Abstract
Background
Despite its role in increasing the number of offspring during the lifetime of an individual animal, controlled ovarian hyperstimulation (COH) may have detrimental effects on oocyte development, embryo quality and endometrial receptivity. Circulating miRNAs in bio-fluids have been shown to be associated with various pathological conditions including cancers. Here we aimed to investigate the effect of COH on the level of extracellular miRNAs in bovine follicular fluid and blood plasma and elucidate their mode of circulation and potential molecular mechanisms to be affected in the reproductive tract.
Method
Twelve simmental heifers were estrous synchronized and six of them were hyperstimulated using FSH. Follicular fluid samples from experimental animals were collected using ovum pick up technique at day 0 of the estrous cycle and blood samples were collected at day 0, 3 and 7 of post ovulation. The expression profile of circulatory miRNAs in follicular fluid and blood plasma were performed using the human miRCURY LNA™ Universal RT miRNA PCR array system. A comparative threshold cycle method was used to determine the relative abundance of the miRNAs.
Results
A total of 504 and 402 miRNAs were detected in both bovine follicular fluid and blood plasma, respectively. Of these 57 and 21 miRNAs were found to be differentially expressed in follicular fluid and blood plasma, respectively derived from hyperstimulated versus unstimulated heifers. Bioinformatics analysis of those circulating miRNAs indicated that their potential target genes are involved in several pathways including TGF-beta signaling pathway, MAPK signaling pathway, pathways in cancer and Oocyte meiosis.
Moreover, detail analysis of the mode of circulation of some candidates showed that most of the miRNA were found to be detected in both exosomal and Ago2 protein complex fraction of both follicular fluid and blood plasma.
Conclusion
Our data provide the consequence of hyperstimulation induced changes of extracellular miRNAs in bovine follicular fluid and blood plasma, which may have a potential role in regulating genes associated not only with bovine ovarian function but also involved in altering various physiological in bovine oocytes, embryos and modulating reproductive tract environment.
Keywords: Extracellular miRNA, Ovarian hyperstimulation, Follicular fluid, Exosome, Ago2
Background
Follicular development is a dynamic process which involves emergence of a group of follicles after an increase of peripheral concentration of follicle stimulating hormone (FSH) [1]. Follicular products from the dominant follicle are responsible for suppressing FSH concentration and leading to atresia of the subordinate follicles [2, 3]. Meanwhile, dominant follicle acquires LH receptors in its granulosa cell and no longer dependent of FSH for further development. Maintenance of elevated level of circulating FSH rescues the subordinate follicles from regression, resulted in the growth of multiple dominant follicles with ovulating capability [4]. Therefore, Controlled ovarian hyperstimulation (COH) is utilized to stimulate the growth of multiple follicles in naturally mono-ovulating species, including cattle and humans, by maintaining supraphysiological level of gonadotrophin.
Although COH is the central component of ART, it is recently became evident that COH may have detrimental effects on oocyte development, embryo quality, endometrial receptivity and perhaps on perinatal outcomes [5]. In vitro studies showed that ovarian stimulation disrupts and delays early embryonic development in mouse [6, 7]. We have previously shown that changes in the bovine oviduct and uterine environment induced by gonadotrophin treatments has resulted in changes in gene expression profile of the resulting blastocysts [8]. Furthermore, an increase in chromosomal abnormalities has also been reported in human embryos after conventional stimulation, mainly resulting from an increased incidence of chromosome segregation errors during the first embryonic divisions [9]. It is well known that COH leads to deviant oocyte development in the follicle [10] as a result of nonphysiological endocrine level in the reproductive organs. Follicular development is a complex coordinated process facilitated by the bidirectional signalling between gonadal and somatic cells in follicular microenvironment. A large number of gene products are involved in this process, which are expressed in cell type and time dependent manner Any deviation in the expression of these genes due to environmental factors could lead to abnormal oocyte development, apoptosis, and poor cellular communication with the companion somatic cells [11]. In this context the role of miRNAs as post transcriptional regulaters of gene expression can not be ruled out as these regulatory molecules are abundantly expressed in follicular cells and in extracellular space in follicular microenvironment. MicroRNAs (miRNAs) are a class of small non-coding RNAs that regulate gene expression at the post-transcriptional level. miRNAs have been found in all mammalian tissues and cell types examined so far and play important roles in a variety of physiological and pathological processes [12]. While majority of miRNAs are detected intracellular, a handful number of miRNAs are also detected in extracellular body fluids such as plasma, serum, urine, saliva and follicular fluid [13–15]. Although the origin of cell-free miRNAs in circulation is not entirely clear, miRNAs are believed to be released to the extracellular space from damaged or apoptotic cells and are also actively secreted by cells via exosomes or exocytosis [13]. Circulating miRNAs have been shown to be useful biomarkers for multiple clinical endpoints including the diagnosis of preeclampsia [16], the diagnosis and monitoring of diabetic retinopathy [17], neuropathy [18] and as diagnostic or prognostic markers for multiple cancers [14, 19–22]. Cellular release of miRNA molecules into the circulation has been shown to be associated with the altered physio-pathological conditions. The function of exosomes and shedding vesicles are believed to be cell-to-cell communication and platforms for multi signalling processes [23, 24].
Circulating miRNAs have been extensively studied since their discovery [13, 14], and they are known to be coupled with vesicles, in particular microvesicles or exosomes [25] or high density lipoproteins (HDL) [24, 26, 27] and protein complexes [28–30] and their presence in the blood has been attributed to release by tissue injury. We have recently demonstrated that the presence of extracellular miRNAs in bovine follicular fluid and their association with the growth status of oocytes [15]. However, effect of gonadotrophin treatments during COH on extracellular miRNAs in follicular fluid and blood plasma has not been investigated yet in mammalian species. Therefore, the aim of this study was to test the hypotheses that controlled ovarian hyperstimulation may change the extracellular miRNA population in follicular fluid and blood plasma of cyclic heifers, which may be involved in various physiological processes. For this a qRT-PCR array based expression profiling was used to identify a panel of differentially expressed circulating miRNAs in follicular fluid samples and blood plasma samples of hyperstimulated compared to unstimulated heifers. Moreover, the bioinformatics analysis of the candidate miRNAs revealed the potential involvement of those miRNAs in regulation of several signaling pathways, which are known to be active in various key reproductive processes.
Methods
Animal treatments and sample collection
All experimental animals were handled according to the animal protection law of Germany. The experiment was approved by the Animal Welfare committee of the University of Bonn with proposition number 84–02.05.20.12.075. Simmental heifers (n =10), aged from 15 to 17 months and weighing between 380 to 450 kg were used in this study. All animals were kept under identical farm conditions within the same herd. Synchronization and ovarian hyperstimulation was performed according to the previously mentioned protocol [8] Briefly, pre-synchronization was performed for all animals by intra-muscular administration of 500 mg of cloprostenol (PGF2a, Estrumatew; Essex Tierarznei, Munich, Germany) twice within 11 days. Two days after each of the PGF2a treatments animals received 10 mg of GnRH (Receptalw; Intervet, Boxmeer, the Netherlands). Of 10 synchronized heifers 6 were used for hyperstimulation in which twelve days after the last GnRH injection, these heifers received the first of eight consecutive FSH-injections over 4 days in decreasing doses (in total 300–400 mg of FSH equivalent according to the body weight; Stimufol, University of Liege, Belgium). Two PGF2a treatments were performed 60 and 72 h after the initial FSH injection. Finally, 48 h after the application of first PGF2a, ovulation was induced by simultaneous administration of 10 mg of GnRH. Afer 60 h of first PGF2a application was considered as onset of oestrus (D0). Follicular contents (follicle 35 mm) were collected by transvaginal, ultrasound-guided follicular aspirations. Follicular fluid was collected using a 12-gauge needle, centrifuged at 1500 × g for 5 min, and later stored at −80 °C, while blood samples were collected from each animal from day 0 (D0), day 3 (D3) and day7 (D7) by tail vein puncture. Blood serum following collection, blood samples were refrigerated at 4 °C for 12–24 h before being centrifuged at 1500 × g at 4 °C for 15 min. Serum was separated and stored at −20 °C until assayed to determine progesterone concentration. Blood plasma for miRNA detection was collected by EDTA Tubes (Carl Roth, Karlsruhe, Germany) from the both group animals and stored at −80 °C until processed for microvesicles/ exosomes, RNA, or protein isolation.
Progesterone assay
Serum progesterone concentration in different time points was determined by time-resolved immunofluorescence using an Auto DELFIA™ Progesterone kit (Perkin Elmer, Wallac Oy, Turku, Finland) which is based on the fluorescence of elements where the assay sensitivity was 0.01 ng/ml. The assay principle combines an enzyme immunoassay competition method with final fluorescent detection. The DELFI test is based on the competition for binding sites on the antibody molecule that occurs between the Europium + 3-labeled hormone and a not-labeled hormone, contained in the sample. The amount of the labeled hormone is constant, whilst the not-labeled hormone content is a function of antibody- labeled hormone complex formation. On this basis, a standard curve was drawn for reading the hormone levels in the sample.
Isolation total RNA and reverse transcription
Total RNA was isolated from follicular fluid and blood plasma, ultracentrifugation pellets and immunoprecipitation pellets using the miRNeasy kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol with some modifications. Briefly, 800 μL of QIAzol buffer was added to 200 μL of plasma or follicular fluid or exosome pellet or Ago2 pellet and incubated at room temperature for 8 min. After that to inactivate RNases activity 200 μL chloroform was added to each sample. At that point, the manufacturer’s protocol was followed. Total RNA concentration and purity was determined using NanoDrop ND-1000 spectrophotometer. Moreover, prior to reverse transcription procedure RNA samples from both plasma and follicular fluid were checked for the presence or absence of PCR inhibitors described elsewhere [15]. Briefly, cDNA was synthesized using different input volume, for instant 0.5 μl, 1 μl, 2 μl and 4 μl of total RNA, in 10 μl of reaction volume and quantify the expression of selected miRNA using qPCR. All RNA samples showed linear amplification of the candidate miRNAs for different levels of RNA input, which showed the absence of PCR inhibitors and good quality RNA (data not shown). A reverse transcription reaction was performed using the miRCURY LNA™ Universal RT microRNA PCR system (Exiqon, Denmark) according to the manufacturer’s instructions. In brief, approximately a total of 100 ng of total RNA, including small RNA, were anchor-tailed with a poly(A) sequence at their 3'end and then reverse transcribed into cDNA using a universal poly(T) primer with a 3'end degenerate anchor and a 5' end universal tagged.
miRNA Profiling and expression analysis
The expression of miRNAs in blood plasma and follicular fluid were profiled using miRNA Ready-to-Use PCR, Human Panel I &II, V2.M qRT-PCR arrays (Exiqon, Vedbaek, Denmark) which contains 748 mature miRNA sequences along with 8 empty wells, 6 intra-plate calibrators and 6 endogenous controls. A SYBR green based real time PCR technology was employed for signal detection during quantification. Prior to real time PCR analysis the cDNA products from each samples were diluted 100-fold and mixed with ready to use SYBR-green master mix. Then the cDNA-master mix was robotically pipette to a 384-well PCR plate containing miRNA specific primers. The real time PCR was run on a ABI-7900HT thermocycler (Applied Biosystems) using the following thermal-cycling parameters: 95 °C for 10 min, 40 cycle of 95 °C for 10 sec, 60 °C for 1 min followed by a melting curve analysis. Raw Ct values were calculated as recommended by Exiqon using the RQ manager software v1.2.1 (Applied Biosystems), and Ct values were calculated by using automated assay-specific baseline and threshold settings. To minimize the potential noise, miRNAs with Ct value greater than 35 in all groups were considered as undetected. The PCR data were analysed using web-based PCR array data analysis software (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php-). The raw miRNA data was normalized using a global normalization method.
Target prediction and pathway analysis
Target Scan 6.2 and miRDB were used to predict the target genes of the differentially expressed miRNAs in both follicular fluid and blood plasma of hyperstimulated and unstimulated groups. The predicted miRNA target genes were analyzed by using the DAVID Bioinformatic Resource (http://david.abcc.ncifcrf.gov/) server for Annotation, Visualization, and Integrated Discovery to identify the pathway distribution. These pathways were presented according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/ kegg/). Fisher’s exact test was used to calculate a p-value determining the probability that each biological function or canonical pathway assigned to the data set. In addition, the significance of the association between the data set and the canonical pathway was calculated as the ratio of the number of genes from the data set that were mapped to the pathway divided by the total number of genes that mapped to the canonical pathway.
Exosome isolation
Exosomes were isolated from 400 μL follicular fluid and blood plasma using differential ultra-centrifugation as previously described [15] with some modification. Briefly, samples were subjected to centrifugation at 4,000 × g for 10 min to remove cell and cell debris. Cell free plasma samples were then filtered through 0.22 μm screen to remove particles larger than 200 nm. Purified plasma were diluted into 4 mL of DPBS (life technologies, paisley, UK) solution and mixed gently. Samples were ultracentrifuged in 5-mL (13 × 51 mm) polyallomer tubes (no. 326819; Beckman Coulter) at 120,000 × g for 70 min at 4 °C in a swinging-bucket SWTi 55 rotor. The resulting supernatant was removed and exosomes pellets were resuspended in DPBS solution and centrifuged again at 120,000 × g for 70 min at 4 °C. Finally, the resulting pellet was resuspended in 200 μL DBPS and stored for further use.
Immunoprecipitation Ago2 protein complex
A total of 400 μL of plasma was used from each group to immunoprecipitate Ago2 protein in order to isolate Ago2 coupled miRNAs. Each plasma sample was diluted with 400 μL of DPBS (pH 7.4) resulting 800 μL of diluted plasma. Following this, 400 μL of Magna Bind goat anti-mouse IgG Magnetic Bead slurry (Thermo Scientific, Rockford, USA) were washed in DPBS and incubated with 10 μg of mouse monoclonal anti-Ago2 mouse normal IgG (Santa Cruz Biotechnology Inc, USA) antibodies for 2 h at 4 °C. The pre-incubated beads and antibody were then added to the 800 μL of diluted plasma and incubated overnight at 4 °C. Beads were washed three times with 1 % Nonidet P-40 buffer (1 % Nonidet P-40, 50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA (BD Bioscience, Heidelberg, Germany). After the final wash 800 μL of QIAzol lyses buffer was added directly to the Ago 2 pellet and processed for RNA isolation.
Western blot analysis
Exosomal and Ago2 immunoprecipitate proteins were isolated from organic phenol part during total RNA isolation using miRNeasy mini kit (Qiagen, Hilden, Germany) and resuspended in 8 M Urea. Approximately 20 μg of protein from each sample were resolved in 12 % SDS-PAGE polyacrylamide gel (Bio-Rad, Corp., Hercules, CA, USA) and immune reactive proteins were visualized with a Chemidoc XRS (Bio-Rad) instrument. CD63 (ExoAB Antibody, SBI, CA, USA) was used to check the efficiency of exosomes isolation, while Ago2 antibody (Santa Cruz Biotechnology Inc, USA) was employed to detect the efficiency of Ago2 immunoprecipitation.
Individual qRT-PCR Assays
Based on their enrichment in follicular fluid and blood plasma from hyperstimulated heifers, 11 candidate miRNAs were selected for further study. Moreover, high merit was given to those miRNAs which has sequence similarities between human and bovine during the selection of candidate miRNAs. Primer sets of individual qRT-PCR assays for miRNAs: miR-212, miR-182, let-7 g, miR-100, miR-877, miR-200c, miR-221, miR-103, miR-134, miR-147 and miR-127-3p were obtained from Exiqon (Vedbaek, Denmark). Reverse transcription reaction products were combined with SYBR Green master mix (Exiqon, Vedbaek, Denmark) and loaded into the 96-well plates preloaded with individual miRNA primers. Quantitative PCR was run in a Step one plus real time PCR instrument (Applied Biosystems). Relative expression of each miRNA was analysed using a comparative Ct (2-ΔΔCT) method and global normalization strategy was employed to normalize the data.
Statistical analysis
All experiments were performed a minimum of three times. When two groups were compared (i.e., synchronised vs. super ovulated) a Student’s t-test was used to detect differences between treatment groups. A P value of ≤ 0.05 was considered to be significant. Data are expressed as mean ± SD of replicates.
Results
Progesterone profile of experimental heifers
Blood serum progesterone (P4) concentration at day 7 of the experimental heifers was used to identify animals with improper stimulation phenotype. Of the 6 hyperstimulated heifers, while 4 have shown a significantly elevated P4 level (>24 ng/mL) and considered as properly stimulated, 2 heifers (P4 concentration < 23 ng/mL) considered as improperly stimulated and excluded from the experiment. The progesterone concentration in blood serum of the selected hyperstimulated and unstimulated heifers at days 0 & 7 is presented in Fig. 1. The mean progesterone concentration at day 7 was 25.17 ± 1.01 and 2.21 ± 1.19 ng/ml (p < 0.001) in the hyperstimulated and unstimulated heifers, respectively.
Fig. 1.
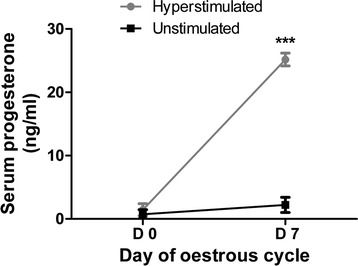
Progesterone profile of experimental heifers: Serum progesterone concentrations (ng/mL) for unstimulated controls and hyperstimulated heifers. Data is presented as mean ± SD and *** designated a significance level of p < 0.001
Detection of miRNAs in bovine follicular fluid and blood plasma
Here, we examined the abundance of mature miRNAs in follicular fluid and blood plasma of hyperstimulated and unstimulated groups using ready-to-use Human miRNA Panels (I + II) (Exiqon, Denmark) covering 748 mature human miRNAs. After comparative analysis between mature human miRNA sequences used in PCR panels and bovine mature miRNA sequence available in miRBase version 19 (http://www.mirbase.org/), around 241 miRNAs were found to be completely identical, while 126 miRNAs showed differences in size due to addition or deletion of 1–5 nucleotides and 58 miRNA sequences contained mismatches. However, no bovine homologous sequences were matched for 323 miRNAs in the present miRBase databank. A miRNA was considered to be detected when its Ct value is less than 35 and present in at least in 75 % of the samples. Of the 748 miRNA investigated, while 504 miRNAs were detected in follicular fluid, 402 miRNAs were detected in blood plasma (Fig. 2). Among the detected miRNAs 373 miRNAs were commonly detected in both follicular fluid and blood plasma. However, 131 and 29 miRNAs were detected exclusively in follicular fluid and blood plasma, respectively.
Fig. 2.
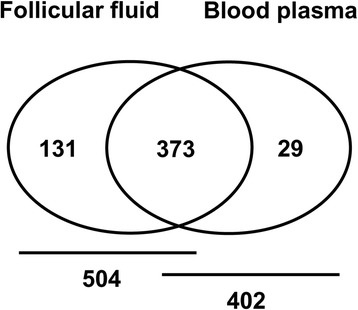
Venn diagram showing the number of detected miRNAs in follicular fluid and blood plasma. Out of 748 miRNAs investigated in the PCR panel I + II, a total of 504 and 402 miRNAs were detected (with threshold cycle value of ≤35 in real time PCR analysis) in follicular fluid and blood plasma, respectively
Differential expression of extracellular miRNAs in follicular fluid from hyperstimulated and unstimulated heifers
After excluding 244 miRNAs (31.8 %) with a Ct value above 35 from the dataset, 504 informative miRNAs were analysed in follicular fluid samples. In order to compare miRNA expression differences between follicular fluid from hyperstimulated (n = 4) and unstimulated (n = 4) animals, first the raw data was normalized using global normalization method (correction with the sum of the expression levels of detected miRNAs), as it was successfully used in previous study compared to spiked-in miRNA or mammalian U6 [31]. Expression analysis of the detected miRNAs in follicular fluid revealed that 57 miRNAs are differentially expressed between two groups (with a fold change of ≥2 & p ≤ 0.05). Of these, 30 miRNAs (52.6 %) were found to be up-regulated, while 27 miRNAs (47.4 %) were down-regulated in hyperstimulated animals compared to the unstimulated ones. Among the up-regulated miRNAs, miR-212 and miR-148b-5p showed the highest fold change regulation. On the other hand, miR-100 and miR-877 showed the greatest fold change regulation among the downregulated miRNAs (Table 1).
Table 1.
Differential expression of extra miRNAs in follicular fluid and blood plasma obtained from hyperstimulated and unstimulated heifers
| Follicular fluid (Hyp vs. Unst) | Blood plasma (Hyp vs. Unst) | ||||
|---|---|---|---|---|---|
| MicroRNA Name | Fold change | P value | MicroRNA Name | Fold change | P value |
| miR-643 | 51.36 | 0.032678 | miR-708-3p | 8.15 | 0.038772 |
| miR-212 | 40.19 | 0.007121 | miR-26b-3p | 4.92 | 0.031749 |
| miR-148b* | 33.13 | 0.009156 | miR-598 | 3.69 | 0.002565 |
| miR-548j | 23.84 | 0.031322 | miR-423-3p | 2.77 | 0.045389 |
| miR-132 | 22.45 | 0.033763 | miR-130b-5p | 2.73 | 0.022855 |
| miR-224* | 13.98 | 0.008991 | miR-576-5p | 2.69 | 0.024159 |
| miR-129-3p | 11.4 | 0.005949 | miR-221 | 2.66 | 0.016328 |
| miR-33b | 10.57 | 0.000110 | miR-1468 | 2.63 | 0.016305 |
| miR-202* | 6.53 | 0.000805 | miR-18a-3p | 2.55 | 0.031849 |
| miR-155* | 6.51 | 0.039305 | miR-181c | 2.40 | 0.033951 |
| miR-374a | 5.2 | 0.040696 | let-7 g | 2.00 | 0.008124 |
| miR-1207-5p | 5.11 | 0.000203 | miR-103 | 2.29 | 0.002404 |
| miR-182 | 4.81 | 0.047938 | miR-34a | 2.13 | 0.023235 |
| miR-103 | 4.49 | 0.006078 | miR-125b | −3.07 | 0.013367 |
| let-7 g | 4.23 | 0.012259 | miR-410 | −3.2 | 0.038659 |
| miR-106b | 3.96 | 0.034579 | miR-127-3p | −3.22 | 0.000817 |
| miR-191 | 3.95 | 0.021379 | miR-494 | −3.23 | 0.008614 |
| miR-542-5p | 3.78 | 0.005687 | miR-147 | −3.63 | 0.047214 |
| miR-505* | 3.57 | 0.006276 | miR-99a* | −5.54 | 0.044064 |
| miR-550a | 3.19 | 0.046925 | miR-134 | −7.03 | 0.043678 |
| miR-107 | 3.03 | 0.034726 | miR-153 | −7.21 | 0.006251 |
| miR-595 | 2.78 | 0.030403 | |||
| miR-33a* | 2.74 | 0.042360 | |||
| miR-374b | 2.65 | 0.038757 | |||
| miR-30b | 2.62 | 0.039581 | |||
| miR-31 | 2.54 | 0.029654 | |||
| miR-106b* | 2.43 | 0.005030 | |||
| miR-940 | 2.37 | 0.014177 | |||
| miR-495 | 2.27 | 0.010936 | |||
| miR-494 | 2.06 | 0.046091 | |||
| miR-300 | −2.08 | 0.020863 | |||
| miR-24 | −2.09 | 0.000273 | |||
| miR-452 | −2.15 | 0.020850 | |||
| miR-151-5p | −2.22 | 0.033121 | |||
| miR-320a | −2.58 | 0.027659 | |||
| miR-125a-3p | −2.78 | 0.008997 | |||
| miR-99a | −2.94 | 0.048804 | |||
| miR-449a | −2.97 | 0.018677 | |||
| miR-206 | −3.19 | 0.010009 | |||
| miR-23a | −3.26 | 0.007032 | |||
| miR-27b | −3.44 | 0.013330 | |||
| miR-139-5p | −3.46 | 0.006195 | |||
| miR-125b-2* | −3.6 | 0.029345 | |||
| miR-378 | −3.72 | 0.017333 | |||
| miR-125b | −3.86 | 0.017793 | |||
| miR-141 | −4.03 | 0.038193 | |||
| miR-23b | −4.11 | 0.033330 | |||
| miR-190 | −4.2 | 0.004785 | |||
| miR-382 | −4.45 | 0.035549 | |||
| miR-92a | −4.53 | 0.007967 | |||
| miR-145 | −4.78 | 0.000015 | |||
| miR-361-5p | −5.25 | 0.029699 | |||
| miR-200a* | −6.41 | 0.040837 | |||
| miR-200c | −6.48 | 0.048195 | |||
| miR-659 | −8.42 | 0.037679 | |||
| miR-877 | −12.16 | 0.009853 | |||
| miR-100 | −14.02 | 0.004324 | |||
Expression profile of circulatory miRNAs in blood plasma derived from hyperstimulated vs. unstimulated heifers
The same PCR array platform was used to compare the pattern of circulatory miRNA expression in blood plasma at day 7 of oestrous cycle between two experimental groups. By employing the same criteria for data filtration, a total of 402 (54.8 %) informative miRNAs were identified in blood plasma samples of both groups. Among the detected miRNAs, 21 miRNAs were identified as differentially expressed (p < 0.05), of which 13 were overexpressed and 8 were underexpressed in the plasma of hyperstimulated heifers (Table 1). The miRNA expression in the hyperstimulated plasma compared to the unstimulated control was increased by 2 to11-fold but decreased by 2.8to 7-fold (Table 1). Among the overexpressed miRNAs miR-20b-3p and miR-708-3p showed the highest fold change regulation, while miR-153 and miR-134 exhibit highest fold change among the downregulated miRNAs.
Cluster of miRNAs affected by ovarian hyperstimulation in follicular fluid and blood plasma
MiRNA cluster represent group of miRNA genes located within 10 Kb of distance on the same chromosome. To test the hypothesis that controlled ovarian hyperstimulation may affect the expression of individual or cluster miRNAs, the genomic location of the 57 and 21 miRNAs that were differentially expressed in follicular fluid and blood plasma of hyperstimulated heifers, respectively, were determined. Subsequently, we found that approximately 18 % of the follicular fluid derived differentially expressed miRNAs to belong to miRNA clusters and the rest 82 % were individual miRNAs (Fig. 3). Of those in blood plasma differentially expressed miRNAs, 15 % were found to be present in clusters. Variable numbersof miRNAs were present in each cluster with minimum of two. For example, the miRNA cluster miR-410 ~ 134, which is found to be differentially expressed between hyperstimulated and unstimulated heifers blood plasma, contained 3 miRNAs. Since miRNAs within the cluster may share a common gene promoter and may be regulated as a single transcriptional unit, it could be of interest to determine whether any entire cluster changed in the same direction following treatment. Subsequently, we found that all miRNAs within all 6 clusters (Fig. 3) showed the same direction of expression tended to change in the same direction as the other members in that cluster.
Fig. 3.
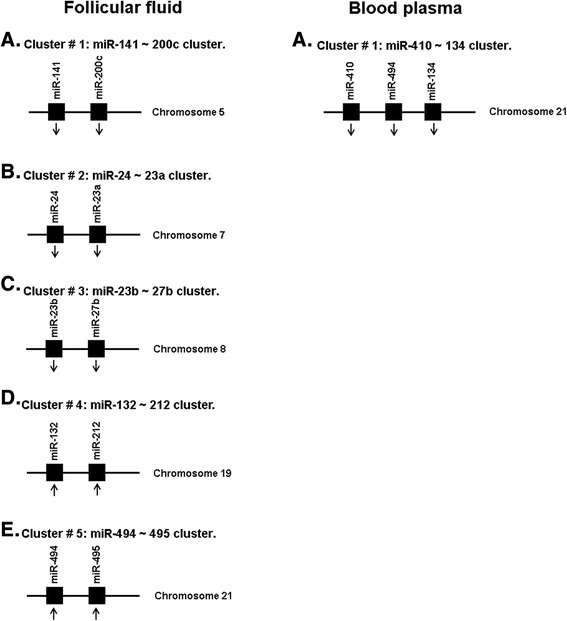
The number of miRNAs in each cluster and their distribution in bovine chromosomes. While clusters differentially expressed in follicular fluid (a-e) comprise two miRNAs, clusters in blood plasma shown to comprise three miRNAs. Multiple miRNA clusters are affected in the follicular fluid and blood plasma due to ovarian hyperstimulation. The number of miRNA genes in each cluster ranged from 2-3. In all clusters, the expression of miRNA genes in a particular cluster was in the same direction (the arrow ↑ for increased expression, and the arrow ↓ for decreased expression). Please note that differentially expressed miRNAs (≥2 fold and p < 0.05) in follicular fluid and blood plasma of hyperstimulated heifers compared to the un stimulated ones are listed in Table 1
miRNA target prediction and pathway analysis
As the number of experimentally validated targets of miRNA is limited, we used the widely used TargetScan and miRDB algorithm to obtain the list of genes predicted to be targeted by the differentially expressed miRNAs obtained from follicular fluid and blood plasma of the experimental animals. Accordingly, 8311 and 8050 genes were predicted to be targeted by up- and down-regulated miRNAs, respectively, between follicular fluid from hyperstimulated vs. unstimulated heifers. To overcome the limitation of DAVID Bioinformatic Resource (limited processing ability: 3,000 genes), only top 100 genes were considered for each miRNA for further bioinformatics analysis. Accordingly, after screening of the long list of genes, 3000 and 2700 genes were identified as potential targets of up- and down-regulated miRNAs, respectively, in follicular fluid samples. Similarly, a total of 6267 and 4982 genes were found to be potentially targeted by up- and down-regulated miRNAs in blood plasma samples of the experimental animals. Following screening, a total of 1300 and 800 genes were identified as potential targets of up- and down-regulated miRNAs, respectively, in blood plasma samples of hyperstimulated versus unstimulated heifers. Finally the screened list of genes from both follicular fluid and blood plasma samples were subjected to a pathway analysis using NCBI DAVID Bioinformatic Resource 6.7 to identify the significantly enriched canonical pathway (P < 0.05).
As shown in Table 2 and Table 3, among the several list of pathways, MAPK signalling pathway and Wnt signaling pathway are the most enriched pathways potentially regulated by the differentially expressed miRNAs. In addition, pathways involved in various signal transductions and cell-to-cell interaction including ErbB signaling pathway, Axon guidance, Neurotrophin signaling pathway, Endocytosis, Oocyte meiosis, TGF-beta signaling pathway and Focal adhesion were among the most enriched pathways for genes potentially targeted by miRNAs derived from both follicular fluid and blood plasma of hyperstimulated heifers. Taken together, these bioinformatics exploratory analyses suggest that variation in the level of circulatory miRNAs in follicular fluid and blood plasma might affect critical pathways involved in follicular development and oocyte maturation. As miRNAs target prediction algorithms are known to contain both false positives and false negatives, and our in silico pathway enrichment analysis is based on mRNA genes predicted to be targeted by extracellular miRNAs, a full understanding of the potential functional role of extracellular miRNAs can only be established using functional experiments.
Table 2.
List of enriched pathways (p <0.05), in which genes predicted to be targeted by differentially expressed miRNAs (P <0.05) between follicular fluid from hyperstimulated vs. unstimulated heifers
| Pathway | miRNAs involved | P value | Top genes |
|---|---|---|---|
| Overexpressed miRNAs (hyp vs. unst) | |||
| TGF-beta signaling pathway | miR-106b, −132, −148b-5p, −182, −212, −374a, −548j | 1.75E-07 | TGFBR2, TGFB2, SMAD2, SMAD3, SMAD4, BMPR2 |
| Axon guidance | miR-22-5p, −30b, −31, −33a-3p, −182, −132, −550a | 2.85E-06 | EFNB1, DCC, EPHB4, EPHA3, PLXNA1, PAK4 |
| MAPK signaling pathway | miR-30b, −106b, −132, −182, −212, −548j, −202-5p | 6.43E-05 | MAP3K1, MAP3K5, KRAS, MRAS, GRB2, FGF7 |
| Endocytosis | miR-33a-3p, −106b, −182, −374a, −374b, −202-5p | 3.83E-04 | RAB11FIP4, RAB11FIP2, EEA1, IGF1R, EPS15, EPN |
| Colorectal cancer | miR-30b, −33a-3p, −106b, −132, −212, −384, −494 | 5.94E-04 | SOS1, FZD3, SMAD2, DCC, MAPK1, BAX |
| Pathways in cancer | miR-33a-3p, −107, −132, −212, −494, −495, −548j | 5.95E-04 | E2F1, FGF18, WNT16, FGF7, PTEN, MITF |
| Wnt signaling pathway | miR-132, −212, −33a-3p, −494, −940, −495, −548j, −107 | 6.73E-04 | LRP5, LRP6, TCF7, PLCB4, DVL3, WNT1, WNT5A |
| Neurotrophin signaling pathway | miR-106b, −30b, −940, −182, −212, −107, | 0.001267 | PIK3R2, NTRK2, NTRK3, RPS6KA6, IRS1, RAC1 |
| Oocyte meiosis | miR-212, −132, −940, 495, −595, −107 | 0.003369 | CDC27, CPEB1, PRKACB, FBXW11, MAPK1, RPS6KA3 |
| GnRH signaling pathway | miR-940, −495 | 0.007085 | PRKCA, SRC, MAP3K2, MAPK14, GRB2, MAP3K3 |
| Underexpressed miRNAs (Hyp vs. Unst) | |||
| Pathways in cancer | miR-659, −141, −190, −449a, −200c, −361-5p, −145 | 9.51E-12 | KRAS, PTEN, VEGFA, STAT5B, MITF, BCL2 |
| MAPK signaling pathway | miR-23a, −23b, −141, −145, −200c, −92a, −125b, −24 | 2.94E-11 | MKNK2, MEF2C, FGF, SOS1, RAS, MAerkPK2 |
| Wnt signaling pathway | miR-125b, −449a, −302e, −145, −23a, −29b-2-5p, −659, −200c | 2.76E-08 | LRP5, LRP6, TCF7, PLCB4, DVL3, WNT1, WNT5A |
| ErbB signaling pathway | miR-23a, −23b, −125b, −206, −302e, −513c | 3.19E-07 | ERBB4, GRB2, SHC1, PIK3R3, PIK3CD, AKT3 |
| Colorectal cancer | miR-659, 23a, −23b, −141, −190, −449a, −200c, −361-5p, −145 | 4.48E-07 | DCC, APPL1, TGFBR2, SMAD3, SMAD4, APC |
| Axon guidance | miR-23a, −23b, −200c, −145 | 7.23E-07 | PLXNC1, EFNB2, PAK4, DCC, SEMA6A, PAK7, MET |
| Neurotrophin signaling pathway | miR-302e, −361-5p, −452, −525-5p, −449a, −659 | 1.82E-06 | NTRK2, NGFR, KRAS, MAP3K1, PIK3R3, PLCG1 |
| Renal cell carcinoma | miR-659, −141, −190, −449a, −200c, −361-5p, −145 | 3.75E-05 | GAB1, MET, SOS1, GRB2, VEGFA, NRAS |
| Focal adhesion | miR-24, −27b, −125a-3p, −139-5p, −206, −200c | 5.04E-05 | ITGB3, PXN, SHC3, ACTB, SRC, PAK2 |
| Regulation of actin cytoskeleton | miR-200c, −145, −27b, −92a, −300, −206 | 8.29E-05 | RAC1, ROCK2, GIT1, PIK3R3, ITGA3, LIMK1 |
Table 3.
List of enriched pathways (P < 0.05), in which genes predicted to be targeted by differentially expressed miRNAs (P < 0.05) in blood plasma of hyperstimulated vs. unstimulated heifers
| Pathway name | miRNA involved | P value | Top genes |
|---|---|---|---|
| Overexpressed miRNAs (hyp vs. unst) | |||
| Pathways in cancer | let-7 g, miR- 20b-3p, −22, −26b-3p, −34a, −221, −181c | 1.44E-09 | AKT3, PIK3R3, RASSF1, KRAS, MEK, DCC |
| Wnt signaling pathway | let-7 g, miR-221, −32 | 4.04E-08 | WNT1, SMAD2, DKK2, DVL3, VANGL2, CCND1 |
| Neurotrophin signaling pathway | let-7 g, miR-22, −181c, −221 | 6.73E-08 | NTRK2, IRS2, AKT3, NGF, PIK3CD, NRAS |
| Axon guidance | miR-34a, −181c, −221 | 2.19E-07 | PLXNC1, DPYSL2, PAK1, MET, KRAS, EFNB1 |
| Endocytosis | miR-22, −34a, −130b-3p, −576-5p | 1.23E-06 | TGFBR1, RAB5A, RAB11FIP4, PDCD6IP, VPS37A |
| MAPK signaling pathway | let-7 g, miR-22, −32, −34a, −221 | 2.69E-06 | RRAS, MAP3K1, MAPK1, MAPK9, SRF, MAP2K4 |
| Colorectal cancer | let-7 g, miR-22, −26b-3p, −24-2-5p | 4.59E-06 | DCC, TGFBR1, KRAS, MAP2K1, AKT3, CASP3 |
| Chronic myeloid leukemia | miR-22, −181c, −221 | 4.98E-06 | NRAS, AKT2, BCR, MAPK1, GAB2, BCL2L1 |
| TGF-beta signaling pathway | miR-26b-3p, −221, −22, −32 | 9.63E-06 | ACVR2A, TGFBR2, SMAD2, SMAD4, BMP7 |
| ErbB signaling pathway | miR-22, −181c, −221 | 2.96E-05 | ERBB4, GAB1, MAP2K1, PAK4, PIK3CD |
| Underexpressed miRNAs (Hyp vs. Unst) | |||
| Pathways in cancer | miR-125b, −153, −410, −494 | 9.51E-12 | DCC, GRB2, SMAD2, SOS1, E2F3 |
| MAPK signaling pathway | mir-125b, −153, −494 | 2.94E-11 | MEF2C, MAPKAPK2, MAP3K1, TGFB2, FGFR2 |
| Wnt signaling pathway | miR-147, −153, −410, −494 | 2.76E-08 | LRP6, WNT5A, DVL3, DKK2, PLCB1, ANGL2 |
| ErbB signaling pathway | miR-125b, −410 | 3.19E-07 | ERBB4, SOS1, MAPK1, NRG3, GAB1, MAP2K7 |
| Colorectal cancer | miR-125b, −410, −153, −494 | 4.48E-07 | DCC, BCL2, SMAD4, RAF1, SMAD2, PIK3R3 |
| Axon guidance | miR-147, −153, −410, −494 | 7.23E-07 | PLXNA2, ROCK2, EFNA3, NFAT5, MAPK1 |
| Neurotrophin signaling pathway | miR-125b, −134, −147, −494 | 1.82E-06 | NTF3, MAP3K1, SOS1, SORT1, BCL2 |
| Melanogenesis | miR-134, −99a-3p | 4.52E-06 | MAPK1, WNT5A, KRAS, GNAI3, CREB1 |
| Melanoma | miR-125b, −494 | 1.70E-05 | E2F2, FGFR1, IGF1R, FGF7, RAF1 |
| Prostate cancer | miR-125b, −147, −153, −410, −494 | 1.82E-05 | FGFR2,, IGF1R, MAPK1, IGF1, BCL2 |
Temporal expression of candidate circulatory miRNAs in blood plasma across oestrus cycle
In order to explore the temporal changes in the expression of circulatory miRNA especially in blood plasma of hyperstimulated heifers, the expression of candidate miRNAs was investigated at different days during the estrous cycle (Days 0, 3 and 7). As shown in Fig. 4 expression analysis of candidate miRNAs (miR-221, miR-103, let-7 g, miR-134, miR-147 and miR-127-3p) using qRT-PCR revealed the presence of temporal changes in the pattern of expression depending on the time during the estrous cycle as shown in Fig. 4. While four of the candidates namely. miR-221, miR-103, miR-134 and miR-127-3p showed a significant increase in abundance at day 7 of the estrous cycle compared to days 0 and 3, miR-147 showed a decreasing pattern from day 0 to day 7. No significant difference in the expression of let-7 g miRNA has been observed between the days during estrous cycle.
Fig. 4.
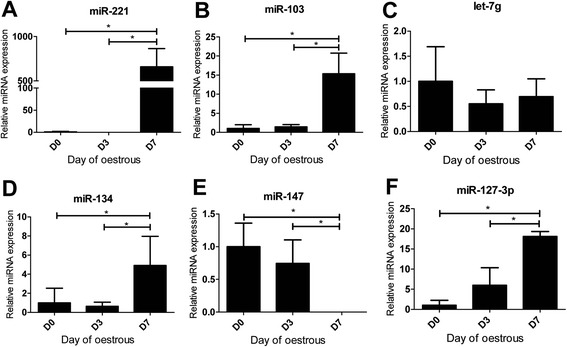
Temporal expression of candidate circulatory miRNAs in blood plasma across estrous days. Relative expression of candidate circulatory miRNAs in blood plasma of hypersitimulated heifers across different days during estrous (a-f) Expression of miR-221, miR-103, let-7g, miR-134, miR-147 and miR-127-3p was determined by RT-qPCR and each miRNA expression level was normalized using global normalization method. The error bars showed the SD. The significance of differences for miRNA expression was calculated using two-tailed T-test and *, p < 0.05. D0: onset of estrous; D3: day 3 of estrous; D7: day 7 of estrous
Validation of exosomes and Ago2 protein complexes isolated from follicular fluid and blood plasma
To determine the mode of circulation of candidate miRNAs in follicular fluid and blood plasma, the exosomal and Ago2 protein fractions of the biological fluid samples were isolated using differentially ultracentrifugation and immunoprecipitation procedures respectively. The specificity of both isolation procedures was validated using a western blot analysis of exosomes and Ago2 specific marker proteins. Immunoblotting of CD63 protein showed the presence of strong band in the exosome fraction of both follicular fluid and blood plasma samples, while the Ago2 protein band was evident in immunoprecipitated portion of the samples (Fig. 5). Despite ignorable detection of Ago2 protein in exosome fractions, strong bands for CD63 and Ago2 proteins in exosomes and Ago2 fraction, respectively suggest the efficient recovery of exosomes and Ago2 protein complexes from follicular fluid and blood plasma.
Fig. 5.
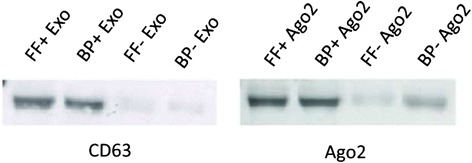
Specificity of isolation of exosomes and Ago2 protein complex from follicular fluid and blood plasma. Exosome and Ago2 proteins were isolated from organic-phenol fraction during total RNA isolation using miRNeasy kit and resolved in 8M urea. Protein concentrations were quantified using Bradford assay and total of twenty microgram protein from each group were separated in 12 % SDS-PAGE, transferred nitrocellulose membrane and incubated with specific antibody (CD63 and Ago2). Followed by HRP-conjugated secondary antibody and detected using chemiluminescent substrate. Western-blot results indicate the specificity of exosome and Ago2 protein isolation from follicular fluid (FF) and blood plasma (BP) as indicated in the corresponding figures
Detection of candidate circulatory miRNAs in exosomes and Ago2 fraction of follicular fluid
Here we investigate the expression pattern of selected candidate miRNAs in both exosomes and Ago2 fractions of follicular fluid derived from hyperstimulated versus unstimulated heifer groups. For this a total of six miRNAs representing those whose expression was induced (miR-212, miR-182 & let-7 g) or suppressed (miR-100, miR-877 and miR-200c) due to hyperstimulation were selected for detecting their expression in exosome and Ago2 fractions. The level of detection of candidate miRNAs was determined by real time PCR and data are shown as raw Ct value in Fig. 6. As shown in this Fig. 6, except miR-182, all candidate miRNAs were detected in both exosomal and Ago2 fraction of follicular fluid of both hyperstimulated and unstimulated animals. Mir-182 was not detected in Ago2 fraction (with raw Ct value more than 35) in both hyperstimulated and unstimulated heifers.
Fig. 6.
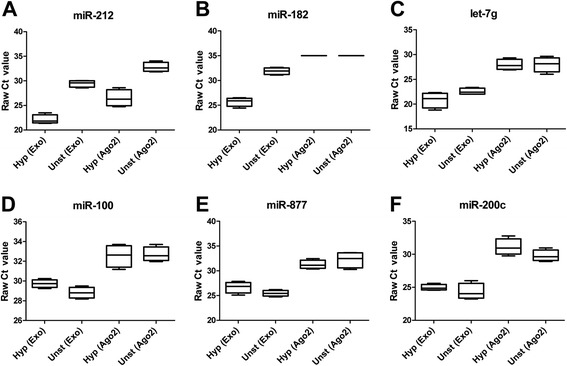
Detection of candidate extracellular miRNAs in exosomes and Ago2 fraction of follicular fluid derived from hyperstimulated and unstimulated heifers. The boxplots on panels a through f represent the detection of candidate extracellular miRNAs in exosomes (Exo) or Ago2 protein complex (Ago2) fraction of follicular fluid samples derived from hyperstimulated (Hyp) and unstimulated (Unst) heifers. The error bars show the SD. Data are presented as raw Ct value and Ct value of more than 35 was considered as undetected
Detection of candidate circulatory miRNAs in exosomes and Ago2 fraction of blood plasma
Here we have tried to detect candidate miRNAs, which were enriched (miR-221, miR-103 & let-7 g) or suppressed (miR-134, miR-147 & miR-127-3p) in blood plasma samples of hyperstimulated animals compared to the unstimulated ones. As shown in Fig. 7, all candidate miRNAs, except miR-103 and miR-127-3p, were detected in both exosomal and Ago2 fraction of blood plasma of both hyperstimulated and unstimulated heifers. Mir-103 and miR-127-3p were not detected in the Ago2 protein fraction of both treatment groups.
Fig. 7.
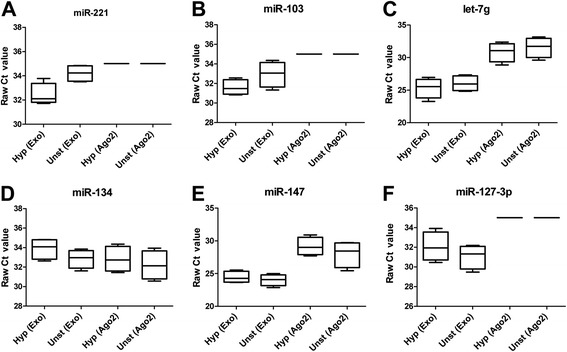
Detection of candidate circulating miRNAs in exosomes and Ago2 fraction of blood plasma derived from hyperstimulated and unstimulated heifers. The boxplots on panels a through f represent the detection of candidate extracellular miRNAs in exosomes (Exo) or Ago2 protein complex (Ago2) fraction of blood plasma samples from hyperstimulated (Hyp) and unstimulated (Unst) heifers. The error bars show the SD of 4 replicates. Data are presented as raw Ct value and Ct value of more than 35 was considered as undetected
Discussion
Controlled ovarian hyperstimulation (COH) facilitate growth of multiple follicles in monoovulatory animals. COH involves stimulation of the ovaries by supraphysiological levels of gonadotropins and also associated with very low level of luteinizing hormone during the luteal phase, the peri-implantation and implantation period (Kolibianakis et al., 2006). Thus, the goal of COH is multifollicular recruitment with retrieval of multiple oocytes in an effort to compensate for the “inefficiencies” of the outcome of IVF laboratory. However, several reports showed that, COH itself has adverse effects on oocyte development, embryo quality, endometrial receptivity and perhaps also perinatal outcomes [6, 7, 32] The effect of COH on oviductal environment and subsequently on the transcriptome of the resulting embryos has been evidenced [8].
Recently miRNAs have been detected in extracellular environment mainly in different bio-fluids of various species [15, 25, 33, 34]. Given the emerging association of circulating miRNA species with a spectrum of patho-physiological conditions, we have systematically explored the changes in circulatory miRNAs in follicular fluid and blood plasma induced by controlled ovarian hyperstimulation. The results of this study clearly uphold our primary hypothesis that introduction of supra-physiological level of gonadotropins during the process of COH may induce the alteration of expression of extracellular miRNAs in follicular fluid as well as in blood plasma.
Since their discovery [13, 14], extracellular miRNAs got a research priority as they have been found to be differentially expressed in altered physiological conditions [13]. Recently, it has been shown that a large number of miRNAs are present in bovine follicular fluid and their expression patterns differs depending on the growth status of oocyte [15]. In the present study we profiled extracellular miRNAs in bovine follicular fluid and blood plasma derived from hyperstimulated and unstimulated heifers using Human miRNome PCR array platform. A specific miRNA transcript was considered as detected when its Ct value <35 and present in at least 75 % of the replicates. This selection criteria was set based on the previous experiences in our lab with a similar study design [15]. Similar studies on detection of miRNAs in plasma samples also use the same threshold level to determine the detection level of transcripts [35], even as high as ≤37 cycle as a threshold to determine the level of detection of miRNAs in equine follicular fluid [36]. Despite using a heterologous approach, the quantitative real time PCR analysis revealed a wide array of miRNAs to be present in both follicular fluid and blood plasma, indicating the cross species conservation feature of miRNAs between human and bovine as it has been observed between wide range of species [37]. Of 748 miRNA investigated in the PCR array panel, a total of 504 and 402 miRNAs were detected in follicular fluid and blood plasma, respectively. Comparable detection rate was also reported for follicular fluid [15] and blood plasma [38]. Among the detected miRNAs, 373 were commonly found in both follicular fluid and blood plasma. Results from this study and our previous findings [15] confirmed that, relatively higher number of extracellular miRNAs are present in follicular fluid than blood plasma, which may be attributed to the condensed nature of the former.
By comparing hyperstimulated vs. unstimulated heifers, 57 and 21 miRNAs were found to be differentially regulated in follicular fluid and blood plasma, respectively. Out of 57 differentially expressed miRNAs, 30 miRNAs were found to be up-regulated in follicular fluid of hyperstimulated heifers. Several of these up-regulated miRNAs have been reported to play an important role in follicular development and other physiological processes. For instance, miR-212 & miR-132 were reported to be involved in regulating follicular development [39, 40]. Moreover, these two miRNAs (miR-212, −132) were found to be significantly up-regulated in preovulatory mouse mural granulosa cells following LH/hCG induction [41] indicating their potential involvement in follicular development in those species. The miR-148*, a minor form of miR-148, was also significantly up-regulated in follicular fluid of hyperstimulated heifers. Although, the role of this miRNA in follicular development is unclear, miR-148/152 family was found to be important for growth and development of normal tissue [42]. Interestingly, miRNAs like miR-643, miR-548j, miR-224* and miR-33b, which were found to be highly abundant in follicular fluid of hyperstimulated heifers, are also reported to be highly associated with cancer [43–46]. Among the down-regulated miRNAs, miR-361-5p [47] and miR-27b (homologous to miR-27a, sharing 20 out of 21 nts) [48] were reported as tumor suppressors. Despite accumulated evidences on the hormonal regulatory role of cellular miRNAs and vice versa [41], the present study is the first of its kind reporting the effect of suprarphysiological level of gonadotropins on the level of circulating miRNAs in bovine follicular fluid and blood plasma. However, the consequence of this alteration on the resulting oocytes, embryos and further ovarian function needs further investigation.
In blood plasma, 21 miRNAs were found to be differentially expressed between hyperstimulated and unstimulated heifers where 13 miRNAs were up-regulated and 8 miRNAs were down regulated. Among the up-regulated miRNAs miR-708-3p, which showed the highest fold change regulation, was reported in promoting the development of bladder carcinoma via direct repression of Caspase-2 [49]. Several studies have shown the association of miR-26b, which was induced in expression due to hyperstimulation, with pregnancy-associated disorder like preeclampsia [50]. The potential association of this miRNA in reproductive disorder due to hyperstimulated ovaries cannot be ruled out. Interestingly, miR-26b was also reported to be elevated in plasma from severe preeclamptic pregnancies [51]. MiR-221, which is commonly known as oncomir, was also significantly elevated in blood plasma of hyperstimulated heifers. In the present study we also found a panel of down regulated miRNAs including miR-153 and miR-134 which are commonly known as tumor suppressor miRNA [52, 53].
Among the differentially expressed miRNAs in follicular fluid and blood plasma of hyperstimulated heifers, 13 miRNAs belong to 6 miRNA clusters suggesting the importance of understanding the structure and function of these miRNAs as clusters. miRNA clusters, which are groups of tandem miRNA genes that are closely located in the genome, are abundantly and widely distributed in animal genomes. It has been revealed that about 50 % of the miRNA genes in Drosophila [54] and over 30 % of the miRNA genes in human, mouse, rat and chicken are located in the genome as clusters [55]. The length of the transcript typically varies from a couple of kb to over 10 kb. It has been reported that each miRNA cluster usually has up to 10 miRNA genes [56]. Many of the miRNA clusters may have one core promoter region and transcriptional start site shared by all miRNA genes within that cluster and are ultimately expressed within a single RNA transcript [57]. Co-expression experiments of clustered miRNA genes showed that one miRNA cluster is usually transcribed as a single transcriptional unit [58] as a result of some kind of regulatory coordination within the clustered miRNAs. In the present study, we found that the miR-134 ~ 410 cluster which comprise 3 miR genes, while rest of the clusters has 2 miR genes per cluster. Although several miRNA clusters have been reported to be associated with pathological and physiological conditions in human and mice, the overall expression pattern of extracellular miRNA clusters during follicular development has not been extensively studied. We have recently identified several classes of cluster miRNAs in bovine follicular granulosa cells derived from subordinate and dominant follicles during folliculogenesis [59, 60]. Since many miRNA genes within a cluster share a common promoter or utilize common regulatory machinery, examination of their expression pattern and regulatory mechanism could enhance our understanding of the role of extracellular miRNAs in the process of follicular development.
Another interesting observation from our study is that the number of differentially expressed miRNAs in follicular fluid is higher (57) than blood plasma (21) of hyperstimulated heifers. This is may be due to the fact that gonadotropins action is specific to the target organs [61]. It has been well accepted that miRNAs are involved in hormone regulation or vice versa [41]. FSH controls the development of granulosa cells during folliculogenesis by stimulating their proliferation and differentiation, and by promoting the formation of the follicular antrum. In particular, FSH enhances the biosynthesis of female sex hormones estradiol and progesterone, which control the estrous cycle and reproduction [62, 63]. Numerous studies have shown that the LH surge initiates the transcriptional upregulation and downregulation of genes, including cytokines, transcription factors, and matrix-remodeling proteins, within periovulatory granulosa cells [64, 65]. Therefore, the higher number of differentially regulated miRNAs in follicular fluid of hyperstimulated heifers may indicate the higher transcriptional activity of follicular cells within follicular microenvironment during the process of COH.
In addition to their expression level, we have investigated the biological relevance of these differentially regulated miRNAs. For this the target genes of differentially expressed miRNAs were predicted bioinformatically and the dominant biological pathways were determined. The most significant pathways enriched by predicted targets for up-regulated miRNAs in follicular fluid of hyperstimulated heifers including TGF-beta signaling pathway, Axon guidance, neurotrophin signaling, MAPK signaling colorectal cancer and pathways in cancer. All these pathways are known to be involved in ovarian follicular growth, different developmental processes and also in several pathological conditions. MiRNAs differentially expressed in plasma samples were found to be involved several pathways including cancer, WNT signaling pathway Axon guidance, MAPK signaling pathways colorectal cancer etc. Although the miRNA expression profiles were different from each other, in silico predicted biological pathways were similar between follicular fluid and blood plasma. This is consistent with the observation that the course of ovarian hyperstimulation may be characterized by certain common pathways in the whole physiology of the animal. However, it is important to note that our observations for pathways are exploratory in nature and need further validation using wet-lab studies to elucidate miRNA-mRNA interaction and causal association of differentially expressed miRNAs and the predicted pathways.
So far, it has been established that miRNAs are expressed spatio-temporally in different developmental process [66] and also in adult tissues [67]. A study conducted with sheep ovarian tissues demonstrated differential expression of miRNA in follicles and corpora lutea during the ovine oestrous cycle [68]. This study demonstrated that eight miRNAs (miR-503, miR-21, miR-29b, miR-142-3p, miR-34a, miR-152, miR-25 and miR-130a) were highly expressed, while nine miRNAs (miR-125a, miR-199a-3p, miR-125b, miR-99a, let-7c, miR-145, miR-31, miR-202 and miR-27b) were expressed at lower level between the follicular and luteal stages in ovine ovarian tissues. In the present study temporal expression of miRNAs was observed in blood plasma of hyperstimulated heifers across estrous cycle. For instance, miR-221, miR-103, miR-134 and miR-127-3p show significantly higher expression at day 7 of estrous cycle compared to day 0 or day 3. In contrast, miR-147 has significantly lower expression at day 7 of estrous. However, there is no significant difference in the expression of let-7 g across the estrous days.
Several studies have demonstrated that circulating miRNAs may be coupled either with exosomes [21, 25] or Ago2 protein complex [28] to bypass the high RNases activity in blood stream or in follicular fluid. To proof the mode of circulation of those miRNAs in follicular fluid and blood plasma we have detected the expression of candidate miRNAs in exosomal and Ago2 protein fractions of both follicular fluid and blood plasma of heifers in both treatment groups. The specificity of isolation of exosome and Ago2 fraction was confirmed by western blot analysis of CD63 and Ago2 protein [15, 69]. Candidates including miR-182 from follicular fluid (Fig. 4) and miR-221, miR-103 and miR-127-3p from blood plasma (Fig. 5) were found to be detected only in exosomal fraction and completely undetected in the Ago2 fraction. In both follicular fluid and blood plasma samples, all miRNA which were detected in both fractions, showed higher abundance in exosomal fraction compared to the Ago2 protein complex. This is in agreement with our previous findings showing that exosome mediated release of miRNAs to be the dominant pathway in follicular fluid [15]. Interestingly, the hyperstimulation treatment had no effect on the detection of candidate miRNAs either in exosomal or Ago2 protein complexes.
Conclusion
The present study demonstrated the consequence of ovarian hyperstimulation in inducing changes in the relative abundance of extracellular miRNAs, which are potentially involved in regulation of genes involved in several physiological pathways. Moreover, the release of miRNAs into extracellular space in both follicular fluid and blood plasma was not affected by superstimulation treatment.
Acknowledgement
The authors are indebted to all members of Life and Brain institute of University of Bonn for their careful assistance during the qPCR array work.
Footnotes
Sina Seifi Noferesti and Md. Mahmodul Hasan Sohel contributed equally to this work.
Competing interests
The authors declare no conflict of interest.
Authors’ contributions
SSN participated in sample collection, carried out the molecular analysis, drafting the manuscript. MHS participated in molecular analysis, in data analysis, drafting the manuscript. MH participated in study design and collected samples from experimental animals. DSW participated in the study design, data analysis. ET participated in study design and statistical analysis. CL participated in reading the draft manuscript. FR participated in sample collection and handling works. CN participated in study design and draft manuscript reading. KS participated in study design and supervision of the experiment. DT made the study design, supervised the experiment, and participated in the preparation of the manuscript. All authors read and approved the final manuscript.
References
- 1.Adams GP, Jaiswal R, Singh J, Malhi P. Progress in understanding ovarian follicular dynamics in cattle. Theriogenology. 2008;69(1):72–80. doi: 10.1016/j.theriogenology.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 2.Adams GP, Matteri RL, Kastelic JP, Ko JC, Ginther OJ. Association between surges of follicle-stimulating hormone and the emergence of follicular waves in heifers. J Reprod Fertil. 1992;94(1):177–188. doi: 10.1530/jrf.0.0940177. [DOI] [PubMed] [Google Scholar]
- 3.Berfelt DR, Lightfoot KC, Adams GP. Ovarian synchronization following ultrasound-guided transvaginal follicle ablation in heifers. Theriogenology. 1994;42(6):895–907. doi: 10.1016/0093-691X(94)90113-W. [DOI] [PubMed] [Google Scholar]
- 4.Mapletoft RJ, Steward KB, Adams GP. Recent advances in the superovulation in cattle. Reprod Nutr Dev. 2002;42(6):601–611. doi: 10.1051/rnd:2002046. [DOI] [PubMed] [Google Scholar]
- 5.Santos MA, Kuijk EW, Macklon NS. The impact of ovarian stimulation for IVF on the developing embryo. Reproduction. 2010;139(1):23–34. doi: 10.1530/REP-09-0187. [DOI] [PubMed] [Google Scholar]
- 6.Ertzeid G, Storeng R. Adverse effects of gonadotrophin treatment on pre- and postimplantation development in mice. J Reprod Fertil. 1992;96(2):649–655. doi: 10.1530/jrf.0.0960649. [DOI] [PubMed] [Google Scholar]
- 7.Van der Auwera I, D'Hooghe T. Superovulation of female mice delays embryonic and fetal development. Hum Reprod. 2001;16(6):1237–1243. doi: 10.1093/humrep/16.6.1237. [DOI] [PubMed] [Google Scholar]
- 8.Gad A, Besenfelder U, Rings F, Ghanem N, Salilew-Wondim D, Hossain MM, et al. Effect of reproductive tract environment following controlled ovarian hyperstimulation treatment on embryo development and global transcriptome profile of blastocysts: implications for animal breeding and human assisted reproduction. Hum Reprod. 2011;26(7):1693–1707. doi: 10.1093/humrep/der110. [DOI] [PubMed] [Google Scholar]
- 9.Baart EB, Macklon NS, Fauser BJ. Ovarian stimulation and embryo quality. Reprod Biomed Online. 2009;18(Suppl 2):45–50. doi: 10.1016/S1472-6483(10)60448-8. [DOI] [PubMed] [Google Scholar]
- 10.Hyttel P, Greve T, Callesen H. Ultrastructure of oocyte maturation and fertilization in superovulated cattle. Prog Clin Biol Res. 1989;296:287–297. [PubMed] [Google Scholar]
- 11.Toloubeydokhti T, Bukulmez O, Chegini N. Potential regulatory functions of microRNAs in the ovary. Semin Reprod Med. 2008;26(6):469–478. doi: 10.1055/s-0028-1096127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 14.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sohel MM, Hoelker M, Noferesti SS, Salilew-Wondim D, Tholen E, Looft C, et al. Exosomal and Non-Exosomal Transport of Extra-Cellular microRNAs in Follicular Fluid: Implications for Bovine Oocyte Developmental Competence. PLoS One. 2013;8(11):e78505. doi: 10.1371/journal.pone.0078505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freeman DJ, Tham K, Brown EA, Rumley A, Lowe GD, Greer IA. Fetal corticotrophin-releasing hormone mRNA, but not phosphatidylserine-exposing microparticles, in maternal plasma are associated with factor VII activity in pre-eclampsia. J Thromb Haemost. 2008;6(3):421–427. doi: 10.1111/j.1538-7836.2007.02882.x. [DOI] [PubMed] [Google Scholar]
- 17.Shalchi Z, Sandhu HS, Butt AN, Smith S, Powrie J, Swaminathan R. Retina-specific mRNA in the assessment of diabetic retinopathy. Ann N Y Acad Sci. 2008;1137:253–257. doi: 10.1196/annals.1448.008. [DOI] [PubMed] [Google Scholar]
- 18.Sandhu HS, Butt AN, Powrie J, Swaminathan R. Measurement of circulating neuron-specific enolase mRNA in diabetes mellitus. Ann N Y Acad Sci. 2008;1137:258–263. doi: 10.1196/annals.1448.044. [DOI] [PubMed] [Google Scholar]
- 19.Kopreski MS, Benko FA, Kwak LW, Gocke CD. Detection of tumor messenger RNA in the serum of patients with malignant melanoma. Clin Cancer Res. 1999;5(8):1961–1965. [PubMed] [Google Scholar]
- 20.Dasi F, Lledo S, Garcia-Granero E, Ripoll R, Marugan M, Tormo M, et al. Real-time quantification in plasma of human telomerase reverse transcriptase (hTERT) mRNA: a simple blood test to monitor disease in cancer patients. Lab Invest. 2001;81(5):767–769. doi: 10.1038/labinvest.3780285. [DOI] [PubMed] [Google Scholar]
- 21.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110(1):13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 22.Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10(1):42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 23.Halicka HD, Bedner E, Darzynkiewicz Z. Segregation of RNA and separate packaging of DNA and RNA in apoptotic bodies during apoptosis. Exp Cell Res. 2000;260(2):248–256. doi: 10.1006/excr.2000.5027. [DOI] [PubMed] [Google Scholar]
- 24.Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19(2):43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 26.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285(23):17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13(4):423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108(12):5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39(16):7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laterza OF, Lim L, Garrett-Engele PW, Vlasakova K, Muniappa N, Tanaka WK, et al. Plasma MicroRNAs as sensitive and specific biomarkers of tissue injury. Clin Chem. 2009;55(11):1977–1983. doi: 10.1373/clinchem.2009.131797. [DOI] [PubMed] [Google Scholar]
- 31.Cui W, Ma J, Wang Y, Biswal S. Plasma miRNA as biomarkers for assessment of total-body radiation exposure dosimetry. PLoS One. 2011;6(8) doi: 10.1371/journal.pone.0022988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raga F, Casan EM, Kruessel J, Wen Y, Bonilla-Musoles F, Polan ML. The role of gonadotropin-releasing hormone in murine preimplantation embryonic development. Endocrinology. 1999;140(8):3705–3712. doi: 10.1210/endo.140.8.6899. [DOI] [PubMed] [Google Scholar]
- 33.Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, et al. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3(9):e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008;3(11):e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui XS, Zhang DX, Ko YG, Kim NH. Aberrant epigenetic reprogramming of imprinted microRNA-127 and Rtl1 in cloned mouse embryos. Biochem Biophys Res Commun. 2009;379(2):390–394. doi: 10.1016/j.bbrc.2008.12.148. [DOI] [PubMed] [Google Scholar]
- 36.da Silveira JC, Veeramachaneni DN, Winger QA, Carnevale EM, Bouma GJ. Cell-secreted vesicles in equine ovarian follicular fluid contain miRNAs and proteins: a possible new form of cell communication within the ovarian follicle. Biol Reprod. 2012;86(3):71. doi: 10.1095/biolreprod.111.093252. [DOI] [PubMed] [Google Scholar]
- 37.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37(7):766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 38.van Schooneveld E, Wouters MC, Van der Auwera I, Peeters DJ, Wildiers H, Van Dam PA, et al. Expression profiling of cancerous and normal breast tissues identifies microRNAs that are differentially expressed in serum from patients with (metastatic) breast cancer and healthy volunteers. Breast Cancer Res. 2012;14(1):R34. doi: 10.1186/bcr3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schauer SN, Sontakke SD, Watson ED, Esteves CL, Donadeu FX. Involvement of miRNAs in equine follicle development. Reproduction. 2013;146(3):273–282. doi: 10.1530/REP-13-0107. [DOI] [PubMed] [Google Scholar]
- 40.Tripurani SK, Wee G, Lee KB, Smith GW, Wang L, Jianboyao MicroRNA-212 post-transcriptionally regulates oocyte-specific basic-helix-loop-helix transcription factor, factor in the germline alpha (FIGLA), during bovine early embryogenesis. PLoS One. 2013;8(9):e76114. doi: 10.1371/journal.pone.0076114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fiedler SD, Carletti MZ, Hong X, Christenson LK. Hormonal regulation of MicroRNA expression in periovulatory mouse mural granulosa cells. Biol Reprod. 2008;79(6):1030–1037. doi: 10.1095/biolreprod.108.069690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen L, Li Y, Fu Y, Peng J, Mo MH, Stamatakos M, et al. Role of deregulated microRNAs in breast cancer progression using FFPE tissue. PLoS One. 2013;8(1):e54213. doi: 10.1371/journal.pone.0054213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takwi AA, Li Y, Becker Buscaglia LE, Zhang J, Choudhury S, Park AK, et al. A statin-regulated microRNA represses human c-Myc expression and function. EMBO Mol Med. 2012;4(9):896–909. doi: 10.1002/emmm.201101045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyachi K, Sawada Y, Shida Y, Sugawara A, Hisatomi H. Lipogenic gene expression profile in patients with gastric cancer. Mol Clin Oncol. 2013;1(5):825–827. doi: 10.3892/mco.2013.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Yao X, Li SN, Suo AL, Tian T, Ruan ZP, et al. Detection of prostate cancer related copy number variations with SNP genotyping array. Eur Rev Med Pharmacol Sci. 2013;17(21):2916–2922. [PubMed] [Google Scholar]
- 46.Zhang GJ, Zhou H, Xiao HX, Li Y, Zhou T. Up-regulation of miR-224 promotes cancer cell proliferation and invasion and predicts relapse of colorectal cancer. Cancer Cell Int. 2013;13(1):104. doi: 10.1186/1475-2867-13-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu D, Tao T, Xu B, Chen S, Liu C, Zhang L, et al. MiR-361-5p acts as a tumor suppressor in prostate cancer by targeting signal transducer and activator of transcription-6(STAT6) Biochem Biophys Res Commun. 2014;445(1):151–156. doi: 10.1016/j.bbrc.2014.01.140. [DOI] [PubMed] [Google Scholar]
- 48.Vimalraj S, Miranda PJ, Ramyakrishna B, Selvamurugan N. Regulation of breast cancer and bone metastasis by microRNAs. Dis Markers. 2013;35(5):369–387. doi: 10.1155/2013/451248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song T, Zhang X, Zhang L, Dong J, Cai W, Gao J, et al. miR-708 promotes the development of bladder carcinoma via direct repression of Caspase-2. J Cancer Res Clin Oncol. 2013;139(7):1189–1198. doi: 10.1007/s00432-013-1392-6. [DOI] [PubMed] [Google Scholar]
- 50.Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28(10):1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 51.Wu L, Zhou H, Lin H, Qi J, Zhu C, Gao Z, et al. Circulating microRNAs are elevated in plasma from severe preeclamptic pregnancies. Reproduction. 2012;143(3):389–397. doi: 10.1530/REP-11-0304. [DOI] [PubMed] [Google Scholar]
- 52.Zhao S, Deng Y, Liu Y, Chen X, Yang G, Mu Y, et al. MicroRNA-153 is tumor suppressive in glioblastoma stem cells. Mol Biol Rep. 2013;40(4):2789–2798. doi: 10.1007/s11033-012-2278-4. [DOI] [PubMed] [Google Scholar]
- 53.Tahiri A, Leivonen SK, Luders T, Steinfeld I, Ragle Aure M, Geisler J, et al. Deregulation of cancer-related miRNAs is a common event in both benign and malignant human breast tumors. Carcinogenesis. 2014;35(1):76–85. doi: 10.1093/carcin/bgt333. [DOI] [PubMed] [Google Scholar]
- 54.Lai EC, Tomancak P, Williams RW, Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4(7):R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Megraw M, Sethupathy P, Corda B, Hatzigeorgiou AG. miRGen: a database for the study of animal microRNA genomic organization and function. Nucleic Acids Res. 2007;35(Database issue):D149–155. doi: 10.1093/nar/gkl904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hertel J, Lindemeyer M, Missal K, Fried C, Tanzer A, Flamm C, et al. The expansion of the metazoan microRNA repertoire. BMC Genomics. 2006;7:25. doi: 10.1186/1471-2164-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282(4):2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- 58.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11(3):241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salilew-Wondim D, Ahmad I, Gebremedhn S, Sahadevan S, Hossain MD, Rings F, et al. The expression pattern of microRNAs in granulosa cells of subordinate and dominant follicles during the early luteal phase of the bovine estrous cycle. PLoS One. 2014;9(9):e106795. doi: 10.1371/journal.pone.0106795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gebremedhn S, Salilew-Wondim D, Ahmad I, Sahadevan S, Hossain MM, Hoelker M, et al. MicroRNA Expression Profile in Bovine Granulosa Cells of Preovulatory Dominant and Subordinate Follicles during the Late Follicular Phase of the Estrous Cycle. PLoS One. 2015;10(5):e0125912. doi: 10.1371/journal.pone.0125912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hazum E, Conn PM. Molecular mechanism of gonadotropin releasing hormone (GnRH) action. I. The GnRH receptor. Endocr Rev. 1988;9(4):379–386. doi: 10.1210/edrv-9-4-379. [DOI] [PubMed] [Google Scholar]
- 62.Amsterdam A, Selvaraj N. Control of differentiation, transformation, and apoptosis in granulosa cells by oncogenes, oncoviruses, and tumor suppressor genes. Endocr Rev. 1997;18(4):435–461. doi: 10.1210/edrv.18.4.0306. [DOI] [PubMed] [Google Scholar]
- 63.Amsterdam A, Rotmensch S, Ben-Ze'ev A. Coordinated regulation of morphological and biochemical differentiation in a steroidogenic cell: the granulosa cell model. Trends Biochem Sci. 1989;14(9):377–382. doi: 10.1016/0968-0004(89)90012-1. [DOI] [PubMed] [Google Scholar]
- 64.Richards JS, Russell DL, Robker RL, Dajee M, Alliston TN. Molecular mechanisms of ovulation and luteinization. Mol Cell Endocrinol. 1998;145(1–2):47–54. doi: 10.1016/S0303-7207(98)00168-3. [DOI] [PubMed] [Google Scholar]
- 65.Russell DL, Robker RL. Molecular mechanisms of ovulation: co-ordination through the cumulus complex. Hum Reprod Update. 2007;13(3):289–312. doi: 10.1093/humupd/dml062. [DOI] [PubMed] [Google Scholar]
- 66.Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9(10):1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McBride D, Carre W, Sontakke SD, Hogg CO, Law A, Donadeu FX, et al. Identification of miRNAs associated with the follicular-luteal transition in the ruminant ovary. Reproduction. 2012;144(2):221–233. doi: 10.1530/REP-12-0025. [DOI] [PubMed] [Google Scholar]
- 69.Logozzi M, De Milito A, Lugini L, Borghi M, Calabro L, Spada M, et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS One. 2009;4(4):e5219. doi: 10.1371/journal.pone.0005219. [DOI] [PMC free article] [PubMed] [Google Scholar]


