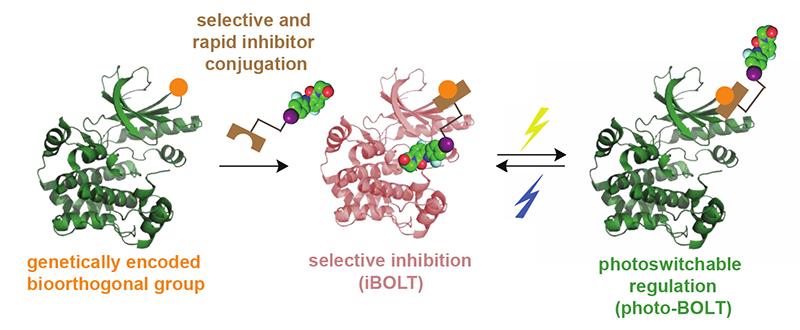
Abstract
The rapid and selective regulation of a target protein within living cells that contain closely related family members is an outstanding challenge. Here we introduce genetically directed bioorthogonal ligand tethering (BOLT) and demonstrate selective inhibition (iBOLT) of protein function. In iBOLT, inhibitor-conjugate/target protein pairs are created where the target protein contains a genetically encoded unnatural amino acid with bioorthogonal reactivity and the inhibitor conjugate contains a complementary bioorthogonal group. iBOLT enables the first rapid and specific inhibition of MEK isozymes, and introducing photoisomerizable linkers in the inhibitor conjugate enables reversible, optical regulation of protein activity (photo-BOLT) in live mammalian cells. We demonstrate that a pan kinase inhibitor conjugate allows selective and rapid inhibition of the lymphocyte specific kinase, indicating the modularity and scalability of BOLT. We anticipate that BOLT will enable the rapid and selective regulation of diverse proteins for which no selective small molecule ligands exist.
Graphical abstract
Introduction
Small molecules allow the regulation of protein function, and provide powerful tools for dissecting biological systems. Small-molecule-mediated regulation is commonly rapid, may be temporally controlled, is dose-dependent, and allows the inhibition of proteins where gene disruption is developmentally lethal or leads to adaptive compensation1,2. Small molecules can modulate a particular function of a protein, such as its catalytic activity, while maintaining the protein’s other interactions and functions. The phenotypes elicited by small-molecule regulators may therefore be distinct from those resulting from genetic perturbation.2
Modern drug discovery approaches have developed some remarkably potent inhibitors and created new therapeutics3. However, the selective inhibition of a target protein in the cell with small molecules remains incredibly challenging. Many proteins have closely related family members with similar sequences and structures, making it difficult to selectively target the desired protein inside a cell without affecting other members of the protein’s family. As a result, it is often challenging to dissect the distinct roles that are executed by closely related proteins using small-molecule regulators. Several approaches have been developed to address this challenge.
A ‘bump-hole’ strategy4 has been used to make selective inhibitors of protein kinases2, providing a broadly applicable and powerful tool to study signaling5. While this approach has had a tremendous impact, not all kinases are amenable to this approach6, and extending it to other protein families has proved challenging.
A strategy that may increase the affinity, and potentially the specificity, of a non-covalent ligand for its target involves creating molecules like X (Fig. 1a), for covalently tethering ligands to their targets. The utility of these approaches depends critically on both the selectivity and rate of the chemical reactions used to target the protein of interest and therefore on the nature of the chemical groups involved.
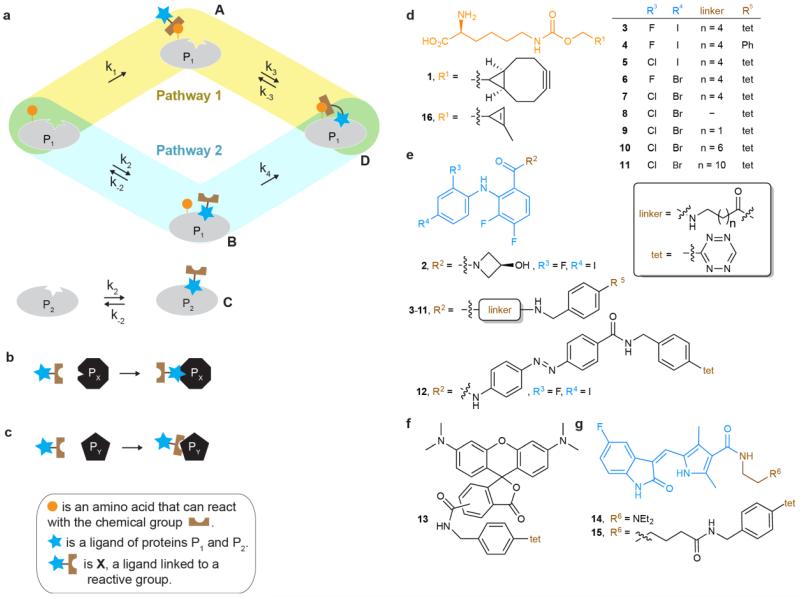
Figure 1. Pathways for tethering a ligand conjugate to a protein, and the structures of compounds used in this study.
a, A protein P1 may bind a ligand conjugate X through two pathways. In the first pathway, an intermolecular chemical reaction first takes place between X and protein P1 to form adduct A; non-covalent binding of the tethered ligand to its binding site then occurs. In the second pathway, the ligand first binds reversibly to protein P1 to form the non-covalent complex B; a tethered chemical reaction then takes place between the protein P1 and X to form the covalently tethered ligand bound complex D. The relative contributions of these pathways to the formation of D depend on the indicated rate constants. Protein P2 has an identical ligand binding pocket to P1 but does not have the reactive group. At inhibitor conjugate concentrations where C is minimally populated, D may be formed and inhibit P1. b, Off-target ligand binding of the ligand conjugate to other biomolecules (PX) decreases specificity. c, Off-target reaction of the reactive group on the ligand conjugate with other biomolecules (PY) decreases specificity. d, Unnatural amino acids 1 and 16. e, MEK1/2 specific inhibitor 2 and its derivatives 3-12. f, TAMRA tetrazine conjugate 13. g, Sunitinib 14 and its tetrazine derivative 15.
A variety of ligand-reactive group conjugates have been developed to tether ligands to proteins for distinct purposes, including affinity labeling, traceless protein conjugation, fragment-based drug discovery and modulation of activity7-9. Because these approaches commonly employ tethering via reactions with natural amino acid side chain nucleophiles (commonly cysteine thiols) present on endogenous proteins, the specificity for a target protein is commonly derived from the intrinsic specificity of the ligand. As a result, these approaches do not differentially target proteins that are common targets of the parent ligand.
Cysteines may be engineered into protein targets to direct tethered ligand binding10,11 and engineered cysteine tethering and bump hole approaches have been combined to create more selective covalent inhibitors12. Tethering ligands, attached through photoswitchable linkers, to cysteine residues in cell surface receptors has enabled receptor state photoswitching, and rapid control of protein function13-15, but this approach is limited to cell surface proteins.
Approaches that take advantage of the reactivity of natural amino acid side chains can increase the fraction of ligand bound to its target9. However, the reactive group that is linked to the ligand to enable tethering may react with endogenous nucleophiles, including thiols and cysteines in the proteome, and selectivity for a target protein, which depends on the relative rates of ligand binding and off-target reactions (Fig. 1b,c) may be low.
Bioorthogonal reactions16 have been reported, with rate constants spanning nine orders of magnitude (from 10−5 M−1s−1 to 104 M−1s−1; refs 17,18). We and others have shown that bioorthogonal groups, including strained alkenes and alkynes for rapid and specific inverse electron demand Diels-Alder reactions with tetrazines, can be genetically encoded in cells19-24. This has enabled the site-specific fluorescent labelling of proteins on the cell surface19,20 and within cells23 and animals24,25. Here we describe genetically encoded bioorthogonal ligand tethering (BOLT), in which an unnatural amino acid bearing a bioorthogonal group is directed into a protein of interest and a ligand for the proteins is connected, via a linker, to a bioorthogonal group (to create a bioorthogonal version of X, Fig. 1) that can react with the group introduced into the protein. The rate constant for small molecules binding to target enzymes (k2, Fig. 1a) is commonly26 104 to 106 M−1 s−1, and a very fast covalent tethering reaction (large k1) is therefore required for pathway 1 to contribute substantially to the rate of formation of D (assuming k4 is at least equal to k1)27. For most intermolecular bioorthogonal reactions, pathway 2 (Fig. 1a) will be the major contributor to the formation of D via BOLT (Supplementary Tables 1-3). The formation of D depends on the rate constant k4, for the irreversible conversion of B to D via a bioorthogonal reaction. Notably, if we apply the inhibitor conjugate at a concentration much lower than k-2/k2 (KD, dissociation constant of the inhibitor conjugate) so that most inhibitor conjugate is not bound to P1, only trace amounts of the non-covalent complex B are formed, but once B is formed it is irreversibly converted to D and all protein P1 is tethered. The extent of tethered inhibition depends on the intramolecular ligand binding equilibrium (k3/k−3) between D and A, and the increase in the effective molarity of tethered ligands28,29 in the proximity of the inhibitor binding site may result in complete inhibition for optimal sites of tethering. Notably, at concentrations of inhibitor much lower than the KD for P1 only trace amounts of the inhibitor conjugate will bind P2 (a protein that does not have a bioorthogonal group installed, but may otherwise be identical to P1, Fig. 1a) and P2 will not be inhibited, allowing selective inhibition of protein P1 without inhibiting protein P2. The timescale of selective inhibition is critically dependent on k4, which in turn depends on the rate of the bioorthogonal reaction, and relatively rapid tethered bioorthogonal reactions are required to achieve selective inhibition on the timescales commonly associated with small-molecule inhibition (Supplementary Table 1).
Here we demonstrate that inhibition via BOLT (iBOLT) can be used to selectively inhibit either MEK1 or MEK2 in live mammalian cells. Although the parent inhibitors are non-selective between the two MEK isozymes, inhibition of the targeted protein by the inhibitor conjugate proceeds without inhibition of the other isozyme. We demonstrate that iBOLT is exquisitely selective and is sensitive to inhibitor structure, linker and amino acid tether, and that inhibition is rapid and quantitative. Introduction of a photoswitch into the inhibitor conjugate linker enables the activity of the protein to be reversibly toggled on and off with light (photo-BOLT) in live mammalian cells. Furthermore, defining amino acid positions that can be targeted by iBOLT with a pan kinase inhibitor in MEK isozymes enables LCK, a protein kinase that contains just 26% sequence identity to MEK1, to be targeted, indicating that the strategy we develop is scalable and modular.
Results
Design of MEK inhibitor-tetrazine conjugate
MEK1 and MEK2 are key kinases in the MAP kinase signalling pathway, which regulates growth and development. Misregulation of this pathway is observed in 30% of human tumors, including melanoma and Hodgkin lymphoma30-32. Selective inhibition of MEK1 or MEK2 by small molecules has not been achieved, and is challenging because the two kinases share 82% sequence identity33. Knockout of MEK1 is embryonic lethal34 and knockdown of MEK1 takes 16 h, enabling the cell to adapt and compensate for the loss35. Efforts to generate inhibitor-sensitive alleles of MEK1 by the ‘bump-hole’ strategy abolished its kinase activity6.
Our first task was to design and synthesize an inhibitor conjugate for iBOLT (Supplementary Fig. 1). Many of the MEK1/2 inhibitors reported to date do not compete with adenosine triphosphate (ATP) binding36, and several have structures that are similar to 2 (Fig 1)37. Inhibitor 2 has an IC50 (concentration of inhibitor where activity goes to half of its maximal value) of 44 nM against MEK138 and inhibits the activity of both MEK1 and MEK2 with comparable potency in cell culture (Supplementary Fig. 2). Inhibitor 2 binds MEK1 with the amide moiety facing the opening of the binding pocket38, suggesting that this would be a good position to attach a linker for iBOLT (Fig. 2a). We therefore designed and synthesized inhibitor conjugate 3 (Fig. 1). Compound 2 completely inhibits MEK1 at 10 μM, but compound 3 does not inhibit it at the same concentration (Supplementary Fig. 3). Thus, the changes between 2 and 3 lead to a decrease in the potency in untethered inhibitor.
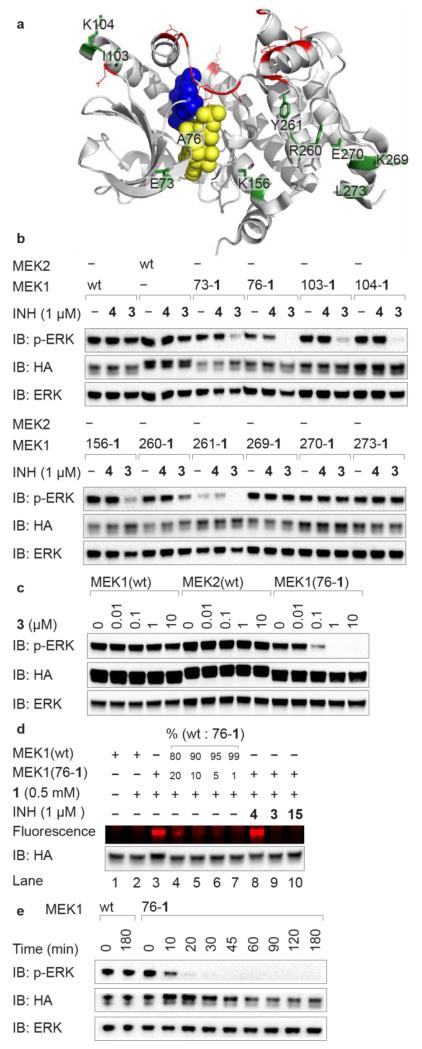
Figure 2. iBOLT of MEK1(XXX-1) variants by 3.
a, Sites targeted for site-directed mutagenesis to amino acid 1 in MEK1. Permissive (green with black labels) and non-permissive (red) sites are highlighted. ATP and inhibitor 2 are shown as yellow and blue spheres, respectively. The figure was created using Pymol (www.pymol.org) and PDB ID 3ZLS. b, Several MEK1(XXX-1) variants can be selectively inhibited by 3, but not 4 in HEK293ET cells. Cells were incubated with the indicated inhibitor (INH) for 3 h before western blot analysis. c, Concentration dependence of MEK1(76-1) inhibition by 3 after 3 h. d, Fluorescent labeling of cell lysates by TAMRA tetrazine conjugate 13 is consistent with the quantitative formation of iBOLT product. e, Following inhibition of MEK1(76-1) by 1 μM 3 as a function of time. EGFP-ERK2 is detected by immunoblot (IB) for ERK and phosphorylated EGFP-ERK2 is detected by immunoblot for pERK. All MEK variants are human influenza hemagglutinin (HA) tagged, contain DD mutations making them constitutively active, and are detected by immunoblot for HA.
Identifying active MEK1(XXX-1) variants
Our next task was to discover MEK1 variants that incorporate unnatural amino acid 1 (Fig. 1) and retain activity (Supplementary Fig. 1). We identified 22 solvent-exposed amino acid residues that are within 40 Å of the carbonyl group of the amide in the structure of inhibitor 2 bound to the kinase (Fig. 2a)38 as targets for substitution by 1. We transiently expressed MEK1(XXX-1) variants in human cells, where XXX refers to each of the positions identified. We discovered ten MEK1(XXX-1) variants that retain the ability to phosphorylate EGFP-ERK2 (Fig. 2a and Supplementary Fig. 4a), an EGFP tagged version of the MEK substrate ERK. Both the expression of these variants in cells containing MEK1(XXX-TAG) and the BCNRS/tRNACUA pair, and the resulting EGFP-ERK2 phosphorylation, were strictly dependent on the addition of 1 to cells (Supplementary Fig. 4b).
iBOLT of MEK1 (XXX-1) variants by 3
We tested the inhibition of the ten active MEK1 variants by tetrazine-inhibitor conjugate 3 (1 μM) in cells (Fig. 2b, full blots for all Figures in Supplementary Fig. 5). We observed clear inhibition of EGFP-ERK2 phosphorylation for six MEK1 variants (73-1, 76-1, 103-1, 104-1, 156-1, 261-1), and for three of these MEK1 variants (76-1, 104-1, 261-1) we detected little or no phosphorylated EGFP-ERK2. Neither wild-type MEK1 nor wild-type MEK2 were inhibited by 3 (1 μM), indicating that the presence of the unnatural amino acid 1 at the genetically defined site is critical for inhibition. Furthermore, an analogue of 3 in which the tetrazine is replaced by a phenyl group (4) (Fig. 1) did not inhibit wild-type MEK1/2 or any MEK1(XXX-1) variant tested (Fig. 2b), indicating that the tetrazine moiety is also essential for inhibition.
iBOLT of MEK1(76-1) by 3 is selective, quantitative, and rapid
We observed substantial inhibition of MEK1(76-1) by 100 nM 3 (Fig. 2c, See Supplementary Fig. 6 for quantification). In contrast, 10 μM 3 (the solubility limit) does not inhibit either wild-type MEK1 or MEK2 (Fig. 2c), indicating that 3 selectively inhibits MEK1(76-1) over wild type MEK1/2 by a factor of at least 100. The parent inhibitor (2) completely inhibits MEK1 at 10 μM (Supplementary Figs 2 and 3a), but 3 does not inhibit MEK1 at 10 μM (Fig. 2c and Supplementary Fig. 3a). The specificity of 3 for MEK1(76-1) results from a decrease in inhibition of wild-type MEK1 by 3 (with respect to 2) and more potent inhibition of MEK1(76-1) by 3, than MEK1 by 2.
To demonstrate that the attachment of 3 to MEK1(76-1) is quantitative and correlated with inhibition, we labelled the lysate from cells containing MEK1(76-1), and the lysate from cells containing MEK1(76-1) treated with 3, with tetrazine fluorophore 13 (Fig. 1). Lysates from cells expressing MEK1(76-1) were strongly labelled with 13 (Fig. 2d, lane 3, and Supplementary Fig. 7A). In contrast, when we incubated lysates from cells expressing MEK1(76-1) that had been treated with inhibitor 3, with 13 we did not observe fluorescent labelling of MEK1(76-1) (Fig. 2d, lane 9) and the extent of labeling and inhibition are correlated (Supplementary Fig. 7b). Control experiments demonstrate that wild-type MEK1 is not labelled by 13 (Fig. 2d, lanes 1 and 2) and that lysates from cells expressing MEK1(76-1) treated with 4 are efficiently labelled with 13 (Fig. 2d, lane 8). These experiments demonstrate that the quantitative attachment of 3 to MEK1(76-1) leads to its inhibition by BOLT and that this inhibition is specific, quantitative and requires both the genetically directed incorporation of 1 into MEK1 and the presence of the tetrazine reaction partner in the inhibitor conjugate 3.
One key advantage of small-molecule-mediated inhibition over genetic methods is the speed of inhibition (Supplementary Fig. 3c). On adding 3 (1 μM) to cells we observed complete inhibition of MEK1(76-1) within 30 minutes (Fig. 2e, Supplementary Figs 3d and 8 for quantification). These data indicate that the inhibition of MEK1(76-1) by 3 is rapid and is consistent with the rate constants for labelling 1 on proteins with similar tetrazines (k1 of ~102 M−1 s−1), and kinetic simulations of iBOLT inhibition (Supplementary Table 1).
MEK1(76-1) is selectively labeled with respect to the proteome of mammalian cells
Potential considerations for BOLT, as well as other approaches that use unnatural amino acid incorporation at the amber stop codon to selectively label a target protein in the cell, are (1) the selectivity of the bioorthogonal reaction and (2) the extent to which the unnatural amino acid is incorporated in response to the amber stop codons of endogenous genes and leads to labeling of off-target proteins.
Labeling lysates from cells expressing MEK1(76-TAG) and the BCNKRS/tRNACUA pair fed 1 with 13 led to strong fluorescent labelling of MEK1(76-1) with 13, but minimal background fluorescence from the rest of the lysate (Supplementary Fig. 7a). MEK1(76-1) is not detectable as an overexpressed band in the lysate, and is therefore expressed at a level comparable to many endogenous proteins, demonstrating that the selective labelling reflects the selectivity of the chemical reaction rather than protein overexpression. These data demonstrate that the fluorescent labelling we observe results from the selective incorporation of the unnatural amino acid into the protein of interest and its labelling in a bioorthogonal reaction that is selective with respect to the soluble proteome. The minimal labelling of endogenous proteins, some of which are encoded by genes that terminate in amber stop codons (Supplementary Fig. 7a), is similar to previous observations in Escherichia coli19, while the selectivity of tetrazine labeling with respect to the proteome (Supplementary Fig. 7a) is consistent with previous reports39.
Inhibitor potency and linker length affect iBOLT
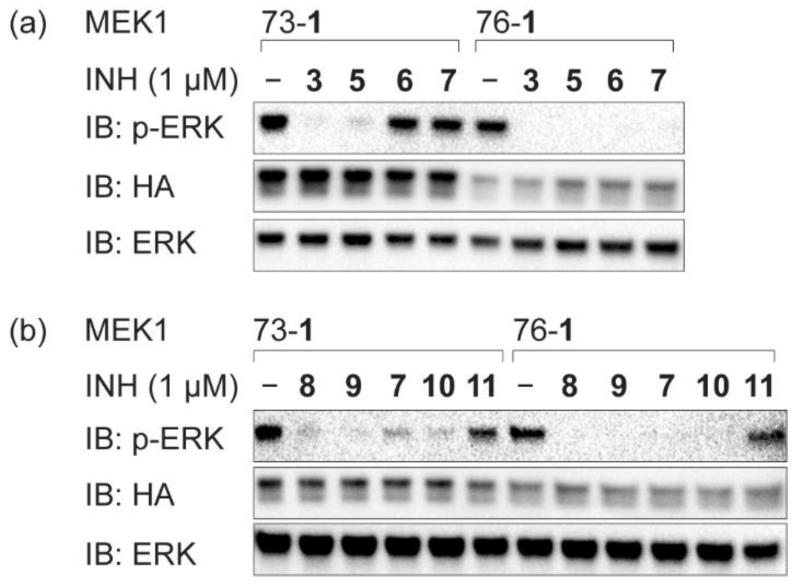
To further investigate how the precise structure of the inhibitor impacts on the potency of the tetrazine inhibitor conjugate we varied the halogen pattern of 3 to create compounds 5-7 (Fig. 1). Based on the reported IC50 values for derivatives of 2 bearing the same halogen patterns as those found in 5-7, which are 50–1,000-fold less active than the parent inhibitor40, the potency of the inhibitors should decrease in the order 3 > 5 > 6 > 7. MEK1(73-1), MEK1(103-1) and MEK1(156-1), for which we observed partial inhibition by 3, showed a clear decrease in inhibiton level in going from inhibitor 3 to inhibitor 7 (Fig. 3a and Supplementary Fig. 9a). These data indicate that the level of inhibition observed for iBOLT can be exquisitely sensitive to the precise structure of the inhibitor and that structure-activity relationships determined for the parent inhibitor qualitatively translate to iBOLT. This provides additional evidence that the mode of tethered inhibitor binding is very similar to the mode of parent inhibitor binding. In contrast, we observed complete inhibition of MEK1(76-1), MEK1(104-1) and MEK1(261-1) with 1 μM 3, 5, 6, and 7, consistent with the increase in potency provided by tethering outweighing any weakening of the inhibitor complex as a result of alterations in the inhibitor structure.
Figure 3. iBOLT is sensitive to inhibitor potency and linker length in inhibitor tetrazine conjugates.
a, Inhibition of MEK1(73-1), but not MEK1(76-1), depends on the potency of the inhibitor. b, Inhibition of MEK1(73-1) and MEK1(76-1) depends on the linker length in the inhibitor conjugate. Cells were incubated with the indicated inhibitor (INH) for 3 h before western blot analysis. EGFP-ERK2 is detected by immunoblot (IB) for ERK and phosphorylated EGFP-ERK2 is detected by immunoblot for pERK. All MEK variants are HA tagged, contain DD mutations making them constitutively active, and are detected by immunoblot for HA. Results of other MEK1 variants are shown in Supplementary Fig. 9.
To investigate the effect of linker length on inhibition we synthesized several derivatives of 7, which is the weakest tetrazine-containing inhibitor in the n = 4 series (Fig. 1) and may therefore be the most sensitive to linker length. The derivatives, 8-11, have the same halogen substitution pattern as 7 and linker lengths up to n = 10. We find that shorter linkers are always as good or better than longer linkers, and that the n = 10 linker leads to a decrease in inhibition with many MEK1(XXX-1) variants (Fig. 3b and Supplementary Fig. 9b). This observation may result from an increased entropic cost of binding the ligand with a long linker as there is no observable difference in tethering 7-11 to MEK1(76-1) (Supplementary Fig. 9c).
Reversible optical switching of kinase activity in live mammalian cells by photo-BOLT
Inhibition of MEK1 by iBOLT is sensitive to the position of tethering, linker length and the potency of the inhibitor. Based on these observations, we investigated whether the introduction of a photoisomerizable group into the linker in compound X (Fig. 1a), to create photo-X (Fig. 4a) might enable the reversible switching of the tethered linker between isomeric forms and enable the activity of the protein to be reversibly toggled on and off in response to light of distinct wavelengths.
Figure 4. Reversible optical toggling of protein function in live mammalian cells via photo-BOLT.
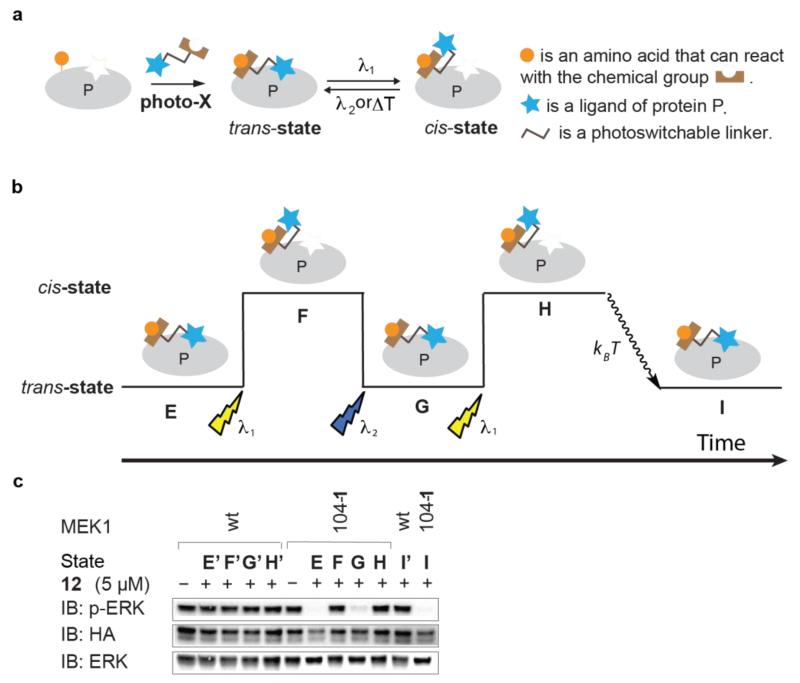
a, General mechanism for toggling the activity of a target protein with light by photo-BOLT. In the inhibitor conjugate the reactive group and the ligand are connected via a photoswitchable linker. After incubation with photo-X the activity of the target protein may be inhibited. Illumination with light at one wavelength (λ1) changes the linker geometry and leads to recovery of function. Further illumination at a second wavelength (λ2) or thermal relaxation (kBT) restores the original geometry, allowing reversible control of protein activity. While we depict the trans-state as inhibitory and the cis-state as activated, other permutations are possible. b, The sequential illuminations implemented in c. c, HEK293ET cells expressing MEK1(104-1) were incubated with 12 for 1 h to achieve state E. The cells were then illuminated with light (λ1 = 360 nm) to achieve state F. Cells were illuminated (λ2 = 440 nm) to achieve state G. Cells were further illuminated (λ1 = 360 nm) to achieve state H. Cells were incubated for an additional 3 h at 37°C without illumination to allow thermal relaxation to state I. HEK293ET cells expressing wild-type MEK1 were treated under identical conditions to those described for MEK1(104-1), leading to the corresponding states: E′, F′, G′, H′ and I′. EGFP-ERK2 is detected by immunoblot (IB) for ERK and phosphorylated EGFP-ERK2 is detected by immunoblot for pERK. All MEK variants are HA tagged, contain DD mutations making them constitutively active, and are detected by immunoblot for HA.
We designed an azobenzene containing linker, as azobenzenes have a defined trans/cis geometry, exhibit good photostability (making them switchable over many cycles), display high extinction coefficients and quantum yields41, and have been used as photocontrol elements in a range of biological systems14,15, 42,43. We synthesized compound 12 (Fig. 1) in which the carbon linker of compound 3 is replaced by an azobenzene-containing linker, and tested its photophysical properties in vitro. The UV-Vis spectra of trans-12 shows a maximum peak of absorption at 373 nm (Supplementary Fig. 10a) which disappears after irradiation and isomerization to cis-12 with UV light (360 nm) (Supplementary Fig. 10b). Irradiation of cis-12 with blue light (440 nm) leads to the recovery of the UV spectra of trans-12. In the absence of illumination, cis-12 also thermally relaxes at room temperature to trans-12 in 3.5 h (Supplementary Fig. 10c).
We screened the active MEK1(XXX-1) variants for inhibition by trans-12 in cells. We identified six kinase variants, MEK1 (73-1, 76-1, 103-1, 104-1, 261-1 and 269-1) that were inhibited by trans-12 (data not shown). For one of these variants, MEK1(104-1), we were able to reversibly toggle kinase activity in mammalian cells with light (Fig. 4b,c, see Supplementary Fig. 11 for quantification). The addition of 12 to HEK293ET cells expressing MEK1(104-1) led to inhibition of EGFP-ERK2 phosphorylation (state E, Fig. 4b,c). Illumination of the cells at 360 nm re-established EGFP-ERK2 phosphorylation (state F, Fig. 4b,c), consistent with the isomerization of 12 from trans to cis relieving inhibition by the tethered inhibitor. Subsequent illumination of cells at 440 nm converted the tethered cis-12 to trans-12 (state G, Fig. 4b,c), re-establishing inhibition of EGFP-ERK2 phosphorylation. Additional illumination of cells at 360 nm relieved inhibition of EGFP-ERK2 phosphorylation by the tethered inhibitor (state H, Fig. 4b,c). Finally, thermal relaxation of state H to state I restored inhibition of EGFP-ERK2 phosphorylation within 3 h. These data demonstrate that photo-BOLT enables the reversible toggling of protein activity in live mammalian cells.
Selective inhibition of MEK2 by iBOLT
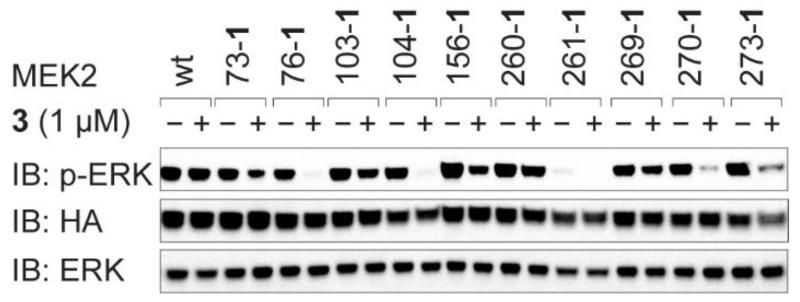
To investigate whether the strategy we have developed for selective MEK1 inhibition can be extended for selective MEK2 inhibition we created ten MEK2(XXX-1) variants (Supplementary Fig. 12a), where XXX represents positions in MEK1(XXX-1) that lead to activity (we use MEK1 numbering for all MEK2 positions to aid comparison between experiments). We tested the inhibition of MEK2(XXX-1) variants by 3 in cells (Fig. 5). For most MEK2(XXX-1) variants we observed inhibition of EGFP-ERK2 phosphorylation and two of these MEK2 variants (76-1 and 104-1) were effectively inhibited by 3. Tethering of 3 to MEK2(76-1) is quantitative (Supplementary Fig. 7a) and 1 μM 3 inhibits MEK2(76-1) within 30 minutes (Supplementary Fig. 12b,c). Comparing the inhibition of MEK1 and MEK2 variants by 3 (Figs 2b and 5) reveals that tethering 3 at positions 76 and 104 lead to effective inhibition of both MEK1 and MEK2, while tethering 3 at position 270 or 273 leads to partial inhibition of MEK2, but not MEK1. Interestingly, a major site of sequence difference between MEK1 and MEK2 is in the kinase insert domain from residues 260 to 308 (ref. 33) and the distinct sequence of the two proteins in this region may explain the distinct effects of tethering 3 at position 270 or 273 on the function of the two proteins.
Figure 5. iBOLT of MEK2(XXX-1) variants.
Cells were incubated with 3 for 3 h before western blot analysis. EGFP-ERK2 is detected by immunoblot (IB) for ERK and phosphorylated EGFP-ERK2 is detected by immunoblot for pERK. All MEK variants are HA tagged, contain DD mutations making them constitutively active, and are detected by immunoblot for HA.
iBOLT of MEK1 and MEK2 with 15
To further extend the scope of iBOLT we designed and synthesized a new conjugate (15, Fig. 1) in which the clinically approved ATP competitive pan kinase inhibitor sunitinib (SU011248, 14) is linked to a tetrazine44 at a position on the inhibitor that protrudes from the well-conserved ATP binding pocket of protein kinases.45
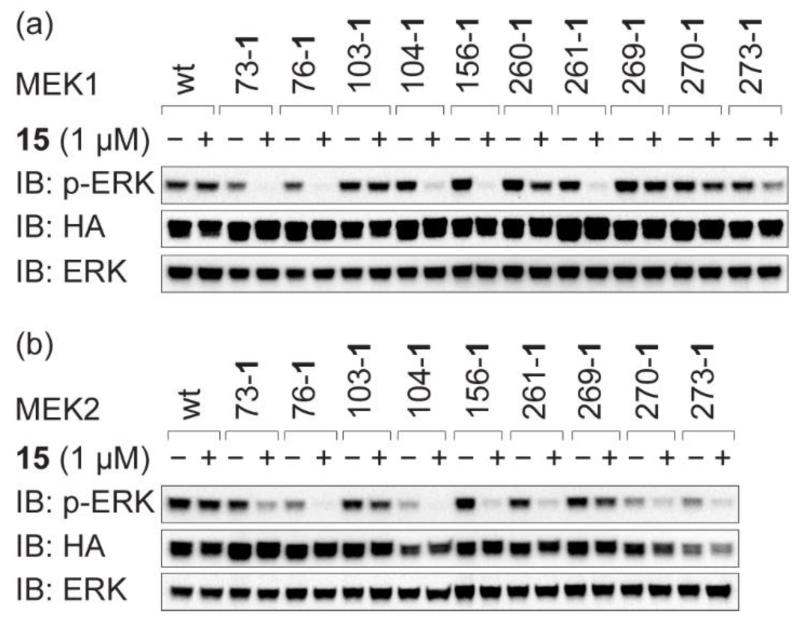
Neither sunitinib tetrazine conjugate 15 nor unmodified sunitinib (14) inhibited wild-type MEK1/2 at the solubility limit (10 μM) (Supplementary Fig. 13a). However, 15 (1 μM) inhibited several MEK1(XXX-1) and MEK2(XXX-1) variants (Fig. 6). We observed differences in inhibition profile between 3 and 15. For example, we observed near-complete inhibition of MEK1(156-1) and MEK2(156-1) by 15, although they are only partially inhibited by 3. In contrast, MEK1(103-1), which was partially inhibited by 3, shows no inhibition by 15. Interestingly, MEK1(76-1) and MEK2(76-1) are inhibited by both 3 and 15. This is consistent with the proximity of position 76 to both the binding site for 2 (the parent compound for 3) and the ATP binding site, bound by sunitinib (14). Fluorescent labeling of cell lysates with 13 indicate that 15 is quantitatively tethered to MEK1(76-1) and to MEK2(76-1) upon addition to cells (Supplementary Fig. 7a). Inhibition of MEK1(76-1) is rapid, and complete inhibition is observed after 30 minutes (Supplementary Fig. 13b). Additional experiments demonstrate that the 1,3-disubstituted cyclopropene containing amino acid 16 (ref. 24; Fig. 1) can be site-specifically incorporated into MEK1(XXX-TAG) variants and used to direct rapid and selective iBOLT with 3 and 15 (Supplementary Fig. 14).
Figure 6. Inhibition of MEK1(XXX-1) and MEK2(XXX-1) variants with sunitinib tetrazine conjugate 15.
Cells were incubated with 15 for 3 h before western blot analysis. EGFP-ERK2 is detected by immunoblot (IB) for ERK and phosphorylated EGFP-ERK2 is detected by immunoblot for pERK. All MEK variants are HA tagged, contain DD mutations making them constitutively active and, are detected by immunoblot for HA.
Inhibition of lymphocyte specific kinase (LCK) with BOLT
We next asked whether the approach developed to selectively inhibit isozymes of the threonine, tyrosine kinase MEK, can be extended to inhibit a distinct protein kinase. We chose the lymphocyte specific kinase LCK, a T-cell-specific, SRC family tyrosine kinase that initiates signalling from the T-cell antigen receptor (TCR) complex at the plasma membrane of T cells46,47, as a target for inhibition. LCK can phosphorylate the TCR at the plasma membrane47, leading to binding of an otherwise cytoplasmic tyrosine kinase ZAP7048, which is phosphorylated by LCK. LCK has only 26% sequence identity to MEK1, and therefore provides a stringent test of the scope and potential scalability of iBOLT.
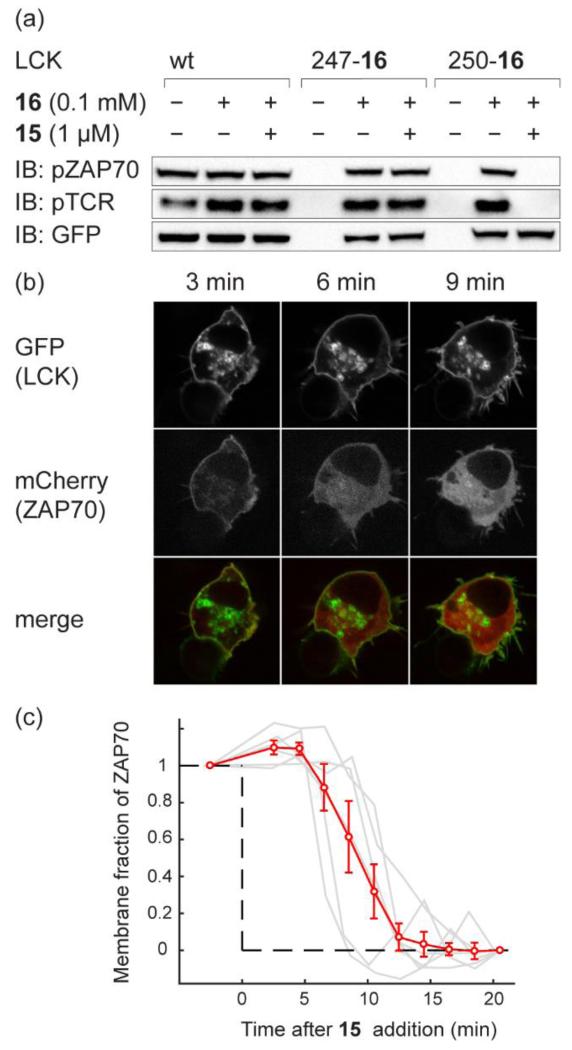
Superposition of the structures of MEK1 and LCK revealed that position 250 of LCK corresponds to position 73 in crystal structures of MEK1 (Supplementary Fig. 15). We asked whether LCK(250-16)-GFP, like MEK1(76-16), can be inhibited by 15. Expression of LCK(250-16) and ZAP70-mCherry in human cells that express the TCR (HEK-TCR) led to phosphorylation of both the TCR and ZAP70-mCherry. Similar results were obtained for other LCK(XXX-16) variants (Supplementary Fig. 15b,c). Phosphorylation of both TCR and ZAP70-mCherry by LCK(250-16)-GFP was inhibited by 1 μM conjugate 15 in HEK-TCR cells (Fig. 7a), and control experiments demonstrate that 15 does not inhibit wild-type LCK (Fig. 7a and Supplementary Fig. 16). Inhibition was rapid, with TCR phosphorylation disappearing within 20 minutes and ZAP70-mCherry phosphorylation disappearing within 30 minutes (Supplementary Fig. 17). LCK(250-16)-GFP and ZAP70-mCherry had the expected localization at the plasma membrane (Fig. 7b), and upon addition of 15, we observed re-localization of ZAP70-mCherry from the plasma membrane to the cytosol of cells as a result of LCK(250-16)-GFP inhibition by BOLT (Fig. 7b,c, Supplementary Movie 1). These data clearly demonstrate that the strategies we have described for one kinase can be extended to rapidly and selectively inhibit other distantly related kinases in mammalian cells.
Figure 7. iBOLT of LCK(250-16)-GFP using conjugate 15 inhibits ZAP70-mCherry and TCR phosphorylation and elicits rapid ZAP70-mCherry dissociation from the membrane.
a, Inhibition of LCK(250-16)-GFP by conjugate 15. Wild type LCK-GFP and LCK(XXX-TAG)-GFP variants were expressed in the presence or absence of 16 in HEK-TCR cells. Cells were then incubated with 1 μM 15 for 3 h before western blot analysis. b, Confocal microscope images of ZAP70-mCherry dissociation from the membrane upon addition of 1 μM 15 to HEK-TCR cells expressing LCK(250-16)-GFP. c, Membrane fraction of ZAP70-mCherry after addition of 1 μM 15 into HEK-TCR expressing LCK(250-16)-GFP. The red curve shows the mean data (± standard error of mean) from six analysed cells. Data for individual cells are plotted in grey lines. The fraction of membrane bound ZAP70-mCherry before adding 15 and after 20 minutes is normalised between 1 and 0, respectively, to allow comparison between datasets.
Conclusion
We have exemplified BOLT with genetically directed competitive and non-competitive inhibitors (iBOLT). We have realized the rapid and selective inhibition of specific isozymes of MEK in live mammalian cells. Identifying parameters for selectively inhibiting one member of an enzyme family enabled the selective inhibition of both a closely related family member and a kinase with low sequence identity to MEK. These results demonstrate that BOLT is a modular and scalable approach to generate highly selective inhibitor-protein pairs. By introducing photoswitchable linkers we demonstrate the first optical toggling of protein function for tethered ligands (photo-BOLT) in live mammalian cells.
It will be possible to apply BOLT to other classes of proteins for which no selective small-molecule modulators exist, to enable selective inhibition, activation or optical modulation of protein function with tethered ligands in cells. We note that it will be possible to spatiotemporally control the optical toggling of protein activity by photo-BOLT, allowing the specificity of BOLT to be combined with a level of spatial control over regulation not commonly achieved by small molecules. The combination of increasingly efficient methods for site-specifically incorporating unnatural amino acids into proteins in mammalian cells49 and genome engineering50 may allow the introduction of ‘phenotypically-silent’, bioorthogonally reactive unnatural amino acids mutants into genomically encoded proteins. This will enable the selective and rapid modulation of protein function by BOLT, providing a route to selectively and rapidly dissect the functions of proteins for which no specific small-molecule modulator exists.
Methods
Detailed procedures for the synthesis of all compounds and their characterization, as well as details of plasmid construction procedures, are provided in the Supplementary Information. Blots show a representative of at least two replicates.
Expression and inhibition
HEK293ET cells were grown at 37 °C in a 5% CO2 atmosphere in DMEM + GlutaMAX medium (Gibco) supplemented with 10% fetal bovine serum (FBS) for 24 hours before transfection. All MEK constructs include the S218D and S222D mutations (unless otherwise noted). These mutations (referred to herein as DD mutations) mimic phosphorylation by RAF rendering MEK1 constitutively active51. Endogenous MEK1/2 has minimal activity in low serum media without external stimulation by epidermal growth factor (EGF). For expression of MEK1 or MEK2 mutants, the growth media from HEK293ET cells was replaced with fresh DMEM (0.5 ml for 24-well, 2 ml for 6-well plate) supplemented with 0.1% FBS and 0.5 mM 1 or 0.1 mM 16, as indicated. Cells (2×105/well, 24-well plate; 106/well, 6-well plate) were transiently transfected with the relevant MEK1 or MEK2 plasmid (pEF1a-MEK1 (XXX_TAG)-218D222D-HA-4xU6-PylTU25C, the corresponding MEK2 variant or control plasmids that do not contain the amber stop codon), pEF1a-FLAG-MbBCNRS-4xU6-PylTU25C and pCMV-EGFP-ERK2 plasmids using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s protocol. Cells were grown for 24 h before analysis. For inhibition assays, cells were washed with media (0.5 ml for 24-well, 2 ml for 6-well plate; DMEM with 0.1% FBS) four times (3 × 1 min, 1 × 30 min) before the addition of inhibitor conjugates. Inhibitor conjugates were used as 200× stock solutions in DMSO. For photo-BOLT, cells were incubated with 12 for 45 min and washed twice (1 × 1 min, 1 × 15 min) with media (0.5 mL, phenol red free DMEM with 0.1% FBS) before illumination at 360 nm (3 × 30 s, 15 min incubation between each illumination, 80 mW cm−2) to isomerize 12 from trans to cis relieving inhibition by the tethered inhibitor conjugate. Subsequent illumination of cells at 440 nm (120 s, 20 min incubation, 20 mW cm−2) converted the tethered cis-12 to trans-12, re-establishing inhibition of EGFP-ERK2 phosphorylation. HEK TCR cells were handled as HEK293ET cells, except that the DMEM contained 10% FBS throughout. Cells were transiently transfected with the relevant LCK plasmid, pEF1a-FLAG-PylRS-4xU6-PylTU25C and pHR-ZAP70-mCherry (where indicated) using GeneJuice (Merck Millipore) according to the manufacturer’s protocol. Inhibition assays were performed as described above. Imaging experiments were performed using a spinning-disk confocal microscope47.
Western blot
Inhibitor conjugate mediated inhibition was followed by western blot. Cells (2 × 105/well of a 24-well plate) were washed with 0.5 ml phosphate buffered saline (PBS), then lysed with RIPA buffer (0.1 mL per well, Sigma, R0278) supplemented with protease inhibitor (Roche, 11873580001) and phosphatase inhibitor (1×, Sigma, P5726) at 4°C for 10 min. The lysates were pelleted (20,000g, 10 min, 4 °C), and the supernatant was added to LDS sample buffer (Life Technologies). The samples were heated (95 °C, 10 min), loaded on NuPAGE 4-12% bis-tris protein gel (Life Technologies) and transferred to a nitrocellulose membrane (iBlot2, Life Technologies). Each membrane was incubated (4°C, overnight) with a primary rabbit anti-p-ERK1/2(pT202/pY204) antibody (Cell Signaling, #4370, 1:1,000 dilution) and a primary rat anti-HA antibody (Roche, #12158167001, 1:4,000 dilution) in 20 ml PBST (PBS with 0.1% Tween 20) supplemented with 5% milk powder. The membrane was washed three times (20 ml PBST, 5 min per wash). All subsequent washing steps use this procedure. A secondary anti-rabbit antibody (Cell Signaling, #7074, 1:2,000 dilution) was incubated (23 °C, 1 h), the membrane was washed and the signal was developed by the addition of 1 ml Luminata Forte Western HRP substrate (Millipore). After imaging on a Bio-Rad ChemiDoc XRS+ system, the membrane was washed and incubated (23 °C, 1 h) with secondary anti-rat antibody (Santa Cruz Biotech, sc-2032, 1:2,000 dilution). The membrane was washed again, the signal developed and imaged as described above. A second membrane loaded with the same sample was incubated with rabbit anti-ERK1/2 antibody (Cell Signaling, #4695, 1:1,000 dilution) and incubated (4°C, overnight) before washing. A secondary anti-rabbit antibody (Cell Signaling, #7074, 1:2,000 dilution) was incubated (23 °C, 1 h), before being washed and processed as described above. For some experiments membranes were washed and re-probed with a second primary antibody. Procedures for LCK experiments were analogous to those described above for MEK, but LDS buffer was also supplemented with 1 mM dithiolthreitol (DTT). Three membranes were prepared. The first was incubated with primary rabbit anti-ZAP70(pY319) antibody (Cell Signaling, #2717, 1:1,000 dilution), the second with rabbit anti-LCK(pY394) (Cell Signaling, #2101, 1:1,000 dilution) and the third was cut in two pieces. The top half of this membrane was incubated with mouse anti-GFP antibody (Roche, #11814460001, 1:2,000 dilution) and the bottom half with mouse anti-p-TCR antibody (BD Biosciences, #558402, 1:2,000 dilution). The blots were developed as described above.
Fluorescence imaging
Tethering of inhibitor conjugates to MEK1/2(XXX) variants was determined by treating cells with a fluorescent tetrazine conjugate. Fluorescent labelling is inhibited if the tetrazine is ligated to the inhibitor conjugate. HEK293ET cells (106/well, 6-well plate) expressing the relevant MEK1/2(XXX) variant and previously treated (or not) with an inhibitor conjugate were washed with PBS (2 ml), then lysed in PBS containing 0.1% Triton X-100 (0.2 ml per well, 4 °C, 10 min). The lysate was pelleted (20,000g, 10 min, 4 °C) and the supernatant collected in an amber tube (Eppendorf, 0030120.191). TAMRA-tetrazine conjugate 13 was added to a final concentration of 10 μM. The mixture was incubated (4 °C, 4 h) before the addition of 4× LDS sample buffer (Life Technologies), heated (95 °C, 10 min) and loaded onto a to NuPAGE 4-12% bis-tris protein gel (Life Technologies). Fluorescent images were obtained by using a Typhoon Trio Variable Mode Imager (GE Healthcare Life Sciences) with excitation at 532 nm (O580 filter). The proteins were then transferred to a nitrocellulose membrane (iBlot2, Life Technologies), and incubated with Ponceau S (Santa Cruz Biotech, sc-301558) to visualize total protein content, before MEK expression was visualized as described above by anti-HA western blot.
Supplementary Material
Acknowledgements
This work was supported by the UK Medical Research Council (grants nos. U105181009 and UD99999908) and the European Research Council. Y.-H.T. was supported by an EMBO Long-Term Fellowship and S.E by a DFG Research Fellowship. J.R.J. is a Sir Henry Dale fellow of The Wellcome Trust and The Royal Society. The authors thank K. Wang, L. Davis and M. Mohan for discussions, S. Elsaesser for plasmids and T. Elliot for compound 16.
Footnotes
Supplementary information and chemical compound information are available in the online version of the paper.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Shogren-Knaak MA, Alaimo PJ, Shokat KM. Recent Advantages in Chemical Approaches to the Study of Biological Systems. Annu. Rev. Cell Dev. Bio. 2001;17:405–433. doi: 10.1146/annurev.cellbio.17.1.405. [DOI] [PubMed] [Google Scholar]
- 2.Knight ZA, Shokat KM. Chemical Genetics: Where Genetics and Pharmacology Meet. Cell. 2007;128:425–430. doi: 10.1016/j.cell.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 3.Schreiber SL. Organic synthesis toward small-molecule probes and drugs. Proc. Natl. Acad. Sci. 2011;108:6699–6702. doi: 10.1073/pnas.1103205108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belshaw PJ, Schoepfer JG, Liu K-Q, Morrison KL, Schreiber SL. Rational Design of Orthogonal Receptor-Ligand Combinations. Angew. Chem. Int. Ed. 1995;34:2129–2132. [Google Scholar]
- 5.Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, Wood JL, Morgan DO, Shokat KM. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 6.Zheng H, Al-Ayoubi A, Eblen ST. MAP Kinase Signaling Protocols. Vol. 661. Springer; 2010. [Google Scholar]
- 7.Erlanson DA, Wells JA, Braisted AC. Tethering: fragment-based drug discovery. Annu. Rev. Biophys. Biomol. Struct. 2004;33:199–223. doi: 10.1146/annurev.biophys.33.110502.140409. [DOI] [PubMed] [Google Scholar]
- 8.Cravatt BF, Wright AT, Kozarich JW. Activity-Based Protein Profiling: From Enzyme Chemistry to Proteomic Chemistry. Annu. Rev. Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 9.Takaoka Y, Ojida A, Hamachi I. Protein Organic Chemistry and Applications for Labeling and Engineering in Live-Cell Systems. Angew. Chem. Int. Ed. 2013;52:4088–4106. doi: 10.1002/anie.201207089. [DOI] [PubMed] [Google Scholar]
- 10.Chen Z, Jing C, Gallagher SS, Sheetz MP, Cornish VW. Second-Generation Covalent TMP-Tag for Live Cell Imaging. J. Am. Chem. Soc. 2012;134:13692–13699. doi: 10.1021/ja303374p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Q, Sabnis Y, Zhao Z, Zhang T, Buhrlage Sara J., Jones Lyn H., Gray NS. Developing Irreversible Inhibitors of the Protein Kinase Cysteinome. Chem. Biol. 2013;20:146–159. doi: 10.1016/j.chembiol.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blair JA, Rauh D, Kung C, Yun C-H, Fan Q-W, Rode H, Zhang C, Eck MJ, Weiss WA, Shokat KM. Structure-guided development of affinity probes for tyrosine kinases using chemical genetics. Nat. Chem. Biol. 2007;3:229–238. doi: 10.1038/nchembio866. [DOI] [PubMed] [Google Scholar]
- 13.Volgraf M, Gorostiza P, Numano R, Kramer RH, Isacoff EY, Trauner D. Allosteric control of an ionotropic glutamate receptor with an optical switch. Nat. Chem. Biol. 2006;2:47–52. doi: 10.1038/nchembio756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fehrentz T, Schönberger M, Trauner D. Optochemical Genetics. Angew. Chem. Int. Ed. 2011;50:12156–12182. doi: 10.1002/anie.201103236. [DOI] [PubMed] [Google Scholar]
- 15.Szymański W, Beierle JM, Kistemaker HAV, Velema WA, Feringa BL. Reversible Photocontrol of Biological Systems by the Incorporation of Molecular Photoswitches. Chem. Rev. 2013;113:6114–6178. doi: 10.1021/cr300179f. [DOI] [PubMed] [Google Scholar]
- 16.Sletten EM, Bertozzi CR. Bioorthogonal Chemistry: Fishing for Selectivity in a Sea of Functionality. Angew. Chem. Int. Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lang K, Chin JW. Bioorthogonal reactions for labeling proteins. ACS Chem. Biol. 2014;9:16–20. doi: 10.1021/cb4009292. [DOI] [PubMed] [Google Scholar]
- 18.Lang K, Chin JW. Cellular Incorporation of Unnatural Amino Acids and Bioorthogonal Labeling of Proteins. Chem. Rev. 2014;114:4764–4806. doi: 10.1021/cr400355w. [DOI] [PubMed] [Google Scholar]
- 19.Lang K, et al. Genetically encoded norbornene directs site-specific cellular protein labelling via a rapid bioorthogonal reaction. Nat. Chem. 2012;4:298–304. doi: 10.1038/nchem.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plass T, et al. Amino Acids for Diels-Alder Reactions in Living Cells. Angew. Chem. Int. Ed. 2012;51:4166–4170. doi: 10.1002/anie.201108231. [DOI] [PubMed] [Google Scholar]
- 21.Kaya E, et al. A Genetically Encoded Norbornene Amino Acid for the Mild and Selective Modification of Proteins in a Copper-Free Click Reaction. Angew. Chem. Int. Ed. 2012;51:4466–4469. doi: 10.1002/anie.201109252. [DOI] [PubMed] [Google Scholar]
- 22.Seitchik JL, et al. Genetically Encoded Tetrazine Amino Acid Directs Rapid Site-Specific in Vivo Bioorthogonal Ligation with trans-Cyclooctenes. J. Am. Chem. Soc. 2012;134:2898–2901. doi: 10.1021/ja2109745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lang K, et al. Genetic Encoding of Bicyclononynes and trans-Cyclooctenes for Site-Specific Protein Labeling in Vitro and in Live Mammalian Cells via Rapid Fluorogenic Diels-Alder Reactions. J. Am. Chem. Soc. 2012;134:10317–10320. doi: 10.1021/ja302832g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elliott TS, et al. Proteome labeling and protein identification in specific tissues and at specific developmental stages in an animal. Nat. Biotech. 2014;32:465–472. doi: 10.1038/nbt.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bianco A, Townsley FM, Greiss S, Lang K, Chin JW. Expanding the genetic code of Drosophila melanogaster. Nat. Chem. Biol. 2012;8:748–750. doi: 10.1038/nchembio.1043. [DOI] [PubMed] [Google Scholar]
- 26.Krishnamurthy VM, et al. Carbonic Anhydrase as a Model for Biophysical and Physical-Organic Studies of Proteins and Protein-Ligand Binding. Chem. Rev. 2008;108:946–1051. doi: 10.1021/cr050262p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vauquelin G. Simplified models for heterobivalent ligand binding: when are they applicable and which are the factors that affect their target residence time. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2013;386:949–962. doi: 10.1007/s00210-013-0881-0. [DOI] [PubMed] [Google Scholar]
- 28.Gargano JM, Ngo T, Kim JY, Acheson DWK, Lees WJ. Multivalent Inhibition of AB5 Toxins. J. Am. Chem. Soc. 2001;123:12909–12910. doi: 10.1021/ja016305a. [DOI] [PubMed] [Google Scholar]
- 29.Krishnamurthy VM, Semetey V, Bracher PJ, Shen N, Whitesides GM. Dependence of Effective Molarity on Linker Length for an Intramolecular Protein-Ligand System. J. Am. Chem. Soc. 2007;129:1312–1320. doi: 10.1021/ja066780e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hilger RA, Scheulen ME, Strumberg D. The Ras-Raf-MEK-ERK Pathway in the Treatment of Cancer. Oncol. Res. Treat. 2002;25:511–518. doi: 10.1159/000068621. [DOI] [PubMed] [Google Scholar]
- 31.Sebolt-Leopold JS, Herrera R. Targeting the mitogen-activated protein kinase cascade to treat cancer. Nat. Rev. Cancer. 2004;4:937–947. doi: 10.1038/nrc1503. [DOI] [PubMed] [Google Scholar]
- 32.Solit DB, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brott BK, et al. Mek2 Is a Kinase Related to Mek1 and Ts Differentially Expressed in Murine Tissues. Cell Growth. Differ. 1993;4:921–929. [PubMed] [Google Scholar]
- 34.Giroux S, et al. Embryonic death of Mek1-deficient mice reveals a role for this kinase in angiogenesis in the labyrinthine region of the placenta. Curr. Biol. 1999;9:369–376. doi: 10.1016/s0960-9822(99)80164-x. [DOI] [PubMed] [Google Scholar]
- 35.Ussar S, Voss T. MEK1 and MEK2, Different Regulators of the G1/S Transition. J. Biol. Chem. 2004;279:43861–43869. doi: 10.1074/jbc.M406240200. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Y, Adjei AA. The clinical development of MEK inhibitors. Nat. Rev. Clin. Oncol. 2014;11:385–400. doi: 10.1038/nrclinonc.2014.83. [DOI] [PubMed] [Google Scholar]
- 37.Tecle H, et al. Beyond the MEK-pocket: Can current MEK kinase inhibitors be utilized to synthesize novel type III NCKIs? Does the MEK-pocket exist in kinases other than MEK? Bioorg. Med. Chem. Lett. 2009;19:226–229. doi: 10.1016/j.bmcl.2008.10.108. [DOI] [PubMed] [Google Scholar]
- 38.Rice KD, et al. Novel Carboxamide-Based Allosteric MEK Inhibitors: Discovery and Optimization Efforts toward XL518 (GDC-0973) ACS Med. Chem. Lett. 2012;3:416–421. doi: 10.1021/ml300049d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yao JZ, et al. Fluorophore Targeting to Cellular Proteins via Enzyme-Mediated Azide Ligation and Strain-Promoted Cycloaddition. J. Am. Chem. Soc. 2012;134:3720–3728. doi: 10.1021/ja208090p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barett SD, Biwersi C, Kaufman M, Tecle H, Warmus JS. International Patent, WO 02/06213 A2 Oxygenated esters of 4-iodo phenylamino benzhydroxamic acid. 2002
- 41.Hamon F, Djedaini-Pilard F, Barbot F, Len C. Azobenzenes-synthesis and carbohydrate applications. Tetrahedron. 2009;65:10105–10123. [Google Scholar]
- 42.Renner C, Moroder L. Azobenzene as Conformational Switch in Model Peptides. ChemBioChem. 2006;7:868–878. doi: 10.1002/cbic.200500531. [DOI] [PubMed] [Google Scholar]
- 43.Beharry AA, Wong L, Tropepe V, Woolley GA. Fluorescence Imaging of Azobenzene Photoswitching In Vivo. Angew. Chem. Int. Ed. 2011;50:1325–1327. doi: 10.1002/anie.201006506. [DOI] [PubMed] [Google Scholar]
- 44.Tang PC, et al. US Patent, US 6,573,293 B2 Pyrrole substituted 2-indolinone protein kinase inhibitors. 2003
- 45.Gajiwala KS, et al. KIT kinase mutants show unique mechanisms of drug resistance to imatinib and sunitinib in gastrointestinal stromal tumor patients. Proc. Natl. Acad. Sci. 2009;106:1542–1547. doi: 10.1073/pnas.0812413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palacios EH, Weiss A. Function of the Src-family kinases, Lck and Fyn, in T-cell development and activation. Oncogene. 2004;23:7990–8000. doi: 10.1038/sj.onc.1208074. [DOI] [PubMed] [Google Scholar]
- 47.James JR, Vale RD. Biophysical mechanism of T-cell receptor triggering in a reconstituted system. Nature. 2012;487:64–69. doi: 10.1038/nature11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Au-Yeung BB, et al. The structure, regulation, and function of ZAP-70. Immunol. Rev. 2009;228:41–57. doi: 10.1111/j.1600-065X.2008.00753.x. [DOI] [PubMed] [Google Scholar]
- 49.Schmied WH, Elsässer SJ, Uttamapinant C, Chin JW. Efficient Multisite Unnatural Amino Acid Incorporation in Mammalian Cells via Optimized Pyrrolysyl tRNA Synthetase/tRNA Expression and Engineered eRF1. J. Am. Chem. Soc. 2014;136:15577–15583. doi: 10.1021/ja5069728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mansour S, et al. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.