Abstract
Trichoderma atroviride is a mycoparasite of a number of plant pathogenic fungi thereby employing morphological changes and secretion of cell wall degrading enzymes and antibiotics. The function of the tmk1 gene encoding a mitogen-activated protein kinase (MAPK) during fungal growth, mycoparasitic interaction, and biocontrol was examined in T. atroviride. Δtmk1 mutants exhibited altered radial growth and conidiation, and displayed de-regulated infection structure formation in the absence of a host-derived signal. In confrontation assays, tmk1 deletion caused reduced mycoparasitic activity although attachment to Rhizoctonia solani and Botrytis cinerea hyphae was comparable to the parental strain. Under chitinase-inducing conditions, nag1 and ech42 transcript levels and extracellular chitinase activities were elevated in a Δtmk1 mutant, whereas upon direct confrontation with R. solani or B. cinerea a host-specific regulation of ech42 transcription was found and nag1 gene transcription was no more inducible over an elevated basal level. Δtmk1 mutants exhibited higher antifungal activity caused by low molecular weight substances, which was reflected by an over-production of 6-pentyl-α-pyrone and peptaibol antibiotics. In biocontrol assays, a Δtmk1 mutant displayed a higher ability to protect bean plants against R. solani.
Keywords: Trichoderma atroviride, Mycoparasitism, Mitogen-activated protein kinase (MAPK), Chitinase, Antifungal metabolites, Peptaibols
1. Introduction
Members of the genus Trichoderma are potent mycoparasites as they attack and parasitize plant pathogens, and therefore they are commercially applied as biocontrol agents (Hjeljord and Tronsmo, 1998). What currently is defined as biocontrol is a combination of different mechanisms like formation of infection structures (e.g. coiling), production of hydrolytic enzymes, secretion of antifungal metabolites, and induction of defense responses in plants, that work synergistically to achieve disease control (Harman et al., 2004; Howell, 2003).
After recognizing the presence of a potential host fungus, Trichoderma inhibits or kills the plant pathogen by parasitizing its hyphae, thereby employing hydrolytic enzymes like chitinases and glucanases to degrade the host’s cell wall (Chet et al., 1998; Kubicek et al., 2001). Sensing of the host’s presence may involve a variety of signal transduction pathways resulting in the expression of mycoparasitism-related genes. The current model is that both enzyme production and infection structure formation are induced responses triggered by molecules released from the host fungus (e.g. degradation products from its cell wall) or located on its surface (e.g. lectins). Recent findings suggest that additionally the production of antimicrobial metabolites could be enhanced by the presence of the host, as 6-pentyl-α-pyrone production in T. harzianum is elicited by Rhizoctonia solani (Serrano-Carreon et al., 2004).
Highly conserved mitogen-activated protein kinase (MAPK) cascades found in animals, plants, and fungi are involved in the transmission of extracellular and intracellular signals, thereby often regulating transcription factors by MAPK-mediated phosphorylation (Schaeffer and Weber, 1999). The study of fungal MAPKs revealed their involvement in several essential developmental processes such as sporulation, mating, hyphal growth, and pathogenicity (Gustin et al., 1998; Xu, 2000; Zeilinger, 2004b). In the model pathogenic fungus Magnaporthe grisea three MAPK-encoding genes have been characterized, among which pmk1 (an ortholog of FUS3/KSS1 of Saccharomyces cerevisiae) and mps1 (an ortholog of S. cerevisiae SLT2) were found to be essential for pathogenicity-related processes like appressoria formation, host tissue colonization, and penetration of the host cuticle (Dixon et al., 1999; Xu and Hamer, 1996; Xu et al., 1998). All examined Pmk1 homologs from other phytopathogenic fungi were also shown to be involved in pathogenicity, and some of them to regulate the induction of secreted plant cell wall-degrading enzymes (Di Pietro et al., 2001; Jenczmionka and Schaeffer, 2005; Lev and Horwitz, 2003; Zheng et al., 2000).
Elucidation of signaling pathways of Trichoderma affecting mycoparasitism recently started and confirmed the involvement of conserved signaling components. In T. atroviride and T. virens, α-subunits of heterotrimeric G proteins were demonstrated to play important roles in the antagonism of plant pathogens (Mukherjee et al., 2004; Reithner et al., 2005; Rocha-Ramirez et al., 2002; Zeilinger et al., 2005), and in T. virens in addition a MAP kinase was found to affect mycoparasitism-related processes (Mendoza-Mendoza et al., 2003; Mukherjee et al., 2003) as well as plant systemic resistance (Viterbo et al., 2005). Although both species are closely related, examination of G protein signaling revealed significant differences among these biocontrol agents. Whereas in T. atroviride the subgroup I Gα protein Tga1 affects chitinase and antifungal metabolite production (Reithner et al., 2005), and was shown to be indispensable for the overgrowth of the host R. solani (Rocha-Ramirez et al., 2002), T. virens TgaA is involved in antagonism against Sclerotium rolfsii but not R. solani and ΔtgaA loss-of-function mutants sporulate and coil similar to the parental strain (Mukherjee et al., 2004).
In this study, we describe the characterization of the T. atroviride MAP kinase-encoding gene tmk1 according to its influence on mycoparasitism-related processes and plant protection. Δtmk1 mutants showed reduced mycoparasitism in direct mycoparasite–host interactions on plates and a host-specific regulation of ech42 gene transcription. In addition, they exhibited a 10-times elevated production of peptaibols and displayed a higher ability to protect bean plants against R. solani infection. To our knowledge, this is the first report on increased secondary metabolite synthesis in a fungal MAPK deletion mutant.
2. Materials and methods
2.1. Strains and culture conditions
Trichoderma atroviride strain P1 (formerly T. harzianum, ATCC 74058) was used for this study and grown on potato-dextrose agar (PDA; Merck, Germany) at 28 °C until sporulation. Botrytis cinerea and R. solani were used as pathogens and were obtained from the collection of the Institute of Plant Pathology, Università degli Studi di Napoli Federico II, Naples, Italy. Escherichia coli JM 109 was the host for plasmid amplification.
Trichoderma atroviride was grown in liquid synthetic medium (SM) as described previously (Brunner et al., 2003). For induction experiments, the fungus was pre-cultivated for 36 h in SM containing 1% (w/v) glycerol as carbon source, harvested by filtration, washed with sterile tap water and transferred to fresh SM medium containing 1% (w/v) colloidal chitin or 1% (w/v) N-acetyl-β-d-glucosamine.
For the determination of conidia production T. atroviride was incubated on PDA at 28 °C for 7 days with daily exposure to sunlight for 30 min. Spores were collected and counted in a counting chamber. The results are representing the total amount of conidia produced on one plate determined in four replicates.
2.2. Cloning of tmk1
A ~750-bp product was obtained by PCR amplification of genomic DNA from T. atroviride P1 with degenerate oligonucleotide primers based on conserved regions of MAPK sequences (MAPK-F: 5′-GCNTAYGGNRTNGTNTG-3′ and MAPK-R: 5′-CATYTCNGCNARDATRCANCC-3′). The product was subcloned into pGEM-T (Promega, Madison, WI) and sequenced. For screening a genomic λ BlueStar library of T. atroviride P1, the tmk1-containing PCR fragment was radioactively labeled with [α32P]dCTP by random priming and used as a probe. The genomic sequence of the isolated clones carrying the tmk1 gene and its flanking regions was determined by primer walking using the following internal primers: Tmk1intF1: 5′-GTCCGTTGGCTGTATC-3′ (bp 815–830), Tmk1intF2: 5′-GTATTCTGGTTCACTAC-3′ (bp 1603–1619), Tmk1intR1: 5′-CTTACCAAACAACACCGTAG-3′ (bp 98–117), and Tmk1intR2: 5′-CAAGGCAATA ATTC AGGAG-3′ (bp 688–670).
2.3. Fungal transformation
For obtaining Δtmk1 mutants, Agrobacterium-mediated transformation of T. atroviride was carried out as previously described (Zeilinger, 2004a). Briefly, after co-cultivating a conidial suspension (107 spores ml−1) for 24 h with an Agrobacterium tumefaciens strain containing the disruption construct pTSZ-Δ tmk1, in which the entire tmk1 coding region is replaced by an hph (hygromycin B phosphotransferase-encoding) expression cassette (Zeilinger, 2004a), fungal transformants were selected on PDA containing 200 μg/ml hygromycin B. Transformants were recovered and purified to mitotic stability by repeated transfer to selective medium and by two rounds of single spore isolation.
For complementation assays, an ~3.5-kb fragment bearing the tmk1 gene and its 5′ and 3′ regulatory regions from the parental strain T. atroviride P1 was reintroduced into the Δtmk1-12 mutant by co-transformation with plasmid p3SR2 (Kelly and Hynes, 1985), a vector containing the Aspergillus nidulans amdS gene. Transformants were selected on acetamide-containing SM medium without any nitrogen source and 2% (w/v) of Agar-Nobel (Difco, BD, Germany) and purified to mitotic stability by two rounds of single spore isolation. Re-integration of the tmk1 gene was verified by Southern analysis and a transformant strain named tmk1-re carrying one copy of tmk1 was selected for further experiments.
2.4. DNA and RNA manipulations
Genomic DNA was isolated as previously described (Gruber et al., 1990); Southern hybridizations were carried out as described by Sambrook et al. (Sambrook et al., 1989). RNA was isolated as described by Chomczynski and Sacchi (1987).
2.5. Real-time reverse transcriptase-polymerase chain reaction
Real-time reverse transcriptase PCR of total RNA was carried out as described previously (Zeilinger et al., 2005). For amplification of a ~170-bp fragment of the actin-encoding gene act1 as reference gene, real-time RT-PCR was carried out with primers actinP1F (5′-GCACGGAATCGCTCGTTGC-3′) and actinP1R (5′-TTCTCCACCACCGCCAAGC-3′) in the same run than the transcripts of interest. A ~90-bp fragment of nag1 was amplified using primers Fnag1Taq (5′-TGTCCTACAGCCTCTGCTGCAAAAGTTC-3′) and Rnag1Taqkurz (5′-CATCTCCTCACAGACAAGCGGTGAAAG-3′) and the following program: 40 cycles consisting of 95 °C for 15 s, 60 °C for 15 s, and 72 °C for 20 s. A fragment of the endochitinase-encoding gene ech42 was amplified with primers ech42 P1 forward (5′-CGCAACTTCCAGCCTCAGAACC-3′) and ech42 P1 reverse (5′-TCAATACCATCGAAACCCCAGTCC-3′) using 40 cycles of 95 °C for 20 s, 60 °C for 20 s, and 72 °C for 30 s. A ~140-bp fragment of the condensation domain-encoding region of the peptide synthetase Tex1 was amplified with primers tex1_Ta_f (5′-GGTACACGTCTCTGCCGCTATGCTATC-3′) and tex1_Ta_r (5′-CATTTCGGTGCCAGCGTACGCGG-3′) using 40 cycles consisting of 95 °C for 15 s, 60 °C for 15 s, and 72 °C for 20 s. All determinations were performed three times from two biological replicates with three different sample dilutions. The efficiency of the real-time RT-PCR was calculated from the given threshold cycles in Bio-Rad iCycler (Bio-Rad) according to the equation: E = 10−1/slope (Rasmussen et al., 2001). The relative transcription ratio was determined according to the mathematical model published by Pfaffl (Pfaffl, 2001).
2.6. Transmitting light microscopy
Microscopic studies were mainly performed as previously described by Lu et al. (2004). Glass slides, on which 500 μl of PDA was spread onto, were inoculated and incubated on a moistened filter paper (Gel-blotting paper; Roth, Karlsruhe, Germany) at 28 °C in a Petri dish sealed with parafilm. Additionally, sterile nylon 66 fibers (approximate diameter 14 μm; Nilit, Migdal-Haemek, Israel) were placed on the media before inoculation. For mycoparasitic studies of the antagonistic behavior, T. atroviride and R. solani were inoculated on opposite sides of the glass slides. After 48–72 h, the fungal hyphae were viewed under a Leitz Aristoplan (Wetzlar, Germany) microscope, and pictures were taken using an Olympus DP 10 (Olympus America Inc., Melville, NY) camera.
2.7. Antagonistic and biocontrol assays
Plate confrontation assays with R. solani or B. cinerea as hosts were performed as previously described (Lorito et al., 1996; Zeilinger et al., 1999). Plant experiments for in vivo biocontrol tests were conducted with Phaseolus vulgaris L. (var. nanus L.) (Austrosaat, Vienna, Austria) and R. solani-infested soil as previously described (Brunner et al., 2003). The height of surviving plants was investigated for a period of 6 days after germination and related to the number of growing beans. The vitality was calculated from the relative plant height with Trichoderma compared to a control experiment without Trichoderma. Additional control experiments were conducted without R. solani and a total of 14 bean seeds per attempt.
2.8. Antibiotic activity assays
As described previously (Graeme-Cook and Faull, 1991), an agar plug with Trichoderma was placed on a PDA plate covered with a cellophane or a dialysis tubing cellulose membrane (cut off 12 kDa; Sigma–Aldrich, St. Louis, MO), and cultivated for 3 days. The area covered by the colony was marked and the membrane carrying the fungus was removed. A spore suspension containing 107 B. cinerea spores was spread over the surface of the plate which was further incubated for 7 days at room temperature (RT). The antibiotic activity secreted by the Trichoderma colony was detected as a zone of inhibition of germination of B. cinerea spores. The inhibition index (an extent for inhibition) was calculated as (diameter of the zone of inhibition)/(diameter of the colony growth). These assays were set up in triplicate.
2.9. Combined antagonistic and antibiotic activity assays
Plate confrontation assays were carried out on PDA plates covered with cellophane or dialysis tubing cellulose membranes and incubated for 4 days at 28 °C. The areas covered by the fungal colonies as well as the confrontation zone were marked on the plate and the membrane was removed. A lawn of 107 B. cinerea spores was spread over the surface of the plate and incubated at RT for 7 days. The inhibition index for this assay was calculated as (area of the zone of inhibition)/(area of fungal growth).
2.10. Enzyme assays
Chitinase activity in the culture filtrates was determined in triplicate. Enzyme assays were performed as previously described (Harman et al., 1993) using the substrates p-nitrophenyl N-acetyl-β-d-glucosaminide for determination of N-acetyl-glucosaminidase and 4-nitrophenyl β-d-N,N′,N″-triacetylchitotriose (all from Sigma) for determination of endochitinase activity as substrates. Enzyme activity was measured as U/ml (one unit is defined as the release of 1 μmol of nitrophenol per minute) and related to biomass by determination of the mycelial dry weight resulting in expression of enzyme activity as U/g mycelial dry weight.
2.11. Quantitative 6-pentyl-α-pyrone analysis
Quantification of 6-pentyl-α-pyrone (6-PP) from PDA plates previously inoculated with Δtmk1-12 strain and T. atroviride parental strain, respectively, was performed as described (Reithner et al., 2005).
2.12. Qualitative analysis of peptaibol production
Sterilized synthetic medium composed of (0.5% w/v) glucose, (0.08% w/v) KH2PO4, (0.07% w/v) KNO3, (0.02% w/v) CaCl2, (0.05% w/v) MgSO4·7H2O, (0.001% w/v) MnSO4·5H2O, (0.0005% w/v) CuSO4·5H2O, and (0.0001% w/v) FeSO4·7H2O was inoculated with conidial suspensions of T. atroviride P1 and the Δtmk1-12 mutant. The stationary cultures were incubated at 28 °C for 10 days. Five microliters of a mixture containing 0.5 ml of the culture filtrate and 1 ml of mobile phase A (acetonitrile/water/formic acid; 28:72:0.1) were used for LC-MS/MS analysis (QTrap 4000 LC-MS/MS system; Applied Biosystems, Foster City, CA, USA). The chromatographic separation was carried out on a BDS Hypersil® C-18 column (5 μm, 150 × 2.1 mm; Thermo Electron, Woburn, MA, USA) using a linear gradient elution starting with mobile phase A and ending with mobile phase B (acetronitrile/water/formic acid; 100:0:0.1). The LC flow rate was set to 0.3 ml/min at a column oven temperature of 25 °C. The ESI interface was used in positive ion mode at 450 °C with the following settings: nitrogen curtain gas (CUR) at 69 kPa, nebuliser gas (GS1) nitrogen at 345 kPa, auxiliary gas (GS2) nitrogen at 345 kPa, ion spray voltage (IS) +4000 V, collision-activated dissociation gas (CAD) “high”, interface heater (ihe) on. Enhanced product ion scan (EPI) was used as a hybrid triple quadrupole-linear ion trap (LIT) scan mode, involving selection of the precursor in quadrupole 1, fragmentation by collision with CAD gas molecules in quadrupole 2, trapping and scanning product ions in quadrupole 3 operated as LIT. For screening of atroviridins, three sequential EPI experiments in a single period with precursor ions corresponding to [M+2H]2+ species of atroviridins A–C (Oh et al., 2002) were used: atroviridin A: m/z 982.2, scan range 150–1965 amu; atroviridin B: m/z 989.2, scan range 150–1980 amu; atroviridin C: m/z 996.2, scan range 150–1995 amu. For all experiments the following settings were kept constant: the average of three successive mass spectra were recorded (CE 10, 30, and 50 V); scan rate 4000 amu/s, step size 0.12 amu, Q1 unit resolution, Q3 entry barrier 8 V, declustering potential (DP) +50 V.
3. Results
3.1. Cloning and characterization of tmk1
PCR amplification with degenerate primers designed according to conserved regions of fungal MAPKs resulted in a 747-bp product. The PCR product was used to screen a T. atroviride P1 genomic γ BlueStar library, where multiple clones carrying inserts of 10–15 kb were obtained. Sequencing with internal primers Tmk1intF1, F2, R1, and R2 showed that T. atroviride tmk1 (GenBank Accession No. AF452096) consists of an open reading frame of 1335-bp interrupted by three putative introns at positions conserved in other fungal MAPK homologs, e.g. M. grisea Pmk1 (Xu et al., 1998), and Fusarium oxysporum Fmk1 (Di Pietro et al., 2001). The deduced protein sequence of the tmk1 gene comprises 355 amino acids and is 99% identical to T. virens TmkA and 97% identical to M. grisea Pmk1 with most of the changes located in the N-terminal parts of the proteins. Analysis of Tmk1 by the PROSITE website (http://us.expasy.org/prosite/; (Falquet et al., 2002) revealed a consensus sequence typical for MAP kinases between residues 57 and 159 (TXY+F-X (19)-R-E-X (72, 86) R-D-X-K-X (9)-C) and a nuclear localization sequence at aa 256–272. The catalytic domain is located between aa 23 and 311 with residues 29–53 being responsible for ATP binding. Based on Southern analysis with genomic DNA of T. atroviride digested with several restriction enzymes and the 747-bp PCR product as a probe, tmk1 is present as single copy in the genome of T. atroviride (data not shown).
3.2. Generation and characterization of tmk1 mutants
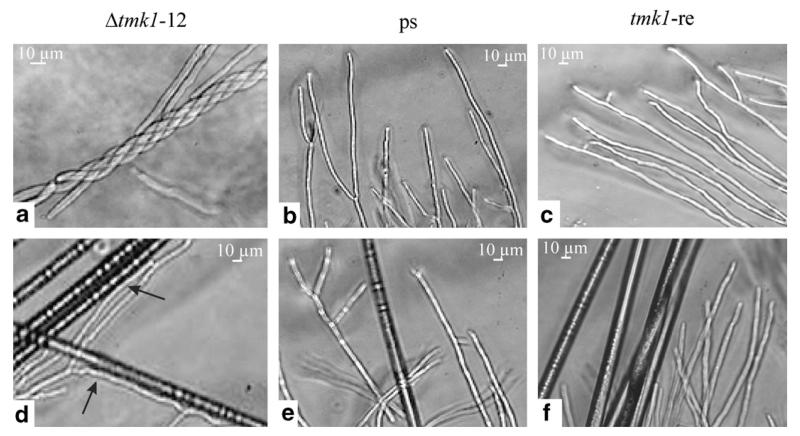
tmk1 gene deletion by Agrobacterium-mediated transformation resulted in seven transformants with single-copy integration of the hph marker cassette at the homologous tmk1 gene locus (Zeilinger, 2004a). Because of identical phenotypes of the obtained Δtmk1 mutants, two of them (Δtmk1-11 and Δtmk1-12) were chosen for further studies and compared to the parental strain and a respective rescued transformant (tmk1-re). tmk1-re was derived from mutant Δtmk1-12 by replacing the integrated deletion cassette by a single functional copy of the tmk1 gene. Δtmk1 mutants grew at about 80–85% the rate of the parental strain and tmk1-re on PDA plates (Zeilinger, 2004a), but produced ~3-times more conidia under these conditions. They began to sporulate at the center of the colony—similar to the phenotype described for T. virens ΔtmkA mutants (Mukherjee et al., 2003). Conidia of T. atroviride Δtmk1 mutants appeared to be pale green and, in contrast to the parental and the tmk1-re strain, were produced light-independently. Microscopic analysis of Δtmk1 mycelia showed an increased tendency of the mutant’s hyphae to coil around themselves compared to the parental and the tmk1-re strain (Fig. 1a–c). To further elucidate this abnormal hyper-coiling, the behavior of the Δtmk1 mutants in the presence of autoclaved, uncoated nylon fibers was examined. The fibers should mimic the physical appearance of a host hyphae without the presence of a contact stimulant, e.g. lectins. As shown in Fig. 1d, the Δtmk1-12 mutant still grew towards and along the nylon fibers and attached to them. The same effect was observed with Δtmk1-11 (data not shown). As expected, neither directed growth nor attachment to the plain nylon fibers was noticed with the parental strain T. atroviride P1 (Fig. 1e) or the tmk1-re strain (Fig. 1f).
Fig. 1.
Light microscopic examination of the Δtmk1-12 mutant, the T. atroviride parental strain P1 (ps), and the rescued transformant tmk1-re after growth for 48–72 h on PDA without (a–c) and with (d–f) plain nylon 66 fibers (approximate diameter 14 μm). Arrows mark the attachment to and the growth along the nylon fibers; the thick, dark structures in panels (d–f) represent the nylon fibers.
The results presented above show the involvement of the Tmk1 MAP kinase in the production of morphological changes as tmk1 gene deletion causes permanent coiling around hyphae or attachment to hyphal-like structures in the absence of a host-derived signal.
3.3. Mycoparasitic abilities of Δtmk1 strains
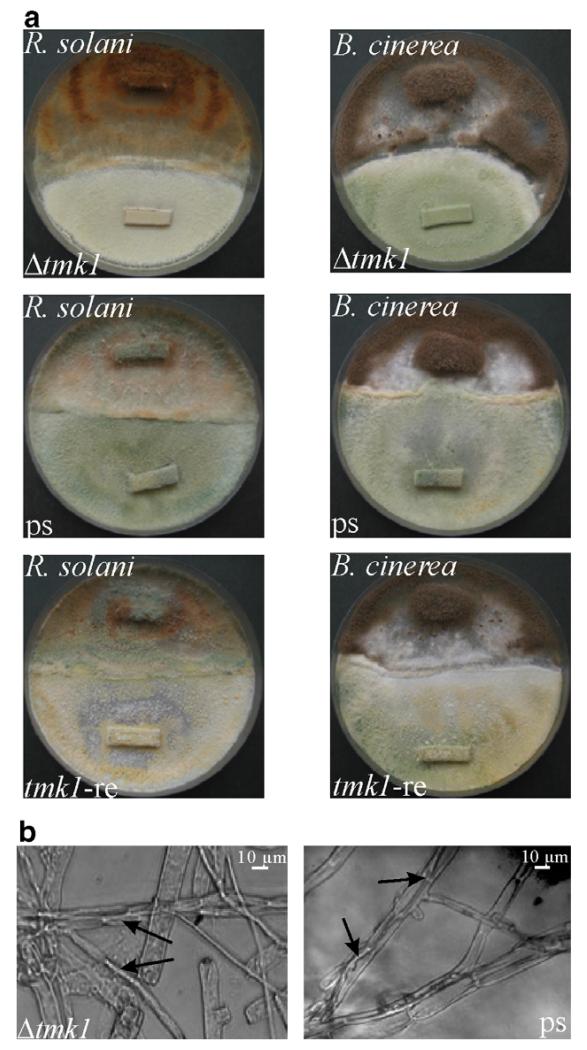
In order to investigate whether this release from host-derived signals for initiating primary steps of mycoparasitism is reflected by an increased host attack, we performed direct plate confrontation assays against R. solani and B. cinerea as fungal hosts. In contrast to the parental strain T. atroviride P1 and the rescued transformant tmk1-re, which already overgrew and lysed R. solani 7 days after inoculation of the mycoparasite and the host on opposite sides of the plate (data not shown), the Δtmk1 mutant just contacted and started to overgrow the host fungus during this period. After 7 more days the hyphae of Rhizoctonia were almost completely lysed by T. atroviride P1 and the tmk1-re strain, while the Δtmk1 mutant did not completely overgrow and lyse the host fungus even under prolonged incubation times (reflected by still intact hyphae of the host fungus; Fig. 2a). Although the mycoparasitic activity of the Δtmk1 mutant was reduced, typical morphological changes associated with mycoparasitism (e.g. attachment to the host’s hyphae and growth alongside these hyphae) could be detected microscopically (Fig. 2b).
Fig. 2.
(a) Plate confrontation assays of Δtmk1 mutant tmk1-11 and the rescued transformant tmk1-re in comparison to the parental strain T. atroviride P1 (ps) against R. solani and B. cinerea for analyzing the mycoparasitic ability. Pictures were taken 14 days after inoculation of the two fungi on opposite sides of the plate. (b) Microscopic analyses of the confrontation zone between T. atroviride (thin hyphae) and R. solani (thick hyphae). The interaction of Trichoderma hyphae with the host is marked with arrows.
When the Δtmk1 mutants were inoculated opposite to B. cinerea on PDA plates, no formation of a zone with direct contact could be observed with the naked eye, while the control strains (parental strain and rescued transformant) touched and started to overgrow the host (Fig. 2a). Nevertheless, microscopic analysis revealed single Δtmk1 hyphae attaching to and growing alongside B. cinerea hyphae (data not shown).
3.4. nag1 and ech42 gene transcription in Δtmk1 mutants
Besides attaching to and coiling around the host’s hyphae, the mycoparasitic interaction of Trichoderma also involves the secretion of cell wall-degrading enzymes preceded by the induction of the respective genes (Mach et al., 1999; Zeilinger et al., 1999). The transcription of two mycoparasitism-related genes coding for the Ech42 chitinase and the Nag1 N-acetyl-glucosaminidase was examined in the Δtmk1-11 mutant when grown in liquid culture under chitinase-inducing conditions and in dual plate cultures when confronted with R. solani or B. cinerea as hosts.
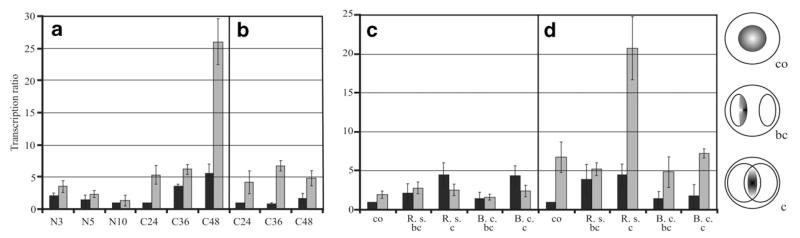
Three hours after transfer to liquid growth medium containing N-acetyl-β-d-glucosamine, nag1 transcript formation in the Δtmk1 mutant and the parental strain was at its maximum and decreased again in both strains 5 h after transfer to induction medium (Fig. 3a). Upon transfer to liquid growth medium containing colloidal chitin, nag1 transcript levels were at their maximum at 48 h in both the Δtmk1 mutant and the parental strain. Under both conditions, the Δtmk1 mutant attained higher nag1 transcript levels compared to the parental strain T. atroviride P1. When analyzing ech42 gene transcription, the Δtmk1 mutant again displayed enhanced mRNA levels compared to the parental strain at all time points tested (Fig. 3b).
Fig. 3.
Relative transcription ratios of the chitinase-encoding genes nag1 (a and c) and ech42 (b and d) in the Δtmk1-11 mutant (grey bars) compared to the parental strain T. atroviride P1 (black bars). (a and b) Real-time RT-PCR analyses were performed 3, 5, and 10 h after transfer of mycelia to liquid growth medium containing 1% N-acetylglucosamine (N) and 36 and 48 h after transfer to 1% colloidal chitin (C). The samples of the parental strain showing the lowest mRNA levels within one PCR run were arbitrarily assigned the factor 1 (N10, C24). (c and d) Direct confrontation assays with R. solani (R.s.) and B. cinerea (B.c.) as host fungi. Samples were collected from a control (co) where T. atroviride was grown alone, from an early stage before direct contact between the two fungi (bc), and from a later stage of direct contact—if present—with an interaction zone of 1.0 cm (c) and subjected to real-time RT-PCR. The control sample of the parental strain was arbitrarily assigned the factor 1.
In direct plate confrontation assays, induction of nag1 gene transcription reached its maximum in the parental strain T. atroviride P1 upon direct contact with the host fungi R. solani and B. cinerea. Interestingly, in the Δtmk1 mutant neither host fungus was able to significantly enhance nag1 gene transcription over the basal level detected when the fungus was grown alone on PDA plates (Fig. 3c).
When analyzing ech42 gene transcription at different mycoparasitic stages, we found enhanced mRNA levels in the parental strain already before contact with the host fungi, whereas in the Δtmk1 mutant ech42 gene transcription was only significantly induced upon direct contact with R. solani but remained un-induced upon confrontation with B. cinerea (Fig. 3d).
Interestingly, the Δtmk1 mutant produced elevated basal nag1 and ech42 mRNA levels in the control condition (growth on PDA in the absence of a host fungus) when compared to the parental strain, indicating a de-regulated production of these cell wall-degrading enzymes under non-inducing conditions.
To summarize, tmk1 gene deletion results in a de-regulated transcription of the two mycoparasitism-related genes studied as even under non-inducing conditions elevated mRNA levels were found in the Δtmk1 mutant. Nevertheless, nag1 gene transcription could not be induced over the basal level by the presence of a host fungus, whereas induction of ech42 gene transcription was triggered in a host-specific manner in the Δtmk1 mutant.
3.5. Extracellular chitinase activities in Δtmk1 mutants
In the culture supernatants of the parental strain T. atroviride P1 increasing amounts of extracellular chitinases (N-acetyl-glucosaminidases (NAGase) as well as endochitinases) were detected during induction with N-acetylglucosamine or colloidal chitin in liquid culture (Table 1). Upon induction with N-acetyl-glucosamine the Δtmk1-11 mutant produced similar NAGase activities as the parental strain for the first 5 h of induction. After 10 h under these conditions NAGase production in the Δtmk1-11 mutant was 1.6-times higher compared to the parental strain. When induced with colloidal chitin, the Δtmk1-11 mutant showed slightly increased NAGase activities compared to the parental strain after 36 h; at 48 h after induction Δtmk1-11 showed an about 1.8-fold higher extracellular activity than the parental strain. Similar to the NAGase activities, 48 h after induction with colloidal chitin endochitinase activities of the Δtmk1-11 mutant were increased ~1.5-fold. Enzyme activities measured for cultures of the rescued transformant tmk1-re were comparable to those of the parental strain (data not shown). These results suggest that Tmk1 negatively influences the production of extracellular chitinases during later points of induction.
Table 1.
Extracellular N-acetyl-glucosaminidase (NAGase) and endochitinase activities in culture filtrates of T. atroviride parental strain and the Δtmk1-11 mutant
| Carbon source | Strain | Induction time [h] | NAGase-activity [U/g]a | Endochitinase-activity [U/g]a |
|---|---|---|---|---|
| N-Acetylglucosamine | Parental strain | 3 | 11.3 ± 3.0 | n.d.b |
| 5 | 25.8 ± 2.0 | n.d. | ||
| 10 | 47.2 ± 2.1 | n.d. | ||
| Δtmk1-11 | 3 | 11.0 ± 2.1 | n.d. | |
| 5 | 23.7 ± 3.0 | n.d. | ||
| 10 | 77.8 ± 1.1 | n.d. | ||
| Colloidal chitin | Parental strain | 36 | 82.1 ± 0.4 | 17.3 ± 0.1 |
| 48 | 132.7 ± 5.9 | 30.4 ± 0.3 | ||
| Δtmk1-11 | 36 | 92.5 ± 4.5 | 14.2 ± 1.4 | |
| 48 | 242.5 ± 18.5 | 46.3 ± 0.9 |
Values are related to the mycelial dry weight and are means ± standard deviations of three independent experiments.
Not determined.
3.6. Production of antifungal metabolites
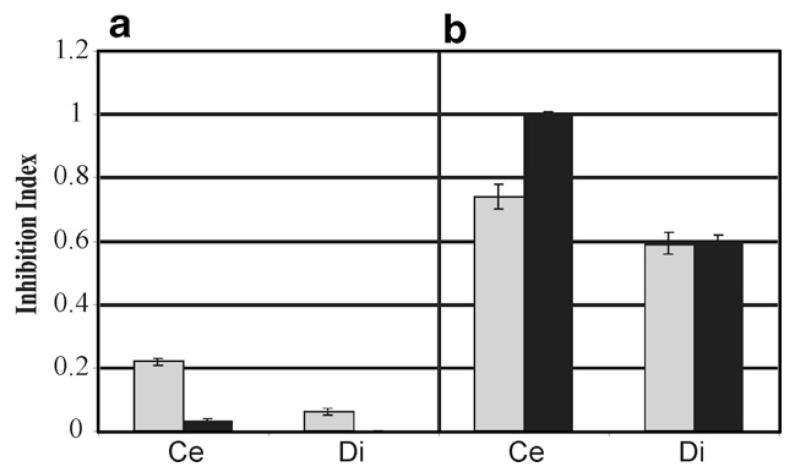
Since the mycoparasitic action of Trichoderma is not merely an effect of hydrolytic enzymes and direct contact to the host fungus but also due to the secretion of antimicrobial substances (Howell, 1998), the antifungal activity of one Δtmk1 mutant was determined. In the B. cinerea spore germination assay the Δtmk1-12 mutant showed higher biological activity (inhibition index of 0.22 when pre-cultivated on cellophane and 0.06 when pre-cultivated on dialysis tubing cellulose membrane; standard deviation for the experiments are displayed in Fig. 4) compared to the parental strain (inhibition index of 0.06 on cellophane and no detectable inhibition index on dialysis tubing membrane; Fig. 4a). When the inhibition indices were calculated after plate confrontation assays against B. cinerea, the Δtmk1-12 mutant and the parental strain secreted similar amounts of inhibitory substances into the media (inhibition index of 0.74 and 0.59, respectively), independent from the membrane type used (Fig. 4b). These data indicate that a fraction of secreted metabolites smaller than 12 kDa is significantly increased in the deletion strains when grown alone, whereas production of antifungal substances remains inducible by the presence of a host fungus to a similar extent in all strains tested. Higher molecular weight antifungal substances were secreted to an even higher extent by the parental strain when confronted with the host (Fig. 4b).
Fig. 4.
Inhibition indices for T. atroviride P1 (∎) and the Δtmk1-12 mutant ( ) obtained from B. cinerea spore germination assays. The strains were grown on PDA for 7 days on plates covered by a cellophane (Ce) or a dialysis tubing cellulose membrane (Di) either alone (a) or were confronted against B. cinerea (b). Each bar represents the average of at least three experiments; error bars indicate standard deviation which is less than 15% for all experiments.
) obtained from B. cinerea spore germination assays. The strains were grown on PDA for 7 days on plates covered by a cellophane (Ce) or a dialysis tubing cellulose membrane (Di) either alone (a) or were confronted against B. cinerea (b). Each bar represents the average of at least three experiments; error bars indicate standard deviation which is less than 15% for all experiments.
Since one of the major antimicrobial substances produced by Trichoderma is 6-pentyl-α-pyrone (6-PP) (Claydon et al., 1987; Ghisalberti and Sivasithamparam, 1991; Simon et al., 1988), we were interested in its production in the Δtmk1 mutants. Quantitative analysis of 6-PP revealed 1.6-fold elevated amounts secreted into PDA plates by the Δtmk1-12 mutant (1.23 ± 0.16 g 6-PP/g mycelial dry weight) compared to the parental strain (0.75 ± 0.11 g 6-PP/g mycelial dry weight).
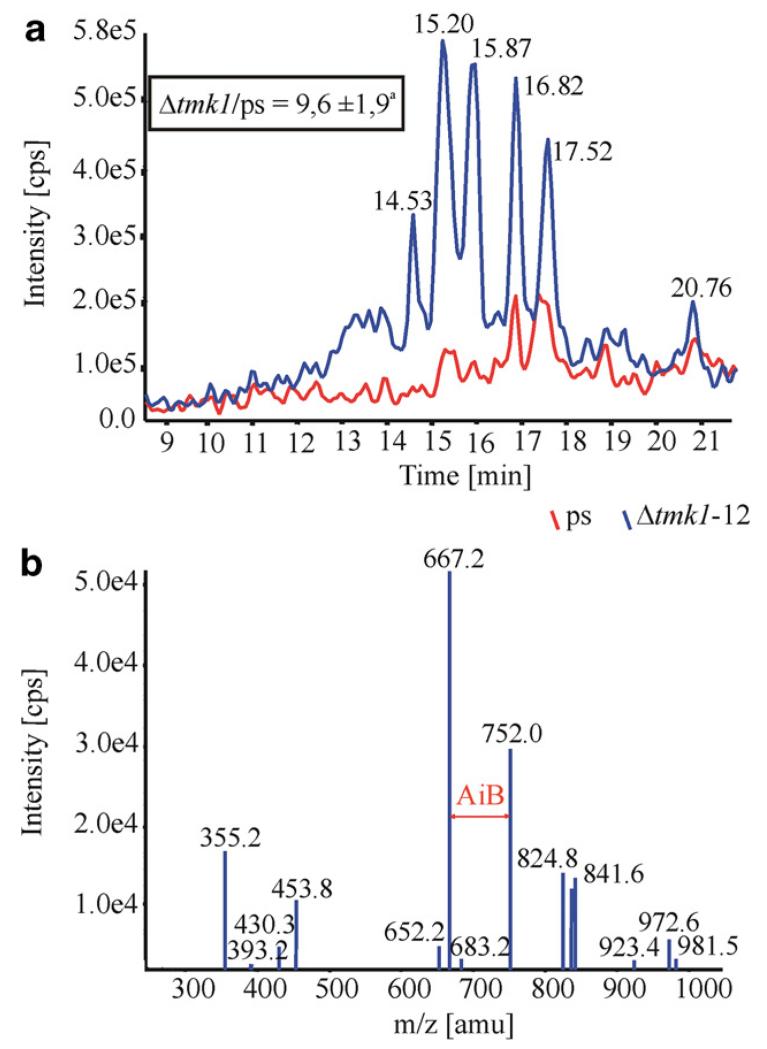
To obtain insights into the production of peptaibols, another important class of antifungal metabolites (Correa et al., 1995), the transcription ratio of the coding region of the peptaibol-specific α-aminoisobutyric acid (AiB) condensation domain of a non-ribosomal peptide synthetase of T. atroviride P1 (GenBank Accession No. DQ973296), with high similarity to T. virens Tex1, compared to act1 as reference gene was determined by real-time RT-PCR. After growth under peptaibol-producing conditions for 10 days, the transcription ratio of the Δtmk1-12 mutant was 2.5-fold ( ± 0.1) higher induced compared to the parental strain. The culture filtrates, obtained from the same cultivations, were screened for the presence of peptaibols by LC-MS/MS analysis. The chromatograms revealed that the peptaibol production pattern and peak intensities differed between the Δtmk1-12 mutant and the parental strain (Fig. 5a). When the peak intensities of the Δtmk1-12 mutant and T. atroviride P1 were related to the mycelial dry weight of the cultures and compared with each other, it was found that the production of peptaibols was approximately 10-times higher in the mutant. However, when the [M+2H]2+ species were taken as precursors for MS/MS fragmentation, the spectra showed the presence of AiB indicated by the fragments differing in 85 amu (Fig. 5b). It shall be noted that the tandem mass spectra did not match with those expected for the atroviridins and neoatroviridins. Thus we conclude that these peptaibols were absent in all samples tested. Nevertheless, the presence and position of AiB strongly suggests significant over-production of yet uncharacterized peptaibols of the trichorzianin-family in our Δtmk1-12 mutant.
Fig. 5.
LC-MS/MS screening for peptaibols (a) of culture filtrates from the Δtmk1-12 mutant (blue graph) and the parental strain (red graph). The m/z ratio 982.2 of the peak with the retention time of 15.87 min suggests the presence of atroviridins; nevertheless fragmentation of this peak related these peptaibols to the trichorzianin family (b). aThe inset indicates the ratio of the peak-intensities related to the mycelial dry weight of the strains tested.
3.7. In vivo biocontrol abilities of Δtmk1 mutants
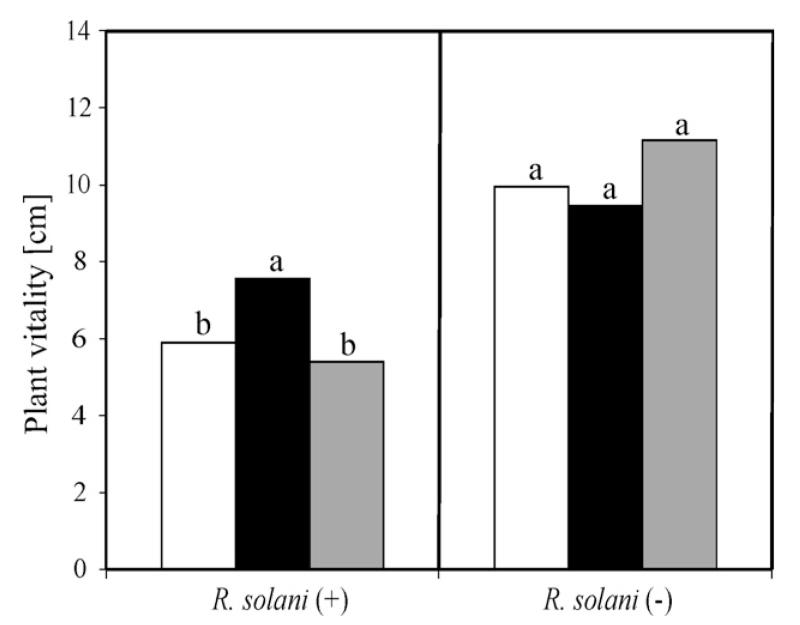
To test the in vivo biocontrol abilities of our Δtmk1 mutants, assays with beans planted in R. solani-infected soil were performed. In these experiments the vitality of the beans coated with spores of the Δtmk1-12 mutant was 30% higher than of beans protected by T. atroviride P1 when considering plant growth in the control experiment with uncoated beans (Fig. 6). To approve the exclusion of a possible influence of the deletion of tmk1 on the vitality of beans, control assays without addition of R. solani to the soil were conducted. It was found that neither Δtmk1-12 mutant nor the parental strain had a statistically significant influence on the plant vitality (Fig. 6). Thus we conclude that in contrast to a reduced mycoparasitic activity, plant protection is increased in Δtmk1 mutants. This enhanced protection is clearly not due to a growth promoting effect on the plants but to a direct (mycoparasite–host interaction) or indirect (increased plant resistance) effect.
Fig. 6.
Effect of T. atroviride parental strain P1 and the Δtmk1 mutant on the vitality of beans infected with R. solani (+). Bean seeds were either coated with spores of the parental strain (◻), the Δtmk1-12 mutant (∎) or were uncoated (without Trichoderma) as a control ( ). Additional control experiments were carried out without pathogen (−). Vitality of the plants is given in plant height (cm) 6 days after germination. Columns marked with the same letter differed significantly according to the Mann–Whitney test at a significance level of 10%.
). Additional control experiments were carried out without pathogen (−). Vitality of the plants is given in plant height (cm) 6 days after germination. Columns marked with the same letter differed significantly according to the Mann–Whitney test at a significance level of 10%.
4. Discussion
In phytopathogenic fungi, signal transduction via MAPK cascades is, among other processes, involved in the parasitic interaction (Banuett and Herskowitz, 1994; Di Pietro et al., 2001; Horwitz et al., 1999; Lev et al., 1999; Takano et al., 2000; Xu and Hamer, 1996; Xu et al., 1998). So far at least three different MAPKs were found to be functional in most of these fungi. MAPKs homologous to T. atroviride Tmk1 like M. grisea Pmk1 and S. cerevisiae FUS3/KSS1 were previously shown to be involved in the regulation of mating and fusion (Madhani et al., 1997), as well as appressorium formation and invasive growth (Xu and Hamer, 1996).
Tmk1 function was analyzed by generating and characterizing respective T. atroviride gene deletion mutants in which tmk1 was replaced by the hph (hygromycin B phosphotransferase-encoding) gene (Zeilinger, 2004a). To clearly relate all observed alterations to tmk1 gene deletion, a rescued transformant again exhibiting the characteristics of the parental strain was generated. Thus the observed phenotypic aberrations like reduction of the growth rate and light-independent formation of conidia in Δtmk1 mutants are truly due to tmk1 deletion, which is in accordance to the fact that similar phenotypes were also reported for T. virens ΔtmkA and Δtvk1 mutants (Mendoza-Mendoza et al., 2003; Mukherjee et al., 2003). Further investigations on the growth behavior of Δtmk1 mutants demonstrated a permanent induction of the coiling response in the absence of host-derived signals as the mutant coiled around its own hyphae and around plain nylon fibers in the absence of a host fungus.
To reveal the role of T. atroviride Tmk1 during mycoparasitism, mycoparasitism-related processes like chitinase gene expression, infection structure formation (coiling), and the secretion of antifungal substances were examined in Δtmk1 mutants. T. atroviride Δtmk1-11 mutant showed enhanced transcription of the mycoparasitism-related genes ech42 and nag1 and increased production of cell wall-degrading chitinases when cultivated in liquid culture under inducing conditions. Similar findings were also described for T. virens Δtvk1 strains (Mendoza-Mendoza et al., 2003).
Results from direct plate confrontation assays against R. solani und B. cinerea as hosts suggested that Tmk1 affects the host specificity of T. atroviride as the Δtmk1 mutant still could parasitize R. solani (although less effectively than the parental strain) whereas it was no longer able to overgrow B. cinerea, although attachment to and growth alongside of Botrytis hyphae could still be observed microscopically (data not shown). This host-dependent behavior of the Δtmk1 mutant was also reflected by the fact that ech42 gene transcription was inducible upon direct contact with R. solani, whereas it remained un-induced when the Δtmk1 mutant was confronted with B. cinerea. These findings are similar to the host-specific loss of the biocontrol potential of T. virens tmkA mutants which were as effective against R. solani as the parental strain whereas they failed to colonize S. rolfsii sclerotia (Mukherjee et al., 2003).
As the production of secondary metabolites was reported to be associated with the formation of conidiospores (Bu’Lock, 1961) and the inhibition of sporulation was shown to be accompanied by an inhibition of aflatoxin production in Aspergillus parasiticus (Reiss, 1982), we hypothesized that the light-independent conidiation of our Δtmk1 mutants may correlate with changes in the production of antimicrobial substances. Qualitative and quantitative analysis of the secreted metabolites demonstrated an increased production of 6-pentyl-α-pyrone and peptaibols upon deletion of tmk1; additionally also the pattern of the secreted peptaibols differed compared to the parental strain. Nevertheless, the secretion of antifungal substances could still be induced during direct confrontation with B. cinerea compared to the constitutive level during sole growth on PDA. Summarizing, while tmk1 gene deletion resulted in a de-regulation of the production of antifungal metabolites, their formation still was inducible by the presence of a host fungus. Therefore, although the lack of Δtmk1 mutants to overgrow B. cinerea would suggest a loss of their ability to appropriately react on signals derived from this host, the enhanced secretion of antifungal substances in the presence of this pathogen evidences a still remaining host–pathogen cross-talk. Thus we speculate that deletion of tmk1 at least not completely impairs sensing of this host fungus.
The finding that mycoparasitism-related processes like infection structure formation (coiling) as well as chitinase and antifungal metabolite production (Chet et al., 1998; Kubicek et al., 2001) were unaltered or even enhanced upon tmk1 gene deletion somehow contrasts the reduced/lost ability of the Δtmk1 mutants to control host fungi in direct confrontation assays. On the other hand, in greenhouse experiments the examined Δtmk1-12 mutant was able to protect bean plants against R. solani infection even better than the parental strain indicating a negative regulatory role of Tmk1 on Trichoderma-triggered plant resistance. Similar results were reported by Mendoza-Mendoza et al. (Mendoza-Mendoza et al., 2003) for T. virens Δtvk1 mutants which showed a greater capacity to control and reduce damage by R. solani and Pythium ultimum on cotton plants, whereas for T. virens ΔtmkA mutants attenuated mycoparasitism and a failure to induce full systemic resistance against the bacterial leaf pathogen Pseudomonas syringae pv. lacrymans, but similar effectivity as the wild-type in biocontrol activity against R. solani on bean plants was found (Viterbo et al., 2005). These findings strongly suggest the presence of further, still unknown, mycoparasitism-related factors which are missing in our Δtmk1 mutants and which are therefore affected by a signaling pathway involving Tmk1.
Acknowledgments
This work was supported by grants from the Fonds zur Förderung Wissenschaftlicher Forschung (FWF P15483 and P18109) and the DOC-FFORTE Program of the Austrian Academy of Sciences.
We thank Ilan Chet and Ada Viterbo (Weizmann Institute of Science, Rehovot, Israel) for their helpful support with the nylon fibers and Robert Mach for fruitful discussions.
References
- Banuett F, Herskowitz I. Identification of fuz7, a Ustilago maydis MEK/MAPKK homolog required for a-locus-dependent and -independent steps in the fungal life cycle. Genes Dev. 1994;8:1367–1378. doi: 10.1101/gad.8.12.1367. [DOI] [PubMed] [Google Scholar]
- Brunner K, Peterbauer CK, Mach RL, Lorito M, Zeilinger S, Kubicek CP. The Nag1 N-acetylglucosaminidase of Trichoderma atroviride is essential for chitinase induction by chitin and of relevance to biocontrol. Curr. Genet. 2003;43:289–295. doi: 10.1007/s00294-003-0399-y. [DOI] [PubMed] [Google Scholar]
- Bu’Lock JD. Intermediary metabolism and antibiotic synthesis. Adv. Appl. Microbiol. 1961;3:293–342. doi: 10.1016/s0065-2164(08)70514-8. [DOI] [PubMed] [Google Scholar]
- Chet I, Benhamou N, Haran S. Mycoparasitism and lytic enzymes. In: Harman GE, Kubicek CP, editors. Trichoderma and Gliocladium. Vol. 2. Taylor and Francis Ltd.; London, UK: 1998. pp. 153–172. [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Claydon N, Allan M, Hanson JR, Avent AG. Antifungal alkyl pyrones of Trichoderma harzianum. Trans. Br. Mycol. Soc. 1987;88:503–513. [Google Scholar]
- Correa A, Rebuffat S, Bodo B, Roquebert MF, Dupont J, Bettucci L. In vitro inhibitory activity of trichorzianines of Sclerotium rolfsii Sacc. Crypt. Mycol. 1995;16:185–190. [Google Scholar]
- Di Pietro A, Garcia-MacEira FI, Meglecz E, Roncero MI. A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 2001;39:1140–1152. [PubMed] [Google Scholar]
- Dixon K, Xu J-R, Smirnoff N, Talbot NJ. Independent signaling pathways regulate cellular turgor during hyperosmotic stress and appressorium-mediated plant infection by Magnaporte grisea. Plant Cell. 1999;11:2045–2058. doi: 10.1105/tpc.11.10.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falquet L, Pagni M, Bucher P, Hulo N, Sigrist CJ, Hofmann K, Bairoch A. The PROSITE database, its status in 2002. Nucleic Acids Res. 2002;30:235–238. doi: 10.1093/nar/30.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisalberti EL, Sivasithamparam K. Antifungal antibiotics produced by Trichoderma spp. Soil Biol. Biochem. 1991;23:1011–1020. [Google Scholar]
- Graeme-Cook KA, Faull JL. Effect of ultraviolet-induced mutants of Trichoderma harzianum with altered antibiotic production on selected pathogens in vitro. Can. J. Microbiol. 1991;37:659–664. doi: 10.1139/m91-112. [DOI] [PubMed] [Google Scholar]
- Gruber F, Visser J, Kubicek CP, de Graaff LH. The development of a heterologous transformation system for the cellulolytic fungus Trichoderma reesei based on a pyrG-negative mutant strain. Curr. Genet. 1990;18:71–76. doi: 10.1007/BF00321118. [DOI] [PubMed] [Google Scholar]
- Gustin MC, Albertyn J, Alexander M, Davenport K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1998;62:1264–1300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman GE, Howell CR, Viterbo A, Chet I, Lorito M. Trichoderma species—opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2004;2:43–56. doi: 10.1038/nrmicro797. [DOI] [PubMed] [Google Scholar]
- Harman GE, Hayes CK, Lorito M, Broadway RM, Di Pietro A, Peterbauer CK, Tronsmo A. Chitinolytic enzymes of Trichoderma harzianum: purification of chitobiosidase and endochitinase. Mol. Plant Pathol. 1993;83:313–318. [Google Scholar]
- Hjeljord L, Tronsmo A. Trichoderma and Gliocladium in biological control: an overview. In: Harman GE, Kubicek CP, editors. Trichoderma and Gliocladium. Vol. 2. Taylor and Francis Ltd.; London, UK: 1998. pp. 131–152. [Google Scholar]
- Horwitz BA, Sharon A, Lu SW, Ritter V, Sandrock TM, Yoder OC, Turgeon BG. A G protein alpha subunit from Cochliobolus heterostrophus involved in mating and appressorium formation. Fungal Genet. Biol. 1999;26:19–32. doi: 10.1006/fgbi.1998.1094. [DOI] [PubMed] [Google Scholar]
- Howell CR. The role of antibiosis in biocontrol. In: Harman GE, Kubicek CP, editors. Trichoderma and Gliocladium. Vol. 2. Taylor & Francis Ltd.; London, UK: 1998. pp. 173–184. [Google Scholar]
- Howell CR. Mechanisms employed by Trichoderma species in the biological control of plant diseases: the history and evolution of current concepts. Plant Dis. 2003;87:4–10. doi: 10.1094/PDIS.2003.87.1.4. [DOI] [PubMed] [Google Scholar]
- Jenczmionka NJ, Schaeffer W. The Gpmk1 MAP kinase of Fusarium graminearum regulates the induction of specific secreted enzymes. Curr. Genet. 2005;47:29–36. doi: 10.1007/s00294-004-0547-z. [DOI] [PubMed] [Google Scholar]
- Kelly JM, Hynes MJ. Transformation of Aspergillus niger by the amdS gene of Aspergillus nidulans. EMBO J. 1985;4:475–479. doi: 10.1002/j.1460-2075.1985.tb03653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek CP, Mach RL, Peterbauer CK, Lorito M. Trichoderma: from genes to biocontrol. J. Plant Pathol. 2001;83:11–23. [Google Scholar]
- Lev S, Horwitz BA. A mitogen-activated protein kinase pathway modulates the expression of two cellulase genes in Cochliobolus heterostrophus during plant infection. Plant Cell. 2003;15:835–844. doi: 10.1105/tpc.010546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev S, Sharon A, Hadar R, Ma H, Horwitz BA. A mitogen-activated protein kinase of the corn leaf pathogen Cochliobolus heterostrophus is involved in conidiation, appressorium formation, and pathogenicity: diverse roles for mitogen-activated protein kinase homologs in foliar pathogens. Proc. Natl. Acad. Sci. USA. 1999;96:13542–13547. doi: 10.1073/pnas.96.23.13542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorito M, Woo SL, D’Ambrosio M, Harman GE, Hayes CK, Kubicek CP, Scala F. Synergistic interaction between cell wall degrading enzymes and membrane affecting compounds. Mol. Plant Microbe Interact. 1996;9:206–213. [Google Scholar]
- Lu Z, Tombolini R, Woo S, Zeilinger S, Lorito M, Jansson JK. In vivo study of Trichoderma-pathogen-plant interactions, using constitutive and inducible green fluorescent protein reporter systems. Appl. Environ. Microbiol. 2004;70:3073–3081. doi: 10.1128/AEM.70.5.3073-3081.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach RL, Peterbauer CK, Payer K, Jaksits S, Woo SL, Zeilinger S, Kulling CM, Lorito M, Kubicek CP. Expression of two major chitinase genes of Trichoderma atroviride (Trichoderma harzianum P1) is triggered by different regulations signals. Appl. Environ. Microbiol. 1999;65:1858–1863. doi: 10.1128/aem.65.5.1858-1863.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Styles CA, Fink GR. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- Mendoza-Mendoza A, Pozo MJ, Grzegorski D, Martínez P, García JM, Olmedo-Monfil V, Cortés C, Kenerley C, Herrera-Estrella A. Enhanced biocontrol activity of Trichoderma through inactivation of a mitogen-activated protein kinase. Proc. Natl. Acad. Sci. USA. 2003;100:15965–15970. doi: 10.1073/pnas.2136716100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee PK, Latha J, Hadar R, Horwitz BA. TmkA, a mitogen-activated protein kinase of Trichoderma virens, is involved in biocontrol properties and repression of conidiation in the dark. Eukaryot. Cell. 2003;2:446–455. doi: 10.1128/EC.2.3.446-455.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee PK, Latha J, Hadar R, Horwitz BA. Role of two G-protein alpha subunits, TgaA and TgaB, in the antagonism of plant pathogens by Trichoderma virens. Appl. Environ. Microbiol. 2004;70:542–549. doi: 10.1128/AEM.70.1.542-549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SU, Yun BS, Lee SJ, Kim JH, Yoo ID. Atroviridins A–C and neoatroviridins A–D, novel peptaibol antibiotics produced by Trichoderma atroviride F80317. I. Taxonomy, fermentation, isolation and biological activities. J. Antibiot. (Tokyo) 2002;55:557–564. doi: 10.7164/antibiotics.55.557. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen R. Quantification on the LightCycler. In: Meuer S, et al., editors. Rapid Cycle Real-Time PCR Methods and Applications. Springer Press; Heidelberg: 2001. pp. 21–34. [Google Scholar]
- Reiss J. Development of Aspergillus parasiticus and formation of aflatoxin B1 under the influence of conidiogenesis affecting compounds. Arch. Microbiol. 1982;133:236–238. doi: 10.1007/BF00415008. [DOI] [PubMed] [Google Scholar]
- Reithner B, Brunner K, Schuhmacher R, Peissl I, Seidl V, Krska R, Zeilinger S. The G protein alpha subunit Tga1 of Trichoderma atroviride is involved in chitinase formation and differential production of antifungal metabolites. Fungal Genet. Biol. 2005;42:749–760. doi: 10.1016/j.fgb.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Rocha-Ramirez V, Omero C, Chet I, Horwitz BA, Herrera-Estrella A. Trichoderma atroviride G-protein alpha-subunit gene tga1 is involved in mycoparasitic coiling and conidiation. Eukaryot. Cell. 2002;1:594–605. doi: 10.1128/EC.1.4.594-605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed Cold Spring Harbor Laboratory Press; New York: 1989. [Google Scholar]
- Schaeffer HJ, Weber MJ. Mitogen-activated protein kinases: specific messages from ubiquitous messengers. Mol. Cell Biol. 1999;19:2435–2444. doi: 10.1128/mcb.19.4.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Carreon L, Flores C, Rodriguez B, Galindo E. Rhizoctonia solani, an elicitor of 6-pentyl-alpha-pyrone production by Trichoderma harzianum in a two liquid phases, extractive fermentation system. Biotechnol. Lett. 2004;26:1403–1406. doi: 10.1023/B:BILE.0000045640.71840.b5. [DOI] [PubMed] [Google Scholar]
- Simon A, Dunlop RW, Ghisalberti EL, Sivasithamparam K. Trichoderma koningii produces a pyrone compound with antibiotic properties. Soil Biol. Biochem. 1988;20:263–264. [Google Scholar]
- Takano Y, Kikuchi T, Kubo Y, Hamer JE, Mise K, Furusawa I. The Colletotrichum lagenarium MAP kinase gene CMK1 regulates diverse aspects of fungal pathogenesis. Mol. Plant Microbe Interact. 2000;13:374–383. doi: 10.1094/MPMI.2000.13.4.374. [DOI] [PubMed] [Google Scholar]
- Viterbo A, Harel M, Horwitz BA, Chet I, Mukherjee PK. Trichoderma mitogen-activated protein kinase signaling is involved in induction of plant systemic resistance. Appl. Environ. Microbiol. 2005;71:6241–6246. doi: 10.1128/AEM.71.10.6241-6246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J-R. MAP kinases in fungal pathogens. Fungal Genet. Biol. 2000;31:137–152. doi: 10.1006/fgbi.2000.1237. [DOI] [PubMed] [Google Scholar]
- Xu J-R, Hamer JE. MAP Kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporte grisea. Genes Dev. 1996;10:2696–2706. doi: 10.1101/gad.10.21.2696. [DOI] [PubMed] [Google Scholar]
- Xu J-R, Staiger CJ, Hamer JE. Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc. Natl. Acad. Sci. USA. 1998;95:12713–12718. doi: 10.1073/pnas.95.21.12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeilinger S. Gene disruption in Trichoderma atroviride via Agrobacterium -mediated transformation. Curr. Genet. 2004a;45:54–60. doi: 10.1007/s00294-003-0454-8. [DOI] [PubMed] [Google Scholar]
- Zeilinger S. Signal transduction in fungi: signalling cascades regulating virluence in filamentous fungi. In: Arora DK, editor. Handbook of Fungal Biotechnology. 2nd ed Marcel Dekker, Inc; New York: 2004b. pp. 181–192. [Google Scholar]
- Zeilinger S, Reithner B, Scala V, Peissl I, Lorito M, Mach RL. Signal transduction by Tga3, a novel G protein alpha subunit of Trichoderma atroviride. Appl. Environ. Microbiol. 2005;71:1591–1597. doi: 10.1128/AEM.71.3.1591-1597.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeilinger S, Galhaup C, Payer K, Woo SL, Mach RL, Fekete C, Lorito M, Kubicek CP. Chitinase gene expression during mycoparasitic interaction of Trichoderma harzianum with its host. Fungal Genet. Biol. 1999;26:131–140. doi: 10.1006/fgbi.1998.1111. [DOI] [PubMed] [Google Scholar]
- Zheng L, Campbell M, Murphy J, Lam S, Xu JR. The BMP1 gene is essential for pathogenicity in the gray mold fungus Botrytis cinerea. Mol. Plant Microbe Interact. 2000;13:724–732. doi: 10.1094/MPMI.2000.13.7.724. [DOI] [PubMed] [Google Scholar]