Abstract
The JmjC oxygenases catalyse the N-demethylation of Nε-methyl lysine residues in histones and are current therapeutic targets. A set of 2-oxoglutarate analogues were screened using a unified assay platform for JmjC demethylases and related oxygenases. Results led to the finding that daminozide (N-(dimethylamino)succinamic acid, 160 Da), a plant growth regulator, selectively inhibits the KDM2/7 JmjC subfamily. Kinetic and crystallographic studies reveal daminozide chelates the active site metal via its hydrazide carbonyl and dimethylamino groups.
Introduction
Histone modifications are central to the regulation of eukaryotic gene expression. The dynamic methylation of lysine and arginine residues in histones has diverse transcriptional outcomes, with different methylation sites and states being associated with promotion or repression of transcription. There are two classes of histone lysine demethylases (KDMs), the largest of which uses 2-oxoglutarate (2OG) as a cosubstrate (the JmjC enzymes) and comprises ~18 human enzymes grouped into 5 subfamilies (Figure 1). Several JmjC demethylases are targets for the treatment of diseases including leukaemia, breast and prostate cancers1,2 and inflammation.3 However, only limited inhibition data has been reported for the JmjC enzymes with few reports of selective inhibitors.4-8
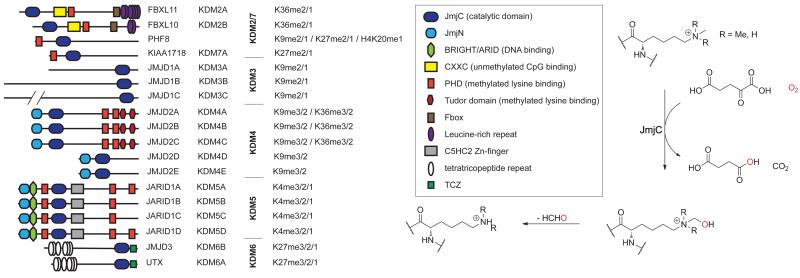
Figure 1. Subfamilies of the human 2-oxoglutarate (‘JmjC’) histone demethylases, showing their domain architecture.
Not all proposed family members are shown. Each demethylation reaction is coupled to 2OG decarboxylation and formaldehyde production. KIAA1718 has recently been assigned as KDM7A, however its JmjC domain shows high sequence and structural similarity with the KDM2 subfamily members.
2OG oxygenases have been targeted for therapeutic and agricultural applications9: an inhibitor of γ-butyrobetaine hydroxylase, BBOX1, is used clinically as a ‘cardioprotectant’, inhibition of the collagen prolyl hydroxylases has been investigated for treating fibrotic disease, and inhibitors of the hypoxia inducible factor hydroxylases are in clinical trials for the treatment of anaemia. These examples provide evidence that 2OG oxygenases are viable targets for therapeutic inhibition by small molecules in vivo. 2OG oxygenases involved in the biosynthesis of gibberellins, plant hormones involved in growth regulation, have also been targeted using small molecule inhibitors for both agricultural and horticultural applications. Here we report that daminozide, N-(Dimethylamino)succinamic acid, which was once widely used as a plant growth retardant but later withdrawn due to toxicity concerns, is a highly selective inhibitor of the KDM2/7 family of human JmjC histone demethylases.
Results
To enable the identification of selective JmjC demethylase subfamily inhibitors, we developed a unified screening platform containing representatives of each of the five human demethylase subfamilies (KDM2A/FBXL11, KDM3A/JMJD1A, KDM4E/JMJD2E, KDM5C/JARID1C, KDM6B/JMJD3) (Supporting Information). Active enzymes were expressed using bacterial or eukaryotic expression systems and purified to near homogeneity (Supporting Information). Kinetic parameters for 2OG and histone fragment substrates were determined (Table 1), and activity AlphaScreens (amplified luminescence proximity homogeneous assays) were developed (Figure S1).10 This assay is based on immunodetection of the lysine methylation state of biotin-conjugated histone peptide product, using Streptavidin- and Protein A- conjugated beads for quantification. For counter-screening, we selected three clinically relevant hypoxic response oxygenases, PHD2, FIH (prolyl and asparaginyl hydroxylases acting on Hypoxia Inducible Factor, respectively) and BBOX1 (γ-butyrobetaine hydroxylase) for which AlphaScreen or fluorescence assays11 were also developed. This screening platform is a significant improvement on previous methods for human 2OG oxygenases9; it enables the oxygenases to be used at low nanomolar concentrations. Standardisation of the analytical methods for enzymes employing different types of substrates enabled quantitative comparisons of inhibitor potencies to be made.
Table 1. Inhibition data (IC50) for daminozide and its analogues across KDMs and other 2OG oxygenases.
Data are reported as IC50 values in μM. AlphaScreen was used for all IC50 determinations except where values are given in brackets where MALDI assay was used; all BBOX assays were run using a fluorescence based assay.11 AlphaScreen assays were optimised to run at linear range of the reaction. Assays were performed at concentrations of 2OG near or below experimentally determined 2OG Km values. Where Kmapp values for 2OG for enzymes were unknown, the kinetic parameter was determined by using the FDH assay: KDM2A (12.5 ± 1.4 μM), PHF8 (16.6 ± 1.8 μM), KDM3A (5.3 ± 2.2 μM), KDM5C (41.7 ± 6.3 μM) and KDM6B (5.4 ± 0.4 μM).
In work aimed at identifying inhibitor scaffolds for the JmjC histone demethylases, we assembled a set of 2OG analogues known to inhibit 2OG oxygenases (Figure 2), and screened these against the 2OG oxygenase panel using the assay platform. As expected, known ‘generic’ 2OG oxygenase inhibitors9 including pyridine 2,4-dicarboxylic acid (compound 11), and N-oxalylglycine (compound 12) showed inhibition across the 2OG oxygenases tested, validating this assay platform. Interestingly, the clinically used histone deacetylase (HDAC) inhibitor Vorinostat/SAHA (suberoylanilide hydroxamic acid, compound 15) inhibited several histone demethylases (Figure 2).
Figure 2. Heatmap of JmjC demethylase inhibition by a set of 2OG analogues.
Daminozide 19 is selective for KDM2A. Tricarboxylic acid cycle intermediates succinate 1 and fumarate 2 were generally poor demethylase inhibitors, though KDM4E7 was an exception to this trend. (R)- and (S)-2-hydroxyglutarate enantiomers, produced by gain-of-function mutations to isocitrate dehydrogenase were inhibitors of the KDM2, KDM3 and KDM4 histone demethylases.12 The catechols 16 and 17, bipyridyl 18 and 8-hydroxyquinoline 21 inhibited all demethylases screened. By contrast, 5 most potently inhibited PHD2 (prolyl hydroxylase domain enzyme isoform 2). Each compound was screened in an AlphaScreen assay and is represented as % inhibition of the enzyme at 20 μM compound. Details of the assays are in the Supporting Information.
Unexpectedly, the plant growth regulator daminozide (compound 19) stood out as a selective inhibitor of KDM2A (IC50 = 1.5±0.7 μM), which is selective for dimethyl lysine residues. Daminozide was identified as a plant growth retardant in the 1960s13 and used to control stem growth, plant size and fruit ripening for over 20 years.14 In vivo, daminozide is proposed to inhibit 2OG oxygenases involved in gibberellin, and possibly ethylene, biosynthesis in plants.15,16 However, daminozide usage in food crops was curtailed due to the potential carcinogenicity of its metabolite, 1,1-dimethylhydrazine (although is still used for ornamental plants).17-19 A formaldehyde dehydrogenase (FDH) coupled assay monitoring formaldehyde production confirmed the potency of daminozide against KDM2A (IC50 = 1.5±0.4 μM). To test further for subfamily selectivity, daminozide was screened against two other members of the KDM2/7 subfamily (PHF8 and KIAA1718, Table 1) which are structurally highly related to KDM2A and KDM2B, and are both also selective for demethylation of dimethyl lysine residues, but have different sequence specificities.20-22 The results indicate that daminozide is at least 100-fold selective as an inhibitor of the KDM2/7 subfamily over the other demethylase subfamily members tested, with IC50s of 2 μM or less against KDM2A, PHF8, and KIAA1718 and IC50s of 127 μM for KDM3A (a different subfamily of dimethyl lysine demethylase) or greater (mM range) against other demethylases (Table 1). No inhibition was observed (at 1 mM) against other biologically important 2OG oxygenases that catalyse hydroxylation, i.e. PHD2, FIH and BBOX1. Given its simple achiral structure and low molecular weight (160 Da), the degree of selectivity exhibited by daminozide is remarkable.
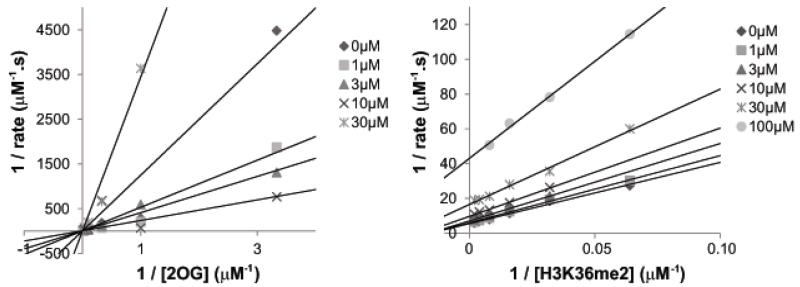
Kinetic analyses revealed that daminozide is predominantly a competitive inhibitor with respect to 2OG (Ki = 1.97 μM) for KDM2A, but shows mixed inhibition with respect to peptide substrate, binding predominantly to the enzyme-peptide complex (Ki = 85 μM, α = 0.13). (Figure 3). The latter observation is notable given that daminozide contains a dimethylamino group as do the KDM2/7 subfamily dimethyl lysine substrates, and might thus be expected to compete with the dimethylated lysine substrate. The pKa of the daminozide hydrazide amine is 2.8, whilst that of its carboxylate is 4.623, suggesting that it may bind the active site iron; NMR analyses show daminozide complexes to Fe(II) in solution (Figure S2).
Figure 3. Mode of inhibition of the KDM2/7 subfamily by daminozide.
Inhibition of KDM2A by daminozide is competitive with 2OG but not peptide substrate.
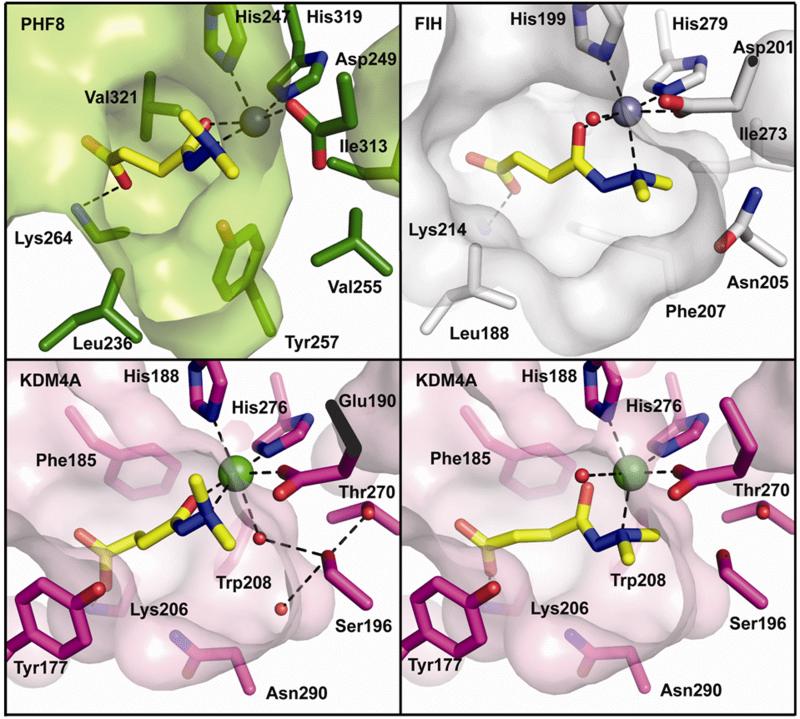
We then investigated the structural basis of the selective inhibition by daminozide on the KDM2/7 subfamily. We obtained structures of daminozide complexed with two demethylases, PHF8 and KDM4A, and a hydroxylase, FIH (Figure 4). The structures support the proposed general mode of inhibition by daminozide, i.e. by binding in the 2OG binding pocket and chelating to the active site metal via its acyl hydrazide carbonyl and dimethylamino groups (Figure 4). Whilst the coordinate position of the daminozide carbonyl was invariant, i.e. in all cases trans to the Asp/Glu protein-based ligand, in the KDM4A structure the dimethylamino group was observed to complex either trans to His276 or trans to His188 in the two different molecules present in the asymmetric unit. Although in the case of PHF8 and FIH the daminozide dimethyl amino group was only observed to bind trans to the histidine equivalent to His276 of KDM4A, previous work24 on complexes of 2OG oxygenases with 2OG and the cosubstrate analogue 12 demonstrate that the 1-carboxylate can bind in either of the two conformational positions, in an analogous manner to that observed for the dimethylamino group in the KDM4A structure. We propose that the selectivity of daminozide for the KDM2/7 subfamily could, at least in part, arise from a ‘snug fit’ obtained via binding in the position trans to His247 wherein its two methyl groups would be accommodated in a tight hydrophobic pocket (formed by Val255, Ile313, and Tyr257), which is conserved in the KDM2/7subfamily (Figure 4). This pocket might bind the daminozide methyl groups less tightly in other demethylases/oxygenases because it is either more hydrophilic (as demonstrated by crystallographic analysis) or predicted to be more hydrophilic (by structure based on sequence alignments), for all the other 2OG oxygenases tested (Figure 1) (e.g. for KDM4A: Ser196, Thr270 and Asn198, KDM6B: Ser225, Ile291 and Asn227, FIH: Asn205, Ile273 and Phe257).
Figure 4. Crystal structures reveal the mode of inhibition of the KDM2/7 subfamily by daminozide.
PHF8 (with Zn(II) substituting for Fe(II),) and KDM4A (with Ni(II) substituting for Fe(II)) and FIH (with Zn(II) substituting for Fe(II)) (Table S3). Daminozide binds the metal through its hydrazide amine lone pair and carbonyl oxygen. Two distinct orientations of daminozide are observed in the two KDM4A molecules in the asymmetric unit; both are shown (see Table S2 for electron density maps). Selectivity of daminozide for the KDM2/7 subfamily may arise because they possess a hydrophobic region (Tyr257, Val255 and Ile313) into which the two daminozide methyl groups may bind. In contrast, the equivalent regions of KDM4A, FIH and the other tested demethylases/oxygenases are more hydrophilic.
We also synthesised and tested analogues of daminozide (Table 1 and Table S3). The trimethylated analogue 22, and compound 27, both of which lack the terminal amine lone pair, displayed little/no KDM inhibition, consistent with the proposed mode of daminozide inhibition involving chelation by its terminal hydrazide amine. The monomethylated 23 and unmethylated analogues 24 were more potent than daminozide against KDM2A.
However, when tested against other 2OG subfamilies, these compounds were substantially less selective than daminozide (and were somewhat unstable in aqueous solution). The succinyl hydroxamic acid 25 and dioxoheptanoic acid 28, in which the acyl-hydrazinamide of daminozide is replaced by metal-chelating hydroxamic acid and malonyl groups, respectively were also relatively potent but non-selective inhibitors (Table 1). Notably, compound 26, in which the amide nitrogen of daminozide is N-methylated, is also selective for the KDM2/7demethylases, albeit with reduced potency. Like daminozide, 26 has two methyl groups on the acyl hydrazide amine, suggesting that their presence confers selectivity. The other tested analogues were less active (Table S3).
Conclusions
Overall, we have demonstrated how the development of a screening platform employing multiple human 2OG oxygenases can enable the discovery of compounds selective for particular subfamilies. In the long term, we aim to help to develop screens for all human 2OG oxygenases and see them applied to the discovery of medicinally useful inhibitors. Using a relatively focused set of 2OG analogues, the screening platform led to the discovery that daminozide is a selective inhibitor of the KDM2/7demethylase subfamily. Whilst daminozide itself is unlikely to be of medicinal use, the work has revealed a new class of 2OG oxygenase inhibitor, which employs alkylamino iron chelation. Of particular note is the high degree of selectivity exhibited by daminozide, especially given its achiral nature and low molecular weight. The precise mode of action of daminozide as a plant growth regulator15,16, and its potential human toxicity (if any) under physiologically relevant conditions are unknown. We have no evidence that our results are directly relevant to the toxicity issues relating to daminozide. However, the results do suggest a potential of daminozide or its derivatives to exert biological effects via the inhibition of 2OG oxygenases involved in epigenetics in animals and, potentially, in other organisms including plants. Given the link between JmjC enzymes and diseases including cancer2,3, further work on the biological effects of daminozide are of interest.
Experimental Section
Chemical synthesis of potential inhibitors. Reagents and solvents were from Aldrich, Alfa Aesar or Acros. Reactions were monitored by TLC, which was performed on precoated aluminum-backed plates (Merck, silica 60 F254). Melting points were determined using a Leica Galen III hot-stage melting point apparatus and microscope. Infrared spectra were recorded from Nujol mulls between sodium chloride discs, on a Bruker Tensor 27 FT-IR spectrometer. NMR spectra were acquired using a Bruker DPX500 NMR spectrometer. Chemical shifts (δ) are given in ppm, and the multiplicities are given as singlet (s), doublet (d), triplet (t), quartet (q), multiplet (m), broad (br). Coupling constants J are given in Hz (± 0.5 Hz). High resolution mass spectra (HRMS) were recorded using a Bruker MicroTOF spectrometer. The purity of all compounds synthesized were ≥95% as determined by analytical reverse-phase HPLC (Ultimate 3000). Daminozide (Alar™) and compound 28 are commercially available. The synthesis and characterisation of compounds 2225, 2526, 2727, 3628, 3729 and 3826 has been reported. The synthesis of compounds 31-35, 39-41 and 13C NMR spectra for 22, 23, 24, 26, 31-35 are given in the Supporting Information.
4-(2,2,2-Trimethylhydrazinyl)-4-oxobutanoate 22. The synthesis of compound 22 was as reported25, thus reaction of daminozide (500mg, 3.1 mmol) with methyl iodide (700mg, 0.31 mL, 5.0 mmol) gave 22 as a white solid (75% yield), mp: 137-138 °C (lit.1 137-138.5 °C); 1H NMR (500 MHz, MeOD): δ 2.40 (t, J = 6.5 Hz, 2H), 2.51 (t, J = 6.5 Hz, 2H), 3.56 (s, 9H); 13C NMR (125 MHz, MeOD): δ 28.5, 29.1, 56.1, 170.4, 173.4; IR (neat) υ/cm−1: 3405, 3312, 1729, 1693; HRMS (m/z): [M]+ calcd. for C7H15N2O3, 175.1077; found, 175.1081.
General procedure for the coupling of hydrazine to succinic anhydride. To a stirred solution of the appropriate hydrazine (1 equiv.) in acetonitrile (5 mL) was added dropwise a solution of succinic anhydride (200 mg, 2.0 mmol, 1 equiv.) in acetonitrile (5 mL). The mixture was stirred at room temperature for 24 h, after which the solvent was evaporated in vacuo and the resulting crude purified using semipreparative reverse-phase HPLC, performed on a phenomenex C18 column (150 mm × 4.6 mm). Separation was achieved using a linear gradient of solvent A (water + 0.1% CF3CO2H) and solvent B (acetonitrile + 0.1% CF3CO2H), eluting at a flow rate of 1 mL/min and monitoring at 220 nm: 0% B to 40% B over 30 min.
4-(2-Methylhydrazinyl)-4-oxobutanoic acid 23. Compound 23 is a colourless oil (63% yield), 1H NMR (500 MHz, DMSO-d6): δ 2.35 (t, J = 7.0 Hz, 2H), 2.68 (t, J = 7.0 Hz, 2H), 2.98 (s, 3H), 4.76 (s, 1H), 7.74 (s, 1H); 13C NMR (125 MHz, DMSO-d6): δ 28.3, 29.1, 170.1, 173.6; IR (neat) υ/cm−1: 33 3219, 3057, 1708, 1632; HRMS (m/z): [M+Na]+ calcd. for C5H10N2NaO3, 169.0584; found, 169.0577.
4-Hydrazinyl-4-oxobutanoic acid 24. Compound 24 is a colourless oil (87% yield), 1H NMR (500 MHz, DMSO-d6): δ 2.34 (t, J = 7.0 Hz, 2H), 2.60 (t, J = 7.0 Hz, 2H), 5.86 (s, 1H), 8.99 (s, 1H); 13C NMR (125 MHz, DMSO-d6): δ 28.2, 29.1, 170.8, 173.9; IR (neat) υ/cm−1: 3303, 3290, 3199, 1712, 1624; HRMS (m/z): [M-H]- calcd. for C4H7N2O3, 131.0462; found, 131.0468.
4-Oxo-4-(1,2,2-trimethylhydrazinyl)butanoic acid 26. Compound 26 is a white solid (56% yield), mp: 97-98 °C, 1H NMR (500 MHz, DMSO-d6): δ 2.37 (t, J = 7.0 Hz, 2H), 2.66 (t, J = 7.0 Hz, 2H), 2.74 (s, 3H), 11.98 (s, 1H); 13C NMR (125 MHz, DMSO-d6): δ 28.2, 29.8, 43.4, 48.7, 173.5, 175.0; IR (neat) υ/cm−1: 2958, 1723, 1615; HRMS (m/z): [M+Na]+ calcd. for C7H14N2NaO3, 197.0897; found, 197.0895.
4-((Dimethylamino)oxy)-4-oxobutanoic acid 29. N,N-Dimethylhydroxylamine (39 mg, 0.63 mmol, 1.1 equiv. ) was added to a solution of 4-(tert-butoxy)-4-oxobutanoic acid (100 mg, 0.57 mmol, 1 equiv.), hydroxybenzotriazole (100 mg, 0.74 mmol, 1.3 equiv.), 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide ( 140 mg, 0.74 mmol, 1.3 equiv.) and diisopropylethylamine ( 0.2 mL, 1.14 mmol, 2.0 equiv.) in CH2Cl2 (10 mL). The reaction was stirred at room temperature overnight, washed with water, HCl 1N, brine, dried on MgSO4. The organic phase was evaporated in vacuo and purified by chromatography (MeOH/CH2Cl2 0.5/9.5) to obtain 110 mg of tert-butyl 4((dimethylamino)oxy)-4-oxobutanoate (90% yield). CF3CO2H (0.04 ml, 0.37 mmol, 4 equiv.) was added to a solution of tert-butyl 4((dimethylamino)oxy)-4-oxobutanoate (20 mg, 0.09 mmol, 1 equiv.) in CH2Cl2 (1.5 ml). The reaction was stirred at room temperature for 4h and evaporated in vacuo to give 14 mg of 29 (yield 95%). 1H NMR (500 MHz, CD3OD) δ 2.59 (s, 6H), 2.57 (s, 4H); 13C NMR (500 MHz, CD3OD) δ 176.2, 172.0, 48.5, 29.9; IR (neat) 3341, 2485,1717, 1120, 1026, 975 cm−1; HRMS (m/z):[M+]calcd. for C6H11NO4 161.0688; found 161.0923.
N‘1, N‘1, N‘4, N‘4-Tetramethylsuccinohydrazide 30. A solution of succinic acid (100 mg, 0.85 mmol, 1 equiv.), hydroxybenzotriazole (350 mg, 2.11 mmol, 2.3 equiv.), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (421 mg, 2.11 mmol, 2.3 equiv.), diisopropylethylamine (0.6 mL, 3.4 mmol, 4 equiv.) and 1,1-dimethylhydrazine (0.16 mL, 2.04 mmol, 2.2 equiv.) in CH2Cl2 (20 mL) was stirred at room temperature overnight. CH2Cl2 (10 mL) was added and the reaction mixture was washed with water, a saturated solution of NaHCO3, brine and dried on MgSO4. The organic phase was evaporated in vacuo and purified by chromatography (MeOH/ CH2Cl2 1/9) to give 77 mg of 30 (45% yield). 1H NMR (500 MHz, CD3OD) δ 2.87 (s, 6H), 2.65 (s, 2H); 13C NMR (500 MHz, CD3OD) δ 178.2, 43.8, 27.6; IR (neat) 3356, 2485, 2071, 1695, 1120, 1027, 974 cm−1; HRMS (m/z):[(M-2CH3)−]calcd. for C6H14N4O2, 174.1117; found, 174.1022.
Supplementary Material
ACKNOWLEDGMENT
This research was supported in part by the The Wellcome Trust, The Commonwealth Scholarship Commission in the United Kingdom, the Biotechnology and Biological Research Council (U.K.) and the European Research Council. The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research and the Wellcome Trust.
ABBREVIATIONS
- 2OG
2-oxoglutarate
- BBOX
γ-butyrobetaine hydroxylase
- FBXL
F-box, leucine-rich repeat protein
- FIH
factor inhibiting hypoxia inducible factor
- HDAC
histone deacetylase
- JARID
jumonji, AT rich interactive domain containing protein
- JmjC
jumonji-C domain
- JMJD
jumonji-C domain containing protein
- PHD2
human prolyl hydroxylase
- PHF
plant homeodomain containing protein
Footnotes
Associated Content
Supporting Information Available: Assay methods, synthetic procedures, characterization of all synthesised compounds, 1H NMR spectra of daminozide and analogues with iron(II), crystalllographic data collection and refinement statistics. This material is provided free of charge via the Internet at http://pubs.acs.org. PDB ID Codes: 4AI9, 4AI8, 4DO0
REFERENCES
- (1).He J, Nguyen AT, Zhang Y. KDM2b/JHDM1b, an H3K36me2-specific demethylase, is required for initiation and maintenance of acute myeloid leukemia. Blood. 2011;117(14):3869–3880. doi: 10.1182/blood-2010-10-312736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Yang J, Jubb AM, Pike L, Buffa FM, Turley H, Baban D, Leek R, Gatter KC, Ragoussis J, Harris AL. The histone demethylase JMJD2B is regulated by estrogen receptor alpha and hypoxia, and is a key mediator of estrogen induced growth. Cancer Res. 2010;70(16):6456–6466. doi: 10.1158/0008-5472.CAN-10-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The Histone H3 Lysine-27 Demethylase Jmjd3 Links Inflammation to Inhibition of Polycomb-Mediated Gene Silencing. Cell. 2007;130(6):1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- (4).Chang KH, King ON, Tumber A, Woon EC, Heightman TD, McDonough MA, Schofield CJ, Rose NR. Inhibition of histone demethylases by 4-carboxy-2,2′-bipyridyl compounds. ChemMedChem. 2011;6(5):759–764. doi: 10.1002/cmdc.201100026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Hamada S, Suzuki T, Mino K, Koseki K, Oehme F, Flamme I, Ozasa H, Itoh Y, Ogasawara D, Komaarashi H, Kato A, Tsumoto H, Nakagawa H, Hasegawa M, Sasaki R, Mizukami T, Miyata N. Design, synthesis, enzyme-inhibitory activity, and effect on human cancer cells of a novel series of jumonji domain-containing protein 2 histone demethylase inhibitors. J Med Chem. 2010;53(15):5629–5638. doi: 10.1021/jm1003655. [DOI] [PubMed] [Google Scholar]
- (6).Luo X, Liu Y, Kubicek S, Myllyharju J, Tumber A, Ng S, Che KH, Podoll J, Heightman TD, Oppermann U, Schreiber SL, Wang X. A selective inhibitor and probe of the cellular functions of Jumonji C domain-containing histone demethylases. J Am Chem Soc. 2011;133(24):9451–9456. doi: 10.1021/ja201597b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Rose NR, Ng SS, Mecinovic J, Liénard BMR, Bello SH, Sun Z, McDonough MA, Oppermann U, Schofield CJ. Inhibitor Scaffolds for 2-Oxoglutarate-Dependent Histone Lysine Demethylases. J Med Chem. 2008;51(22):7053–7056. doi: 10.1021/jm800936s. [DOI] [PubMed] [Google Scholar]
- (8).Sekirnik R, Rose NR, Mecinovic J, Schofield CJ. 2-Oxoglutarate oxygenases are inhibited by a range of transition metals. Metallomics. 2010;2(6):397–399. doi: 10.1039/c004952b. [DOI] [PubMed] [Google Scholar]
- (9).Rose NR, McDonough MA, King ON, Kawamura A, Schofield CJ. Inhibition of 2-oxoglutarate dependent oxygenases. Chem Soc Rev. 2011;40(8):4364–4397. doi: 10.1039/c0cs00203h. [DOI] [PubMed] [Google Scholar]
- (10).Kawamura A, Tumber A, Rose NR, King ON, Daniel M, Oppermann U, Heightman TD, Schofield C. Development of homogeneous luminescence assays for histone demethylase catalysis and binding. Anal Bioch. 2010;404(1):86–93. doi: 10.1016/j.ab.2010.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Rydzik A, Leung IKH, Kochan GT, Thalhammer A, Opperman U, Claridge TDW, Schofield CJ. Development and application of a fluoride detection based fluorescence assay for gamma-butyrobetaine hydroxylase. ChemBioChem. doi: 10.1002/cbic.201200256. in press. [DOI] [PubMed] [Google Scholar]
- (12).Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IKH, Li XS, Woon EC, Yang M, McDonough MA, King ON, Clifton IJ, Klose RJ, Claridge TDW, Ratcliffe PJ, Schofield CJ, Kawamura A. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12(5):463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Riddell JA, Hageman HA, J’Anthony CM, Hubbard WL. Retardation of Plant Growth by a New Group of Chemicals. Science. 1962;136(3514):391. doi: 10.1126/science.136.3514.391. [DOI] [PubMed] [Google Scholar]
- (14).Rademacher W. Growth Retardants: Effects on Gibberellin Biosynthesis and Other Metabolic Pathways. Ann Rev Plant Physiol Plant Mol Biol. 2000;51(1):501–531. doi: 10.1146/annurev.arplant.51.1.501. [DOI] [PubMed] [Google Scholar]
- (15).Brown RGS, Kawaide H, Yang Y-Y, Rademacher W, Kamiya Y. Daminozide and prohexadione have similar modes of action as inhibitors of the late stages of gibberellin metabolism. Physiol Plant. 1997;101:309–313. [Google Scholar]
- (16).Looney NE. Inhibition of Apple Ripening by Succinic Acid, 2,2-Dimethyl Hydrazide and its Reversal by Ethylene. Plant Physiol. 1968;43:1133–1137. doi: 10.1104/pp.43.7.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Hasegawa R, Cabral R, Hoshiya T, Hakoi K, Ogiso T, Boonyaphiphat P, Shirai T, Ito N. Carcinogenic potential of some pesticides in a medium-term multi-organ bioassay in rats. Int J Cancer. 1993;54(3):489–493. doi: 10.1002/ijc.2910540322. [DOI] [PubMed] [Google Scholar]
- (18).Mott L. Alar Again: Science, the Media, and the Public’s Right to Know. Int J Occup Environ Health. 2000;6(1):68–70. doi: 10.1179/oeh.2000.6.1.68. [DOI] [PubMed] [Google Scholar]
- (19).Toth B, Wallcave L, Patil K, Schmeltz I, Hoffmann D. Induction of tumors in mice with the herbicide succinic acid 2,2-dimethylhydrazide. Cancer Res. 1977;37(10):3497–3500. [PubMed] [Google Scholar]
- (20).Horton JR, Upadhyay AK, Qi HH, Zhang X, Shi Y, Cheng X. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat Struct Mol Biol. 2010;17(1):38–43. doi: 10.1038/nsmb.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia-Bassets I, Aggarwal AK, Desai A, Dorrestein PC, Glass CK, Rosenfeld MG. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466(7305):508–512. doi: 10.1038/nature09272. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Qi HH, Sarkissian M, Hu GQ, Wang Z, Bhattacharjee A, Gordon DB, Gonzales M, Lan F, Ongusaha PP, Huarte M, Yaghi NK, Lim H, Garcia BA, Brizuela L, Zhao K, Roberts TM, Shi Y. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature. 2010;466(7305):503–507. doi: 10.1038/nature09261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Schoenherr J, Bukovac MJ. Dissociation Constants of Succinic Acid 2,2-Dimethylhydrazide. J Agr Food Chem. 1972;20(6):1263–1265. [Google Scholar]
- (24).Zhang Z, Ren J, Harlos K, McKinnon CH, Clifton IJ, Schofield CJ. Crystal structure of a clavaminate synthase–Fe(II)–2-oxoglutarate–substrate–NO complex: evidence for metal centred rearrangements. FEBS Letters. 2002;517(1–3):7–12. doi: 10.1016/s0014-5793(02)02520-6. 2002. [DOI] [PubMed] [Google Scholar]
- (25).Wawzonek S, Kellen JN. Thermolysis of triethylamine with N-p-toluenesulfonylarylhydrazidoyl chlorides. J Org Chem. 1973;38:2058–2061. [Google Scholar]
- (26).Mecinovic J, Loenarz C, Chowdhury R, Schofield CJ. 2-Oxoglutarate analogue inhibitors of prolyl hydroxylase domain 2. Bioorg Med Chem Lett. 2009;19:6192–6195. doi: 10.1016/j.bmcl.2009.09.005. [DOI] [PubMed] [Google Scholar]
- (27).Alain V, Dominique C, Frédéric Z, Roger L. Atypical Oxidation Reaction by Thionyl Chloride: Easy Two-Step Synthesis of N-Alkyl-1,4-dithiines. Synthetic Commun. 2006;36:3591–3597. [Google Scholar]
- (28).Speiser PP, Joshi RK. US 5149695 A Fumaric acid derivatives, process for the production thereof and pharmaceutical compositions containing same. 1968
- (29).Hageman HA, Hubbard WL. US 3257414 A N-disubstituted amino maleimides and succinimides. 1968
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.