolution of 30 000 in a mass range from 400 to 1,800 m/z.
After the performance of the mass spectrometric analyses, the raw files were searched against a composite protein FASTA file using the Sequest algorithm. The composite protein FASTA file was created by fusing the following databases. For M. truncatula: (1) Uniprot UniRef100 Medicago; origin: www.uniprot.org. The search was performed on May 7, 2013; 54 246 entries. (2) IMGA; origin: http://medicago.org/, 64 123 entries. (3) DCFI; origin: http://compbio.dfci.harvard.edu/; 59 601 entries. For G. max: (1) Uniprot UniRef100 G. max; origin: www.uniprot.org. The search was performed on May 15, 2013; 70 318 entries. (2) Gmax_189_protein_annotated.fasta; from phytozome/v9.0/Gmax/assembly/JGI assembly with 73 320 entries. (3) DCFI; origin: http://compbio.dfci.harvard.edu/. Uniprot accession or gene names missing in the originally used version of the FASTA file were added for those proteins presented in the tables in cases when this was available in May 2015.
All nucleotide FASTA files were translated into amino acid sequences selecting only the longest ORF per accession number using “sixpack” from the Jemboss 1.5 toolbox.19 The three M. truncatula protein FASTA files described above, as well as the common contaminants, were combined, producing a new FASTA containing 130 824 entries, which will henceforth be referred to as MT-fasta. Similarly, the three G. max FASTA files described above were combined, producing a new FASTA containing 209 273 entries, which will be referred to as GM-fasta.
Protein sequences that were 100% identical in sequence and length were combined by subsequently adding one header after the other, separating them using the following characters “ __***__ “ (regardless of whether the redundancies originated from one or multiple FASTA files). All other entries were simply added to the new file. The first accession number of the header was repeatedly written to the very beginning of the header line, separated by a “|” to consistently view and parse the accession numbers. The weighted order of headers (different database accessions and annotations) agreed with the above-mentioned order, being (1) Uniprot UniRef100, followed by other entries.
A decoy database enabled a false-positive rate analysis. Only high-confidence peptides (false-positive rate <0.1%) with better than 5 ppm precursor mass accuracy and at least two distinct peptides per protein passed criteria. This led to a data matrix including the spectral count for relative quantification, generated by the Proteome Discoverer. The spectral count is a semiquantitative measure for tracking changes in protein abundance in complex samples that is based on the cumulative sum of ion fragments that are recorded in a MS/MS spectrum.20 To visualize the data for functional category enrichment, the relative abundance of protein across all IDs was calculated using the normalized spectral abundance factor, NSAF21 (Figure 1).
Figure 1.
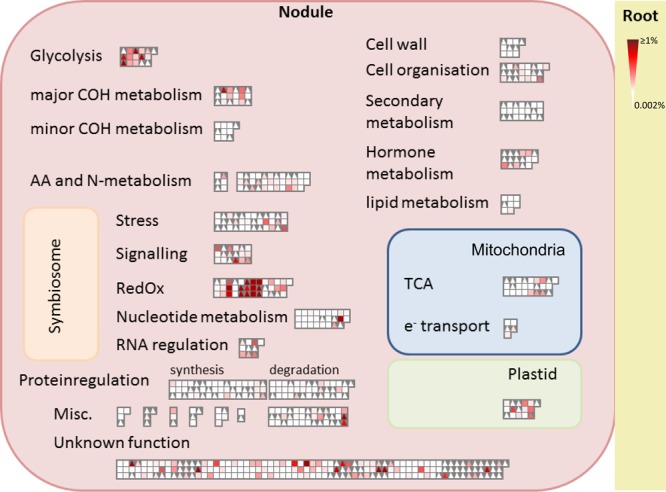
Mapman overview. Functional distribution and relative abundance of 304 M. truncatula and 341 G. max plant nodule proteins. Triangles stand for M. truncatula nodule proteins, and squares represent G. max nodule proteins. The strength of the color indicates the abundance of the protein (NSAF; normalized spectral abundance factor, n = 5).
A new Mapman mapping file was created from the newly generated FASTA files of the two species, using Mercator.22 The two mapping files were then added together to make a single, combined mapping where the two species were distinguished by assigning MT accessions as “P” (usually used for proteins) and GM protein entries as “T” (normally used to display transcripts).
BLAST Table Generation
MT-fasta was blasted against GM-fasta and vice versa. The latter task was performed using the standalone NCBI blastp version 2.2.26+ with the default matrix (Blosum 62). Using these results, tables for Medicago-against-Glycine and Glycine-against-Medicago results were created. Only hits below an e value of 0.001 were considered; if subsequent hits yielded the same e value as the previous one, they were included; a column was introduced indicating if the query and hit pair of one results table was equivalent in the other. With the exception of BLAST, all data was processed using Python version 2.7.3 employing in-house scripts.
Statistical Analysis
For physiological measurements, the normal distribution of the samples was checked via Shapiro–Wilk tests and the homogeneity of variances via Levene’s test. Significant differences between treatments were determined using one-way ANOVA. If significant differences between means were obtained, then comparisons between each treatment and its control were performed using the LSD test. Differences were considered to be significant at P ≤ 0.05.
Relative changes in protein abundance were calculated for the proteomic data. Using the average spectral count values from five biological replicates, the log2 ratios (treatment/control) were calculated. Proteins that were found in at least one treatment and in at least three replicates were used for quantification. Those proteins showing more than a twofold change in relative abundance and significance differences (Student’s t test, P ≤ 0.05) between treatments were used in the Venn diagram and to illustrate relationships between the various treatments.
Results
SRS experiments were performed to examine the response of one side of the root system to the water deprivation experienced by the other side and to identify both local and systemic changes of the nodule proteome in response to drought stress. Any changes occurring in the untreated side of the root system result from the altered water status of the other root fraction, denoting that systemic responses are taking place. A local response occurs in those organs directly exposed to drought conditions.
Physiological Characterization of Plants Subjected to Partial Drought
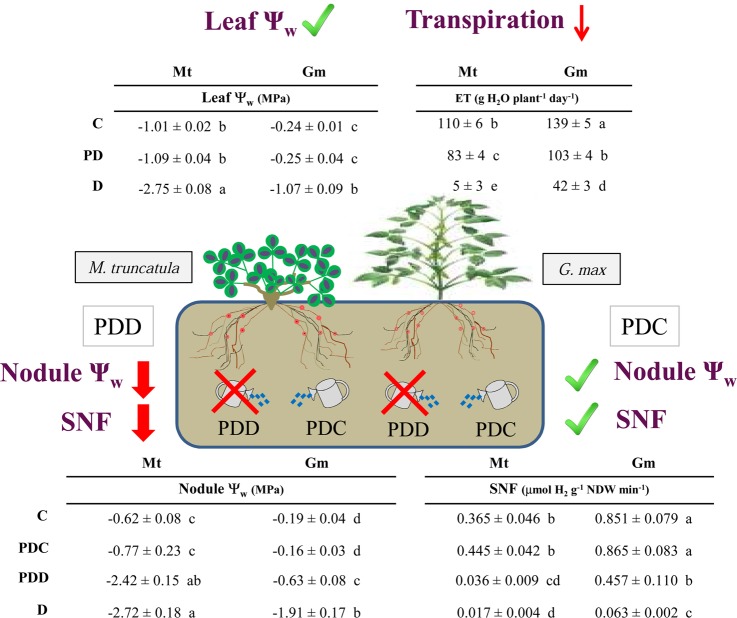
A scheme of the main physiological responses observed in both species in partial drought conditions is presented in Figure 2. The physiological characteristics of both legumes, the amide exporter M. truncatula and the ureide exporter G. max, are discussed together to highlight the similarities and the differences between both plants after 7 days without being irrigated. Detailed time-course studies describing the physiological responses of both legumes showed a local SNF drought response.10,13
Figure 2.
Overview of the effect on leaf water potential, transpiration rate, nodule water potential, and apparent nitrogenase activity in M. truncatula and G. max plants exposed to 7 day partial drought treatment. Values represent mean ± SE (n = 3). For each parameter and species, significant differences (P ≤ 0.05) between treatments were denoted by different letters. Red arrows indicate a decreasing trend of different parameters, and the check mark symbols denote unchanged parameters.
D plants showed a significant decline in Ψleaf, while PD plants maintained their Ψw in the same range as C plants. Just as with leaves, a decline in Ψnodule was also ascertained for D nodules compared to C in both legumes. PDD nodules also suffered a significant decline in Ψnodule reaching similar values to those of the D nodules in M. truncatula, while in soybean plants the drop was slightly less. On the irrigated side of PD plants, however, PDC nodules had similar Ψnodule values to C in both species. It is worth noting that the leaf and nodule Ψw in C conditions were different for each legume, the Ψw of soybean plants being closer to zero. The transpiration rate was higher in soybean plants than in M. truncatula, but the decreasing trend shown in both species was similar. After 7 days of water deprivation, transpiration rates decreased in D plants compared to those in C conditions. However, in PD plants, the decline was less and similar in both (26% in soybean plants and 25% in M. truncatula).
Similar to the pattern observed for Ψnodule, a differential ANA behavior was observed between the PDC and PDD root systems. Drought stress caused a 90% and a 95% reduction of ANA in M. truncatula PDD and D nodules, respectively, when compared to C plants, while PDC had values close to those of C. In soybeans, however, the decline of 93% in D was similar to that seen in M. truncatula, while the 46% decrease seen in PDD was less. PDC maintained similar ANA values to C, just as was observed in M. truncatula.
Nodule Proteome Profile and Functional Classification
The nodule proteins were analyzed to compare the response to drought stress of both legumes. The mass spectra obtained were searched against MT-fasta and GM-fasta. This resulted in a stringent identification of 304 M. truncatula plant nodule proteins and 341 soybean plant nodule proteins. The complete lists of identified proteins and their spectral count values for relative quantification are included in the Supporting Information in tables in the electronic appendix (Table S2 for M. truncatula proteins and Table S3 for soybean proteins).
For an overview of the proteome in control conditions, the Mapman database was employed and proteins were classified into 20 functional groups (Figure 1). The largest groups for number of identified proteins were protein regulation, amino acids and N-metabolism, and redox and stress. In general, the distribution of the proteins in the different functional groups was similar in both legumes with the exception of lipid metabolism and nucleotide metabolism, which were much more abundant in soybeans (Figure 1). In the context of relative protein abundance, both in M. truncatula and G. max, proteins from the redox and glycolysis functional groups were the most abundant (Figure 1). According to the BLAST comparison, from the protein ID list for the two species, 134 proteins were found to have a high degree of similarity (e value ≤0.001) between M. truncatula and G. max (Table S1), with 14 proteins being unique; that is to say that they only matched with one protein from the other species (Table 1). Among the 134 highly similar proteins, 54 changed significantly in the drought comparisons in either M. truncatula or G. max (Table S1 in bold).
Table 1. Extracted List of 14 Proteins (From All Identified Proteins) With High Sequence Similarity (e value ≤ 0.001) between M. truncatula and G. max Databases for Improved Functional Annotationa.
| MEDTR accession | protein description | GLYMA accession | protein description |
|---|---|---|---|
| A2Q2 V1 | citrate lyase a-subunit | I1L0Q8 | uncharacterized protein |
| B7FI39 | 12-oxophytodienoate reductase | I1JAQ7 | uncharacterized protein |
| B7FL28 | similar to putative uncharacterized protein | Q96453 | 14-3-3-like protein D |
| G7I8P7 | 26S proteasome regulatory particle triple-A ATPase protein | I1NJ15 | uncharacterized protein |
| A0A072VE66 | glutamate synthase | I1KAR0 | uncharacterized protein |
| A0A072VT43 | elongation factor EF-2 | I1KU21 | uncharacterized protein |
| G7I9Q9 | uncharacterized protein | Q01915 | ATP synthase subunit α |
| G7IAA2 | adenosine kinase | C6T7F3 | uncharacterized protein |
| G7IAG4 | neutral invertase-like protein | I1LD52 | uncharacterized protein |
| G7INB7 | ABA-responsive protein ABR17 | C6SWY6 | uncharacterized protein |
| G7JNN9 | cysteine synthase | Glyma02g15640.1 | uncharacterized protein |
| G7KXR2 | transaldolase | I1KZJ1 | uncharacterized protein |
| G7L970 | alanine aminotransferase | I1KHJ4 | uncharacterized protein |
| Q1RSH4 | chaperonin CPN60–2 | I1NHW4 | uncharacterized protein |
The full list of highly similar proteins (∼60) is available in Table S1.
Local Changes in the Proteome Profiles of M. truncatula and G. max Subjected to Partial Drought
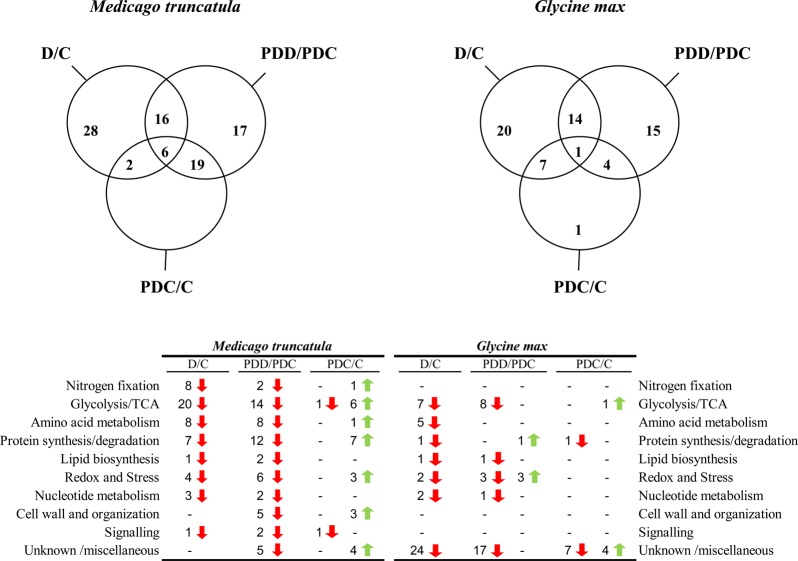
Venn diagrams were employed to examine the drought response of the nodule proteomes (Figure 3). The Gene Ontology Annotation Database from UniProt was utilized to classify proteins into ten functional groups for both M. truncatula and G. max. These diagrams provided an important overview of whether the changes represented specific or common responses. Of the relatively quantified proteins in Medicago, 28 changed significantly and exclusively in the total drought comparison (D/C), 17 proteins were altered exclusively in the partial drought comparison (PDD/PDC), and 16 proteins were shared between both comparisons (Figure 3). In soybean, however, 20 proteins varied significantly and exclusively in the total drought comparison (D/C), 15 were altered in the partial drought comparison (PDD/PDC), and 4 were shared between both comparisons (Figure 3). The main functional groups that changed in the total (D/C) and partial drought (PDD/PDC) comparisons in both legumes were the glycolysis and TCA cycle and amino acid metabolism, followed by redox- and stress-related proteins (Figure 3). It should be noted that in soybean, the proteins with an unknown function represent almost 49% of the studied proteins while in M. truncatula, these are around 6%. The identified proteins that change exclusively in total or partial drought comparisons are listed in Table 2 for M. truncatula and Table 3 for G. max; unknown proteins are not displayed. A common pattern was observed in all the identified proteins: a decrease in the relative content of proteins from the treatments directly subjected to drought (D and PDD) in comparison with their controls (C and PDC) (Tables 2 and 3 and Figure 3). Glycolysis and TCA was the most numerous group that changed in total and partial drought comparisons, and similar proteins were altered significantly in both legumes (mainly sucrose synthase (SuSy), fructose-bisphosphate aldolase (FBPA), phosphoenolpyruvate carboxylase (PEPC), and malate dehydrogenase (MDH) (Tables 2 and 3). Amino-acid metabolism was the second largest protein functional group and included nitrogen assimilation enzymes such as aspartate aminotransferase (AAT), glutamate synthase (GOGAT), glutamine synthetase (GS), and asparagine synthetase (AS); as well as enzymes involved in the metabolism of sulfur-containing amino acids (Tables 2 and 3). In M. truncatula, protein synthesis and degradation and nitrogen fixation-related proteins were also numerous, while this latter functional group was almost absent in soybean plants (Figure 3 and Tables 2 and 3). Additionally in soybean, in contrast to M. truncatula, some exceptions were found to the general pattern of decline in D and PDD nodules. A total of three redox- and stress-related proteins presented a higher relative content in PDD nodules in comparison with PDC (an In2-1 protein and two ATP synthase proteins) (Table 3 and Figure 3). The proteins that showed the highest significant fold-change were two SuSys (P13708; Glyma13g17421.1 and I1L1U4; and Glyma09g08550.5) in soybean plants and one PEPC (G7IH71), two AS (TC180197; O24483; and G7JZK0) and a lipoxygenase (G7LIY0) in M. truncatula.
Figure 3.
Venn diagrams of M. truncatula and G. max plant nodule protein changes (n = 5, P ≤ 0.05 and fold change ≥2). A list is shown with the functional classification of the proteins and number of proteins within each comparison. Green arrows denote increases in the numerator of each comparison, and red arrows indicate decreases.
Table 2. Changes in M. truncatula Nodule Proteins after Drought Treatmenta.
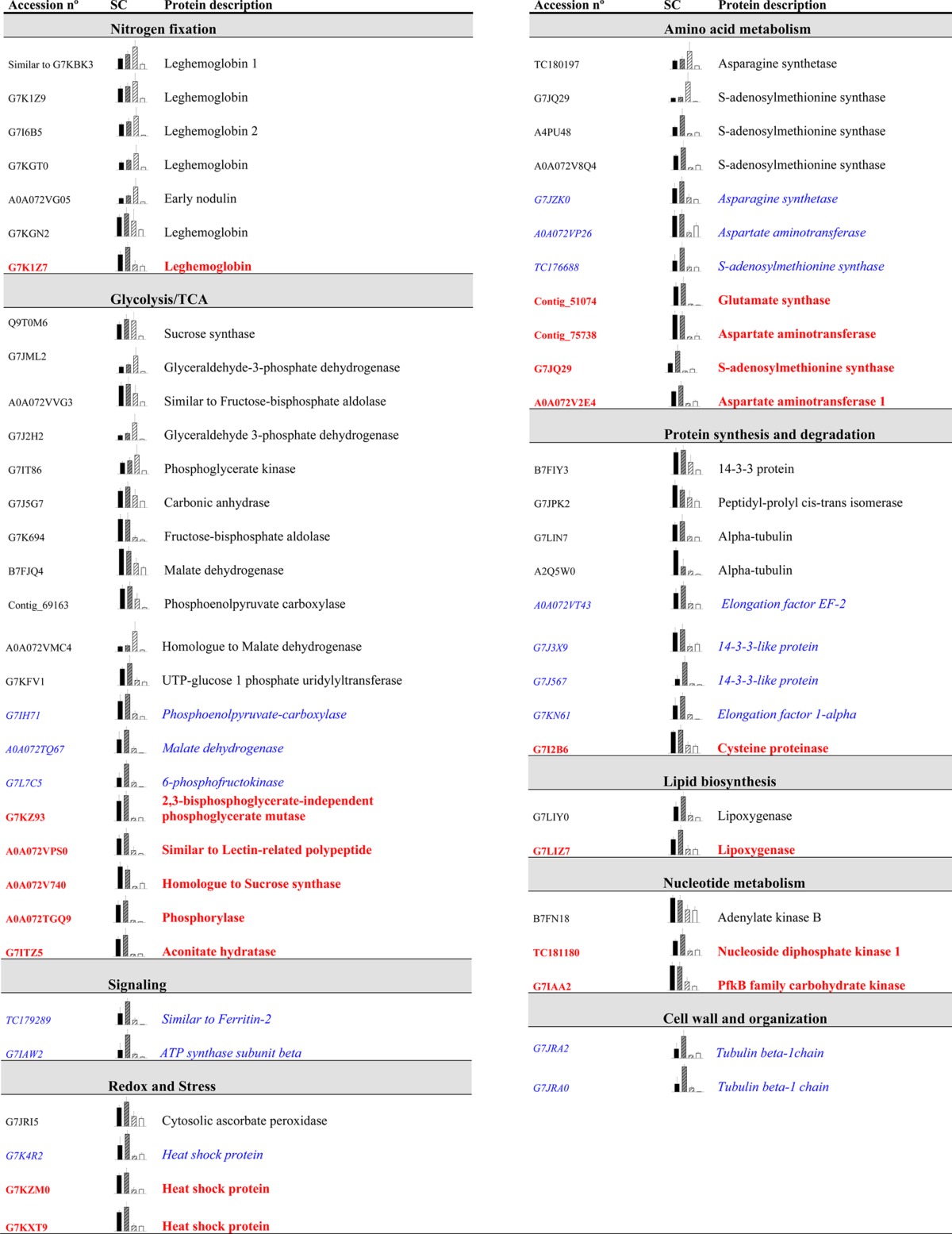
Proteins exhibiting significant changes exclusively in the total (D/C, in normal black) and partial (PDD/PDC, in blue italics) drought comparisons from Venn diagrams are listed below (n = 5, P ≤ 0.05, and fold change ≥ 2). Unknown proteins are not displayed. Proteins shared between both groups are shown in bold red. Protein accessions (UniprotKB) and gene codes are given if available. SC refers to the average spectral counts for each treatment: C is shown in black, PDC is striped grey, PDD is striped white, and the D samples are white.
Table 3. Changes in G. max Nodule Plant Proteins after Drought Treatmenta.

Proteins exhibiting significant changes exclusively in the total (D/C, in normal black) and partial (PDD/PDC, in italics blue) drought comparisons from Venn diagrams are listed below (n = 5, P ≤ 0.05 and fold change ≥ 2). Unknown proteins are not displayed. Proteins shared between both groups are shown in bold red. Protein accessions (UniprotKB) and gene codes are given if available. SC refers to the average of spectral count values for each treatment: C is shown in black, PDC is striped grey, PDD is striped white, and the D samples are white.
Systemic Changes in the Proteome Profiles of M. truncatula and G. max Subjected to Partial Drought
The main feature of the SRS experimental setup is that it enables the identification of systemic changes. Proteins responding systemically have been categorized by comparing the proteins from C nodules with those from PDC nodules. Therefore, significant changes in the PDC nodules in comparison with those in C conditions were understood as systemic changes caused by drought stress. The proteins that changed systemically are listed in Table 4; unknown proteins are excluded. From the relatively quantified proteins, 25% changed systemically in Medicago and 10% in soybean (Figure 3). An overall and marked increase in the relative quantification of PDC proteins compared with C conditions was seen as being a general systemic response in both legumes (Table 4). The only exception to this general pattern was a Medicago FBPA that showed a higher relative content in C nodules than in the other treatments (Table 4). It is worth noting that the proteins changing systemically also changed in the partial and total drought comparisons; in other words, none of the proteins exhibited an exclusively systemic response, with the exception of an uncharacterized soybean protein (I1KK66 and Glyma07g16910.1) (Table S3). The rest of the proteins that varied significantly in the systemic comparison (PDC/C) also altered significantly in the total drought comparison (D/C: 2 proteins in M. truncatula and 7 proteins in soybean), in the partial drought comparison (PDD/PDC: 19 proteins in M. truncatula and 4 proteins in soybean) or were divided among the three comparisons (one protein in soybean and 6 in M. truncatula) (Figure 3).
Table 4. Changes in M. truncatula (Upper Part) and G. max (Lower Part) Nodule Plant Proteins after Drought Treatmenta.
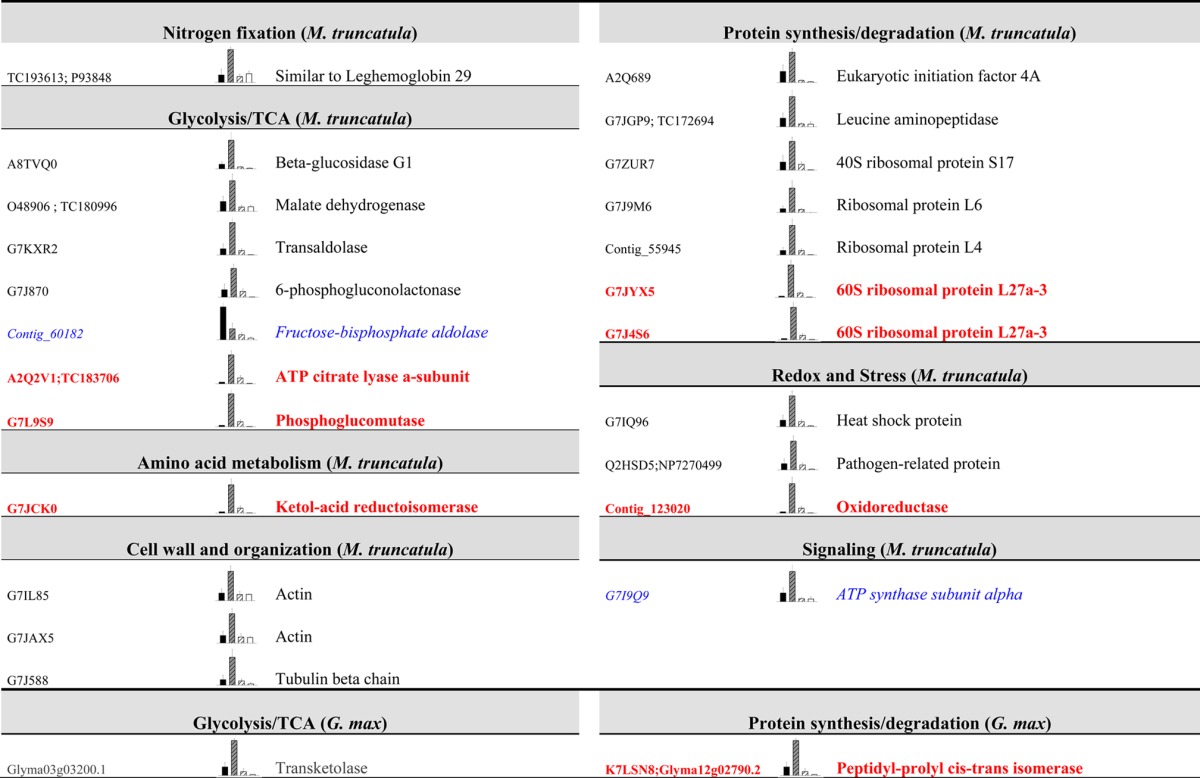
Proteins changed in the systemic comparison (PDC/C, in normal black; most of these are protein shared with the PDD-to-PDC comparison) from Venn diagrams are listed below (n = 5, P ≤ 0.05 and fold change ≥ 2). Proteins shared between the systemic comparison (PDC/C) and the total drought comparison (D/C) are shown in blue italics, and proteins shared among the three comparisons are shown in bold red. Unknown proteins are not displayed. C is shown in black, PDC is striped grey, PDD is striped white and the D samples are white.
The most numerous functional groups in the M. truncatula systemic comparison were glycolysis and TCA cycle and protein synthesis and degradation, which was in agreement with the local response of the proteome (Table 4). Within the set of proteins showing a systemic response in M. truncatula, it is remarkable that three cell-wall- and organization-related proteins changed significantly (two actins: G7IL85 and G7JAX5, and one tubulin β chain: G7J588), while this functional group was not significantly altered in the total drought comparison (D/C) (Figure 3 and Table 4). From the 13 proteins that changed in the systemic comparison (Figure 3) in soybean plants, only two were characterized: a transketolase (Glyma03g03200.1) that also altered in the partial drought comparison and a peptidyl-prolyl cis–trans isomerase (K7LSN8 and Glyma12g02790.2) that was shared between all of the comparisons (Table 4).
Discussion
Drought Tolerance of M. truncatula and G. max
The expansion of water-stressed areas as a result of an increased human population makes it essential to improve legume drought tolerance to ensure sustainable food production in the near future.1 Therefore, comparing the drought response of two economically important legumes is of interest to the research community. In this study, M. truncatula presented lower Ψw values compared to soybean plants in all treatments and in both organs, leaves, and nodules,10,13 indicating a higher tolerance of the amide-exporting legume to water deficit as previously suggested.14 Additionally, M. truncatula D plants showed greater stomatal closure and, consequently, enhanced control of water loss,10,13 something that could ultimately imply increased tolerance to more arid soils compared with soybean.
The decline of Ψnodule in the partial drought treatment showed a similar pattern in both legumes, although the drop in M. truncatula PDD nodules was higher and closer to the D treatment than the decline observed in soybean PDD Ψnodule.10,13 Differences in the nodule anatomy and metabolism of both legumes, determined in soybean and undetermined in M. truncatula, may explain the observed variations. Research on nodule transport under water deficit conditions, although somewhat scarce in the literature, could shed some light on the mechanism behind the observed differences.
Translating the M. truncatula Proteome to That of the Soybean
Despite some encouraging messages,23 it has been questioned whether the advances in M. truncatula “omics” can be applied to crop legumes.24,25 The genome conservation between M. truncatula and crop legumes has been examined26,27 and considerable synteny between M. truncatula and the pea (Pisum sativum)26,28 and between M. truncatula and G. max has been found.26,29,30 Our results show that both nodule proteomes are highly similar (Table 1 and Table S1), and that the distribution of proteins within the 20 functional groups in control conditions was comparable (Figure 1). A total of two main differences were observed in the nodule proteome of soybean plants: a large number of lipid and nucleotide metabolism-related proteins as well as numerous uncharacterized proteins (Figure 1). The large number of lipid metabolism proteins could be partly explained by the high number of lipoxygenases (LOX), proteins that catalyze the oxygenation of polyunsaturated fatty acids.31 In soybean plants, this family contains 19 genes, each encoding one particular subtype of LOX.32 Likewise, the higher number of nucleotide metabolism proteins identified may be related to the fact that soybean is a ureide-exporting legume. Ureide biosynthesis involves the incorporation of amino acids through the purine pathway to ultimately form ureides,33 in contrast to M. truncatula, an amide-exporting legume, that assimilates the reduced N2 into amino acids (for a review, see ref (34)).
Comparing the M. truncatula and soybean databases it can be seen that, although the soybean genome is almost completely sequenced6 and extensive work is being conducted for developing diverse molecular tools,35 the soybean protein databases are not fully annotated. The vast database comparison undertaken in our study and the finding of a high degree of similarity between protein sequences from GM-fasta and the better-annotated MT-fasta could facilitate the functional annotation of several so-far uncharacterized soybean proteins. Moreover, to the best of the authors’ knowledge, this is the first time the nodule proteome of grain and forage legumes has been compared. This could additionally serve to demonstrate the similarities of both proteomes and thus show that the extensive research into M. truncatula, as a model legume for the symbiotic research community,3,4 could be extended to economically important crop legumes such as soybean.
Local Nodule Proteome Regulation in the Drought-Stressed M. truncatula and Soybean
Previous studies showed that the inhibition of SNF in various legumes, including the pea, M. truncatula, and soybean, was local10,12,13 but the origin of the regulation, local or systemic, of diverse metabolic processes is still unknown. This proteomic study is aimed at clarifying whether the changes in the nodule proteome of soybean and M. truncatula are local or systemic in origin, and we discuss the similarities and differences between the two proteomes.
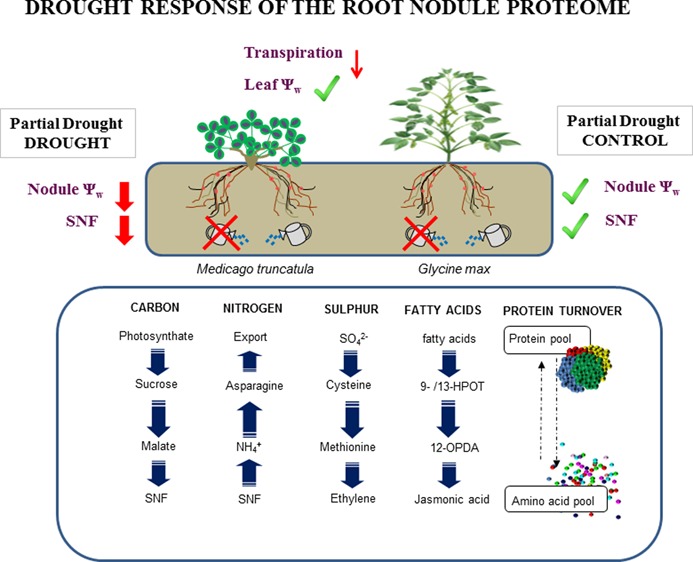
Within the set of proteins found to be highly similar in both species, several changed significantly under drought stress (Table 1 and Table S1). This reinforces the resemblances between the two legumes, not only at the proteome level and in the distribution of proteins among the functional groups (Figure 1) but also in the relative protein abundance changes in both legumes in response to drought. In this sense, in the two legumes there was a general local decrease (in the partial and total drought treatments) in nodule proteins after 7 days of water deprivation. Our research showed that carbon, nitrogen, and sulfur metabolism were the most altered processes in the drought-stressed nodules of M. truncatula and soybean plants. The global decline in the relative content of proteins linked with the glycolysis and TCA cycle in the total and partial drought treatments (Figure 3 and Tables 2 and 3), including SuSy, FBPA, PEPC, and MDH, among others, which catalyze the transformation of sucrose to malate to supply the bacteroids with carbon (for a review, see ref (34)), points to a reduction in carbon metabolism and, therefore, a diminution in the energy source for SNF in the drought-treated nodules of both legumes. The role of carbon metabolism in the regulation of SNF under drought stress has been reviewed elsewhere,36,37 and the decrease in activity of the cited proteins in diverse grain-legume nodules when water-deprived has been widely demonstrated.12,38−42 The proven key role of SuSy in the inhibition of SNF in drought-stressed soybean nodules43,44 seems secondary in forage legumes.16,39 In our results, three SuSys were significantly affected in D and PDD soybean nodules, as well as a further one in M. truncatula (Tables 2 and 3). Similarly, many amino-acid-metabolism-related proteins declined in the PDD and D nodules, denoting a down-regulation in nodule nitrogen assimilation when plants are subjected to drought stress. In addition to a decrease of primary nitrogen assimilation proteins, a local down-regulation of nodule proteins related to sulfur metabolism was recorded in both legumes (Tables 2 and 3). Cysteine biosynthesis in plants marks the connection between nitrogen and sulfur assimilation.45 Sulfur metabolism is known to be very active in legume nodules,46 and its involvement in the response to water deficit in M. truncatula nodules was suggested for the first time by.9 Here, the decrease in relative content of Cysteine synthase and S-adenosylmethionine synthase (SAMS) in D and PDD nodules in both legumes was measured (Tables 2 and 3) because this had been noted previously in diverse legume roots and nodules.9,38,47 SAMS, which responds to various stress conditions in plants,48 catalyzes the adenosylation of methionine to form S-adenosylmethionine, a precursor of polyamines and ethylene.49,50 It could be presumed that ethylene is reduced under drought stress, and that this could have possible implications in SNF signaling and regulation. However, the question of whether ethylene acts as a signaling mechanism to regulate SNF during drought stress remains unanswered.46
Together with the above-cited proteins, the relative content of LOX proteins declines significantly in the D and PDD nodules of both legumes (Tables 2 and 3). Furthermore, in M. truncatula, one of the nodule proteins that decreased the most after the water deprivation treatment was a LOX (G7LIY0) (Table 2 and Table S2). LOX proteins have been identified in numerous legume nodules,32,51−53 and it has been suggested that they play a role in nodule development.54 However, this is the first time LOX proteins have been implicated in the response of the nodule proteome to drought stress. LOXs catalyze the oxygenation of polyunsaturated fatty acids,31 which can be further metabolized into volatile aldehydes and jasmonates in plants.55 These molecules play important signaling roles in defense processes, responses to biotic and abiotic stresses, and in plant growth and development.56,57 It is known that nodules express a variety of LOXs in diverse tissues indicating their possible different functions.58 However, more research needs to be conducted to further understand the implication of LOX in the metabolism of legume nodules when subjected to drought stress.
The major differences in the responses of the two nodule proteomes to drought was the large number of SNF, protein synthesis, and degradation enzymes that changed significantly in M. truncatula and that were absent in soybean plants or were, at least, not annotated. Once again, it should be pointed out that the soybean database had a high number of uncharacterized proteins (Figure 3 and Table S3). The decline in protein biosynthesis components in drought-stressed nodules reinforces the hypothesis that amino acids accumulate in drought-stressed M. truncatula nodules due to their reduced incorporation into proteins.13
New Metabolic Arrangement in Nodules on the Watered Side of Legumes in Partial-Drought Conditions
Along with the local drought-driven proteome changes, the SRS allows the systemic changes occurring at the proteome level after a drought treatment to be revealed. Changes are considered systemic when significant variations in the PDC/C ratios are observed. In our work, there was no systemic reduction of M. truncatula and soybean PDC nodule proteins similar to that found for the local inhibition of the SNF process;10,13 instead, there was a significant increase in the relative content of the proteins in the PDC-treated nodules compared with C (Table 4).
The up-regulation of glycolysis- and TCA-related proteins in PDC nodules could be indicative of a higher energy demand and consumption on that side of the root system. Furthermore, it could be a response to guaranteeing the SNF and, therefore, the nitrogen supply required in the aerial part. Carbon consumption is limited in the drought-treated part of the root system, and this may favor a switch of photosynthate supply to the watered root of PD plants to enhance the PDC root system activity and, therefore, the nitrogen supply to the shoot. The active metabolism in PDC nodules could promote the growth of the root and nodules in response to the nitrogen demand of the aerial part, which can only be supplied by the PDC root system. Although not significant, the nodule dry weight for the PDC treatment was higher than in C conditions in both M. truncatula and soybeans (data not shown). The new metabolic arrangement proposed would require active plant cell growth. This suggestion is in line with the increase in PDC nodules when compared with C conditions of the relative content of diverse ribosomal proteins (G7ZUR7; G7J9M6; Contig_55945; G7JYX5; and G7J4S6), enzymes known to play important roles in protein synthesis, and a peptidyl-prolyl cis–trans isomerase (Glyma12g02790.2), which is related to protein folding and is implicated in stress-response signaling and tolerance59−61 (Table 4). Similarly, the quantity of diverse actin (G7IL85 and G7JAX5) and tubulin β chain proteins (G7J588) increased in PDC nodules when compared with C treatment. These components of the plant cytoskeleton are involved in several subcellular processes, including cell division, cell elongation, cell trafficking, and cell-wall formation.62−64 The increase of the relative content of the above-mentioned proteins in PDC nodules reflects the general adaptive response of the root system to the partial drought treatment applied to the other side of the root, pointing to an enhanced metabolism in the watered side of the PD root system aiming for cell growth.
Taken together, the M. truncatula and soybean proteomes had a high degree of similarity, and comparable responses in the diverse functional groups were identified in the SNF process that showed the same pattern of decline in both legumes, although M. truncatula seemed more tolerant to drought stress. In nodules directly subjected to drought stress, a local down-regulation of the metabolic processes was observed. Carbon, nitrogen, and sulfur metabolism were the most down-regulated processes, and SuSy, in soybean plants, and AS, in M. truncatula, were identified as being the enzymes showing the greatest relative abundance changes. Furthermore, new evidence in the regulation of SNF has come to light, such as the implication of LOX and protein turnover that seem to be important in the response of legume nodules to drought stress. Water deprivation in partial-drought conditions leads to a new metabolic arrangement in PDC nodules, increasing diverse metabolic processes involved in energy supply and cell growth. The high degree of similarity between M. truncatula and soybean proteomes suggests, in our view, that the vast amount of research conducted on M. truncatula could be extended to economically important crop legumes such as soybean.
Acknowledgments
We would like to thank Prof. Tomás Ruiz-Argüeso for providing the B. japonicum strain UPM752 and Xabier Sanz for aiding in nodule harvesting. D.L. and C.S. were funded by the Austrian Science foundation (FWF; P23441–B20). This work was financed by the Spanish Ministry of Economy and Competitiveness (AGL 2011-23738).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.5b00617.
List of highly similar proteins after a BLAST comparison between M. truncatula and G. max nodule proteins. (XLSX)
List of quantified proteins analyzed via shotgun nano-LC-ultra coupled to an Orbitrap XL mass spectrometer in M. truncatula root nodules and their spectral count values. (XLSX)
List of quantified proteins analyzed via shotgun nano-LC-ultra coupled to an Orbitrap XL mass spectrometer in G. max root nodules and their spectral count values. (XLSX)
The authors declare no competing financial interest.
Supplementary Material
References
- Graham P. H.; Vance C. P. Legumes: Importance and Constraints to Greater Use. Plant Physiol. 2003, 131, 872–877 10.1104/pp.017004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey G.; Vodkin L.; Parrott W. A.; Shoemaker R. C. National science foundation-sponsored workshop report. Draft plan for soybean genomics. Plant Physiol. 2004, 135, 59–70 10.1104/pp.103.037903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker D. G.; Bianchi S.; Blondon F.; Dattee Y.; Duc G.; Essad S.; Flament P.; Gallusci P.; Genier G.; Guy P.; Muel X.; Tourneur J.; Denarie J.; Huguet T. Medicago truncatula, a model plant for studying the molecular genetics of the Rhizobium-legume symbiosis. Plant Mol. Biol. Rep. 1990, 8, 40–49 10.1007/BF02668879. [DOI] [Google Scholar]
- Cook D. R. Medicago truncatula -a model in the making!. Curr. Opin. Plant Biol. 1999, 2, 301–304 10.1016/S1369-5266(99)80053-3. [DOI] [PubMed] [Google Scholar]
- Young N. D.; Debelle F.; Oldroyd G. E. D.; Geurts R.; Cannon S. B.; Udvardi M. K.; Benedito V. A.; Mayer K. F. X.; Gouzy J.; Schoof H.; Van de Peer Y.; Proost S.; Cook D. R.; Meyers B. C.; Spannagl M.; Cheung F.; De Mita S.; Krishnakumar V.; Gundlach H.; Zhou S.; Mudge J.; Bharti A. K.; Murray J. D.; Naoumkina M. A.; Rosen B.; Silverstein K. A. T.; Tang H.; Rombauts S.; Zhao P. X.; Zhou P.; Barbe V.; Bardou P.; Bechner M.; Bellec A.; Berger A.; Berges H.; Bidwell S.; Bisseling T.; Choisne N.; Couloux A.; Denny R.; Deshpande S.; Dai X.; Doyle J. J.; Dudez A.-M.; Farmer A. D.; Fouteau S.; Franken C.; Gibelin C.; Gish J.; Goldstein S.; Gonzalez A. J.; Green P. J.; Hallab A.; Hartog M.; Hua A.; Humphray S. J.; Jeong D.-H.; Jing Y.; Jöcker A.; Kenton S. M.; Kim D.-J.; Klee K.; Lai H.; Lang C.; Lin S.; Macmil S. L.; Magdelenat G.; Matthews L.; McCorrison J.; Monaghan E. L.; Mun J.-H.; Najar F. Z.; Nicholson C.; Noirot C.; O’Bleness M.; Paule C. R.; Poulain J.; Prion F.; Qin B.; Qu C.; Retzel E. F.; Riddle C.; Sallet E.; Samain S.; Samson N.; Sanders I.; Saurat O.; Scarpelli C.; Schiex T.; Segurens B.; Severin A. J.; Sherrier D. J.; Shi R.; Sims S.; Singer S. R.; Sinharoy S.; Sterck L.; Viollet A.; Wang B.-B.; Wang K.; Wang M.; Wang X.; Warfsmann J.; Weissenbach J.; White D. D.; White J. D.; Wiley G. B.; Wincker P.; Xing Y.; Yang L.; Yao Z.; Ying F.; Zhai J.; Zhou L.; Zuber A.; Denarie J.; Dixon R. A.; May G. D.; Schwartz D. C.; Rogers J.; Quetier F.; Town C. D.; Roe B. A. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 2011, 480, 520–524 10.1038/nature10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J.; Cannon S. B.; Schlueter J.; Ma J.; Mitros T.; Nelson W.; Hyten D. L.; Song Q.; Thelen J. J.; Cheng J.; Xu D.; Hellsten U.; May G. D.; Yu Y.; Sakurai T.; Umezawa T.; Bhattacharyya M. K.; Sandhu D.; Valliyodan B.; Lindquist E.; Peto M.; Grant D.; Shu S.; Goodstein D.; Barry K.; Futrell-Griggs M.; Abernathy B.; Du J.; Tian Z.; Zhu L.; Gill N.; Joshi T.; Libault M.; Sethuraman A.; Zhang X. C.; Shinozaki K.; Nguyen H. T.; Wing R. A.; Cregan P.; Specht J.; Grimwood J.; Rokhsar D.; Stacey G.; Shoemaker R. C.; Jackson S. A. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Colditz F.; Braun H.-P. Medicago truncatula proteomics. J. Proteomics 2010, 73, 1974–1985 10.1016/j.jprot.2010.07.004. [DOI] [PubMed] [Google Scholar]
- Hossain Z.; Khatoon A.; Komatsu S. Soybean proteomics for unraveling abiotic stress response mechanism. J. Proteome Res. 2013, 12, 4670–4684 10.1021/pr400604b. [DOI] [PubMed] [Google Scholar]
- Larrainzar E.; Wienkoop S.; Weckwerth W.; Ladrera R.; Arrese-Igor C.; Gonzalez E. M. Medicago truncatula root nodule proteome analysis reveals differential plant and bacteroid responses to drought stress. Plant Physiol. 2007, 144, 1495–1507 10.1104/pp.107.101618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Quintana E.; Larrainzar E.; Seminario A.; Diaz-Leal J. L.; Alamillo J. M.; Pineda M.; Arrese-Igor C.; Wienkoop S.; Gonzalez E. M. Local inhibition of nitrogen fixation and nodule metabolism in drought-stressed soybean. J. Exp. Bot. 2013, 64, 2171–2182 10.1093/jxb/ert074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprent J. I.Nodulation in Legumes; Royal Botanic Gardens: London, England, 2001. [Google Scholar]
- Marino D.; Frendo P.; Ladrera R.; Zabalza A.; Puppo A.; Arrese-Igor C.; Gonzalez E. M. Nitrogen fixation control under drought stress: localised or systemic?. Plant Physiol. 2007, 143, 1968–1974 10.1104/pp.107.097139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Quintana E.; Larrainzar E.; Arrese-Igor C.; Gonzalez E. M. Is N-feedback involved in the inhibition of nitrogen fixation in drought-stressed Medicago truncatula?. J. Exp. Bot. 2013, 64, 281–292 10.1093/jxb/ers334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair T. R.; Serraj R. Legume nitrogen-fixation and drought. Nature 1995, 378, 344–344 10.1038/378344a0. [DOI] [Google Scholar]
- Rigaud J.; Puppo A. Indole-3-acetic acid catabolism by soybean bacteroids. J. Gen. Microbiol. 1975, 88, 223–228 10.1099/00221287-88-2-223. [DOI] [Google Scholar]
- Larrainzar E.; Wienkoop S.; Scherling C.; Kempa S.; Ladrera R.; Arrese-Igor C.; Weckwerth W.; Gonzalez E. M. Carbon metabolism and bacteroid functioning are involved in the regulation of nitrogen fixation in Medicago truncatula under drought and recovery. Mol. Plant-Microbe Interact. 2009, 22, 1565–1576 10.1094/MPMI-22-12-1565. [DOI] [PubMed] [Google Scholar]
- Staudinger C.; Mehmeti V.; Turetschek R.; Lyon D.; Egelhofer V.; Wienkoop S. Possible role of nutritional priming for early salt and drought stress responses in Medicago truncatula. Front. Front. Plant Sci. 2012, 3, 285–285 10.3389/fpls.2012.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehenwarter W.; Wienkoop S. Spectral counting robust on high mass accuracy mass spectrometers. Rapid Commun. Mass Spectrom. 2010, 24, 3609–3614 10.1002/rcm.4818. [DOI] [PubMed] [Google Scholar]
- Carver T.; Bleasby A. The design of Jemboss: a graphical user interface to EMBOSS. Bioinformatics 2003, 19, 1837–1843 10.1093/bioinformatics/btg251. [DOI] [PubMed] [Google Scholar]
- Liu H.; Sadygov R. G.; Yates J. R. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004, 76, 4193–4201 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Zybailov B. L.; Florens L.; Washburn M. P. Quantitative shotgun proteomics using a protease with broad specificity and normalized spectral abundance factors. Mol. BioSyst. 2007, 3, 354–360 10.1039/b701483j. [DOI] [PubMed] [Google Scholar]
- Lohse M.; Nagel A.; Herter T.; May P.; Schroda M.; Zrenner R.; Tohge T.; Fernie A. R.; Stitt M.; Usadel B. Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data. Plant, Cell Environ. 2014, 37, 1250–1258 10.1111/pce.12231. [DOI] [PubMed] [Google Scholar]
- Young N. D.; Udvardi M. Translating Medicago truncatula genomics to crop legumes. Curr. Opin. Plant Biol. 2009, 12, 193–201 10.1016/j.pbi.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Sprent J. I.; James E. K. Legume–rhizobial symbiosis: an anorexic model?. New Phytol. 2008, 179, 3–5 10.1111/j.1469-8137.2008.02494.x. [DOI] [PubMed] [Google Scholar]
- Moreau D.; Voisin A.-S.; Salon C.; Munier-Jolain N. The model symbiotic association between Medicago truncatula cv. Jemalong and Rhizobium meliloti strain 2011 leads to N-stressed plants when symbiotic N2 fixation is the main N source for plant growth. J. Exp. Bot. 2008, 59, 3509–3522 10.1093/jxb/ern203. [DOI] [PubMed] [Google Scholar]
- Choi H. K.; Mun J. H.; Kim D. J.; Zhu H. Y.; Baek J. M.; Mudge J.; Roe B.; Ellis N.; Doyle J.; Kiss G. B.; Young N. D.; Cook D. R. Estimating genome conservation between crop and model legume species. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 15289–15294 10.1073/pnas.0402251101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon S. B.; Sterck L.; Rombauts S.; Sato S.; Cheung F.; Gouzy J.; Wang X.; Mudge J.; Vasdewani J.; Schiex T.; Spannagl M.; Monaghan E.; Nicholson C.; Humphray S. J.; Schoof H.; Mayer K. F. X.; Rogers J.; Quetier F.; Oldroyd G. E.; Debelle F.; Cook D. R.; Retzel E. F.; Roe B. A.; Town C. D.; Tabata S.; Van de Peer Y.; Young N. D. Legume genome evolution viewed through the Medicago truncatula and Lotus japonicus genomes. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 14959–14964 10.1073/pnas.0603228103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert G.; Morin J.; Jacquin F.; Loridon K.; Quillet M. C.; Petit A.; Rameau C.; Lejeune-Henaut I.; Huguet T.; Burstin J. Functional mapping in pea, as an aid to the candidate gene selection and for investigating synteny with the model legume Medicago truncatula. Theor. Appl. Genet. 2006, 112, 1024–1041 10.1007/s00122-005-0205-y. [DOI] [PubMed] [Google Scholar]
- Young N. D.; Cannon S. B.; Sato S.; Kim D.; Cook D. R.; Town C. D.; Roe B. A.; Tabata S. Sequencing the Genespaces of Medicago truncatula and Lotus japonicus. Plant Physiol. 2005, 137, 1174–1181 10.1104/pp.104.057034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H. H.; Mudge J.; Kim D. J.; Larsen D.; Shoemaker R. C.; Cook D. R.; Young N. D. Estimates of conserved microsynteny among the genomes of Glycine max, Medicago truncatula and Arabidopsis thaliana. Theor. Appl. Genet. 2003, 106, 1256–1265. [DOI] [PubMed] [Google Scholar]
- Porta H.; Rocha-Sosa M. Plant Lipoxygenases. Physiological and molecular features. Plant Physiol. 2002, 130, 15–21 10.1104/pp.010787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J. H.; Van K.; Kim D. H.; Do Kim K.; Jang Y. E.; Choi B. S.; Kim M. Y.; Lee S. H. The lipoxygenase gene family: a genomic fossil of shared polyploidy between Glycine max and Medicago truncatula. BMC Plant Biol. 2008, 8, 133. 10.1186/1471-2229-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P. M. C.; Atkins C. A. Purine biosynthesis. Big in cell division, even bigger in nitrogen assimilation. Plant Physiol. 2002, 128, 793–802 10.1104/pp.010912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodwig E.; Poole P. Metabolism of Rhizobium bacteroids. Crit. Rev. Plant Sci. 2003, 22, 37–78 10.1080/713610850. [DOI] [Google Scholar]
- Joshi T.; Fitzpatrick M. R.; Chen S.; Liu Y.; Zhang H.; Endacott R. Z.; Gaudiello E. C.; Stacey G.; Nguyen H. T.; Xu D. Soybean knowledge base (SoyKB): a web resource for integration of soybean translational genomics and molecular breeding. Nucleic Acids Res. 2014, 42, D1245–D1252 10.1093/nar/gkt905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrese-Igor C.; Gonzalez E. M.; Gordon A. J.; Minchin F. R.; Galvez L.; Royuela M.; Cabrerizo P. M.; Aparicio-Tejo P. M. Sucrose synthase and nodule nitrogen fixation under drought and other environmental stresses. Symbiosis. 1999, 27, 189–212. [Google Scholar]
- Arrese-Igor C.; González E. M.; Marino D.; Ladrera R.; Larrainzar E.; Gil-Quintana E. Physiological response of legume nodules to drought. Plant Stress 2011, 1, 24–31. [Google Scholar]
- Irar S.; Gonzalez E. M.; Arrese-Igor C.; Marino D. A proteomic approach reveals new actors of nodule response to drought in split root grown pea plants. Physiol. Plant. 2014, 152, 634–645 10.1111/ppl.12214. [DOI] [PubMed] [Google Scholar]
- Naya L.; Ladrera R.; Ramos J.; Gonzalez E. M.; Arrese-Igor C.; Minchin F. R.; Becana M. The response of carbon metabolism and antioxidant defenses of alfalfa nodules to drought stress and to the subsequent recovery of plants. Plant Physiol. 2007, 144, 1104–1114 10.1104/pp.107.099648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez L.; Gonzalez E. M.; Arrese-Igor C. Evidence for carbon flux shortage and strong carbon/nitrogen interactions in pea nodules at early stages of water stress. J. Exp. Bot. 2005, 56, 2551–2561 10.1093/jxb/eri249. [DOI] [PubMed] [Google Scholar]
- Gonzalez E. M.; Galvez L.; Arrese-Igor C. Abscisic acid induces a decline in nitrogen fixation that involves leghaemoglobin, but is independent of sucrose synthase. J. Exp. Bot. 2001, 52, 285–293 10.1093/jexbot/52.355.285. [DOI] [PubMed] [Google Scholar]
- Gonzalez E. M.; Aparicio-Tejo P. M.; Gordon A. J.; Minchin F. R.; Royuela M.; Arrese-Igor C. Water-deficit stress effects on carbon and nitrogen metabolism of pea nodules. J. Exp. Bot. 1998, 49, 1705–1714 10.1093/jxb/49.327.1705. [DOI] [Google Scholar]
- Gordon A. J.; Minchin F. R.; Skot L.; James C. L. Stress-induced declines in soybean N2 fixation are related to nodule sucrose synthase activity. Plant Physiol. 1996, 114, 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez E. M.; Gordon A. J.; James C. L.; Arrese-Igor C. The role of sucrose synthase in the response of soybean nodules to drought. J. Exp. Bot. 1995, 46, 1515–1523 10.1093/jxb/46.10.1515. [DOI] [Google Scholar]
- Hell R. Molecular physiology of plant sulfur metabolism. Planta 1997, 202, 138–148 10.1007/s004250050112. [DOI] [PubMed] [Google Scholar]
- Larrainzar E.; Molenaar J.; Wienkoop S.; Gil-Quintana E.; Alibert B.; Limami A.; Arrese-Igor C.; Gonzalez E. M. Drought stress provokes the down-regulation of methionine and ethylene biosynthesis pathways in Medicago truncatula roots and nodules. Plant, Cell Environ. 2014, 37, 2051–2063 10.1111/pce.12285. [DOI] [PubMed] [Google Scholar]
- Alam I.; Sharmin S. A.; Kim K.-H.; Yang J. K.; Choi M. S.; Lee B.-H. Proteome analysis of soybean roots subjected to short-term drought stress. Plant Soil 2010, 333, 491–505 10.1007/s11104-010-0365-7. [DOI] [Google Scholar]
- Azevedo R. A.; Lancien M.; Lea P. J. The aspartic acid metabolic pathway, an exciting and essential pathway in plants. Amino Acids 2006, 30, 143–162 10.1007/s00726-005-0245-2. [DOI] [PubMed] [Google Scholar]
- Ravanel S.; Gakiere B.; Job D.; Douce R. The specific features of methionine biosynthesis and metabolism in plants. Proc. Natl. Acad. Sci. U. S. A. 1998, 95, 7805–7812 10.1073/pnas.95.13.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roje S. S-Adenosyl-l-methionine: Beyond the universal methyl group donor. Phytochemistry 2006, 67, 1686–1698 10.1016/j.phytochem.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Porta H.; Rueda-Benitez P.; Campos F.; Colmenero-Flores J. M.; Colorado J. M.; Carmona M. J.; Covarrubias A. A.; Rocha-Sosa M. Analysis of lipoxygenase mRNA accumulation in the common bean (Phaseolus vulgaris L.) during development and under stress conditions. Plant Cell Physiol. 1999, 40, 850–858 10.1093/oxfordjournals.pcp.a029614. [DOI] [PubMed] [Google Scholar]
- Perlick A. M.; Albus U.; Stavridis T.; Fruhling M.; Kuster H.; Puhler A. The Vicia faba lipoxygenase gene VfLOX1 is expressed in the root nodule parenchyma. Mol. Plant-Microbe Interact. 1996, 9, 860–863 10.1094/MPMI-9-0860. [DOI] [PubMed] [Google Scholar]
- Gardner C. D.; Sherrier D. J.; Kardailsky I. V.; Brewin N. J. Localization of lipoxygenase proteins and mRNA in pea nodules: identification of lipoxygenase in the lumen of infection threads. Mol. Plant-Microbe Interact. 1996, 9, 282–289 10.1094/MPMI-9-0282. [DOI] [Google Scholar]
- Wisniewski J. P.; Gardner C. D.; Brewin N. J. Isolation of lipoxygenase cDNA clones from pea nodule mRNA. Plant Mol. Biol. 1999, 39, 775–783 10.1023/A:1006184516754. [DOI] [PubMed] [Google Scholar]
- Mosblech A.; Feussner I.; Heilmann I. Oxylipins: Structurally diverse metabolites from fatty acid oxidation. Plant Physiol. Biochem. 2009, 47, 511–517 10.1016/j.plaphy.2008.12.011. [DOI] [PubMed] [Google Scholar]
- Andreou A.; Feussner I. Lipoxygenases – Structure and reaction mechanism. Phytochemistry 2009, 70, 1504–1510 10.1016/j.phytochem.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Wasternack C. Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 2007, 100, 681–697 10.1093/aob/mcm079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S.; Gresshoff P. M.; Kinkema M. Molecular analysis of lipoxygenases associated with nodule development in soybean. Mol. Plant-Microbe Interact. 2008, 21, 843–853 10.1094/MPMI-21-6-0843. [DOI] [PubMed] [Google Scholar]
- Yu Y.; Zhang H.; Li W.; Mu C.; Zhang F.; Wang L.; Meng Z. Genome-wide analysis and environmental response profiling of the FK506-binding protein gene family in maize (Zea mays L.). Gene 2012, 498, 212–222 10.1016/j.gene.2012.01.094. [DOI] [PubMed] [Google Scholar]
- Sharma A. D.; Wajapeyee N.; Yadav V.; Singh P. Stress-induced changes in peptidyl-prolyl cis-trans isomerase activity of Sorghum bicolor seedlings. Biol. Plant. 2003, 47, 367–371 10.1023/B:BIOP.0000023879.74558.48. [DOI] [Google Scholar]
- Sharma A. D.; Singh P. Comparative studies on drought-induced changes in peptidyl prolyl cis-trans isomerase activity in drought-tolerant and susceptible cultivars of Sorghum bicolor. Curr. Sci. 2003, 84, 911–918. [Google Scholar]
- Schwarzerova K.; Vondrakova Z.; Fischer L.; Borikova P.; Bellinvia E.; Eliasova K.; Havelkova L.; Fiserova J.; Vagner M.; Opatmy Z. The role of actin isoforms in somatic embryogenesis in Norway spruce. BMC Plant Biol. 2010, 10, 89–89 10.1186/1471-2229-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassileva V. N.; Kouchi H.; Ridge R. W. Microtubule Dynamics in Living Root Hairs: Transient Slowing by Lipochitin Oligosaccharide Nodulation Signals. Plant Cell 2005, 17, 1777–1787 10.1105/tpc.105.031641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonoike H.; Han I. S.; Jongewaard I.; Doyle M.; Guiltinan M.; Fosket D. E. Hypocotyl expression and light downregulation of the soybean tubulin gene, tubB1. Plant J. 1994, 5, 343–351 10.1111/j.1365-313X.1994.00343.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.