Abstract
Trichoderma harzianum species are well known as biocontrol agents against important fungal phytopathogens. Mycoparasitism is one of the strategies used by this fungus in the biocontrol process. In this work, we analyzed the effect of Epl-1 protein, previously described as plant resistance elicitor, in expression modulation of T. harzianum genes involved in mycoparasitism process against phytopathogenic fungi; self cell wall protection and recognition; host hyphae coiling and triggering expression of defense-related genes in beans plants. The results indicated that the absence of Epl-1 protein affects the expression of all mycoparasitism genes analyzed in direct confrontation assays against phytopathogen Sclerotinia sclerotiorum as well as T. harzianum itself; the host mycoparasitic coiling process and expression modulation of plant defense genes showing different pattern compared with wild type strain. These data indicated the involvement T. harzianum Epl-1 in self and host interaction and also recognition of T. harzianum as a symbiotic fungus by the bean plants.
Chemical pesticides have long been used to protect crops from diseases. Fungicides and fumigants are often applied in greater quantities than herbicides and insecticides in agricultural practices and have adverse effects on the environment and human health. Thus, reduction or elimination of synthetic pesticide in agriculture is highly desirable. Hence, biocontrol agents are being used to control phytopathogenic fungi and to induce natural resistance in plants against these organisms. Further, these agents can be integrated with reduced chemical doses for controlling plant pathogens, resulting in minimal environmental impact1,2.
Proteins of the Cerato-platanin (CP) family are released during the early developmental stages of filamentous fungi. They can act as elicitors and induce defense responses in plants. For example Sm1 or Epl-1 proteins from Trichoderma virens and T. atroviride, respectively3,4,5,6 have been shown to induce phytoalexin production and/or cell death in host and non-host plants.7,8,9
Trichoderma species are widely spread used as biocontrol agents in agriculture and forestry; they can induce a combination of antagonist mechanisms such as antibiosis via the production of secondary metabolites having antifungal activity10,11,12,13; mycoparasitism, which involves the production of cell wall-degrading enzymes, competition for nutrients or space; and induction of resistance in plants via the production and secretion of elicitor molecules14,15,16. Among the Trichoderma species, T. harzianum is considered as one of the most effective biocontrol agents17.
In this study, the role of T. harzianum Epl-1 protein during the interaction with pathogens and plants was analyzed, especially its involvement in the mycoparasitism process. The effect of the absence of this elicitor on the induction of pathogen resistance in bean plants, as well as its role in cell wall protection and host and self-recognition was investigated.
Results
The in silico analyses of T. harzianum Epl-1: Features and probable regulation network
Our in silico analyses, revealed that T. harzianum epl-1 gene (ID: 508110; open reading frame (ORF), 417 bp; intron, 64 bp) encodes a protein of 138 amino acids. According to Expasy program ProtParam18, the protein has a theoretical molecular weight of 14,356.1 Da; and isoeletric point of 6.23 with an instability index of 21.74, suggesting that it is stable. The first 18 amino acid residues correspond to a secretion signal peptide. Analyses of Epl-1using NetOGlyc 3.1 software, indicated a potential O-glyosylation (Supplementary Figure 1), but no potential N-glycosylation sites (data not shown). In addition, seven potential O-(beta)-N-acetyl-glycosylation sites (YinOYang 1.2 software)19 (Supplementary Figure 2) and five phosphorylation sites (Supplementary Figure 3) were observed.
The presence of putative regulatory motifs in T. harzianum epl-1 was investigated by analyzing 2.105 bp region upstream of the ATG initiation codon and 2.000 bp downstream of the ATT termination codon by using SCOPE software20 (Supplementary Figure 4). Analysis of the 5′ region reveled two TATAA Box sites and fourteen CAAT box sites related to transcription initiation21. Only one of the four sequences required for the putative binding of regulatory proteins involved in mycoparasitism (MYC)22 (MYC1 5′-GCTTCA-3′) was found in the T. harzianum epl-1 promoter region. This region also contained six copies of consensus sequence (5′-SYGGRG - 3′) which is associated with the binding of Aspergillus nidulans carbon catabolic repressor CREA23,24. One copy of the motif 5′-HGATAR-3′ which mediates global nitrogen regulation25,26,27; three copies of motif 5′-GCCARG-3′, which is the recognition site for the pH regulatory protein PacC in Aspergillus spp21 and the pentanucleotide CCCCT (C4T) which is related to stress response element regulation in fungi22,28,29 were found in the epl-1 promoter. The analysis of the 3′ region allowed the identification of five putative polyadenylation sites without any TAAATAA motif.
T. harzianum epl-1 gene deletion, recovery and transformant selection
Knockout mutants of epl-1 were produced using double-crossover homologous recombination. Protoplasts of T. harzianum (ALL42) were transformed with a linear DNA sequence containing a hygromycin B expression cassette flanked by two DNA fragments homologous to the epl-1 5′ and 3′ regions. Polymerase chain reaction (PCR) analysis of the epl-1 gene region from mitotically stable transformants showed bands corresponding to the wild-type gene (1.090 bp), complete deletion cassette (3.457 bp) and the gene for hygromycin B resistance (hph 600 bp) (Supplementary Figure 5A, 5B, and 5C, respectively). Six transformants that showed bands only for the complete deletion cassette and hph were selected and one of them was subjected for Southern blot analysis (Supplementary Figure 6). The total DNA isolated and digested with EcoRV endonuclease showed two hybridization patterns for the wild-type strain (800 bp and approximately 1.500 bp; Supplementary Figure 6A,C) and one band pattern for the mutant strain (approximately 4.500 bp; Supplementary Figure 6B,C). The epl-1 deletion (∆epl-1) was also confirmed by real-time quantitative PCR (RT-qPCR) analysis. Subsequently, the mutant T. harzianum ∆epl-1 strain was complemented with the epl-1 ORF fused with the green fluorescent protein (GFP) gene. The T. harzianum epl-1 complemented (RecEpl-1-GFP) transformants were selected by their ability to grow in minimal medium, uridine and uracil free; the GFP signal in fluorescence microscopy images; PCR and electrophoreses of the amplification product of epl-1 encoding gene; and RT-qPCR analysis of the epl-1 relative expression from all Trichoderma strains, as described in Materials and Methods section (Supplementary Figure 7A–D).
Epl-1 protein is not essential for T. harzianum antagonistic potential
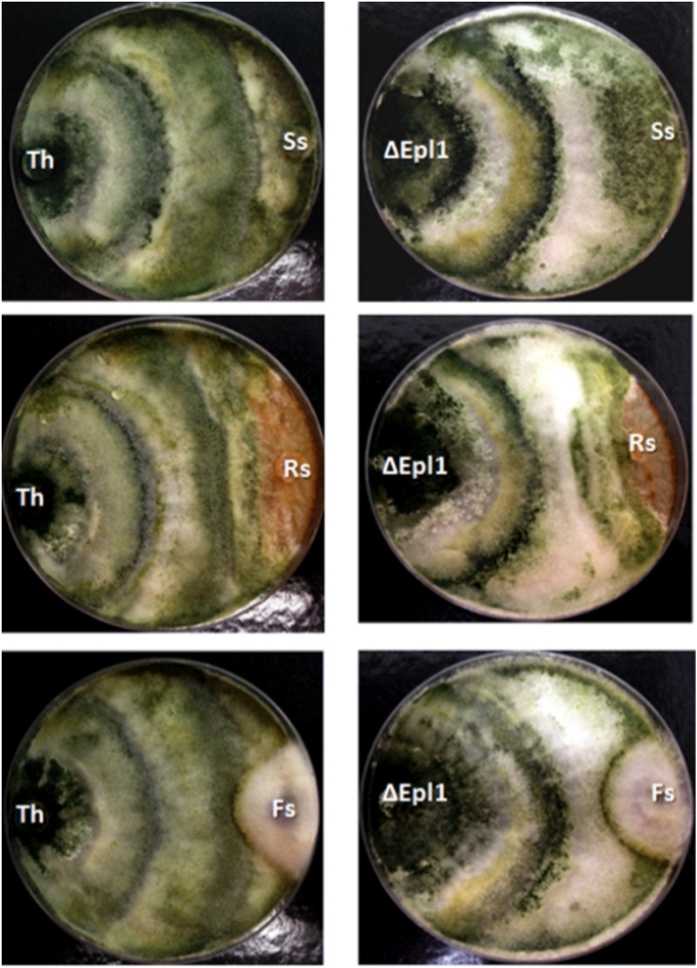
In vitro antagonistic activity of the Trichoderma strains against three plant pathogens (Sclerotinia sclerotiorum, Rhizoctonia solani and Fusarium solani) was analyzed as described by Bell et al.30 (modified method), and Trichoderma strains were classified as “efficient” to antagonize the plant pathogens if the mean score was <3, “moderate” if the mean score was 3 and “inefficient” if the mean score was >3 (Supplementary Figure 8). The deletion of epl-1 did not affect the ability of T. harzianum to antagonize plant pathogens, indicating that Epl-1 was not essential for this function. Wild-type and deletion mutants were efficient against all the pathogen strains tested, showing hyperparasitism signals; further a discrete antagonism improvement was noted in the mutant against R. solani compared to that of the wild-type strain (Fig. 1).
Figure 1. In vitro antagonistic potential assay.
(Th) T. harzianum wild type; (∆Epl1) T. harzianum ∆epl-1; (Ss) S. sclerotiorum; (Rs) R. solani; (Fs) F. solani.
T. harzianum Epl-1 protein is carried by vesicles and predominantly transported to the fungal cell wall
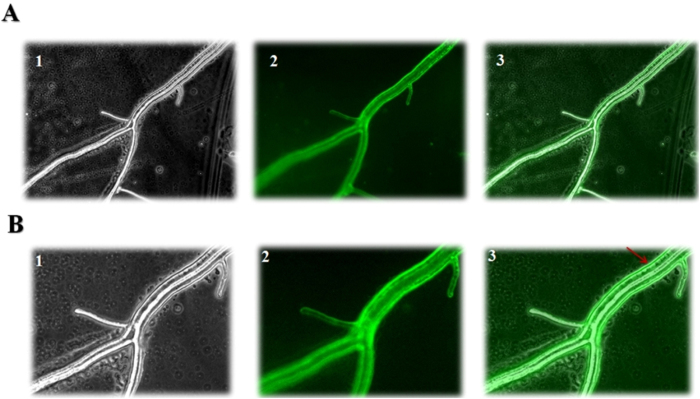
Fluorescence microscopy analyses of T. harzianum strain expressing Epl-1 fused with GFP showed a dynamic flow of vesicles containing Epl-1 through the Trichoderma hyphae in the presence of S. sclerotiorum. The vesicles fused with the plasma membrane and accumulated predominantly in the Trichoderma cell wall (Fig. 2 and Supplementary Videos 1 and 2).
Figure 2. Fluorescence microscopy analyses of Epl-1 fused with GFP.
(A) – Fluorescence microscopy of Trichoderma harzianum RecEpl-1-GFP hyphae in 20× optical magnification. 1 – Light field; 2 – GFP; 3 – Merge. (B) – Fluorescence microscopy of Trichoderma harzianum RecEpl-1-GFP strain hyphae in 40× optical magnification. 1 – Light field; 2 – GFP; 3 – Merge. The arrow indicates the predominance of Epl-1 in the cell wall region and its transport through the hyphae by vesicles.
T. harzianum Epl-1 protein is involved in hyphal recognition and mycoparasitic hyphal coiling process
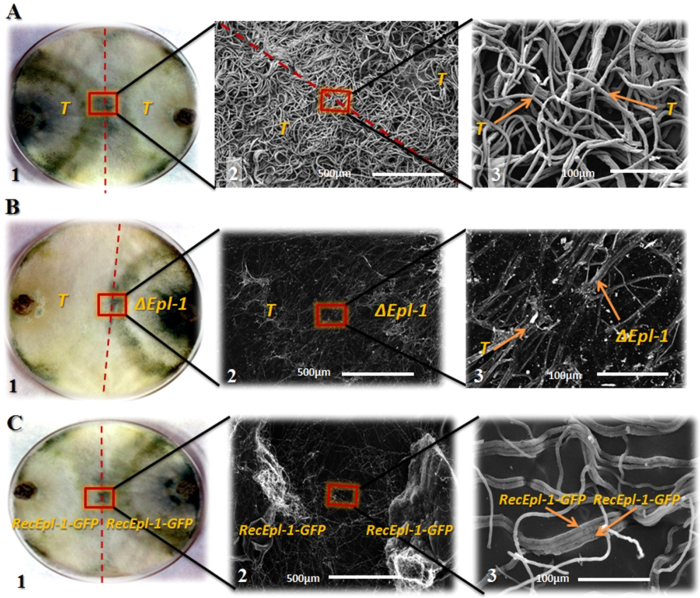
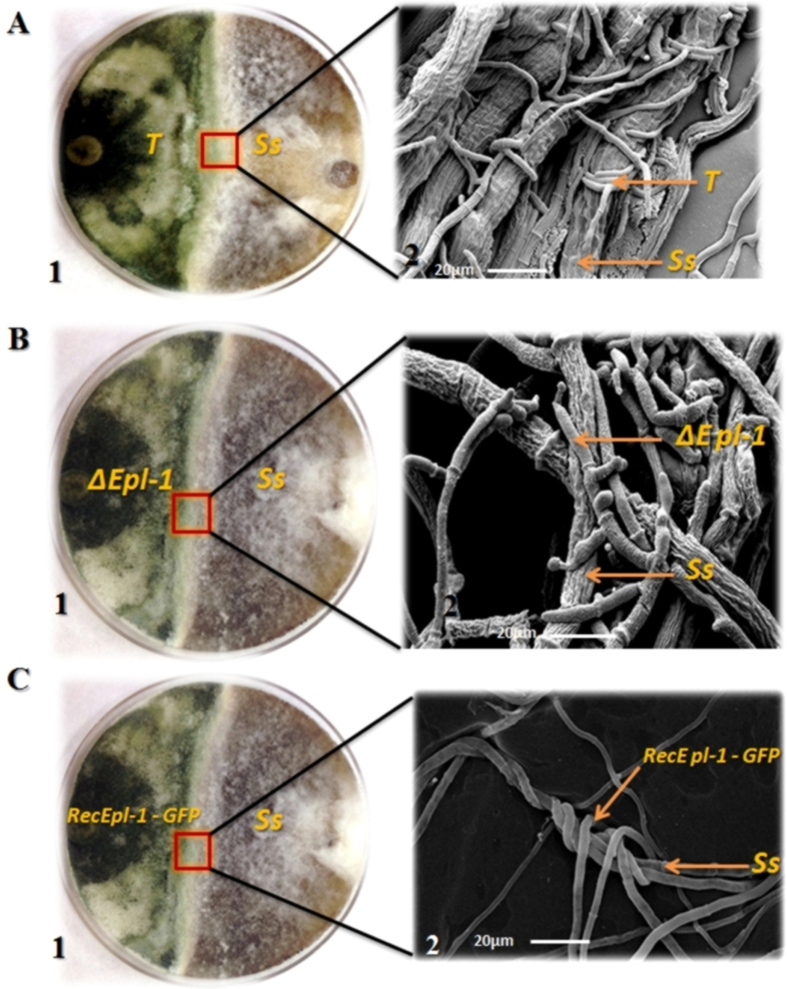
Interaction between T. harzianum strains and S. sclerotiorum was analyzed using scanning electron microscopy (SEM). In the T. harzianum wild-type interaction zone, a homogeneous mycelial mass without a clear division between both the isolates was observed with no hyphal degradation (Fig. 3A). In the interaction zone between wild-type T. harzianum and mutant ∆epl-1 strains, a clear interaction line separating both the strains and debris from probable enzymatic hyphal degradation were noted, indicating non-hyphal recognition between both the isolates (Fig. 3B). The self recognition feature was reestablished in RecEpl-1-GFP complemented strain, which showed parallel hyphal growth between the two strains and no degradation debris was noted in the wild-type interaction (Fig. 3C). In the interaction between wild-type T. harzianum and S. sclerotiorum, the typical Trichoderma mycoparasitism process with hyphal coiling around the host was observed (Fig. 4A). Interestingly, the mycoparasitical interaction process could be observed between T. harzianum ∆epl-1 and S. sclerotiorum; the typical Trichoderma hyphal coiling was not noted (Fig. 4B). Further, the coiling capacity was reestablished in the T. harzianum RecEpl-1-GFP-complemented strain (Fig. 4C), indicating that the Epl-1 interferes with the Trichoderma mycoparasitic process. Similar results were observed during the interaction between T. harzianum strains and R. solani (data not shown).
Figure 3. Scanning electron microscopy analysis (SEM).
(A) - SEM analysis of the interaction between T. harzianum wild type (T) strains. 1 – T. harzianum wild type dual culture plate. 2 – SEM analysis of T. harzianum wild type strains interaction. 2.5 kV acceleration voltage, magnification 250×. 3 – SEM analysis of T. harzianum wild type strains. 2.5 kV acceleration voltage, magnification 500×. (B) - SEM analysis of the interaction between T. harzianum wild type (T) and T. harzianum ∆epl-1 (∆epl-1) strains. 1 – T. harzianum wild type and mutant ∆epl-1 strains dual culture plate. 2 – SEM analysis of T. harzianum wild type and mutant ∆epl-1 interaction. 30 kV acceleration voltage, magnification 350×. 3 – SEM analysis of T. harzianum wild type and mutant ∆epl-1 interaction. 30 kV acceleration voltage, magnification 500×. (C) - SEM analysis of the interaction between T. harzianum RecEpl-1-GFP strains (RecEpl-1-GFP). 1 – T. harzianum RecEpl-1-GFP strains dual culture plate. 2 – SEM analysis of T. harzianum RecEpl-1-GFP strains interaction. 30 kV acceleration voltage, magnification 350×. 3 – SEM analysis of T. harzianum RecEpl-1-GFP strains interaction. 30 kV acceleration voltage, magnification 500×.
Figure 4. Scanning electron microscopy analysis (SEM).
(A) - SEM analysis of the interaction between T. harzianum wild type (T) and S. sclerotiorum (Ss) strains. 1 – T. harzianum wild type and S. sclerotiorum dual culture plate. 2 – SEM analysis of T. harzianum wild type and S. sclerotiorum strains interaction. 5 kV acceleration voltage, magnification 900×. (B) - SEM analysis of the interaction between T. harzianum ∆epl-1 (∆epl-1) and S. sclerotiorum (Ss) strains. 1 – T. harzianum ∆epl-1 and S. sclerotiorum dual culture plate. 2 – SEM analysis of T. harzianum ∆epl-1 and S. sclerotiorum strains interaction. 5 kV acceleration voltage, magnification 900×. C - SEM analysis of the interaction between T. harzianum RecEpl-1-GFP (RecEpl-1-GFP) and S. sclerotiorum (Ss) strains. 1 – T. harzianum RecEpl-1-GFP and S. sclerotiorum dual culture plate. 2 – SEM analysis of T. harzianum RecEpl-1-GFP and S. sclerotiorum strains interaction. 5 kV acceleration voltage, magnification 900×.
Absence of Epl-1 affects expression levels of mycoparasitism genes in direct confrontation
Direct confrontation assays were performed between T. harzianum and S. sclerotiorum strains. Mycelial RNAs from the confrontation area were extracted before (BC), during (C), and after (AC) hyphae contact (Supplementary Figure 9). The relative expression levels of epl-1 and eleven mycoparasitism gene markers were analyzed to determine the effect of Epl-1 on the modulation of mycoparasitism gene expression. In the confrontation between wild-type T. harzianum and mutant ∆epl-1 strains, eight genes were up-regulated, of which exg1, acid phosphatase, chit42, papA, and PRA1 were more prominent (Supplementary Figures 10 and 11). For the confrontation between wild-type and mutant ∆epl-1 Trichoderma strains and S. sclerotiorum, all analyzed genes showed remarkable difference in their expression levels, indicating an altered perception of the host and itself (Supplementary Figures 10 and 11). Gene expression was not significantly different (p > 0.05) between C and AC conditions (data not shown). The qPCR expression data were used to generate heat maps for determine the effects of epl-1 deletion on mycoparasitism gene expression modulation by using Mev MultiExperiment Viewer Software31. In the BC condition, despite the relatively stable gene expressions with slight variations, significant differences were noted in the expression modulation of genes in all the analyzed conditions (Fig. 5A). However, during AC, the difference in gene expression modulation became more evident, as observed between wild-type and mutant Trichoderma strains and Trichoderma vs. Sclerotinia confrontations (Fig. 5B). Another relevant finding was the gene clustering according to their expression pattern. For the BC and AC conditions three distinct groups were formed: Group 1 (PRA1 and sprT), Group 2 (epl-1 and gh92) and Group 3 (nag1 and chit33) for BC (the remaining genes showed different expression profiles; Fig. 5A). Group 1 (epl-1 and gh92); Group 2 (PRA1, acid phosphatase and chit42) and Group 3 (phyAT, papA, mutAW, nag1, and chit33) for AC (Fig. 5B).
Figure 5. Heat map and cluster categorization of T. harzianum mycoparasitism-related genes expression.
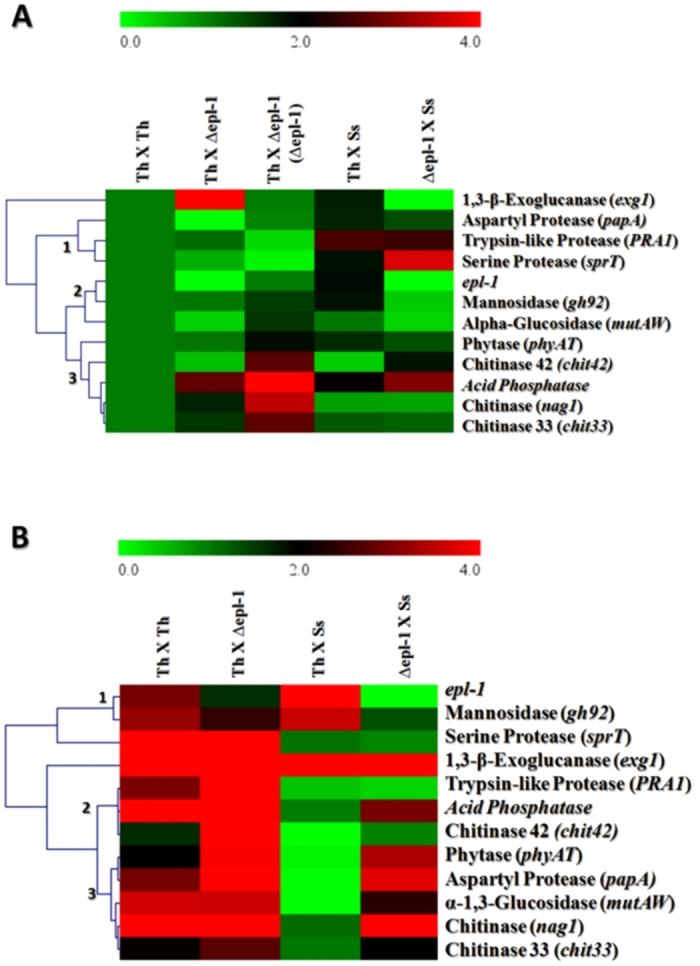
(A) – Heat map of qPCR expression analyzes of mycoparasitism genes before hyphae contact. (B) - Heat map of qPCR expression analyzes of mycoparasitism genes after hyphae contact. Th – T. harzianum wild type; ∆epl-1 – T. harzianum ∆epl-1; Ss – S. sclerotiorum.
Expression analysis of defense-related genes in bean plants was modified by Epl-1
In this study, the effects of T. harzianum Epl-1 in eliciting the expression of five defense-related genes in bean plants was analyzed 24 h after inoculation of both, wild-type and ∆epl-1 mutant T. harzianum strains. Plants without Trichoderma inoculation were used as controls. Plants inoculated with the Δepl-1 mutant and wild-type strains showed the up-regulation of lox and glu genes (19.1- and 6.5-fold respectively; Fig. 6). Trichoderma inoculation significantly induced pod3 gene at 72 h (data not shown).
Figure 6. T. harzianum ∆Epl-1Mutant/Wild Type Fold change analysis of defense-related genes expression (linear) in common bean plant roots 24 hours after T. harzianum strains inoculation.
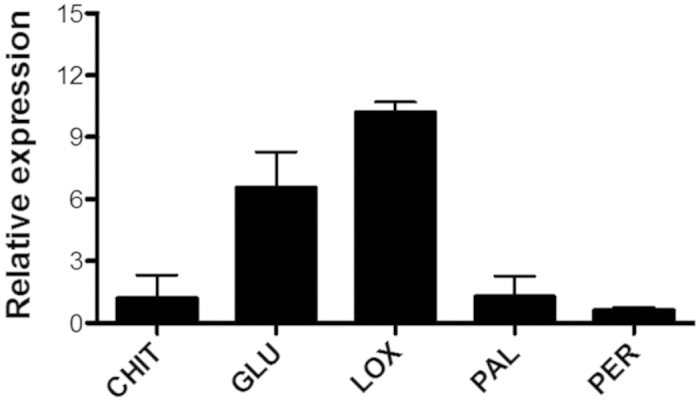
bch1 – chitinase; glu1 - β-1,3-glucanase; lox1 – lypoxygenase; pal – phenylalanine ammonia lyase; pod3 – peroxidase. The data were presented using the 2−ΔΔCt method.
Discussion
The biochemistry and genetics of the mycoparasitism process has been widely studied in several species of Trichoderma, because of their importance as pest biocontrol agents in agriculture. Various enzymes, proteins, and effectors involved in fungi recognition and mycoparasitc responses have been identified32,33,34. In this study, T. harzianum Epl-1 protein, a virulence factor that induces local and systemic defenses in plants, was found to be important during the interaction with itself and with different host organisms and played an essential role in the mycoparasitism process, by affecting T. harzianum self-recognition, regulating mycoparasitism-related gene expression, modulating mycoparasitic hyphal coiling and regulating plant defense-related gene induction.
Proteins of the CP family are involved in fungal growth, development, recognition, adhesion, cell-wall morphogenesis35, antagonism36 and parasitism37. However, data on the primary function of CPs are lacking. The CP member analyzed in this study showed 100% identity with Epl-1 from Trichoderma viride (Hypocrea rufa) and 94% identity with Sm1 from T. harzianum described by Freitas et al.38. Frischmann et al.36 suggested the presence of three epl genes in Trichoderma atroviride genome: epl-1 was predominantly expressed during hyphal growth, epl-2 was mainly expressed during the spore formation and epl3 was not highly expressed during the growth. They suggested that single and double gene knockouts of epl-1 and epl-2 did not remarkably affect the growth and development of the fungus. Their high homology across different species and the presence of some of them in fungal cell wall suggest that they might be associated with the lifestyle of fungi36. The epl-1 gene expression was strongly induced, especially during C condition, consistent with the findings for Epl-1 expression during the confrontation between T. atroviride and R. solani5. Thus, we constructed the T. harzianum ∆epl-1 mutant strain to determine the consequences of the lack of this protein during the interaction process with itself, host pathogens and plants. Furthermore, we constructed a T. harzianum epl-1-complemented strain (RecEpl-1-GFP) by using the Epl-1 protein fused with GFP for complementary analyses and to determine the Epl-1 cell localization and transportation dynamics under direct confrontation conditions.
Comparison of the DNA sequences flanking the promoter region of co-regulated genes might be useful to identify conserved regulatory sequences; such regions could be the binding sites for both; specific proteins and regulators22,39. Thus, we identify motifs and promoter elements that could be involved in epl-1 regulation and/or modification in T. harzianum. Cortés et al.22 described four motifs related to MYC in the promoter of ech42 and prb1 genes in T. atroviride (MYC1, MYC2, MYC3, and MYC4). Lorito et al.40, proposed a model containing the target sequences for carbon-response regulator CreA in Aspergillus nidulans (which correspond to Cre1 in T. harzianum) and showed that specific MYC complex genes overlap with or are close to each other. In this study, in silico analyses of the promoter region of T. harzianum epl-1 gene indicated one of the MYC related-motif (MYC1), which was present in four copies and always close to CreA sequence (Supplementary Figure 4); thus, it could be a target for a MYC regulation. Nonetheless, additional analyses will be necessary to confirm this. Furthermore, the presence of the stress response sequence (C4T)22,28,29 could explain the rapid induction of epl-1 during the direct confrontation between T. harzianum and S. sclerotiorum (Supplementary Figure 4), which corresponds to a stress condition and because of the presence of inducers from the pathogen cell wall, as described for other genes22,41. Another interesting in silico data were the seven O-(beta)-N-acetyl-glycosylation sites in Epl-1 protein (Supplementary Figure 2) proteins from the CP family exhibit a secretion peptide signal (the first 18 peptide residues) and oligosaccharide-binding activity and residues involved in this process are well conserved36,42. This suggested that Epl-1 might play a role in polysaccharide recognition and act directly in the interaction process with different hosts. Accordingly, time lapse fluorescence microscopy indicated the predominant presence of Epl-1 in the Trichoderma cell wall under confrontation conditions. Furthermore, Epl-1 hyphal transportation by vesicles in a highly dynamic protein flow, being strongly induced in the presence of pathogens (Fig. 2 and Supplementary Video 1 and 2). These data are in accordance with the qPCR analyses which indicated an up-regulation of epl-1 expression after hyphae contact in the direct confrontation assay with S. sclerotiorum (Supplementary Figure 10A).
Initially we did not observe phenotypic differences between wild-type and mutant T. harzianumT. harzianum ∆epl-1 strains, with regard to growth (dual culture; Fig. 1) and interaction (antagonistic potential) tests (Fig. 3A1), as was reported for other Trichoderma species36,43. However, microscopically the differences became evident, as showed by SEM analyses. Previous studies indicated that T. harzianum has different host recognition mechanisms, and the mycoparasitism process depend on the pathogen type and its cell wall composition34. The Trichoderma mycoparasitism process involves connection to the host cell wall carbohydrates and host lectins might be involved in the recognition process44. Once connected, the hyphae coil tightly around the host, forming mycoparasitism-related structures (e.g., hooks and appressorium-like bodies); next, cell wall-degrading enzymes are released45. In this study, SEM analyses showed excessive hyphal degradation in the confrontation analyses between wild-type T. harzianum and mutant ∆epl-1 strains, with a very well-defined degradation line in the interaction zone (but not in the wild type-wild type confrontation; Fig. 3A,B). However, hyphal degradation or debris was not observed during the confrontation between complemented strains (RecEpl-1-GFP; Fig. 3C). This finding, could be related to the non-Trichoderma hyphal recognition, which, as suggested by Cortes et al.22, would lead to the strong release of lytic enzymes such as (β-1,3-glucanases [exg1], chitinases [nag1, chit33, and chit42], and proteases [papA, PRA1, and sprT]), resulting in the digestion of the major cell wall components. This evidence can be supported by the fact that most of the analyzed mycoparasitism-related genes were up-regulated during the confrontation between wild-type and mutant ∆epl-1 Trichoderma strains after hyphal contact (Supplementary Figures 10 and 11). The altered expression of these genes became clear from the heat map generated from the qPCR data of T. harzianum strains that had confronted after hyphal contact (Fig. 5B), suggesting an altered perception of Trichoderma after coming in contact with the mutant ∆epl-1 strain and the host pathogen.
Considering the T. harzianum Epl-1 cell localization, probable regulation, and cell wall interaction, its function in mycoparasitism process can be speculated. First, Epl-1 might act as a recognition molecule to identify its own and/or host hyphae while avoiding self-degradation. Alternatively, this protein could have a protection function, such as fish scales, from host and its own degrading enzymes and other metabolites, thereby preserving its own cell wall integrity during confrontation against prey. Gruber & Seidl-Seiboth46 hypothesized that self and non-self fungal cell wall degradation is regulated by substrate accessibility: cell wall protection is noted in healthy hyphae but not during mycoparasitic attack, hyphal ageing, and autolysis. They argued that the accessibility of chitin within the fungal cell wall could be the major determinant, and the accessibility of healthy hyphae might be limited by the protection conferred by cell wall proteins. According to this theory, during mycoparasitism, the prey fungus is weakened by numerous secondary metabolites as well as hydrolytic enzymes which are highly destructive for the host hyphae and leading to partial cell death, thereby increasing the accessibility of prey’s cell wall carbohydrates. This leads to the enzymatic release of oligomers, mainly oligossacharides, which in turn induce the production of even more hydrolytic enzymes and produce greater damage to the host cell wall, as was observed during the confrontation between wild-type T. harzianum and ∆epl-1 mutant strains (Fig. 3B). Hydrophobic cell wall proteins and carbohydrate-binding proteins would be suitable candidates for affording protective functions, such as QID74 from T. harzianum47, which has been shown to be involved in cell protection and adherence to hydrophobic surfaces by acting like hydrophobin. Small cysteines-rich secreted proteins as well as carbohydrate-binding proteins were suggested to also be involved in aspects of the regulation of cell wall degradation and protection by binding to short oligosaccharides influencing the induction of cell wall-degrading enzymes or even by binding to chitin and masking and protecting it from degradation. Thus, Epl-1 might be a small cysteines-rich secreted protein that interacts with the cell wall and protects against enzyme degradation. Thus, the mycoparasitism-related genes could be higher induced during Trichoderma confrontations (Supplementary Figures 10 and 11), because the enzymes released in the interaction region that have more access to the mutant cell wall and degrade more substrate, thereby releasing more products that act as elicitors and induce the production of more enzymes that are important in mycoparasitism process22,48, such as β-1,3-exoglucanase and chitinases.
SEM analyses also showed that T. harzianum wild-type strain perfectly coiled around the S. sclerotiorum hyphae, as previously reported for other phytopathogens34,49, however although T. harzianum ∆epl-1 mutant strain could recognize and parasitize the host, the coiling structures could not be detected (Fig. 4B), indicating that the lack of Epl-1 also affected the mycoparasitism interaction. This is supported by the fact that the coiling ability was recovered by the RecEpl-1-GFP strain (Fig. 4C); however, the mechanism by which Epl-1 affects the mycoparasitic hyphal coiling process is yet unknown.
Numerous signaling molecules of microbial origin that initiate plant defense responses (elicitors) have been characterized50. Plant cells exposed to elicitors, respond with a battery of cellular changes related to defense51,52. Vargas et al.6, showed that Sm1 and Epl-1 monomers could undergo intermolecular interactions forming dimmers, resistant to treatment with SDS/β-mercaptoethanol (and other detergents at various temperatures), suggesting a covalent interaction between these subunits (not disulfide bonds). They also suggested that the monomeric form of Sm1 and Epl-1 proteins could activate defense responses in maize leaves, and no activity was detected for the dimeric form. Thus, the plant was speculated to have specific mechanisms for distinguishing between monomeric and dimeric forms or detecting special features of the monomeric form that are not exposed when the subunits are added. Biochemical characterizations of these proteins showed that two main factors might control their oligomeric state, and thus induce their activities, the first one being glycosylation. Presence of sugar molecules would provide steric hindrance to the protein subunit association, and prevent its dimerization. Thus, the potential O-glycosylation (Supplementary Figure 3) observed in the in silico analyses in this study indicated that T. harzianum Epl-1 could have this kind of dimerization control. Another factor that could affect the oligomeric state of these proteins is the oxidation of a tryptophan residue. Tryptophan oxidation is known to be involved in the aggregation and inactivation of some proteins51. Vargas et al.6 suggested that the dimeric form of the Epl-1 shows a highly conserved site of tryptophan residue in its oxidized state. In this study, the analysis of this conserved site with the alignment of T. harzianum Epl-1 protein sequence showed 100% identity with T. virens Sm-1 (YHWSTQGQIPR) and 91% identity with T. atroviride Epl-1 protein. The accumulation of reactive oxygen species in plant cells was thought to stimulate the oxidation of this site and thus its aggregation, thereby altering the recognition and activation of plant responses6.
The activation of plant defense mechanisms occurs via a succession of events and signals that begin with the plant recognition of pathogenic/nonpathogenic microorganisms and culminates in the formation of physical barriers and the activation of chemical responses, affording protection against invasion49. The plant defense responses include three main mechanisms: (a) hypersensitive response (HR), (b) systemic acquired resistance (SAR) that includes induction of pathogenesis-related (PR) proteins and (c) inducted systemic resistance (ISR), which includes production of compounds such as jasmonic acid and hydrogen peroxide53. The mechanisms used by Trichoderma species to trigger plant defense responses are not completely understood; however, studies concerning differential gene expression and alterations in protein expression patterns of leaves and roots from host plants during association with Trichoderma species have been performed3,54,55. The PR protein synthesis, involving peroxidases, chitinases, β-1,3-glucanases and lipoxygenase, can be considered as one of the most obvious alterations in plant-pathogen interaction. Other parallel infection responses include increased expression of genes related to the synthesis of phytoalexins and phenylalanine ammonia lyase (PAL), along with the deposition of lignin and increased levels of salicylic acid (SA)56. Pereira et al.57 described the interaction between T. harzianum with host Phaseolus vulgaris plants in the presence or absence of the phytopathogenic fungi F. solani and R. solani and showed that the Trichoderma strain could promote the growth of bean plants by increasing their overall size, foliar and root area, and the number of secondary roots, as well as by modifying their root system architecture. The authors also suggested that common bean plants challenged by T. harzianum showed differential expression patterns for defense response genes, compared to those in unchallenged plants and those challenged with F. solani or R. solani alone. In this study, we analyzed the effect of Epl-1 protein in eliciting defense response genes such as bch1, glu1, lox, and pod357, including a phenylalanine ammonia lyase (pal)-encoding gene. Our results also showed the up-regulation of lox and glu1, especially in the first 24 h after plants were challenged with Trichoderma strain49,53,57,58.
Interestingly, the mutant ∆epl-1 showed almost 20-fold higher lox expression than that of the wild-type strain (Fig. 5). Further, a small amount of β-1,3-glucanases is synthesized and secreted into the intercellular space; with the growth of the fungal parasite or symbiont in this space, the enzyme initiates the process of fungal cell wall degradation. When the invading fungus penetrates the plant cells the vacuoles are ruptured leading to the release of enzymes that inhibit the pathogen action59. Interestingly, this gene was strongly induced in mutant ∆epl-1 than in the wild-type strain (Fig. 5), indicating that Epl-1 was a cell wall protection protein. The up-regulation of glu1 in bean plants inoculated with T. harzianum ∆epl-1 compared to that in wild-type strain was consistent with finding in plants challenged with pathogens such as R. solani49,53,57,58. Thus, without the Epl-1 protein, the bean plants could recognize T. harzianum as a pathogen, since PR proteins such as β-1,3-glucanase (PR2) and chitinases (PR3), which are known to disrupt the fungal mycelial wall, can be induced by SA as well as by pathogenic attack3,60 or could also indicate a cross-talk between ISR and SAR defense pathways. The absence of Epl-1 protein in T. harzianum-plant system, found in this study, did not statistically affect the bch1 and pod3 gene expression, although a late pod3 up-regulation was noted, 72 h after T. harzianum strain inoculation (data not shown). Therefore, based on bch1 expression level, we can speculate that other CPs such as Epl-2 and Epl-3, could interact with the chitin and chitin-derived oligomers, where the chitin fragments released by the fungus, because of plant-microbe interaction would be captured by other CPs, according to their affinity61,62, thereby maintaining stable the bch1 expression level. Salas-Marina et al.63 showed that T. atroviride and T. virens could successfully induce systemic disease resistance in tomato accompanied by increased expression levels of SA defense-related genes; however, in our study, this gene was not or slightly affected in plants inoculated with mutant ∆epl-1 strain, suggesting that the induction of plant SA resistance system by T. harzianum strains is Epl-1 independent or that this mechanism acts on the basis of sensitivity rather than occurs because of the accumulation of PAL hormone.
Although the actual role of Epl-1 in Trichoderma physiology is not completely clear, the information available about this protein and the data generated in this study suggest that Epl-1 plays a fundamental role in the successful Trichoderma adaptation to its environment, especially during the interaction with fungal pathogens and plants. Thus, a better understanding of proteins, such as Epl-1 and their role in fungal interaction with themselves and hosts will be necessary for the improvement of strategies to combat plant pathogens and to produce crops that are more resistant to phytopathogenic organisms and to reduce the environmental pollution caused by pesticides and other chemical compounds.
Methods
Microorganisms and culture conditions
T. harzianum (ALL42), F. solani and R. solani isolates were provided by the Department of Biochemistry and Molecular Biology, Biochemistry Laboratory, Federal University of Goiás, Goiânia, Goiás, Brazil. S. sclerotiorum isolate was provided by Embrapa (Brazilian Company of Agronomic Development - Rice and Bean Unit, Santo Antônio de Goiás, Brazil), maintained with periodical sampling on potato dextrose agar (PDA), and stored at 4°C before use.
Generation of ∆epl-1T. harzianum strains
Knockout strains were generated from T. harzianum by using primers that are listed in Table 1. For construction of the epl-1 deletion vector, 950 bp from promoter (Epl-1 P) and 1.000 bp from terminator (Epl-1 T) flanking regions of epl-1 were amplified using genomic DNA as template. The PCR products were digested with the appropriate restriction enzymes and cloned into a pBS (pBluescript SK+) vector (Stratagene, La Jolla, CA) containing a selection hph cassette (hph gene, conferring resistance to hygromycin, under the control of Aspergillus nidulans gpdA promoter and cbh2 terminator)71 (Supplementary Figure 12A,B). The pBShphEpl-1 final vector (as a DNA template) and the more external primer set (Epl1KpnIF and Epl1BamHIR) were used to generate the linear epl-1 deletion cassette. Fungal transformation was performed using protoplast generation as previously described64. Transformants were selected using hygromycin B (100 μg/mL) resistance. The selected transformants were analyzed using PCR and by Southern hybridization (using MSEpl-1 primers set; Table 1) to detect transformants with the pPBShphEpl-1 vector inserted into the epl-1 gene.
Table 1. Primers for T. harzianum Transformation and Transformants Screening.
| PRIMER | TARGET REGION | SEQUENCE | AMPLICON (bp) |
|---|---|---|---|
| Epl1KpnIF | Epl-1 Promoter | ATGGGTACCGCTGTGGCGATGCTTCTCCAAGT | |
| Epl1XhoIR | Epl-1 Promoter | ATGCTCGAGTTTGCCTCCTCCCTTC | 950 |
| Epl1HindIIIF | Epl-1 Terminator | ATGAAGCTTGGTTATTGCATTGCATTGTG | |
| Epl1bBamHIR | Epl-1 Terminator | ATGGGATCCTCCTTTGCCATTGCAGAATC | 1000 |
| MSEpl1F | Epl-1 ORF | TTGGCGATGGCTGGATACTACGAT | WT: 1090 |
| MSEpl1R | Epl-1 ORF | TGCCACGTCGATCAGCTTACAAGA | ∆epl-1: 3457 |
| HphF | Hygromycin ORF | GCGGAGGCCATGGATGCGAT | |
| HphR | Hygromycin ORF | GCGCTTCTGCGGGCGATTTG | 596 |
| RecEpl-1MSF | Epl-1 ORF | ATGCAATTGTCCAGCCTC | |
| RecEpl-1MSF | Epl-1 ORF | CTTGACAGCGACCTGGC | 466 |
Complementation of T. harzianum mutant strain with reintegration of the epl-1 fused with GFP-encoding genes
The Epl-1::GFP cassette was constructed according to Colot et al.; Goldman et al.; Colabardini et al.65,66,67 (Supplementary Figure 7A). The GFP signal was detected in fluorescence microscopy images (×40 magnification) under green fluorescence obtained by illuminating with 480 nm light elicited during the light flash (750 ms duration; Supplementary Figure 7B). PCR of the epl-1 gene using the primer set RecEpl-1MS (Table 1) generated 366-bp fragments from both wild-type and complemented T. harzianum strains (no amplification from T. harzianum ∆epl-1 mutant strain; Supplementary Figure 7C). The RT-qPCR analyses of the epl-1 relative expression were made using the wild-type, mutant ∆epl-1 and RecEpl-1-GFP T. harzianum strains (Supplementary Figure 7D).
Direct confrontation assays
The antagonism of Trichoderma strains against the three phytopathogenic fungi (S. sclerotiorum, R. solani, and F. solani) was detected by taking discs of potato dextrose agar (PDA) medium from the edge of actively growing colonies of fresh fungal cultures and placing 8-cm apart on the surface of a fresh PDA plate. The plates were incubated at 25°C with a photoperiod of 12 h. The evaluation was performed when the pathogen completely covered the control plate without Trichoderma according to the classification proposed by Bell et al.30 with the following modification. Briefly, grade 1-Trichoderma completely overgrew the pathogen and covered the entire medium surface; grade 1.5-Trichoderma occupied 7/8th of the medium surface, grade 2-Trichoderma occupied 6/8th of the medium surface; grade 2.5-Trichoderma occupies 5/8th of the medium surface; grade 3-Trichoderma occupied approximately half of the medium surface; grade 3.5-Trichoderma occupied 3/8th of the medium surface; grade 4-Trichoderma occupied 1/3th of the medium surface; grade 5-the pathogen completely overgrew the Trichoderma strain. The experiment was conducted with three repetitions for each Trichoderma strain. Mycelia from the interacting zones were collected by performing direct confrontation assays on PDA plates covered with sterile cellophane sheets. T. harzianum (wild-Type) and T. harzianum ∆epl-1 were tested against S. sclerotiorum isolate. Agar plugs cut from growing colonies of each fungus were placed on the opposite sides in 9-cm diameter plates and incubated at 28 °C with a 12 h/12 h light/dark cycle68. Mycelia were collected before they contacted each other (after 3 days), when they were just touching (after 4 days), and when the interaction zone was approximately 1-cm wide (after 7 days). The confrontation assays were performed as follows: (1) T. harzianum (wt) × T. harzianum (wt) as control confrontation plate, (2) T. harzianum (wt) × S. sclerotiorum, (3) T. harzianum (wt) × T. harzianum-∆epl-1, and (4) T. harzianum-∆epl-1 × S. sclerotiorum.
Dual culture tests and scanning electron microscopy analysis (SEM)
Mycelial samples and SEM analyses were performed as described in Monteiro et al.69.
Fluorescence microscopy
Fresh spores from T. harzianum RecEpl-1-GFP strain culture and a disc from PDA medium (5-mm diameter) containing S. sclerotiorum mycelium were inoculated in PDA medium for hyphal development. Small pieces of PDA medium containing T. harzianum and S. sclerotiorum mycelium were placed on the opposite sides of the microscope plate covered with a very thin water/agar (2% w/v) film and placed in a chamber for overnight growth at 25°C and 60% humidity. A microscopy plate without S. sclerotiorum was used as a control. The Trichoderma development and the Epl-1-GFP dynamics were monitored in the presence or absence of S. sclerotiorum by using the BioStation IMq (Nikon, Japan) cell incubator and BioStation software version 2.1. The samples were monitored during 24 h at 32 °C and 60% humidity. The images were captured every 10 min at ×20 and ×40 magnification, in the light field and green fluorescence, which was obtained by illuminating with 480 nm light, elicited during the light flash (duration 750 ms). The images were edited, and the movies were obtained using ImageJ Software version 1.48 (National Institute of Health, United States).
DNA/RNA manipulations
All DNA manipulations were performed by standard techniques74. The Southern hybridization was performed as described in Cardoza et al.10. The probe was produced from the PCR-amplified fragment by using MSEpl1 primers set (Table 1). Total fungal RNA was extracted from the mycelia of each sample by using TRIzol® RNA kit (Invitrogen Life Technologies, Carlsbad, CA, USA), according to the manufacturer’s instructions. RNA concentrations were determined by spectrophotometry at OD 260/280, and RNA integrity was verified using 1% agarose gel electrophoresis. Manipulations and analyses of RNA from bean plants was performed as described in Pereira et al.70
qRT-PCR analysis
Expression analysis of T. harzianum mycoparasitism-related genes
Approximately, 0.5 μg of RNA from the direct confrontation assay (described in section 2.3) was isolated and treated with DNase I (Thermo Scientific, Fermentas, Vilnius, Lithuania) and reverse-transcribed to cDNA by using the First strand cDNA kit Maxima™ Synthesis according to manufacturer’s instructions. cDNAs were diluted 1:50 and used for real-time PCR analysis in the Bio-Rad CFX96™ System by using SsoFast™ Eva Green® Supermix (Bio-Rad, San Francisco, CA, USA) for signal detection in accordance with the manufacturer’s instructions. The actin gene (act) was used as an endogenous control (housekeeping)71. Twelve genes were analyzed: epl-1; 1,3-β-exoglucanase (exg1); manosidase (gh92); α-1,3-glucosidase (mutAW); acid phosphatase; phytase (phyAT); chitinase (nag1); chitinase 42 (chit42); chitinase 33 (chit33); aspartyl protease (papA); trypsin-like protease (PRA1); and serine protease (sprT). Primers sequences are described by Steindorff et al.72. The following amplification program was used: 95 °C for 10 min followed by 39 cycles of 95 °C for 10 s, and 60 °C for 30 s followed by a dissociation curve of 60 °C to 95 °C with an increment of 0.5 °C for 10 s. Gene expression values were calculated according to the 2−ΔΔCt method73 by using T. harzianum x T. harzianum (no contact) as a reference sample. Gene expression analyses were performed using three technical and biological replicates. Data analysis was performed using GraphPad Prism v 5.1 software. The results were compared by one-way analysis of variance (ANOVA) with Bonferroni test (α = 5%) to analyze the differences between the related conditions and the reference sample.
Expression analysis of defense genes in bean plants
P. vulgaris growth was assessed in triplicate using 10 plants for each experimental replicate under two conditions (with T. harzianum WT-control condition; with T. harzianum ∆epl-1), totaling 60 plants. Seeds of common bean (P. vulgaris), were surface sterilized by immersion in sodium hypochlorite solution at 1% of active chlorine for 7 min, followed by three autoclaved water washes, 1 min each. After sterilization, the seeds were germinated in sterile glass Petri dishes containing 2% agar and 2% glucose, which were previously autoclaved. The seeds were kept in a germination chamber BOD at 28°C with a photoperiod of 16 h light/8 h dark. After four days, the bean seeds were placed in a hydroponic cultivation system3. Adapted 50 mL conical tubes were placed in 250 mL flasks containing 50 mL of Murashige and Skoog (MS), and Gamborg’s Vitamin Solution prepared according to the manufacturer’s instruction (Sigma-Aldrich, Co., Wisconsin, USA), and sealed with plastic wrap. This system was developed to allow germination, growth, and interaction of bean plants with Trichoderma strains, and the seedling contacted with the medium with partial immersion. Seeds were again grown with a photoperiod of 16 h light for 4 days in a germination chamber BOD with gentle agitation. Subsequently, the medium was replaced by fresh MS medium plus 0.05% sucrose and inoculated with 107 spores/mL of each Trichoderma strain directly on the seedlings roots. The leaves and roots were collected 24, 48, and 72 h after inoculation, and stored at −80 °C until RNA extraction. A group of plants grown in the absence of fungal isolates was used as a control.
Additional Information
How to cite this article: Vieira Gomes, E. et al. The Cerato-Platanin protein Epl-1 from Trichoderma harzianum is involved in mycoparasitism, plant resistance induction and self cell wall protection. Sci. Rep. 5, 17998; doi: 10.1038/srep17998 (2015).
Supplementary Material
Acknowledgments
This work was supported by the State of São Paulo Research Foundation (FAPESP) (Proc. 2012/16895-4).
Thanks to Multiphoton microscopy laboratory and Multiuse electronic microscopy laboratory of cell biology, molecular and pathogenic bioagents department of Ribeirão Preto Medical School – São Paulo University - USP.
Multiuse High Resolution Microscopy (LabMic), Universidade Federal de Goiás - UFG. Dr. Gustavo Henrique Goldman, Faculty of Pharmaceutical Sciences of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, and his group for support in the construction of GFP-Transformant.
Footnotes
Author Contributions E.V.G. performed the experimental design, laboratory experiments, bioinformatics analysis, and drafted the manuscript. M.N.C. and R.R.A. performed the fluorescence microscopy. R.G.P. performed the complementation of T. harzianum mutant strain with reintegration of the epl-1 fused with G.F.P.-encoding genes. F.L.S. and E.F.N. performed the expression analysis of defense genes in bean plants. C.J.U. and V.N.M. performed the SEM analyses, interpreted the data for the work and revised the manuscript. R.E.C. and S.G. performed the southern blot analyses, interpreted the data for the work and revised the manuscript. R.N.S. designed the project, supervised the research study, interpreted the data for the work, drafted/revised the manuscript, and final approved of the version to be published. All the authors have read and approved the final manuscript.
References
- Chet I. & Inbar J. Biological control of fungal pathogens. Appl. Biochem. Biotechnol. 48, 37–43 (1994). [DOI] [PubMed] [Google Scholar]
- Harman G. E. & Kubcek C. P. Enzymes, biological control and commercial applications : Trichoderma and Gliocladium. CRC Press. Taylor & Francis Ltd. London, United Kingdom. 2, 393 (1998). [Google Scholar]
- Djonovi S., Pozo M. J., Dangott L. J., Howell C. R. & Kenerley C. M. Sm1 , a Proteinaceous Elicitor Secreted by the Biocontrol Fungus Trichoderma virens Induces Plant Defense Responses and Systemic Resistance. Mol. Plant. Microbe. Interact. 19, 838–853 (2006). [DOI] [PubMed] [Google Scholar]
- Djonovic S. et al. A proteinaceous elicitor Sm1 from the beneficial fungus Trichoderma virens is required for induced systemic resistance in maize. Plant Physiol. 145, 875–89 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl V., Marchetti M., Schandl R. & Kubicek C. P. Epl1, the major secreted protein of Hypocrea atroviridis on glucose , is a member of a strongly conserved protein family comprising plant defense response elicitors. FEBS J. 273, 4346–4359 (2006). [DOI] [PubMed] [Google Scholar]
- Vargas W. a., Djonović S., Sukno S. a. & Kenerley C. M. Dimerization controls the activity of fungal elicitors that trigger systemic resistance in plants. J. Biol. Chem. 283, 19804–15 (2008). [DOI] [PubMed] [Google Scholar]
- Pazzagli L. et al. Purification, Characterization, and Amino Acid Sequence of Cerato-platanin, a New Phytotoxic Protein from Ceratocystis fimbriata f. sp. platani. J. Biol. Chem. 274, 24959–24964 (1999). [DOI] [PubMed] [Google Scholar]
- Scala A. et al. Cerato-platanin, an early-produced protein by Ceratocystis fimbriata f. sp. platani, elicits phytalexin synthesis in host and non-host plants. J. Plant Pathol. 86, 27–33 (2004). [Google Scholar]
- Boddi S. et al. Cerato-platanin protein is located in the cell walls of ascospores, conidia and hyphae of Ceratocystis fimbriata f. sp. platani. FEMS Microbiol. Lett. 233, 341–6 (2004). [DOI] [PubMed] [Google Scholar]
- Cardoza R. E. et al. Identification of Loci and Functional Characterization of Trichothecene Biosynthesis Genes in Filamentous Fungi of the Genus Trichoderma. Appl. Environ. Microbiol. 77, 4867–4877 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijerino A. et al. Overexpression of the trichodiene synthase gene tri5 increases trichodermin production and antimicrobial activity in Trichoderma brevicompactum. Fungal Genet. Biol. 48, 285–96 (2011). [DOI] [PubMed] [Google Scholar]
- Malmierca M. G. et al. Involvement of Trichoderma trichothecenes in the biocontrol activity and induction of plant defense-related genes. Appl. Environ. Microbiol. 78, 4856–68 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmierca M. G. et al. Relevance of trichothecenes in fungal physiology: disruption of tri5 in Trichoderma arundinaceum. Fungal Genet. Biol. 53, 22–33 (2013). [DOI] [PubMed] [Google Scholar]
- Monteiro V. N. & Ulhoa C. J. Biochemical characterization of a beta-1,3-glucanase from Trichoderma koningii induced by cell wall of Rhizoctonia solani. Curr. Microbiol. 52, 92–6 (2006). [DOI] [PubMed] [Google Scholar]
- Almeida F. B. D. R., Cerqueira F. M., Silva R. D. N., Ulhoa C. J. & Lima A. L. Mycoparasitism studies of Trichoderma harzianum strains against Rhizoctonia solani: evaluation of coiling and hydrolytic enzyme production. Biotechnol. Lett. 29, 1189–93 (2007). [DOI] [PubMed] [Google Scholar]
- Lopes F. A. C. et al. Biochemical and metabolic profiles of Trichoderma strains isolated from common bean crops in the Brazilian Cerrado, and potential antagonism against Sclerotinia sclerotiorum. Fungal Biol. 116, 815–824 (2012). [DOI] [PubMed] [Google Scholar]
- Exiang G. K., Iaoguang L. X., Onghong L. Y., Ianbo Z. T. & Huliang W. S. Potential of Trichoderma harzianum and T. atroviride to Control Botryosphaeria berengeriana f . sp . piricola , the Cause of Apple Ring Rot. J. Phytopathol. 276, 271–276 (2002). [Google Scholar]
- Artimo P. et al. (2012). ExPASy: SIB bioinformatics resource portal. URL: http://www.expasy.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta Ramneek & Brunak S. (2002). YinOYang: predictions for O-β-GlcNAc attachment sites in eukaryotic protein sequences. CBS Prediction Servers. URL: www.cbs.dtu.dk/services/YinOYang/ [Google Scholar]
- Carlson J. M., Chakravarty A., DeZiel C. E. & Gross R. H. (2007). SCOPE: a web server for practical de novo motif discovery. URL: http://genie.dartmouth.edu/scope [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilburn J. et al. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline- expressed genes by ambient pH. EMBO J. 14, 779–790 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes C. et al. The expression of genes involved in parasitism by Trichoderma harzianum is triggered by a diffusible factor. Mol. Gen. Genet. 260, 218–225 (1998). [DOI] [PubMed] [Google Scholar]
- Kulmburg P., Mathieu M., Dowzer C., Kelly J. & Felenbok B. Specific binding-sites in the alcR and alcA promoters of the ethanol regulon for the CREA repressor mediating carbon catabolite repression in Aspergillus nidulans. Mol. Microbiol. 7, 847–857 (1993). [DOI] [PubMed] [Google Scholar]
- Sophianopoulou V., Suarez T., Diallinas G. & Scazzocchio C. Operator derepressed mutations in the proline utilisation gene cluster of Aspergillus nidulans. Mol. Gen. Genet. 236, 209–213 (1993). [DOI] [PubMed] [Google Scholar]
- Ravagnani A. et al. Subtle hydrophobic interactions between the seventh residue of the zinc finger loop and the first base of an HGATAR sequence determine promoter-specific recognition by the Aspergillus nidulans GATA factor AreA. EMBO J. 16, 3974–86 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla B. et al. The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J. 9, 1355–1364 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minehart P. L. & Magasanik B. Sequence and expression of GLN3, a positive nitrogen regulatory gene of Saccharomyces cerevisiae encoding a protein with a putative zinc finger DNA-binding domain. Mol. Cell. Biol. 11, 6216–6228 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler G., Schuller C., Adam G. & Ruis H. A. Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 12, 1997–2003 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N. & Mcentee K. Identification of cis and trans Components of a Novel Heat Shock Stress Regulatory Pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 13, 248–256 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, D. K., Wells, H. D. & Markham C. R. In Vitro Antagonism of Trichoderma species Against Six Fungal Plant Pathogens . Am. Phytopathol. Soc. 72, 379–382 (1982). [Google Scholar]
- Saeed A. I. et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques 34, 374–378 (2003). [DOI] [PubMed] [Google Scholar]
- Kubicek C. P. et al. Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 12, 1–15 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omann M. R., Lehner S., Escobar Rodríguez C., Brunner K. & Zeilinger S. The seven-transmembrane receptor Gpr1 governs processes relevant for the antagonistic interaction of Trichoderma atroviride with its host. Microbiology 158, 107–18 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro V. N. et al. New insights in Trichoderma harzianum antagonism of fungal plant pathogens by secreted protein analysis. Curr. Microbiol. 61, 298–305 (2010). [DOI] [PubMed] [Google Scholar]
- Pazzagli L. et al. Biochimica et Biophysica Acta Characterization of ordered aggregates of cerato-platanin and their involvement in fungus – host interactions. BBA - Gen. Subj. 1790, 1334–1344 (2009). [DOI] [PubMed] [Google Scholar]
- Frischmann A. et al. Self-assembly at air/water interfaces and carbohydrate binding properties of the small secreted protein EPL1 from the fungus Trichoderma atroviride. J. Biol. Chem. 288, 4278–87 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Duan J., Wang B. & Jiang X. The function of snodprot in the cerato-platanin family from Dactylellina cionopaga in nematophagous fungi. Biosci. Biotechnol. Biochem. 76, 1835–42 (2012). [DOI] [PubMed] [Google Scholar]
- Freitas R. S. et al. Cloning and characterization of a protein elicitor Sm1 gene from Trichoderma harzianum. Biotechnol. Lett. 36, 783–8 (2014). [DOI] [PubMed] [Google Scholar]
- Gwynne D. et al. Comparison of the cis- acting control regions of two coordinately controlled genes involved in ethanol utilization in Aspergillus nidulans. Gene 51, 205–216 (1987). [DOI] [PubMed] [Google Scholar]
- Lorito M., Farkas V., Rebuffat S. & Bodo B. Cell wall synthesis is a major target of mycoparasitic antagonism by Trichoderma harzianum. J. Bacteriol. 178, 6382–6385 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores A., Chet I. & Herrera-Estrella A. Improved biocontrol activity of Trichoderma harzianum by over-expression of the proteinase encoding gene prb1. Curr. Genet. 31, 30–37 (1997). [DOI] [PubMed] [Google Scholar]
- De Oliveira A. L. et al. The structure of the elicitor Cerato-platanin (CP), the first member of the CP fungal protein family, reveals a double ψβ-barrel fold and carbohydrate binding. J. Biol. Chem. 286, 17560–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaderer R. et al. Sm2, a paralog of the Trichoderma cerato-platanin elicitor Sm1, is also highly important for plant protection conferred by the fungal-root interaction of Trichoderma with maize. BMC Microbiol. 15, 1–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inbar J. & Chet I. Biomimics of fungal cell-cell recognition by use of lectin-coated nylon fibers. J. Bacteriol. 174(3), 1055–1059 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benítez T., Rincón A. M., Limón M. C. & Codón A. C. Biocontrol mechanisms of Trichoderma strains. Int. Microbiol. 7, 249–60 (2004). [PubMed] [Google Scholar]
- Gruber S. & Seidl-Seiboth V. Self versus non-self: fungal cell wall degradation in Trichoderma. Microbiology 158, 26–34 (2012). [DOI] [PubMed] [Google Scholar]
- Rosado I. V. et al. QID74 Cell wall protein of Trichoderma harzianum is involved in cell protection and adherence to hydrophobic surfaces. Fungal Genet. Biol. 44, 950–64 (2007). [DOI] [PubMed] [Google Scholar]
- Zeilinger S. et al. Chitinase gene expression during mycoparasitic interaction of Trichoderma harzianum with its host. Fungal Genet. Biol. 26, 131–140 (1999). [DOI] [PubMed] [Google Scholar]
- Harman G. E., Howell C. R., Viterbo A., Chet I. & Lorito M. Trichoderma species - opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2, 43–56 (2004). [DOI] [PubMed] [Google Scholar]
- Nimchuk Z., Eulgem T., Holt B. F. & Dangl J. L. Recognition and response in the plant immune system. Annu. Rev. Genet. 37, 579–609 (2003). [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack K. E. & Jones J. D. Resistance gene-dependent plant defense responses. Plant Cell 8, 1773–1791 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Shah J. & Klessig D. F. Signal perception and transduction in plant defense responses. Genes Dev. 11, 1621–1639 (1997). [DOI] [PubMed] [Google Scholar]
- Shoresh M., Harman G. E. & Mastouri F. Induced systemic resistance and plant responses to fungal biocontrol agents. Annu. Rev. Phytopathol. 48, 21–43 (2010). [DOI] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M. & Van Breusegem F. Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498 (2004). [DOI] [PubMed] [Google Scholar]
- Van Loon L. C. & van Strien E. a. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 55, 85–97 (1999). [Google Scholar]
- Verberne M. C., Verpoorte R., Bol J. F., Mercado-Blanco J. & Linthorst H. J. Overproduction of salicylic acid in plants by bacterial transgenes enhances pathogen resistance. Nat. Biotechnol. 18, 779–783 (2000). [DOI] [PubMed] [Google Scholar]
- Pereira Menezes S. et al. The pathogenesis-related protein PR-4b from Theobroma cacao presents RNase activity, Ca(2+) and Mg(2+) dependent-DNase activity and antifungal action on Moniliophthora perniciosa. BMC Plant Biol. 14, 1–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoresh M., Yedidia I. & Chet I. Involvement of Jasmonic Acid/Ethylene Signaling Pathway in the Systemic Resistance Induced in Cucumber by Trichoderma asperellum T203. Phytopathology 95, 76–84 (2005). [DOI] [PubMed] [Google Scholar]
- Mauch F. & Staehelin A. Functional implications of the subcellular localization of ethylene-induced chitinase and β-1,3-glucanase in bean leaves. Plant Cell 1, 447–457 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sticher L., Mauch-Mani B. & Métraux J. P. Systemic acquired resistance. Annu. Rev. Phytopathol. 35, 235–270 (1997). [DOI] [PubMed] [Google Scholar]
- Pazzagli L. et al. Cerato-platanins: Elicitors and effectors. Plant Sci. 228, 79–87 (2014). [DOI] [PubMed] [Google Scholar]
- Kishimoto K. et al. Perception of the chitin oligosaccharides contributes to disease resistance to blast fungus Magnaporthe oryzae in rice. Plant J. 64, 343–54 (2010). [DOI] [PubMed] [Google Scholar]
- Salas-Marina M. a. et al. The Epl1 and Sm1 proteins from Trichoderma atroviride and Trichoderma virens differentially modulate systemic disease resistance against different life style pathogens in Solanum lycopersicum. Front. Plant Sci. 6, 1–13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber F., Visser J., Kubicek C. P. & de Graaff L. H. The development of a heterologous transformation system for the cellulolytic fungus Trichoderma reesei based on a pyrG-negative mutant strain. Curr. Genet. 18, 71–76 (1990). [DOI] [PubMed] [Google Scholar]
- Colot H. V. et al. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA. 103, 10352–10357 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman G. H. et al. Expressed sequence tag analysis of the human pathogen Paracoccidioides brasiliensis yeast phase: Identification of putative homologues of Candida albicans virulence and pathogenicity genes. Eukaryot. Cell 2, 34–48 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colabardini A. C. et al. Molecular characterization of the Aspergillus nidulans fbxA encoding a F-box protein involved in xylanase induction. Fungal Genet. Biol. 49, 130–140 (2012). [DOI] [PubMed] [Google Scholar]
- Carsolio C., Gutiérrez A., Jiménez B. & Van Montagu M. & Herrera-Estrella, a. Characterization of ech-42, a Trichoderma harzianum endochitinase gene expressed during mycoparasitism. Proc. Natl. Acad. Sci. USA. 91, 10903–10907 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro V. N. et al. New insights in Trichoderma harzianum antagonism of fungal plant pathogens by secreted protein analysis. Curr. Microbiol. 61, 298–305 (2010). [DOI] [PubMed] [Google Scholar]
- Pereira J. L. et al. Analysis of Phaseolus vulgaris response to its association with Trichoderma harzianum (ALL-42) in the presence or absence of the phytopathogenic fungi Rhizoctonia solani and Fusarium solani. PLoS One 9, 1–23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger M. G. et al. Transformation system for Hypocrea jecorina (Trichoderma reesei) that favors homologous integration and employs reusable bidirectionally selectable markers. Appl. Environ. Microbiol. 77, 114–21 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steindorff A. S. et al. Identification of mycoparasitism-related genes against the phytopathogen Sclerotinia sclerotiorum through transcriptome and expression profile analysis in Trichoderma harzianum. BMC Genomics 15, 1–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct Method. Methods 25, 402–8 (2001). [DOI] [PubMed] [Google Scholar]
- Sambrook J., Frisch E. F. &Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York. (1989).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.