Abstract
The metastatic spread of cancer cells to distant sites represents the major cause of cancer-related deaths in breast cancer patients, and lungs are one of the most common sites for metastatic colonization. Developing a physiologically relevant tissue culture model to mimic lung colonization of breast cancer is crucial to the investigation of the biology of cancer metastasis and evaluation of drug treatment efficacy. Here, we described an ex vivo lung colonization assay for breast cancer using the native three-dimensional (3D) lung extracellular matrix. The native matrix was isolated from murine lung with a decellularization technique, and the preservation of extracellular matrix (ECM) composition, integrity and mechanical properties was confirmed. We showed that metastatic MDA-MB 231 and 4T1 cells invaded and colonized in the decellularized lung matrix, whereas only a small mass of non-metastatic MCF7 cells survived in the same condition. Furthermore, knockdown of ZEB1, an epithelial-mesenchymal transition (EMT) inducer, significantly reduced invasion and colonization of MDA-MB 231 cells in the decellularized lung, suggesting an important role of EMT in breast cancer metastasis. We conclude that the decellularized lung retains biophysical and biochemical properties of lung ECM and provides a powerful tool to investigate lung colonization of breast cancer.
Introduction
Lungs are one of the most common sites for the metastatic spread of solid tumors.1 Breast cancer metastasis to the lung is a multi-step process in which malignant cells disseminate from the primary tumor, migrate through circulation, and eventually colonize in the lung—the final and the most important step in the lung metastatic process. Currently there is no tissue culture model available to mimic metastasis of breast cancer to lungs. Most research on lung colonization is performed in xenograft models,2 which is time consuming and cannot be used to monitor the dynamic process of metastasis. Therefore, an urgent need exists to develop a physiologically relevant tissue culture model to study lung colonization of breast cancer.
The ‘seed and soil’ theory suggests that some distant organs provide a more favorable environment (soil) than others for facilitating colonization of certain cancer cells (seeds).3 Microenvironmental cues at the metastatic lesion are crucial for cancer cell colonization and metastasis. Lung colonization involves dynamic and reciprocal interactions between cancer cells and the tissue microenvironment. One of the essential components of the tissue microenvironment is ECM.4 We showed previously that mammary epithelial cells rapidly lose their polarized acinus structure and mammary-specific gene expression upon dissociation from primary tissue and placement into two-dimensional (2D) cultures, indicating that a 3D ECM microenvironment is required for maintaining tissue structure and function.5, 6 ECM proteins directly regulate cell adhesion, migration, invasion and colonization during breast cancer progression.7-9 Fibronectin and periostin have been identified as important components of the metastatic niche to support cancer cell colonization.10, 11 Loss of those components dramatically reduced colonization of breast cancer cells in the lung. Therefore, mimicking ECM cues at the metastatic lesion is crucial for the development of a physiologically relevant lung colonization model.
The 3D culture model has been widely used to investigate normal mammary tissue morphogenesis and breast cancer progression.12 Collagen I or laminin-rich ECM (lrECM) gels are the most commonly used ECM substrata in 3D culture assay. However, the ECM microenvironment in vivo is much more complicated than collagen or lrECM gels. Using high-throughput proteomic approaches, Dr. Hynes et al. have analyzed the expression profiles of ECM protein in the lung and colon.13 Hundreds of ECM and ECM-related molecules have been identified in these two tissues.13 Importantly, 59 ECM or ECM-associated molecules were detected only in the lung and 22 proteins were detected only in the colon.13 The lung-specific proteins primarily belong to the following categories: ECM glycoprotein, ECM-affiliated, ECM regulators and ECM secreted factors. Some of these tissue-specific ECM proteins play critical roles in regulating lung physiology.13 These results indicate that the composition and enrichment of ECM molecules are distinct from one organ to another. Therefore, it is important to study proliferation, invasion and lung colonization of breast cancer cells in the native organ matrix.
Recent progress in decellularization techniques provides novel approaches for isolating natural ECM. Decellularized tissue has been successfully utilized for organ reconstruction,14 but has not been used to study tumor progression until recently.15 Native lung ECM has been isolated through the decellularization process, and the isolated ECM retains in vivo structure and composition. It has been shown that composition of lung ECM isolated from different species is very similar, thus decellularized mouse or rat lung may be utilized to study human lung regeneration and lung cancer progression.16
We developed an ex vivo breast cancer lung colonization model using the decellularized lung matrix. By analyzing invasion and colonization of breast cancer cells in this model, we showed that metastatic breast cancer cells invaded and grew in the decellularized lung matrix, while only a small mass of non-metastatic cells survived in the same condition. Moreover, silencing Zeb1, an inducer of EMT, significantly reduced invasion and colonization of MDA-MB 231 cells in the decellularized lung, confirming the critical role of EMT in lung metastasis. Therefore, the ex vivo lung colonization model may bridge the gap between traditional cell-based research and mouse tumor models, especially for investigating the dynamic process of lung metastasis of breast cancer.
Materials and methods
Antibodies and reagents
shZeb1 plasmid was purchased from Sigma. The following antibodies were obtained as indicated: anti-Collagen IV (Santa Cruz); anti-fibronectin (Abcam); anti-Ki67 (Spring Bioscience); anti-active-caspase3 (Millipore); anti-Snail (Cell Signaling); anti-E-cadherin (BD Biosciences); anti-N-cadherin (Millipore); anti-Vimentin (Thermo Scientific); anti-Twist (Santa Cruz); anti-Tubulin (Millipore).
Cell culture and virus preparation
MDA-MB 231-Green cells or MDA-MB 231-Luc cells were propagated in DMEM/F12 (Sigma) with 10% fetal bovine serum (Invitrogen). MCF7-Green cells or MCF7-Luc were propagated in DMEM (Sigma) with 10% fetal bovine serum . HEK293 FT cells were transfected with shRNA vector (Sigma) or shZeb1 plasmid plus packaging lentivector using lipofectamine (Invitrogen). MDA-MB 231 cells were infected with lentivirus and selected by puromycin 48h after infection.
Lung decellularization and cancer cell seeding
The process of lung decellularization was modified from protocols described by Andrew P. Price and Todd Jensen.17, 18 Adult FVB mice were euthanized, the sternum was cut and trachea exposed. To inflate the lungs, a syringe filled with rinse solution (deionized water with 5× Pen/Srep), was attached to an 18-gauge tubing adapter (Becton Dickinson, City, ST) and the tubing adapter was inserted into the trachea; the lungs were inflated with rinse solution. Lungs were excised with the trachea and heart then placed in a sterile 6-well plate and incubated in rinse solution for 1 h at 4oC. Lungs were removed from DI solution and injected with five rinses of 3mL rinse solution through the trachea. Triton solution (3 mL; 0.1% Triton X-100 with 5 × Pen/Srep) was injected through the tracheal aperture and incubated for 8 h at room temperature to lyse cells. Lungs were removed from Triton solution and rinsed 5 times with rinse solution. Deoxycholate solution (3 mL; 2% sodium deoxycholate; SDC with 1× Pen/Srep) was injected through the tracheal aperture and incubated overnight at 4°C. Lungs were removed from the deoxycholate solution and rinsed 5 times with rinse solution. NaCl solution (3 mL; 1M NaCl with 5 × Pen/Srep) was injected through the same hole in the trachea and incubated for 1h at room temperature to lyse residual nuclei. Lungs were removed from NaCl solution and rinsed 5 times with rinse solution. DNase solution (3 mL; 30 mg/mL porcine pancreatic DNase in 1.3mM MgSO4 and 2mM CaCl2 with 5 × Pen/Srep) was injected through the tracheal aperture and incubated for 1h at room temperature to lyse DNA. Lungs were removed from DNase solution and rinsed with PBS solution.
Decellularized lung was cut into 5 mm square cubes and placed in a p-HEMA coated 24-well plate. 0.2×106 MDA-MB 231 cells and MCF7 cells were seeded on the top of the decellularized lung cubes and incubated at 37 °C, 5% CO2 for 2h to allow cells to attach. The unattached cells were removed, and DMEM/F12 medium (.05 mL; 1×B27, 20ng/mL bFGF, 20ng/mL EGF, 0.5 μg/mL hydrocortisone, 5 μg/mL Insulin, 100μg/mL Gentamicin) was carefully added to each well and incubated at 37 °C, 5% CO2. Images were taken using a Nikon microscope on day 9 after seeding. Luciferase activity of seeded MDA-MB 231-Luc cells or MCF7-Luc cells was measured using in vivo imaging system (IVIS, Xenogen IVIS 50 imaging system).
DNA extraction
Equal amounts of mouse native lung tissue and decellularized lung tissue were transferred to a clean microcentrifuge tube and 500 μL extraction buffer (0.1M NaCl, 20mM Trizma Base, 25mM EDTA, disodium and 0.5% SDS) along with 50 μL proteinase-K solution (10 mg/mL) were added. Tissue was crushed in buffer with a clean, sterile scissor until well dispersed and tubes were incubated at 55°C overnight. The incubation was terminated by the addition of 0.75mL Phenol: Chloroform: Isoamyl Alcohol (25:24:1, v/v),the solution was mixed and centrifuged at 12,000 rpm for 6 min. The DNA was precipitated and DNA concentration was determined using a Nanodrop 2000 Spectrophotometer (Thermo Scientific).
Stiffness measurements
Sample preparation: lung tissue and decellularized lung tissue were prepared as described in the section of Lung decellularization and cancer cell seeding. Prior to measurement, a small sample of the tissue was placed on a clean glass cover slip for measurement. Agarose (Promega) and Matrigel (Corning) samples were prepared according to manufacturer's directions and stored in a high-humidity environment until measured—generally within a few hours of preparation.
Mechanical properties of the samples were measured using an Agilent 550 Scanning Probe Microscope. Stiffness of the materials was measured by bringing an AFM tip into contact with the samples and measuring force-displacement curves for each. The displacement corresponds to the vertical movement of the piezo. The point of contact was estimated as the point of displacement where the AFM force became nonzero. Displacment in these measurements is the bulk translation of the AFM cantilever. All samples were measured in an aqueous environment in order to mitigate evaporation effects. AFM tips were 450μm long cantilever tips with 0.2 N/m nominal bending stiffness (Cont AL-G-10 pyramidal with 20 nm radius). In order to account for variability in AFM tip geometry, actual spring constants for each AFM tip was measured prior to use by evaluating the resonant frequency of the cantilever in air.
Immunofluorescence and immunohistochemistry
Decellularized lung cubes with breast cancer cells were fixed in 4% paraformaldehyde (PFA), embedded in paraffin and sectioned. Sections were de-paraffined, hydrated from xylene, 100% ethanol, 95% ethanol, 85% ethanol and 70% ethanol to PBS solution. The tissue sections were blocked with 10% goat serum and incubated with Collagen IV, fibronection or mouse IgG antibody at 4°C overnight. Cell invasion into the decellularized lung tissue was quantified by measuring the invaded depth, invaded area or number of invaded cells of each image.
For immunohistochemical staining, sections were blocked by incubation with 3% H2O2 for 20 min. Antigen retrieval was accomplished by steaming in citrate sodium buffer for 30 min. Slides were incubated with ki67 or active-Caspase 3 antibodies at 4°C overnight, then the sections were incubated with goat anti-rabbit IgG conjugated with horseradish peroxidase at room temperature for 60 min. The conjunct antibody was detected by diaminobenzidine (DAB), and images were taken by Nikon microscope. The ratios of ki67 or active-Caspase 3 positive cells to total cells observed were quantified.
Quantitate RT-PCR and Western blot analysis
Total RNA was extracted from cells using Trizol reagent (Invitrogen). cDNA was synthesized using SuperScript First Strand Synthesis kit (Invitrogen) from 1.0 μg RNA samples. cDNA synthesis was performed with SuperScript III First-Strand Synthesis System according to the manufacturer's instructions. Quantitative RT-PCR reactions were carried out using SYBR Green PCR master mix reagents (Thermo Scientific) on an ABI 7500 Fast Real-Time PCR System (Applied Biosystems, USA). Thermal cycling was conducted at 95°C for 30 s, followed by 40 cycles of amplification at 95°C for 5 s, 55°C for 30 s and 72°C for 15 s. The following primers were used to amplify Zeb1: 5′- TGTGACGCAGTCTGGGTGTAAT-3′ and 5′- TCGCCCATTCACAGGTATCAAG-3′; 18S rRNA: 5′-ACCTGGTTGATCCTGCCAGT-3′ AND 5′-CTGACCGGGTTGGTTTTGAT-3′.
Cells grown on plastic were lysed in situ in 2% SDS in PBS buffer containing phosphatase and protease inhibitor cocktails (Calbiochem). Equal amount of protein lysates were subjected to SDS gel electrophoresis, immunoblotted and detected with an ECL system (Pierce).
Statistical analysis
Experiments were repeated at least twice. Results were reported as mean ± S.E.M. and the significance of difference was assessed by independent Student's t-test. P < 0.05 represented statistical significance and P < 0.01 represented statistical significance.
Live subject statement
All the animal experiments have been approved by the the Division of Laboratory Animal Resources (IACUC) at the University of Kentucky (2011-0862). All procedures were performed within the guidelines of the IACUC. No human subject was used in this study.
Results and discussion
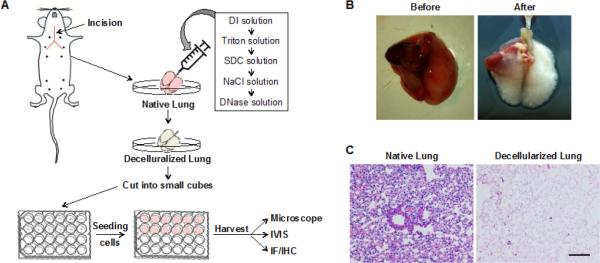
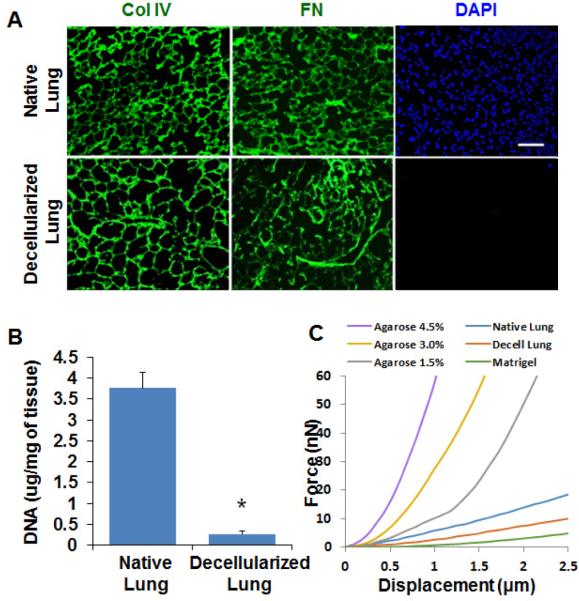
The whole organ decellularization technique has been developed to isolate the native ECM scaffold.14 Using this technique, we generated decellularized lung and examined its biochemical and biophysical properties (Figure 1A, B). The lung-heart block was harvested from adult Balb/c mice, and lung epithelial and stromal cells were removed using the detergent Triton-100 and SDC. Hematoxylin and eosin staining showed that alveolar structure was retained in the decellularized lung, but all epithelial and stromal cells were removed (Figure 1C). The DNA concentration of the decellularized lung was less than 10% of the native lung (Figure 2A). It has been shown that DNA concentration of decellularized lung was reduced to 5% of native lung tissue of rat.19 Immunofluorescence staining with Collagen IV and fibronectin showed intact ECM networks for the airway and alveoli (Figure 2B). These results indicate that most cellular components are removed in the decellularized lung. To examine the biomechanical properties, the stiffness of the decellularized and native lung was measured using atomic force microscopy (AFM). The structural heterogeneity of the native and decellularized lung tissues resulted in some location-dependent variability of force-displacement measurements. Other researchers who have thoroughly studied the mechanics of decellularized lung tissue have noted similar variability and observed similar force-displacement curves.20 We found that decellularized lung has a similar force-displacement curve to that of native lung, with the native lung being slightly stiffer. Also, of the materials measured—Matrigel, decellularized lung, and several concentrations of agarose—the decellularized lung was the most mechanically similar to native lung tissue. All concentrations of agarose tested were stiffer than the native tissue, while Matrigel was more compliant than either native or decellularized lung tissue (Figure 2C). These results indicate that the decellularization process removed the cellular compartment without significant changes to the ECM structure and biomechanical properties.
Figure 1. Decellularization procedure removes the cellular compartment in the lung.
(A) A schematic showing the procedure of decellularization. (B) Images of mouse lungs before and after decellularization process. (C) Hematoxylin and eosin staining of native mouse lung and decellularized mouse lung demonstrate that cellular compartments were removed by the decellularization procedure. Scale bar: 100 μm.
Figure 2. The composition and stiffness are retained in the decellularized lung.
(A) Immunofluorescence images of native and decellularized mouse lung tissue sections. Sections were stained with DAPI (blue) and ECM proteins antibodies: collagen IV (col IV, green) and Fibronectin (FN, green). Cell nuclei were not detected in decellularized lung tissue section. ECM proteins collagen IV and Fibronectin were not affected by decellularization process. Scale bar: 100 μm. (B) DNA concentration of native and decellularized mouse lung. DNA concentration of decellularized lung tissue was significantly reduced compared with native lung tissue; n=3. *, p<0.05. (C) Stiffness of lung tissue, decellularized lung, Matrigel, and agarose gel as measured by AFM. The decellularized lung was slightly softer than the native lung but stiffer than Matrigel.
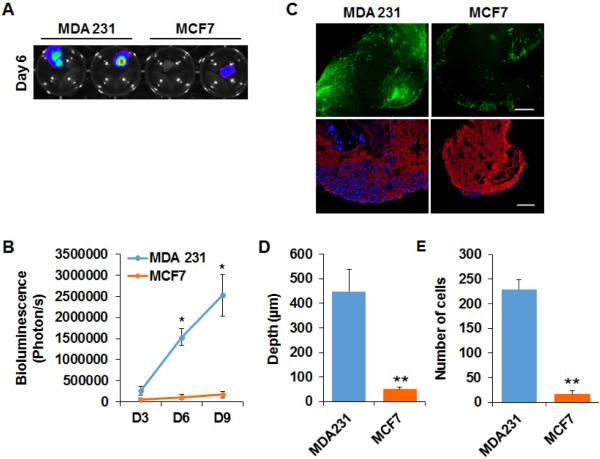
To determine whether decellularized lung matrix retains the functionality of the lung ECM microenvironment and supports the colonization of breast cancer cells, we compared colonization of metastatic breast cancer cell line MDA-MB 231 and non-metastatic MCF-7 cells in decellularized lung. Specifically, decellularized lung tissue was cut into small pieces and plated in non-attached 24 well plates. Luciferase- or GFP-labeled MCF-7 and MDA-MB 231 cells were incubated with the decellularized lung matrix and cultured for 9 days; adhesion, growth and invasion of the cancer cells were monitored by bioluminescence analysis and fluorescence microscope. Equal amounts of Luciferase- or GFP-labeled MCF-7 and MDA-MB 231 cells were incubated with the decellularized lung tissue blocks. After 24 hours, MCF-7 and MDA-MB 231 cells were both able to adhere to the lung matrix, but the number of MCF7 cells attached on the decellularized lung was less than that of MDA-MB 231 cells. The growth curve, assessed by luciferase activity and GFP signal, showed that MDA-MB 231 cells kept growing over the next 8 days (Figure 3A, B). Similar results were obtained with the metastatic cancer cell line 4T1 (Supplemental Figure 1). However, little change in the number of MCF-7 cells placed on decellularized lung matrix was observed (Figure 3A, B). To assess invasion of the cancer cells into the decellularized lung, the lung matrix was fixed, sectioned and examined by fluorescence microscopy. We found that a significant number of MDA-MB 231 cells invaded the lung matrix (Figure 3C, D, E). Only a few MCF-7 cells survived at the surface of the decellularized lung, and the number of invading cells and invasion distance was much less compared to MDA-MB 231 cells (Figure 3C, D, E).
Figure 3. Metastatic MDA-MB 231 cells grow and invade in the decellularized lung matrix.
(A) IVIS Images showed the growth of MDA-MB 231-Luc cells and MCF7-Luc cells in decellularized lung tissue on Day 6. (B) Quantification of the IVIS results showed the number of MDA-MB 231-Luc cells and MCF7-Luc cells on Day 3, Day 6 and Day 9; n=4. *, p<0.05 (C) Upper panel: Fluorescence images showed the growth of GFP-labeled MDA-MB 231 and MCF7 cells in decellularized lung on Day 9. Bottom panel: IF staining of ColIV (red) and DAPI (Blue) staining on sections of decellularized tissue cultured with MDA-MB 231-Green cells and MCF7-Green cells. Scale bar: 200 μm. (D, E) Bar graphs of the invaded depth (D) and number (E) of MDA-MB 231-Green cells and MCF7-Green cells in the decellularized lung tissue; n=5. **, p<0.01.
To understand why MCF-7 cells failed to grow in the decellularized lung matrix, we compared proliferation and apoptosis for MDA-MB 231 and MCF-7 cells. The decellularized lung tissues with MDA-MB 231 or MCF-7 cells were fixed and paraffin embedded at days 3, 6 and 9 after seeding. Ki67 staining showed that both MDA-MB 231 and MCF-7 cells were proliferative in the decellularized lung matrix, although the ratio of Ki67 positive cells was slightly lower in MCF-7 compared to MDA-MB 231 cells (Figure 4A, C). Cell proliferation was active before day 6 in the decellularized lung and was reduced on day 9. These results suggest that the failure of MCF7 cells to colonize in the lung matrix is not due to growth arrest. Cell apoptosis of MDA-MB 231 and MCF7 cells in the decellularized lung was assessed by active caspase 3 staining. We found the ratio of apoptotic cells was much higher for MCF-7 than for MDA-MB 231 cells at days 3 and 6 (Figure 4B, D). These results suggest that the lung ECM microenvironment contains components that induce apoptosis in MCF-7 cells, which may suppress the colonization of the non-metastatic breast cancer cells.
Figure 4. Analysis of cell proliferation and apoptosis in decellularized lung tissue.
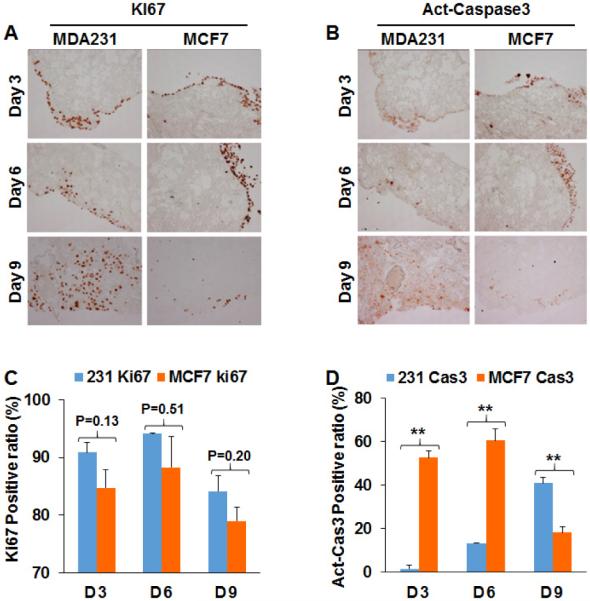
(A) IHC images showed Ki67 positive of MDA-MB 231-Green cells and MCF7-Green cells in decellularized lung matrix on Day3, Day6 and Day9. (B) Active-Caspase3 in MDA-MB 231-Green cells and MCF7-Green cells were analyzed by IHC on Day 3, Day 6 and Day 9. (C) Quantification of Ki67 staining results showed that MDA-MB 231 and MCF7 cells were proliferative in the decellularized lung. The total cell numbers were quantified by hematoxylin staining; n=4; **, p<0.01. (D) Quantification of active-Caspase3 staining showed that apoptosis was induced in MCF7-Green cells in decellularized lung matrix. The total cell numbers was quantified by hematoxylin staining; n=4; **, p<0.01.
The MDA-MB 231 and 4T1 cell lines are highly metastatic and invasive.2 These cells usually colonize in lung after tail vein injection in immune deficient mice. MCF-7 is a low-invasive and non-metastatic breast cancer cell line.21 The results obtained from the decellularized lung matrix are consistent with in vivo phenotypes of MDA-MB 231 and MCF-7 cells with respect to lung metastasis, suggesting that the decellularized lung matrix retains native ECM functionality and is a useful tool to investigate proliferation, invasion and colonization of breast cancer cells in lung.
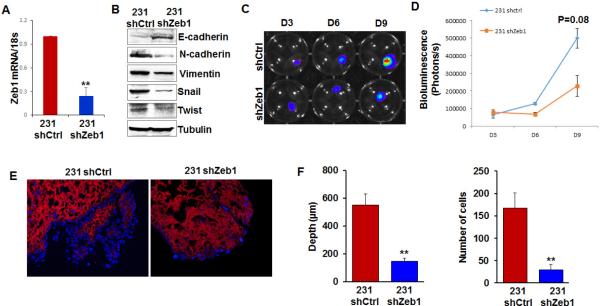
The epithelial-mesenchymal transition (EMT) is a process characterized by loss of epithelial characteristics and acquisition of a mesenchymal phenotype.22 EMT is considered a driving force of breast cancer progression and promotes metastasis by enhancing invasion and colonization.23, 24 Zinc-finger E-box binding homeobox 1 (ZEB1) is an EMT inducer in human breast cancer, and suppresses expression of E-cadherin and basement membrane components.25 Expression of ZEB1 promotes metastasis of tumor cells in a mouse xenograft model.26 MDA-MB 231 cells possess relatively high levels of ZEB1 and display some components of the EMT phenotype.27 To determine whether EMT promotes colonization of MDA-MB 231 cells in the lung matrix, ZEB1 expression was knocked down by shRNA in MDA-MB 231 cells. Quantitative RT-PCR results showed that ZEB1 mRNA level was significantly reduced in shZEB1-expressing cells (Figure 5A). Immunoblotting results confirmed that expression of EMT markers was reduced in ZEB1-silenced cells (Figure 5B). Control and ZEB1-silenced cells were incubated with the lung matrix. We found that silencing ZEB1 had little effect on adhesion of MDA-MB 231 cells on the lung matrix (Figure 5C, D). However, by days 6 and 9, invasion and colonization was significantly reduced in ZEB1-silenced cells compared to control MDA-MB 231 cells (Figure 5E, F), confirming that the EMT process is important for invasion and colonization of breast cancer cells into the lung matrix.
Figure 5. Knockdown of Zeb1 in MDA-MB 231 cells inhibits colonization of MDA-MB 231 cells in decellularized lung tissue.
(A) Quantitate RT-PCR showed knockdown efficiency of Zeb1 shRNA in MDA-MB 231-Luc cells. (B) Western blot results showed the expression of EMT markers in control and Zeb1-silenced MDA-MB 231-Luc cells. (C) IVIS Images showed the growth of control or Zeb1-silenced MDA-MB 231-Luc cells in decellularized lung tissue on Day 3, Day 6, and Day 9. (D) Quantification of the IVIS images showed the number of control or Zeb1-silenced MDA-MB 231-Luc cells on Day 3, Day 6 and Day 9; n=4. (E) Immunofluorescence staining showed invasion of control or Zeb1-silenced MDA-MB 231-Luc cells in decellularized lung matrix. Blue: DAPI, red: collagen IV. (F) Bar graph showed the invaded depth and number of control or Zeb1-silenced MDA-MB 231-luc cells in the decellularized lung tissue; n=5; **, p<0.01.
Biophysical and biochemical cues from the ECM microenvironment play important roles in proliferation and invasion of breast cancer cells. For instance, degradation of lung basement membrane is required for cancer cell invasion in situ. The traditional Transwell invasion assay has been widely used to study cancer cell invasion; however, Matrigel, which is used in this assay, does not have a relevant basement membrane structure and integrity despite a composition of major basement membrane proteins.28 It has been shown that decellularization has little effect on the integrity of basement membranes of airways, alveoli and capillary networks in the lung.16 In addition, we found that factors in the decellularized lung matrix induced apoptosis in the non-metastatic MCF-7 cells. These results suggest that the decellularized lung retains the biophysical and biochemical signals from native lung matrix and provide a physiologically relevant microenvironment to support invasion and colonization of breast cancer cells.
Colonization of breast cancer cells in lung is regulated by cellular components in lung tissue. It has been shown microvasculature of the lung induce dormancy of metastatic breast cancer cells,29 while lung fibroblasts may enhance proliferation of breast cancer cells.30 Therefore, it is important to incorporate these components in the ex vivo lung colonization model to better mimic microenvironmental cues at the metastatic lesion. Breast cancer cells also interact directly or indirectly with lung epithelial cells. Function of the lung epithelial cells in breast cancer metastasis remains unclear, and the decellularized lung matrix may provide a powerful tool to study the crosstalk between the lung epithelial cells and metastatic breast cancer cells.
Conclusions
We have developed an ex vivo lung colonization model of breast cancer using decellularized lung matrix. Utilizing this model, we have examined colonization of breast cancer cells with different metastatic potential in lung ECM, and confirmed the crucial function of EMT in breast cancer lung colonization. This physiologically relevant tissue culture model mimics invasion and colonization of breast cancer cells in lung tissue and provides a powerful tool to investigate the disease and is a potential platform for evaluating drug treatment against breast cancer metastasis.
Supplementary Material
Insight.
Insight: This study demonstrates metastatic breast cancer cells survive in the lung-specific ECM microenvironment, but not non-metastatic MCF7 cells, and EMT facilitates this process.
Innovation: For the first time, we used decellularized lung matrix to study lung colonization of breast cancer. We showed that metastatic MDA-MB 231 and 4T1 cells invaded and colonized in the decellularized lung matrix, whereas only a small mass of non-metastatic MCF7 cells survived in the same condition.
Integration: The native lung matrix isolated by decellularization technology has been used as the 3D culture substrata to investigate lung colonization of human breast cancer. The decellularized lung retains the biophysical and biochemical signals from native lung matrix and provides a physiologically relevant microenvironment to support invasion and colonization of breast cancer cells.
Acknowledgements
We acknowledge the Biospecimen and Tissue Procurement Shared Resource of the University of Kentucky Markey Cancer Center. This study was supported by start-up funding from Markey Cancer Center and funding support from AHA (12SDG8600000 to R. Xu), DOD (W81XWH-15-1-0052). This study was supported in part by NIH Grant P30 GM110787 from the National Institute of General Medical Sciences. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the NIH or the NIGMS.
Footnotes
† Footnotes relating to the title and/or authors should appear here. Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI:10.1039/x0xx00000x
Notes and references
- 1.Lu X, Kang Y. Organotropism of breast cancer metastasis. J Mammary Gland Biol Neoplasia. 2007;12:153–162. doi: 10.1007/s10911-007-9047-3. [DOI] [PubMed] [Google Scholar]
- 2.Fantozzi A, Christofori G. Mouse models of breast cancer metastasis. Breast Cancer Res. 2006;8:212. doi: 10.1186/bcr1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fidler IJ. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 4.Schedin P, Keely PJ. Mammary gland ECM remodeling, stiffness, and mechanosignaling in normal development and tumor progression. Cold Spring Harb Perspect Biol. 2011;3:a003228. doi: 10.1101/cshperspect.a003228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu R, Nelson CM, Muschler JL, Veiseh M, Vonderhaar BK, Bissell MJ. Sustained activation of STAT5 is essential for chromatin remodeling and maintenance of mammary-specific function. J Cell Biol. 2009;184:57–66. doi: 10.1083/jcb.200807021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu R, Spencer VA, Groesser DL, Bissell MJ. Laminin regulates PI3K basal localization and activation to sustain STAT5 activation. Cell Cycle. 2010;9:4315–4322. doi: 10.4161/cc.9.21.13578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson CM, Bissell MJ. Of extracellular matrix, scaffolds, and signaling: tissue architecture regulates development, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2006;22:287–309. doi: 10.1146/annurev.cellbio.22.010305.104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu R, Boudreau A, Bissell MJ. Tissue architecture and function: dynamic reciprocity via extra- and intra-cellular matrices. Cancer Metastasis Rev. 2009;28:167–176. doi: 10.1007/s10555-008-9178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oskarsson T, Acharyya S, Zhang XH, Vanharanta S, Tavazoie SF, Morris PG, Downey RJ, Manova-Todorova K, Brogi E, Massague J. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med. 2011;17:867–874. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malanchi I, Santamaria-Martinez A, Susanto E, Peng H, Lehr HA, Delaloye JF, Huelsken J. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2012;481:85–89. doi: 10.1038/nature10694. [DOI] [PubMed] [Google Scholar]
- 12.Bissell MJ, Weaver VM, Lelievre SA, Wang F, Petersen OW, Schmeichel KL. Tissue structure, nuclear organization, and gene expression in normal and malignant breast. Cancer Res. 1999;59:1757–1763s. discussion 1763s-1764s. [PubMed] [Google Scholar]
- 13.Naba A, Clauser KR, Hoersch S, Liu H, Carr SA, Hynes RO. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol Cell Proteomics. 2012;11:M111014647. doi: 10.1074/mcp.M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crapo PM, Gilbert TW, Badylak SF. An overview of tissue and whole organ decellularization processes. Biomaterials. 2011;32:3233–3243. doi: 10.1016/j.biomaterials.2011.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mishra DK, Thrall MJ, Baird BN, Ott HC, Blackmon SH, Kurie JM, Kim MP. Human lung cancer cells grown on acellular rat lung matrix create perfusable tumor nodules. Ann Thorac Surg. 2012;93:1075–1081. doi: 10.1016/j.athoracsur.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ott HC, Clippinger B, Conrad C, Schuetz C, Pomerantseva I, Ikonomou L, Kotton D, Vacanti JP. Regeneration and orthotopic transplantation of a bioartificial lung. Nat Med. 2010;16:927–933. doi: 10.1038/nm.2193. [DOI] [PubMed] [Google Scholar]
- 17.Price AP, England KA, Matson AM, Blazar BR, Panoskaltsis-Mortari A. Development of a decellularized lung bioreactor system for bioengineering the lung: the matrix reloaded. Tissue Eng Part A. 2010;16:2581–2591. doi: 10.1089/ten.tea.2009.0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen T, Roszell B, Zang F, Girard E, Matson A, Thrall R, Jaworski DM, Hatton C, Weiss DJ, Finck C. A rapid lung decellularization protocol supports embryonic stem cell differentiation in vitro and following implantation. Tissue Eng Part C Methods. 2012;18:632–646. doi: 10.1089/ten.tec.2011.0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mishra DK, Thrall MJ, Baird BN, Ott HC, Blackmon SH, Kurie JM, Kim MP. Human lung cancer cells grown on acellular rat lung matrix create perfusable tumor nodules. Ann Thorac Surg. 2012;93:1075–1081. doi: 10.1016/j.athoracsur.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luque T, Melo E, Garreta E, Cortiella J, Nichols J, Farré R, Navajas D. Local micromechanical properties of decellularized lung scaffolds measured with atomic force microscopy. Acta Biomater. 2013;9:6852–6859. doi: 10.1016/j.actbio.2013.02.044. [DOI] [PubMed] [Google Scholar]
- 21.Lacroix M, Leclercq G. Relevance of breast cancer cell lines as models for breast tumours: an update. Breast Cancer Res Treat. 2004;83:249–289. doi: 10.1023/B:BREA.0000014042.54925.cc. [DOI] [PubMed] [Google Scholar]
- 22.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16:488–494. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 23.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalluri R, Weinberg RA. The basics of epithelialmesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmalhofer O, Brabletz S, Brabletz T. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 2009;28:151–166. doi: 10.1007/s10555-008-9179-y. [DOI] [PubMed] [Google Scholar]
- 26.Spaderna S, Schmalhofer O, Wahlbuhl M, Dimmler A, Bauer K, Sultan A, Hlubek F, Jung A, Strand D, Eger A, Kirchner T, Behrens J, Brabletz T. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res. 2008;68:537–544. doi: 10.1158/0008-5472.CAN-07-5682. [DOI] [PubMed] [Google Scholar]
- 27.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 28.Benton G, Kleinman HK, George J, Arnaoutova I. Multiple uses of basement membrane-like matrix (BME/Matrigel) in vitro and in vivo with cancer cells. Int J Cancer. 2011;128:1751–1757. doi: 10.1002/ijc.25781. [DOI] [PubMed] [Google Scholar]
- 29.Ghajar CM, Peinado H, Mori H, Matei IR, Evason KJ, Brazier H, Almeida D, Koller A, Hajjar KA, Stainier DY, Chen EI, Lyden D, Bissell MJ. The perivascular niche regulates breast tumour dormancy. Nat Cell Biol. 2013;15:807–817. doi: 10.1038/ncb2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Studebaker W, Storci G, Werbeck JL, Sansone P, Sasser AK, Tavolari S, Huang T, Chan MW, Marini FC, Rosol TJ, Bonafé M, Hall BM. Fibroblasts isolated from common sites of breast cancer metastasis enhance cancer cell growth rates and invasiveness in an interleukin-6-dependent manner. Cancer Res. 2008;68:9087–9095. doi: 10.1158/0008-5472.CAN-08-0400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.