Abstract
Covalent modifications to histones play important roles in chromatin dynamics and the regulation of gene expression. The JmjC-containing histone demethylases (HDMs) catalyse the demethylation of methylated lysine residues on histone tails. Here we report the development of homogeneous luminescence based assay methods for measuring the catalytic activity and the binding affinities of peptides to HDMs. The assays use Amplified Luminescent Proximity Homogeneous Assay (ALPHA) technology and are sensitive, robust, and can be used for small molecule inhibitor screening of HDMs. We have profiled known inhibitors of JMJD2E and demonstrate correlation between the inhibitor potencies determined by the ALPHA and other types of assays. Although this study focuses on the JMJD2E isoform, the catalytic turnover and binding assays described here can be used in studies on other HDMs. The assays should be useful for the development of small molecule inhibitors selective for HDM isoforms.
Keywords: Epigenetics, Histone demethylase (HDM), JMJD2, Fe(II) and 2-oxoglutarate (2OG) oxygenases, AlphaScreen
Introduction
An important mechanism for the regulation of gene expression is the post-translational modifications of histones. Covalent modifications to histones identified to date include acetylation, methylation, phosphorylation, ubiquitination, sumoylation and biotinylation [1, 2]. Together with other factors, different combinations of modifications are proposed to regulate gene expression patterns and ultimately lead to phenotypic outcomes in an epigenetic manner [2-4].
Histone methylation plays a role in heterochromatin formation and maintenance, X-chromosome inactivation, transcriptional regulation, DNA repair and genomic imprinting [5]. Abnormal histone Nε-lysine methylation has been linked to diseases, including tumorigenesis and mental retardation [6-8]. Until recently, histone methylation was thought to be an irreversible process [9]. However, the identification of lysine-specific demethylase (LSD1)[10], a flavin-dependent amine oxidase, and more recently, the JumonjiC (JmjC) containing histone lysine demethylases (HDMs) [11, 12] has revealed that methylation / demethylation is a dynamic process.
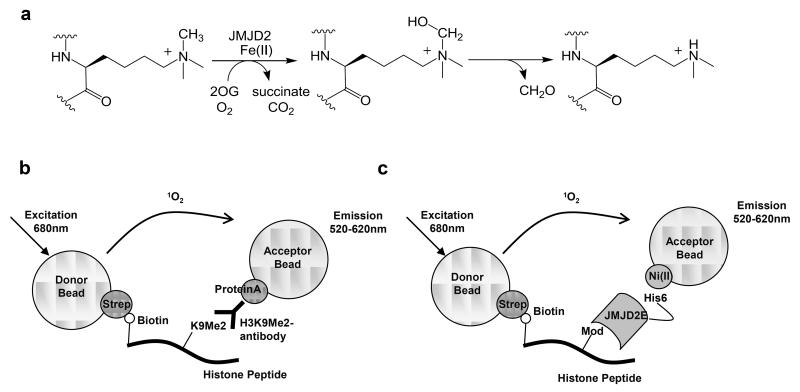
The catalytic JmjC domain comprises a modified double-stranded β-helix fold which is stereotypical of the Fe(II) and 2-oxoglutarate (2OG) dependent oxygenases [6, 12-14]. HDMs can catalyse the demethylation of tri-, di and mono-methylated lysines (Figure 1a). Although the available evidence is that all HDMs follow the same, or closely related, catalytic mechanism, they demonstrate differences in their histone lysine site specificity as well as their ability to demethylate different methylated states of lysines [11, 15, 16]. For instance, whilst the JMJD2 subfamily (JMJD2A, JMJD2B, JMJD2C, JMJD2D, JMJD2E) can demethylate trimethyl lysines (e.g. H3K9Me3)[15], the JHDM1 (KDM2) subfamily can only accept di- and mono- methyl lysines [11].
Figure 1. AlphaScreen assay method.
(a) Jmjc demethylation outline reaction mechanism. JmjC HDMs demethylate tri-, di- and mono-methylated lysines in an oxidative mechanism using Fe(II) and 2OG as co-substrate/co-factor; the hydroxylated intermediate undergoes fragmentation to give formaldehyde and the demethylated lysine product.
(b) HDM peptide turnover AlphaScreen assay setup. The assay utilises a methyl-state specific antibody to detect the demethylated product. Using an AlphaScreen IgG detection kit (Streptavidin-coated donor beads / Protein A-coated acceptor beads), the interaction of the antibody with biotinylated peptide can be detected. In the case of JMJD2, biotinylated-H3K9Me3 peptide is demethylated to its di-methylated state in the presence of Fe(II) and 2OG. An antibody selective for H3K9Me2 (Ab1220) is added to the reaction, and the interaction between the peptide and the antibody is detected by addition of the IgG detection beads.
(c) HDM-peptide binding AlphaScreen assay setup. His-tagged protein and biotinylated-peptide interactions can be detected using AlphaScreen His-detection beads.
Several assay methods have been developed for quantifying the activity of JmjC-containing HDMs. These include immunodetection with bulk histones using methylation status and site specific antibodies [11]; direct detection of histone peptide demethylation by matrix-assisted laser desorption/ionisation-time of flight (MALDITOF) mass spectrometry [11, 15, 17-19]; formaldehyde detection [11, 19-22]; and 2OG turnover assays [8]. The reported HDM assays suffer from drawbacks, including assay throughput, lack of sensitivity that requires relatively high concentrations of HDM to be used placing a lower limit on the IC50 values that can be determined, requirement for expensive equipment or drawbacks associated with the use of assays coupled to the activity of another enzyme (false positives/negatives).
We are interested in identifying selective small molecule inhibitors that can be used as probes for the functions of histone modifying enzymes at biochemical and physiological levels [19, 23]. Here we describe a homogeneous assay for HDMs using the Amplified Luminescent Proximity Homogeneous Assay (ALPHA) technology. The assay is amenable to HTS and ideal for screening of chemical inhibitors of demethylases. In this study, JMJD2E, which demethylates tri and di-methyl-lysine9 (K9) in histone H3, was selected as a model enzyme since it has been kinetically characterised [19, 24] and shows high activity and stability levels. We also demonstrate that the methodology can be used to investigate binding interactions of peptides with HDMs.
Materials and Methods
Reagents
Reagents and solvents were from Aldrich or Alfa Aesar unless otherwise stated. AlphaScreen IgG and Histidine (Nickel Chelate) kits were from PerkinElmer Life and Analytical Sciences. The monoclonal antibody against K9Me2 (Ab1220) was from Abcam.
Peptide synthesis
Peptides with C-terminal amides were synthesised using a CS-Bio automated solid phase peptide synthesiser as described [18]. N-Terminal biotinylation of peptides was carried out on the resin using biotin p-nitrophenyl ester (Merck Chemicals Ltd). Peptides were purified using HPLC (C18 reverse phase column). The synthetic biotinylated and non-biotinylated peptides used in this study are described in Supplementary Table 1. The histone H3 (H3) biotinylated peptide library was from AltaBioscience.
HDM peptide turnover assay using AlphaScreen
The N-terminally His-tagged catalytic domain of recombinant JMJD2E (residues 1-337) was expressed and purified as reported [18, 19]. HDM assays were carried out in 384 well white Proxiplates (PerkinElmer). In a typical assay, JMJD2E was incubated with 100μM ascorbate, 1μM (NH4)2SO4.FeSO4.6H2O, 10μM 2OG and biotin-H3(1-15)K9Me3 peptide in 50mM HEPES (pH7.5) buffer containing 0.01% (v/v) Tween 20 and 0.1% (w/v) BSA (final concentrations), in a 10μL reaction at 22°C. The reaction was quenched by the addition of 5μl of 30mM EDTA. Streptavidin-conjugated donor and Protein-A-conjugated acceptor beads (5μl) were pre-incubated with monoclonal antibody (mAb1220, Abcam), added to the quenched assay and incubated for 1 hr in the dark. The final bead concentration was 20μg/ml and the antibody concentration was 0.3μg/mL in a 20μL reaction volume. For inhibitor studies, assays (10μL) contained compounds dissolved in 0.1% DMSO. The plates were analysed using an Envision (PerkinElmer) plate reader.
HDM peptide binding assay using AlphaScreen
Binding assays were carried out in 384 well white Proxiplates (PerkinElmer) in 20μL reaction volume. His-tagged JMJD2E was incubated with biotinylated peptide for 30 min at 22°C in 50mM HEPES (pH 7.5), 0.01% Tween 20, 0.1% BSA buffer. For inhibition studies, JMJD2E was pre-incubated with inhibitors for 30min prior to the addition of biotinylated peptides. AlphaScreen Streptavidin-conjugated donor and Nickel-chelate conjugated acceptor beads were added to the wells at a final concentration of 10μg/ml and incubated for further 1 hr in the dark at 22°C. The plates were analysed using an Envision (PerkinElmer) plate reader.
Formaldehyde dehydrogenase (FDH) Coupled Inhibition Assay Methods
FDH assays were carried out as described [19] but using a range of histone H3 peptides. In brief, FDH (0.1 U), NAD+ (500 μM), ascorbate (100 μM), Fe(II) (various concentrations), JMJD2E (2 μM), H3 peptide (at various concentrations), 2-OG (100μM) were incubated together at 37°C for 12 min while fluorescence was recorded (355 nm excitation, 460 nm emission) at 30 s intervals. All reagents were used as solutions in HEPES buffer (50 mM, pH 7.5), with the exception of Fe (II) solutions, which were made using (NH4)2Fe(SO4)2 dissolved in 20 mM HCl to make 400 mM stock solutions, which were then diluted to the appropriate concentration using MilliQ water. All solutions were kept on ice until being mixed and incubated at 37 °C. Data were processed using GraphPad Prism 5.0™.
Results
Demethylase turnover assay principle and setup
In AlphaScreen assays, laser excitation (680nm) of a photosensitiser within the “donor bead” converts ambient oxygen to an excited O2 singlet oxygen. If an “acceptor bead” containing thioxene derivates is proximate (~200nm), energy transfer occurs producing a luminescent signal at 520-620nm [25]. In the absence of an acceptor bead, singlet oxygen returns to its ground state and no signal is detected. Both donor and acceptor bead surfaces can be conjugated to biomolecules, giving diversity in the binding partners to be investigated. In the HDM catalytic assay, we used a streptavidin-coated donor and Protein A-coated acceptor bead pair (Figure 1b) to detect the turnover of biotinylated K9Me3 to biotinylated K9Me2 by probing with a K9Me2 selective antibody. JMJD2 demethylase also catalyses the conversion of H3K9Me2 to H3K9Me, but less efficiently (as judged by kcat/Km) than H3K9Me3 to H3K9Me2 [22].
Initially, we assessed the compatibility of donor/acceptor beads with the HDM assay components. The JmjC HDMs require Fe (II) as a cofactor and reducing agent (e.g. ascorbate) for optimal in vitro activity. Because the AlphaScreen methodology relies on the singlet oxygen energy transfer, strong quenchers such as transition metals (e.g. iron) and compounds such as ascorbate (or their reaction products) have the potential to interfere with the assay. The effect of Fe(II) and ascorbate at different concentrations on the luminescence signal produced by biotinylated-IgG was examined (Supplementary Figure 1b,c). Up to 5μM of Fe (II) and 50μM ascorbate (equivalent to 10μM and 100μM in the demethylase assay respectively) had no effect on the luminescence signal.
Secondly, the sensitivity of the assay for JMJD2E demethylase relies on the antibody selectivity for the di-methylated K9 product over the tri-methylated K9 substrate. Monoclonal K9Me2 antibody (Ab1220, 0.3μg/mL) was incubated with a range of concentrations of biotinylated histone H3 peptides with different methylated lysine states (no methylation (K9me0), mono-methyl lysine (K9Me1), di-methyl lysine (K9Me2) and tri-methyl lysine (K9Me3)). Ab1220 was found to be sufficiently selective for binding K9Me2 over K9Me3 (Figure 2a) as well as any other methyl-state K9 peptides (Supplementary Figure 1a). The highest selectivity (>10 fold) for K9Me2 across all methylation states was obtained at 10-30nM, and 30nM substrate was selected for further assay development.
Figure 2. JMJD2E peptide turnover AlphaScreen assay.
(a) Antibody selectivity against different methylation states. Antibody selectivity against biotinylated H3peptides (15mer) with different Lysine (K9) methylation states. Biotinylated peptides were incubated with Ab1220 (0.3μg/ml) and AlphaScreen IgG detection beads (20μg/ml) in 20μl reaction volume at 22°C for 1hr.
(b) AlphaScreen JMJD2E demethylase reaction controls. Increase in AlphaScreen signal was observed only when all the assay components were present in the reaction.
(c) Time-course for JMJD2E enzyme activity. Turnover assay was run at different JMJD2E concentrations and quenched with EDTA at various time intervals. AlphaScreen beads were added to the quenched reactions simultaneously once the final time-point was taken.
(d) Inhibition of JMJD2E by 2,4-PDCA (Compound 1). IC50 value for 2,4-PDCA against JMJD2E was 0.9μM as calculated using non-linear regression using normalised dose-response fit on Prism GraphPad. The final concentration of DMSO in the reaction was 0.1%. Average ± StdError (N=3).
(e) Correlation curve for JMJD2E pIC50 values (FDH vs. AlphaScreen).
The HDM assay was carried out in 10μL reaction volumes in a 384-well plate format. EDTA was used to quench the reaction and was incubated with AlphaScreen beads (20μg/mL final) pre-incubated with Ab1220. The Fe (II) concentration was maintained at 1μM, equivalent to ≥ 100 fold molar excess of the initial enzyme concentrations (≤ 10nM) tested in the assay. Under these assay conditions, the luminescence signal increase was only observed in the presence of all the assay components, and omission of any of the components (JMJD2E, Fe (II), 2OG or biotin-H3(1-15)K9Me3) resulted in no signal increase (Figure 2b).
JMJD2E concentration was titrated in the assay with the biotin-H3(1-15)K9Me3 substrate at 30nM. The time-courses over 60 min show that activity can be measured at JMJD2E concentrations as low as 0.5nM, with the initial rates being proportional to enzyme concentrations (Figure 2c). Signal to noise ratio (S/N) was highest for 5-10nM JMJD2E (S/N ~ 30, 0.6 < Z’ <0.9 at both 10, 20 min time points); approximately 15% biotin-H3(1-15)K9Me3 was demethylated after 15 min at 5nM JMJD2E, as calculated from the biotin-H3(1-15)K9Me2 standard curve. This enzyme concentration was selected for screening of enzyme inhibitors.
Application of the assay to inhibitor studies of JMJD2E
Pyridine carboxylic acids [19], bipyridine carboxylic acids [19], N-oxalyglycine (NOG, compound 7) [26], and N-oxalyl-D-tyrosine derivatives [24] have been identified as inhibitors of JMJD2E, as determined by both FDH and MALDI-TOF mass spectrometry assays, with IC50 values in the micromolar range [19, 27]. We therefore tested the suitability of the luminescence-based turnover assay for inhibitor screening using these chemical scaffolds.
In the inhibition assays, compounds were pre-incubated with JMJD2E (5nM in 10μL final reaction), Fe(II) and ascorbate mixture for 15 min prior to initiating the reaction by addition of biotin-H3(1-15)K9Me3 (30nM in 10μl final reaction) and 2OG. The reaction was quenched with EDTA after 15-20 min, when beads that had been pre-incubated with the K9Me3 antibody (20μg/mL in final 20μL volume) were added.
The inhibition results are summarised in Table 1. Overall, the ALPHA method was able to identify compounds that were inhibitors of JMJD2E across several different chemical scaffolds. A significant correlation was found between the IC50 values determined using ALPHA and FDH assay, (Spearman R = 0.9643, P (two-tailed) = 0.0028), demonstrating that this luminescent-based assay is a valid method for identifying inhibitors of demethylases (Figure 2e). Compound 1 (2,4-PDCA) showed the most potent inhibition, with a clear dose-response curve and an IC50 value of 0.88 μM (Figure 2d), in agreement with IC50 values observed using other assay methods (Table 1). Weak JMJD2E inhibition was observed with compound 6 (NOG), at 225μM. Previously identified potent inhibitor derivatives of N-oxalyl-D-tyrosine were also identified as inhibitors of JMJD2E, validating this assay as a reliable screening method for identifying HDM inhibitors.
Table 1. IC50 values for JMJD2E.
The final concentration of DMSO in the reaction was 0.1%, except NOG where no DMSO was used. SEMs in log(IC50) values were below 10%.
| Compound | R | IC50 (AlphaScreen) /μM | IC50 (FDH) / μM | MALDI-TOF / μM | |
|---|---|---|---|---|---|
| 2,4-PDCA (1) |

|
0.9 | 0.2§-1.4 * | 0.9§ - 1.4* | |
| 2,3-PDCA (2) |
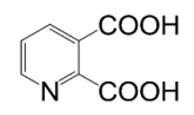
|
> 100 | >5000 * | - | |
| 2,5-PDCA (3) |

|
> 100 | 180 * | - | |
| 3,6-PDCA (4) |

|
> 100 | >1000 * | - | |
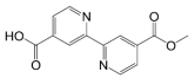
| (5) |
|
10.9 | 6.6* | - | |
| NOG (6) |

|
255 | 78 * | - | |
| (7) |

|

|
110 | 37.1† | 76† |
| (8) |

|
91 | 16.6† | 25† | |
| (9) |

|
20.7 | 14.8† | 59† | |
| (10) |

|
24.9 | 20.1† | - | |
| (11) |

|
>100 | 185† | - |
AlphaScreen enzyme-peptide binding assay principle and setup
AlphaScreen binding assays have been successfully used to investigate protein-protein interactions in epigenetic proteins, including malignant brain tumour (MBT) domains [28], chromo and tudor domains, as well as plant homeodomains [29], all of which are methyl-lysine recognition proteins. Having established that the ALPHA technology can be used to detect HDM activity by peptide product - antibody interaction, we next explored the possibility of using the bead chemistry to study the peptide - HDM (protein-protein) interactions. Such an assay is potentially useful not only for measuring the binding of small molecules at the active site, but also for investigating the binding of peptides and proteins associated with HDMs.
In the binding assay mode, a Streptavidin-coated donor and Ni2+-chelate acceptor bead pair was used (Figure 1c). Upon binding of biotinylated peptide and His6-tagged JMJD2E, the donor and acceptor beads (associated with biotin and His-tags respectively) become proximate, allowing energy transfer and luminescence signal to be detected when the beads are excited.
Initially we investigated the binding of biotin-H3(1-15)K9Me3 to His6-tagged JMJD2E. The H3K9Me3 peptide is a good substrate for JMJD2s and was considered to be the most suitable peptide for development of a binding assay. Biotin-H3(1-15)K9Me3 and JMJD2E were co-incubated at varied concentrations, after which the AlphaScreen beads were added. An increase in luminescence signal was observed with increasing concentrations of enzyme and peptide, indicating peptide-enzyme binding in a dose-dependent manner. There was negligible background signal when either the peptide or the enzyme was omitted, excluding the possibility of bead precipitation by either component. Binding was observed under a wide range of peptide (3 - 1000nM) and enzyme (3 - 1000nM) concentrations. A maximum was observed with the JMJD2E titrations; increases in JMJD2E concentration above 30nM gave a reduction in signal. This “hook effect” is precedented and arises because excess binding molecules beyond the bead binding capacity become inhibitory to the signal production [30] (Figure 3a). No such effect was observed over the range of peptide concentrations tested (Figure 3b). An equimolar JMJD2E/peptide concentration at 10nM, which is below the saturation concentration for JMJD2E, was selected for further binding studies.
Figure 3. JMJD2E protein-protein binding AlphaScreen assay.
(a-b) Titration of JMJD2E and biotin-H3(1-15)K9Me3. Both biotin-H3(1-15)K9Me3 (a) and His-tagged JMJD2E (b) were titrated in order to determine the optimal concentrations for the binding studies. JMJD2E concentration (a) and biotin-K9Me3 concentration (b) was maintained at 10nM for titration studies.
(c-e) Peptide and small molecule affinities against JMJD2E. Assays were all carried out in triplicate, and SEMs in log(IC50) values were below 10%.
*Data from [27] † Highest three concentration data points omitted to calculate EC50. No luminescence signal interference was observed for unlabeled peptides.
HDM-peptide interactions and probing for affinities for other histone marks
To determine the binding affinity of H3(1-15)K9Me3 peptide to JMJD2E, unlabelled H3(1-15)K9Me3 was titrated and pre-incubated with JMJD2E, and subsequently incubated with biotin-H3(1-15)K9Me3 (Figure 3c). The unlabelled H3(1-15)K9Me3 displaced its biotinylated peptide with IC50 value of 1.2μM, demonstrating that the unlabelled peptide competes with biotinylated peptide for JMJD2E binding. This IC50 value is in the similar range to the affinity observed for biotinylated H3K9Me3 and JMJD2A determined using a surface plasmon resonance technique (apparent affinity ~ 1μM)[17]. Competition assays with other unlabelled K9 methyl-state peptide revealed a methylation state dependence on the binding affinities, where higher methylation state had higher affinity, with a rank order of H3(1-15)K9Me3 > H3(1-15)K9Me2 > H3(1-15)K9Me1. This correlates with the ranking of the corresponding peptide Km values observed in the turnover assays determined by the FDH assay (Figure 3e). The H3(6-21)K9Me3 peptide had higher JMJD2E affinity than H3(1-15)K9Me3 in the binding assay, which is in agreement with the lower Km value for this peptide (Figure 3e). These results demonstrate that peptide binding affinities can be detected by AlphaScreen for HDMs and relative binding affinities can be measured for both coupled and un-coupled (e.g. where competing peptide sequence is not the same as the biotinylated peptide sequence) sequences. Moreover, the binding affinities are peptide length and sequence dependent, consistent with peptide kinetic parameters (Figure 3e) and turnover rates (unpublished data).
Small molecule modulators of HDM-peptide binding interactions
The effect of 2OG (compound 1) and NOG (compound 7) on the HDM-peptide interaction was then assessed. JMJD2E was pre-incubated with 2OG or NOG, prior to addition of biotinylated-H3(1-15)K9Me3. An increase in luminescence signal was observed with increasing concentrations of 2OG, a co-substrate of JMJD2E, suggestive of a synergistic effect of 2OG and peptide binding to JMJD2E. The signal maximum is reached at around 10 – 30μM, at approximately the Km of 2OG (14μM)[19]. A similar effect was seen with NOG, a non-reactive 2OG analogue and a competitive inhibitor, where enzyme-peptide binding was enhanced in presence of increasing concentrations of NOG. Whilst NOG inhibits JMJD2E activity (Table 1), the assay demonstrated that NOG does not inhibit the binding of peptide-enzyme interaction. This is in accordance with kinetic data, as well as the binding mode observed in NOG-complexed JMJD2A crystal structures [17, 18, 22]. For 2OG to bind to HDMs, an active site transition metal is required. In these assays, we did not add a metal ion, because JMJD2E co-purifies with at least some Fe(II); Ni(II) may also have been present from the affinity purification step.
2,4-PDCA, and the N-oxalyl-D-tyrosine derivatives described in Table 1, interfered with the bead chemistry and reliable IC50 values could not be measured. The interference is most probably with the Ni2+-chelate acceptor bead, as no interference was observed with Protein-A conjugated acceptor bead in the turnover assays.
The binding assay provides an alternative screening method to the turnover assay for identifying novel JMJD2E inhibitors. Use of both assay modes may allow different types of inhibitors (peptide binding inhibitors / enzyme turnover inhibitors) and novel chemical scaffolds to be identified as modulators of HDMs.
Discussion
The versatility of the ALPHA technology allows its application in a variety of assay formats. We have described two different applications of the luminescent assay for HDMs; an assay for detecting the enzyme activity of HDMs, and an assay for determining the protein-protein binding interactions of HDMs.
In the catalytic turnover mode, the combination of a modified lysine histone peptide substrate with a lysine methylation state selective antibody allowed for successful measurement of HDM enzyme activity. Direct detection of product formation using antibodies not only has the advantage of the high sensitivity of an immunoassay, but also low background due to the specific bead chemistry. Unlike other immunoassays such as western blotting and ELISA, the assay is homogenous and does not require multiple wash steps. We were able to measure JMJD2E enzyme activity at low nM concentrations of enzyme and peptide, avoiding the drawbacks of limited assay range and high protein consumption associated with other reported HDM assay methods where μM enzyme concentrations are required. Because there is a limit to the number of binding sites on the beads and a corresponding upper limit to the peptide concentration that can be used in this assay, the assay cannot be readily used to determine kinetic parameters (e.g. Km, Vmax values) for HDMs; however, we have demonstrated that small-molecule inhibitors can be reliably screened, generating robust IC50 data and with excellent Z’ and S/N values, making the assay amenable to HTS. The assay setup described here is a signal increase type assay, where the product is probed using a product methyl-state selective antibody. However, the assay can also be used in signal decrease assays, where the loss of biotinylated substrate is probed. The assay choice is dependent on the availability of the methyl-state selective antibody and the assay sensitivity that is required. Furthermore, other utilities of ALPHA technology include rapid antibody characterisation and screening for selectivity across various biotinylated peptides, as demonstrated in this paper (Figure 2a).
We are currently extending the catalytic turnover assay for detecting demethylase activities within the JMJD2 subfamily as well as for other HDM subfamilies, by utilising various combinations of appropriate methylated lysine peptide and antibodies.
Previous studies on demethylase binding interactions with histone peptides have employed techniques such as isothermal titration calorimetery (ITC)[20] and surface plasmon resonance [17, 31]. Despite the high sensitivity of these methods, they are limited by low-throughput rates, high enzyme and peptide consumption and complex binding of the protein-peptide interactions, which have made the data analysis difficult. Here, we have provided an alternative method of binding studies for HDMs, which is simple, homogeneous and robust. We were able to demonstrate JMJD2E binding to a variety of modified histone peptide sequences, and the peptide binding affinity determined in our luminescent method was comparable to that of BIAcore studies [17].
The amenability of the assay to high-throughput formatting should make it useful for analysing the combination of binding of histone modifications to HDMs and other histone modifying enzymes.
Supplementary Material
Acknowledgement of Financial Support
This work was supported by the Wellcome Trust. The Structural Genomics Consortium is a registered charity (no. 1097737) funded by the Wellcome Trust, GlaxoSmithKline, Genome Canada, the Canadian Institutes of Health Research, the Ontario Innovation Trust, the Ontario Research and Development Challenge Fund, the Canadian Foundation for Innovation, Vinnova, Knut and Alice Wallenberg foundation and Karolinska Institute. The work was also supported by the Oxford NIHR Biomedical Research Unit.
We thank Owen Chang for providing compound 5, and Martin Philpott and Stan Ng for helpful discussions.
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 4.Karlic R, Chung HR, Lasserre J, Vlahovicek K, Vingron M. Histone modification levels are predictive for gene expression. Proc Natl Acad Sci U S A. 107:2926–31. doi: 10.1073/pnas.0909344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Volkel P, Angrand PO. The control of histone lysine methylation in epigenetic regulation. Biochimie. 2007;89:1–20. doi: 10.1016/j.biochi.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 6.Agger K, Christensen J, Cloos PA, Helin K. The emerging functions of histone demethylases. Curr Opin Genet Dev. 2008;18:159–68. doi: 10.1016/j.gde.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Ng SS, Yue WW, Oppermann U, Klose RJ. Dynamic protein methylation in chromatin biology. Cell Mol Life Sci. 2009;66:407–22. doi: 10.1007/s00018-008-8303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loenarz C, Ge W, Coleman ML, Rose NR, Cooper CD, Klose RJ, Ratcliffe PJ, Schofield CJ. PHF8, a gene associated with cleft lip/palate and mental retardation, encodes for an N{varepsilon}-dimethyl lysine demethylase. Hum Mol Genet. 2009 doi: 10.1093/hmg/ddp480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol. 2001;13:263–73. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- 10.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 11.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–6. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 12.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–18. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 13.Loenarz C, Schofield CJ. Expanding chemical biology of 2-oxoglutarate oxygenases. Nat Chem Biol. 2008;4:152–6. doi: 10.1038/nchembio0308-152. [DOI] [PubMed] [Google Scholar]
- 14.Hewitson KS, Holmes SL, Ehrismann D, Hardy AP, Chowdhury R, Schofield CJ, McDonough MA. Evidence that two enzyme-derived histidine ligands are sufficient for iron binding and catalysis by factor inhibiting HIF (FIH) J Biol Chem. 2008;283:25971–8. doi: 10.1074/jbc.M804999200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–81. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 16.Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–4. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 17.Chen Z, Zang J, Kappler J, Hong X, Crawford F, Wang Q, Lan F, Jiang C, Whetstine J, Dai S, Hansen K, Shi Y, Zhang G. Structural basis of the recognition of a methylated histone tail by JMJD2A. Proc Natl Acad Sci U S A. 2007;104:10818–23. doi: 10.1073/pnas.0704525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng SS, Kavanagh KL, McDonough MA, Butler D, Pilka ES, Lienard BM, Bray JE, Savitsky P, Gileadi O, von Delft F, Rose NR, Offer J, Scheinost JC, Borowski T, Sundstrom M, Schofield CJ, Oppermann U. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature. 2007;448:87–91. doi: 10.1038/nature05971. [DOI] [PubMed] [Google Scholar]
- 19.Rose NR, Ng SS, Mecinovic J, Lienard BM, Bello SH, Sun Z, McDonough MA, Oppermann U, Schofield CJ. Inhibitor scaffolds for 2-oxoglutarate-dependent histone lysine demethylases. J Med Chem. 2008;51:7053–6. doi: 10.1021/jm800936s. [DOI] [PubMed] [Google Scholar]
- 20.Horton JR, Upadhyay AK, Qi HH, Zhang X, Shi Y, Cheng X. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat Struct Mol Biol. 17:38–43. doi: 10.1038/nsmb.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hopkinson RJ, Hamed RB, Rose NR, Claridge TD, Schofield CJ. Monitoring the activity of 2-oxoglutarate dependent histone demethylases by NMR spectroscopy: direct observation of formaldehyde. Chembiochem. 11:506–10. doi: 10.1002/cbic.200900713. [DOI] [PubMed] [Google Scholar]
- 22.Couture JF, Collazo E, Ortiz-Tello PA, Brunzelle JS, Trievel RC. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat Struct Mol Biol. 2007;14:689–95. doi: 10.1038/nsmb1273. [DOI] [PubMed] [Google Scholar]
- 23.Edwards AM, Bountra C, Kerr DJ, Willson TM. Open access chemical and clinical probes to support drug discovery. Nat Chem Biol. 2009;5:436–40. doi: 10.1038/nchembio0709-436. [DOI] [PubMed] [Google Scholar]
- 24.Rose NR, Woon EC, Kingham GL, King ON, Mecinovic J, Clifton IJ, Ng SS, Talib-Hardy J, Oppermann U, McDonough MA, Schofield CJ. Selective Inhibitors of the JMJD2 Histone Demethylases: Combined Nondenaturing Mass Spectrometric Screening and Crystallographic Approaches. J Med Chem. doi: 10.1021/jm901680b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ullman EF, Kirakossian H, Singh S, Wu ZP, Irvin BR, Pease JS, Switchenko AC, Irvine JD, Dafforn A, Skold CN, et al. Luminescent oxygen channeling immunoassay: measurement of particle binding kinetics by chemiluminescence. Proc Natl Acad Sci U S A. 1994;91:5426–30. doi: 10.1073/pnas.91.12.5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elkins JM, Hewitson KS, McNeill LA, Seibel JF, Schlemminger I, Pugh CW, Ratcliffe PJ, Schofield CJ. Structure of factor-inhibiting hypoxia-inducible factor (HIF) reveals mechanism of oxidative modification of HIF-1 alpha. J Biol Chem. 2003;278:1802–6. doi: 10.1074/jbc.C200644200. [DOI] [PubMed] [Google Scholar]
- 27.Sakurai M, Rose NR, Schultz L, Quinn AM, Jadhav A, Ng SS, Oppermann U, Schofield CJ, Simeonov A. A miniaturized screen for inhibitors of Jumonji histone demethylases. Mol Biosyst. 2009;6:357–64. doi: 10.1039/b912993f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wigle TJ, Herold JM, Senisterra GA, Vedadi M, Kireev DB, Arrowsmith CH, Frye SV, Janzen WP. Screening for Inhibitors of Low-Affinity Epigenetic Peptide-Protein Interactions: An AlphaScreenTM-Based Assay for Antagonists of Methyl-Lysine Binding Proteins. J Biomol Screen. 2009 doi: 10.1177/1087057109352902. [DOI] [PubMed] [Google Scholar]
- 29.Quinn AM, Bedford MT, Espejo A, Spannhoff A, Austin CP, Oppermann U, Simeonov A. A homogeneous method for investigation of methylation-dependent protein-protein interactions in epigenetics. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.PerkinElmer . A Practical Guide to working with AlphaScreen. PerkinElmer; 2004. [Google Scholar]
- 31.Tochio N, Umehara T, Koshiba S, Inoue M, Yabuki T, Aoki M, Seki E, Watanabe S, Tomo Y, Hanada M, Ikari M, Sato M, Terada T, Nagase T, Ohara O, Shirouzu M, Tanaka A, Kigawa T, Yokoyama S. Solution structure of the SWIRM domain of human histone demethylase LSD1. Structure. 2006;14:457–68. doi: 10.1016/j.str.2005.12.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.