SUMMARY
RNA quality control pathways get rid of faulty RNAs and therefore must be able to discriminate these RNAs from those that are normal. Here we present evidence that the ATPase cycle of the SF1 Helicase Upf1 is required for mRNA discrimination during Nonsense-Mediated Decay (NMD). Mutations affecting the Upf1 ATPase cycle disrupt the mRNA selectivity of Upf1, leading to indiscriminate accumulation of NMD complexes on both NMD target and non-target mRNAs. In addition, two modulators of NMD -translation and termination codon-proximal poly(A) binding protein - depend on the ATPase activity of Upf1 to limit Upf1-non-target association. Preferential ATPase-dependent dissociation of Upf1 from non-target mRNAs in vitro suggests that selective release of Upf1 contributes to the ATPase-dependence of Upf1 target discrimination. Given the prevalence of helicases in RNA regulation, ATP hydrolysis may be a widely employed activity in target RNA discrimination.
INTRODUCTION
The dynamic interaction of RNA binding proteins (RBPs) with RNA is critical to every aspect of RNA metabolism (Moore, 2005). Yet, how do RNA regulators faithfully distinguish their target RNAs from non-targets in the cell? Most models for RBP-target specificity invoke RBP affinity for target-specific RNA sequences, structures or bound proteins (Ankö and Neugebauer, 2012; Glisovic et al., 2008). However, for RNA quality control pathways, which detect and destroy faulty or non-functional RNAs, target-specific mechanisms for RBP recruitment are harder to envision, as aberrant RNAs have the potential to differ widely in sequence and associated proteins (van Hoof and Wagner, 2011; Porrua and Libri, 2013).
The first mRNA quality control pathway discovered was nonsense-mediated decay (NMD) (Leeds et al., 1991; Losson and Lacroute, 1979; Maquat et al., 1981; Pulak and Anderson, 1993). Conserved in eukaryotes, this translation-dependent pathway degrades transcripts whose termination codons are recognized as premature. In this way, NMD prevents accumulation of truncated polypeptides arising from aberrant mRNAs bearing premature termination codons (PTCs). NMD also affects the accumulation of select naturally occurring mRNAs, impacting up to 10% of protein-coding genes in diverse eukaryotes (Schweingruber et al., 2013).
The key RNA binding regulator in NMD is the Superfamily 1 (SF1) RNA helicase Upf1. Target degradation involves assembly of Upf1 with other NMD factors, such as Upf2, Upf3, and, in most eukaryotes studied to date, Smg1 kinase and one or more of Smg5-7 (Kervestin and Jacobson, 2012; Schweingruber et al., 2013). In humans, Smg1 phosphorylates Upf1 in a manner stimulated by Upf2, Upf3 and the exon junction complex (Kashima et al., 2006). This promotes association of phospho-binding proteins Smg5 and Smg7, as well as general mRNA decay factors (Chakrabarti et al., 2014; Cho et al., 2013; Loh et al., 2013; Okada-Katsuhata et al., 2012). Smg6 is itself an endonuclease, which has both phospho-dependent and -independent interactions with Upf1 (Chakrabarti et al., 2014; Eberle et al., 2009; Nicholson et al., 2014; Okada-Katsuhata et al., 2012). In addition, the ATPase activity of Upf1 has been implicated in a late step of target mRNA degradation: remodeling the mRNP to enhance nuclease access (Franks et al., 2010).
Despite the wealth of information regarding NMD target degradation, a fundamental, yet poorly understood, aspect of NMD is what enables Upf1 to distinguish targets from non-targets in the first place. Well-studied NMD targets share in common the fact that translation termination occurs at an unusual position in the mRNA (Schweingruber et al., 2013). One model for NMD target recognition is that stalled or aberrant termination complexes recruit Upf1 (Kervestin and Jacobson, 2012; Schweingruber et al., 2013). Indeed, ribosome toe-printing in S. cerevisiae extracts and rabbit reticulocyte lysates revealed that ribosome dissociation at an NMD-inducing PTC is stalled aberrantly (Amrani et al., 2004; Peixeiro et al., 2012). Although it remains to be fully elucidated how termination at a PTC becomes aberrant, the absence of proximal mRNP factors that promote normal termination, such as poly(A) binding protein, has been suggested to distinguish NMD targets from non-targets (Cosson and Couturier, 2002; Ivanov et al., 2008; Kervestin and Jacobson, 2012; Uchida et al., 2002). Notably, Upf1 and the ribosome release factors, eRF1 and eRF3, have been found to interact in yeast and human cells (Czaplinski et al., 1998; Ivanov et al., 2008; Kashima et al., 2006; Singh et al., 2008).
However, recent reports cloud the simple view that Upf1 recruitment to aberrantly stalled termination complexes accounts fully for NMD target specificity. For example, Upf1 associates with both target and non-target mRNAs even in the absence of translation, and the manner and degree to which translation seems to affect Upf1-mRNA accumulation varies among different mRNAs (Hogg and Goff, 2010; Kurosaki and Maquat, 2013; Kurosaki et al., 2014; Zünd et al., 2013). Additionally, genome-wide crosslinking-immunoprecipitation (CLIP) studies showed that translation inhibitors cause Upf1 binding sites along mRNAs to expand from a bias for 3’UTRs to be more broadly distributed (Gregersen et al., 2014; Hurt et al., 2013; Zünd et al., 2013). This suggests that translation influences where Upf1 associates along an mRNA’s length, in addition to how strongly it associates with NMD target and non-target mRNAs overall.
Thus, the mechanisms involved in how Upf1 distinguishes differences in target and non-target mRNPs have remained unclear. Here we present evidence for a critical role for Upf1 ATPase activity in NMD target discrimination, with preferential ATPase-dependent release of Upf1 from non-target mRNAs as part of the underlying mechanism.
RESULTS
Upf1 mutants unable to bind or hydrolyze ATP are defective in NMD target discrimination
Previous studies have demonstrated that Upf1 target discrimination is reflected in Upf1-RNA immunoprecipitation (RIP) assays, which yield greater Upf1 copurification of NMD target than non-target mRNAs at steady state (Hwang et al., 2010; Johansson et al., 2007; Johns et al., 2007; Silva et al., 2008). Thus, to examine determinants of human Upf1 target specificity, we established stable cell lines in which endogenous Upf1 could be depleted by RNAi and complemented with siRNA-resistant Flag-Upf1 induced to near-endogenous levels (Figure S1A) for use in RIP assays. We examined the binding specificity of Flag-Upf1 using gene reporters previously shown to be NMD sensitive, due to a premature termination codon (PTC), or NMD insensitive, due to a normal termination codon (NTC) (Singh et al., 2008) (Figure 1A). Non-target mRNAs differing in size were used as internal controls.
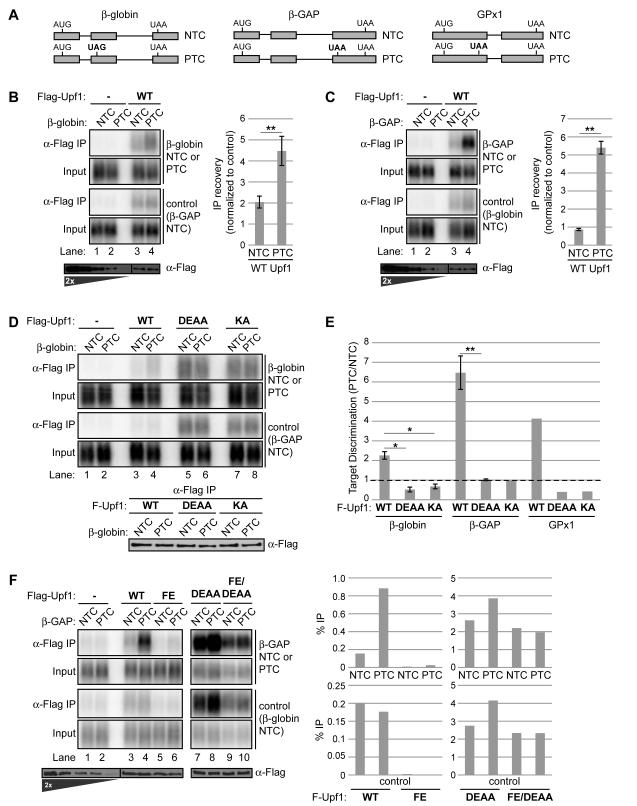
Figure 1. Upf1 ATPase cycle mutants are defective in selective NMD target association.
(A) NMD target (PTC) and non-target (NTC) mRNA reporter pairs used in RNA-IP assays based on human β-globin, β-globin with an insertion from GAPDH (β-GAP), or GPx1. The NMD-inducing termination codon is denoted in bold.
(B) Northerns of β-globin and control mRNAs in input (0.5%) samples or coprecipitated with Flag-Upf1 using anti-Flag antibody (α-Flag IP). Flag-Upf1 recovered in IPs is shown alongside a two-fold titration of input on anti-Flag Westerns below Northerns. The graph on the right represents recovery of β-globin mRNAs (NTC or PTC) with Flag-Upf1 over input normalized to recovery of the internal control (lanes 3,4), after subtraction of background in negative control IPs (lanes 1,2). Data are represented as mean +/− SEM for five biological replicates.
(C) Similar to panel B for RNA-IPs of β-GAP mRNAs.
(D) Similar to panel B, comparing IPs with Flag-Upf1 WT, D637A/E638A (DEAA), or K498A (KA). Western of recovered Flag-Upf1 variants is shown below Northerns.
(E) Graph representing the ratio of normalized IP recovery for NMD target (PTC) to non-target (NTC) mRNAs with the indicated Upf1 variants. A value of one, denoted by the dotted line, reflects an absence of discrimination between target and non-target. Data are represented as mean +/− SEM for three to four biological repeats.
(F) Similar to panel C but without normalization, comparing percent IP recovery of β-GAP mRNAs and the internal control by Flag-Upf1 WT, F192E (FE), DEAA or DEAA/FE mutants.
Asterisks denote P-values: *≤0.05, **≤0.001 (paired student’s t-test, two-tailed).
See also Figure S1
Consistent with expectation, RIP assays performed with Flag-tagged wildtype (WT) Upf1 exhibited a ~2.3- to 6.5-fold greater recovery of PTC mRNAs over their NTC counterparts, as seen in Figures 1B, C and S1B (compare lane 4 to 3 in top panels). This mirrors the range in half-life differences for these PTC/NTC pairs (Singh et al., 2008). Although Upf1 IPs enriched PTC-containing mRNA, both PTC and NTC mRNAs copurified with Upf1 above background (compare lanes 3, 4 to lanes 1, 2), consistent with previous observations that Upf1 can associate with both target and non-target mRNAs (Hogg and Goff, 2010; Hwang et al., 2010; Johns et al., 2007; Silva et al., 2008; Zünd et al., 2013). Importantly, lysate mixing controls (Mili and Steitz, 2004) confirmed that the observed Upf1-mRNA associations reflect interactions occurring in intact cells, rather than after cell lysis (Figure S1C).
Because Upf1 mutations that block the Upf1 ATPase cycle render Upf1 inactive for NMD (Bhattacharya et al., 2000; Cheng et al., 2007; Franks et al., 2010; Kashima et al., 2006), we wanted to examine the impact of these mutations on mRNA association and target discrimination. We therefore repeated our RIP assays with mutant Upf1 defective in two steps of the ATPase cycle, Upf1 K498A (Upf1 KA; deficient in ATP binding) and Upf1 D637A/E638A (Upf1 DEAA; deficient in ATP hydrolysis) (Figure S1A and 1D). In striking contrast to WT Upf1, both mutants exhibited a complete loss in target discrimination and, in fact, recovered increased levels of both target and non-target mRNAs when compared to WT Upf1 (Figure 1D; quantified in Figure 1E). Interestingly, two of the three target/non-target pairs assayed exhibited slightly greater association with NTC mRNA over PTC mRNA with Upf1 DEAA and KA (target discrimination <1 in Figure 1E). This apparent reverse-discrimination was eliminated upon depletion of Smg6 (Figure S1D and S1E), likely reflecting a role for Smg6-mediated cleavage in reducing target mRNA levels in IPs. We conclude that Upf1 mutants defective in ATP binding or ATP hydrolysis exhibit increased association with both target and non-target mRNAs and lack the ability to discriminate between them.
A hyperactive Upf1 ATPase mutant is defective in binding to both target and non-target mRNAs
To examine how mRNA association might be affected in a third Upf1 mutant with a perturbed Upf1 ATPase cycle, we turned to the hyperactive mutant F192E (Upf1 FE). Structural and biochemical studies have shown that the cystidine/histidine-rich (CH) domain partially inhibits Upf1 ATPase, and a F192E mutation within this domain renders bacterially expressed Upf1 hyperactive in its ATPase activity and less stably bound to RNA in vitro (Chakrabarti et al., 2011; Chamieh et al., 2008). Our RIP assays of Upf1 FE with the β-GAP PTC/NTC pair revealed indiscriminate loss in association with both target and non-target mRNA compared to WT Upf1 (Figures 1F and S1G, compare lanes 5, 6 with 3, 4; quantified on the right). Importantly, this loss was predominantly dependent on Upf1 ATPase activity because mRNA association was largely recovered when the FE mutant was rendered deficient in ATP hydrolysis (Upf1 FE/DEAA, Figure 1F and S1G, lanes 7-10; quantified on the right). ATPase assays performed in vitro with Upf1 from our cell lines confirmed Upf1 FE hyperactivity and Upf1 FE/DEAA ATPase deficiency (Figure S1F). Together, these findings for ATPase-hyperactive and -deficient Upf1 indicate that the steady state mRNA association of Upf1 inversely correlates with Upf1 ATPase activity and that Upf1 target discrimination is critically dependent on a normal Upf1 ATPase cycle.
Upf1-mRNA selectivity is lost on a transcriptome-wide level in Upf1 ATP-binding and ATP-hydrolysis mutants
To examine the contribution of Upf1 ATPase activity to mRNA selectivity among endogenous mRNAs, we applied RIP-seq (RIP followed by strand-specific high-throughput sequencing) to Flag-Upf1 WT, DEAA and KA expressed at endogenous levels (Figure 2 and S2A). A parental cell line expressing no exogenous Upf1 was used as a negative control. RIP-seq libraries were sequenced to a mean depth of 23 million reads and approximately twenty thousand genes had >0.1 RPKM per library. Importantly, Upf1 WT and mutant RIPs were all highly enriched for transcripts annotated as protein-coding with a smaller fraction derived from pseudogenes (Table S1), indicating that the ATPase mutations do not disrupt Upf1 preference for mRNAs over non-coding transcripts.
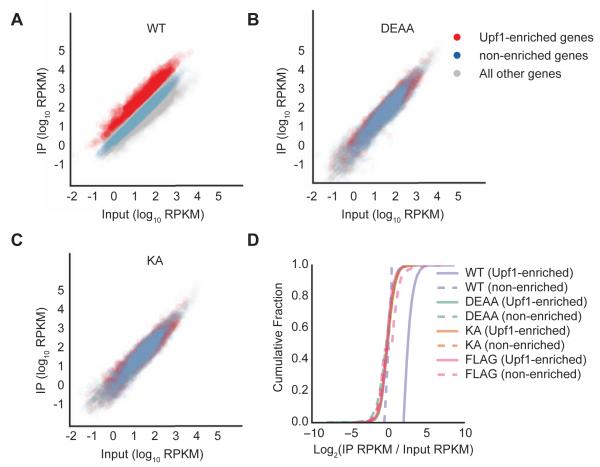
Figure 2. Transcriptome-wide loss in mRNA selectivity for Upf1 ATPase cycle mutants.
(A-C) Scatter plots of reads per kilobase transcript per million mapped reads (RPKM) from RNA-seq of input samples versus IPs for Flag-Upf1 WT (A), DEAA (B) and KA (C). Genes with IP/input ratios for WT Upf1 of greater than 2.05 (cut-off based on 5% false discovery rate (FDR) established using cells expressing Flag epitope only, Figure S2) are shown in red (Upf1-enriched), while genes with log2 (IP/input) between −0.5 and +0.5 are shown in blue (non-enriched). All remaining genes are shown in grey.
(D) Cumulative fraction of Upf1-enriched and non-enriched genes with IP enrichment represented as log2 (IP RPKM/input lysate RPKM) for Flag-Upf1 WT, KA, and DEAA, along with Flag only. Difference between WT Upf1 (Upf1-enriched) curve compared to all other curves was statistically significant (p-value <0.05 for all comparisons, KS-test).
Using the background of negative control IPs to establish a 5% false-discovery rate (Figure S2B), a distinct population of 2,040 Upf1-associated RNAs was found to be enriched by at least 2-fold over input levels (Figure 2A, Upf1-enriched genes indicated in red; solid purple line denote WT in Figure 2D). Based on observations by others (Hogg and Goff, 2010; Kurosaki et al., 2014; Zünd et al., 2013), these RNAs likely include a mixture of NMD sensitive mRNAs and mRNAs that are less sensitive to NMD but limited in downstream steps of the NMD pathway. In striking contrast to WT Upf1, RIPs for Upf1 DEAA and KA did not show enrichment for any RNAs when subjected to the same FDR cutoff (Figure S2B). Accordingly, the population of WT Upf1-enriched RNAs was not enriched in DEAA and KA RIPs over a WT Upf1-non-enriched RNA population defined by 0.97- to 1.03-fold enrichment in WT RIPs over inputs (Figures 2AC, compare red and blue; Figure 2D). These global findings generalize our observations for individual mRNA reporters to the human transcriptome, supporting the conclusion that mRNA selectivity is lost in Upf1 mutants deficient in ATP binding or hydrolysis.
ATP hydrolysis-deficient Upf1 is phosphorylated and assembles Smg5-7 proteins on both target and non-target mRNAs
Our finding that Upf1 mutants blocked in ATP binding and ATPase activity exhibit elevated and indiscriminate mRNA association (Figures 1 and 2) raised the possibility that the Upf1 ATPase cycle plays a critical role in preventing NMD complex assembly on non-target mRNAs. To investigate this, we used RIP assays to compare the phosphorylation levels of WT Upf1 and Upf1 DEAA and their ability to support assembly of Smg5-7 on target and non-target mRNAs.
To examine Upf1 phosphorylation, we used an antibody specific for Upf1 phosphorylated on serine 1116, a Smg1-phosphorylation site shown to be important for efficient NMD (Kurosaki et al., 2014; Yamashita et al., 2001). As seen in Figure 3A and quantified in Figure 3B, α-phospho-Upf1 RIPs from cells treated with okadaic acid prior to harvest to promote recovery of phosphorylated Upf1 (Kurosaki et al., 2014; Yamashita et al., 2001) exhibited a pattern similar to RIPs with a pan-Upf1 antibody: greater recovery of PTC mRNA over NTC mRNA from WT Upf1 expressing cells and equal recovery of both types of mRNA from Upf1 DEAA expressing cells. RIPs with resin only and λ-protein phosphatase pre-treated lysates served as negative controls for α-Upf1 and α-phospho-Upf1, respectively (Figure 3A, panels on the right, and Figure S3). We note that target recovery preference in α-Upf1 RIPs of WT Upf1 exceeds that observed in α-phospho-Upf1 RIPs (Figure 3B, compare top left vs bottom left graphs), perhaps reflecting greater phospho-epitope masking by phospho-Upf1 binding proteins on targets than on non-targets or a population of target-bound Upf1 that has not undergone phosphorylation. Regardless, these observations suggest that indeed, mRNA-associated Upf1 DEAA is phosphorylated and to an extent indistinguishable between target and non-target mRNAs.
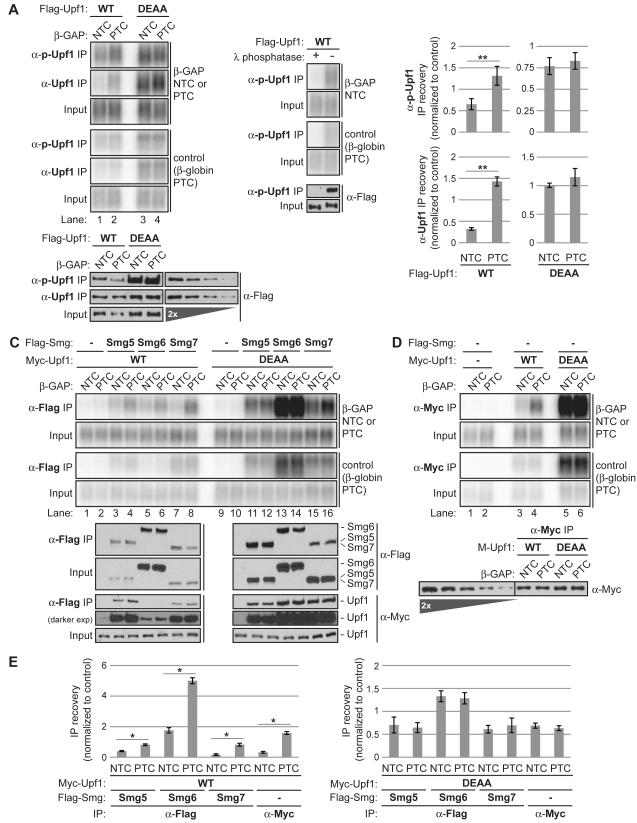
Figure 3. ATP hydrolysis-deficient Upf1 accumulates as a phosphoprotein in complexes with Smg proteins on both target and non-target mRNA.
(A) Northerns of β-GAP and control reporter mRNAs in inputs (0.3%) or RNA-IPs with αphospho-S1116 Upf1 (α-p-Upf1) or α-Upf1 antibodies from okadiac acid-treated cells expressing Flag-Upf1 WT or DEAA. Control RNA-IPs performed with α-phospho-Upf1 from lysates treated with or without λ protein phosphatase are shown on the right. Westerns of Flag-Upf1 recovered in IPs alongside a two-fold titration of Flag-Upf1 WT input are shown below Northerns.
(B) Graphs representing mean IP recovery of β-GAP mRNAs normalized by recovery of the internal control +/− SEM for triplicate biological repeats of α-phospho-Upf1 RNA-IPs and duplicates of α-Upf1 RNA-IPs.
(C) Northerns of β-GAP and control mRNAs in inputs (0.6%) or coprecipitated with Flag-Smg5, -Smg6, or -Smg7 in cells coexpressing Myc-Upf1 WT or DEAA. Westerns of Flag-Smg proteins recovered in IPs, along with copurifying Myc-Upf1 are shown below Northerns.
(D) Similar to panel C for RNA-IPs with Myc-Upf1 WT or DEAA, with 2% inputs loaded. Western of Myc-Upf1 recovered in IPs alongside a two-fold titration of Myc-Upf1 WT input is shown below Northerns.
(E) Quantifications, similar to those in panel B, for RNA-IPs shown in panels C, D with SEM for triplicate (Smg5, Smg7, Upf1) or duplicate (Smg6) biological repeats. Asterisks denote P-values: *≤0.05, **≤0.01 (paired student’s t-test, two-tailed).
See also Figure S3
We had previously observed that Upf1 DEAA supports assembly of Smg5-7 proteins on NMD target mRNA (Franks et al., 2010). Examining whether this occurs also on non-target mRNAs, we observed that all three Smg5-7 proteins coprecipitated PTC mRNA to a greater extent than NTC mRNA in the presence of Upf1 WT (Figure 3C, lanes 1-8), exhibiting a degree of target discrimination equal to or slightly less than that exhibited by Upf1 WT alone (Figure 3D; quantified in Figure 3E, left). In contrast, target discrimination was completely lost for Smg5-7 in the presence of Upf1 DEAA, with equal recovery of PTC and NTC mRNA (Figure 3C, lanes 9-16; quantified in Figure 3E, right). Taken together, these observations suggest that Upf1 ATPase activity serves a critical role in limiting the assembly of NMD complexes containing phosphorylated Upf1 on non-target mRNAs.
Translation prevents Upf1 accumulation on non-target mRNAs in a Upf1 ATPase-dependent manner
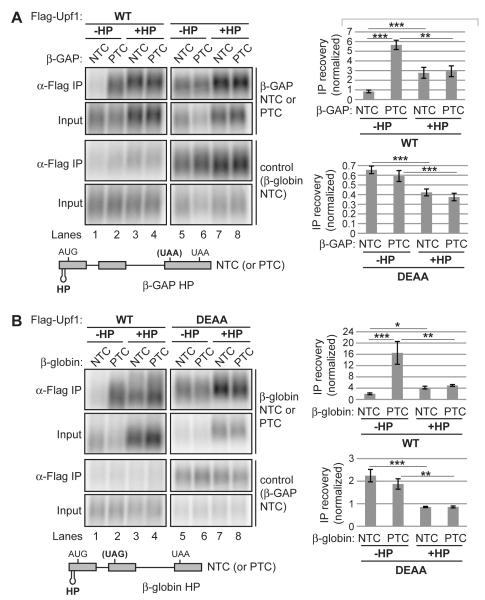
Active translation is critical for NMD and recent reports have suggested a role for translation in reducing the overall level of Upf1 association with non-target mRNAs (Zünd et al., 2013), while promoting or leaving unchanged the level of Upf1 association with target mRNAs (Hogg and Goff, 2010; Kurosaki et al., 2014). Consistent with this, we observed increased Upf1 recovery of NTC mRNAs and decreased recovery of a PTC mRNA upon global repression of translation elongation with cycloheximide or puromycin (Figure S4). To test whether these effects of translation are dependent on Upf1 ATPase activity, we introduced a stable hairpin (HP) into the 5′ UTRs of our PTC/NTC reporter pairs that inhibits translation (Kozak, 1989), efficiently inhibiting polysome formation (Franks and Lykke-Andersen, 2007) and preventing NMD (Belgrader et al., 1993; G. Singh and J.L-A. unpublished observations). RIP assays revealed that HP-induced translational repression reduced PTC mRNA association with both WT Upf1 and Upf1 DEAA, with the magnitude of reduction slightly smaller for DEAA than for WT (Figures 4A and 4B; quantified on the right). This suggests that for target mRNAs, translation promotes Upf1 accumulation in a manner largely independent of Upf1 ATPase activity, consistent with models in which ribosomes contribute to Upf1 recruitment (Czaplinski et al., 1998; Ivanov et al., 2008; Kashima et al., 2006; Min et al., 2013). In contrast, on NTC mRNAs, HP-induced translational repression increased mRNA association with WT Upf1, while reducing Upf1 DEAA association to a similar extent as on PTC mRNAs (Figures 4A and 4B; compare quantifications for NTC mRNAs +/− HP on the right). Therefore, translation limits Upf1 accumulation on non-target mRNAs by a mechanism that is dependent on Upf1 ATPase activity.
Figure 4. Translation prevents Upf1 accumulation on non-target mRNA in a Upf1 ATPase-dependent manner.
(A) Northerns for β-GAP and control reporter mRNAs in inputs (0.5%) or coprecipitated with Flag-Upf1. Schematic of the β-GAP mRNAs used in RNA-IPs is shown below Northerns. HP denotes a stable RNA hairpin in the 5′ UTR that blocks translation. Graphs on the right represent mean IP recovery over input of β-GAP mRNA normalized by recovery of the internal control after subtraction of background from negative control IPs, +/− SEM for triplicate biological repeats.
(B) Similar to panel A for β-globin RNA-IPs, except performed in the presence of Smg6 depletion to prevent Smg6-mediated cleavage of the β-globin PTC mRNA (see Figure S1).
Asterisks denote P-values: *≤0.1, **≤0.05, *** ≤0.01 (paired student’s t-test, two-tailed).
See also Figure S4
PTC-proximal poly(A) binding protein limits Upf1-mRNA accumulation in a manner dependent on Upf1 ATPase activity
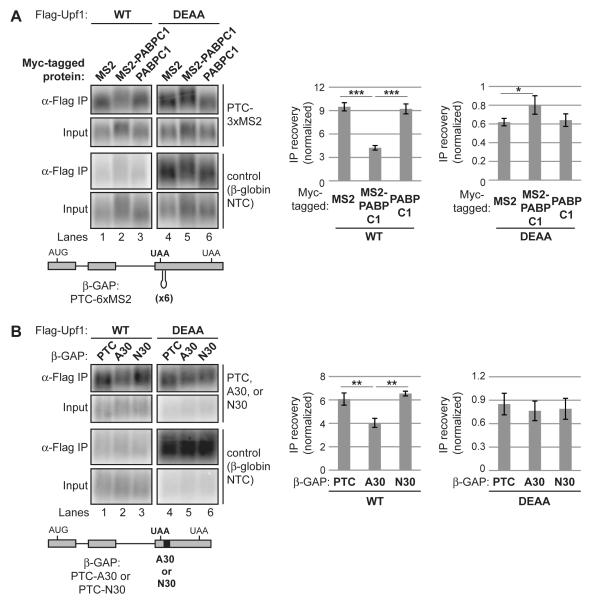
MRNA reporters subject to NMD have been found to partially evade NMD when modified to bring PABP proximal to the PTC (Amrani et al., 2004; Behm-Ansmant et al., 2007; Eberle et al., 2008; Fatscher et al., 2014; Ivanov et al., 2008; Joncourt et al., 2014; Silva et al., 2008; Singh et al., 2008). To test if PTC-proximal PABP affects Upf1-mRNA association, and, if so, requires Upf1 ATPase activity to do so, we repeated our Upf1 RIP assays with two reporters previously shown to evade NMD due to PTC-proximal PABP (Singh et al., 2008) (Figures 5A and 5B).
Figure 5. PTC-proximal poly(A) binding protein prevents Upf1 accumulation on mRNA in a Upf1 ATPase-dependent manner.
(A) Northerns for β-GAP and control reporter mRNAs in inputs (0.3%) or coprecipitated with Flag-Upf1 WT or DEAA from cells coexpressing Myc-tagged proteins as indicated. MS2 denotes fusion with MS2 coat protein. Schematic of the β-GAP PTC-6xMS2 mRNA used is shown below Northerns. Graphs on the right represent mean IP recovery of reporter mRNA normalized by recovery of the internal control after subtraction of background from negative control IPs, +/− SEM for triplicate biological repeats.
(B) Similar to panel A for β-GAP PTC reporter mRNAs containing 30 adenosines (A30) or a 30-nucleotide degenerate sequence (N30) instead of MS2 coat protein binding sites (schematic shown below Northerns), with 0.5% inputs loaded on Northerns and IPs performed in the absence of MS2 fusion proteins.
Asterisks denote P-values: *≤0.1, **≤0.05, *** ≤0.01 (paired student’s t-test, two-tailed).
See also Figure S5
As seen in Figure 5A (left panels), tethering MS2-PABPC1 to a β-GAP PTC variant containing six MS2 binding hairpins (PTC-6xMS2) downstream of the PTC reduced mRNA reporter association with WT Upf1 relative to tethering MS2 alone or when the reporter was coexpressed with untethered PABPC1 (quantified on the right), despite similar levels of IP recovery of tethered and untethered PABP (Figure S5). Significantly, this reduction is dependent on Upf1 ATPase activity as it was not observed in Upf1 DEAA RIPs (Figure 5A, lanes 4-6; quantified on the right). Similar observations were seen for an mRNA bearing a PTC-proximal tract of 30 adenosines predicted to recruit PABPC1 (Figure 5B). These observations suggest that termination-proximal PABPC1 reduces Upf1 accumulation on mRNA in a Upf1 ATPase-dependent manner.
ATP binding- and ATPase-deficient Upf1 accumulate on mRNA 3’UTRs and are enriched near termination codons and 3′ ends
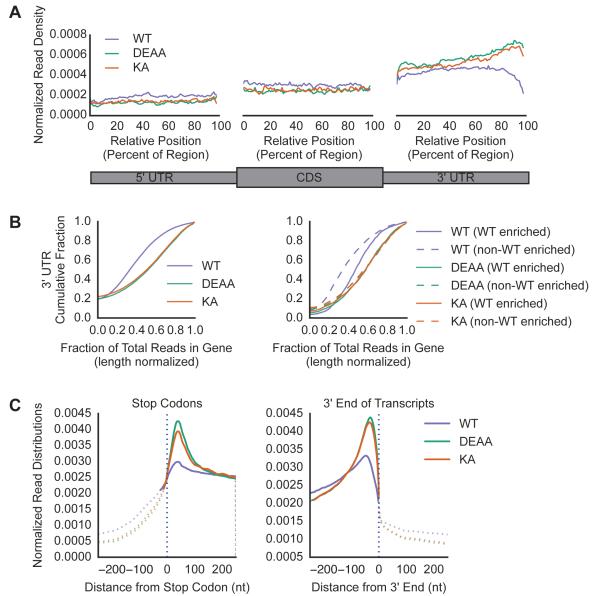
Our observations suggest that Upf1 ATPase activity is required for preventing Upf1 from accumulating and promoting NMD complex formation on translated non-target mRNAs. Recent studies employing UV cross-linking and Upf1 IP followed by high throughput sequencing (CLIP-seq) have reported that while Upf1 binding sites can be found all along the length of mRNAs, the overall distribution has a distinct 3’UTR bias (Gregersen et al., 2014; Hurt et al., 2013; Zünd et al., 2013). To gain insight into how a disrupted Upf1 ATPase cycle might impact Upf1 binding site distribution, we performed CLIP-seq on Flag-Upf1 WT, DEAA and KA expressed at near endogenous levels (Figures S6A, B, C).
Notably, the distributions of Upf1 DEAA and KA binding were nearly identical to each other and similar to WT Upf1 in 3’UTR bias (Figures 6A, and S6D, E) and preferential binding to mRNAs over non-coding RNAs (Table S2). However, Upf1 mutants exhibited an even greater average accumulation in 3’UTRs than WT Upf1 (Figure 6A; compare read densities in 3’UTRs for DEAA, KA with WT) across all genes, as seen by the increased fraction of 3’UTR derived reads per mRNA normalized to length in DEAA and KA CLIPs compared to WT (Figure 6B, left graph, see right-shifted curves for DEAA, KA compared to WT; p-value < 0.05, KS test). Comparison of read distributions for WT Upf1-enriched and non-enriched RNA subpopulations identified by RIP-seq (Figure 2) revealed that for WT Upf1, a greater fraction of enriched mRNAs had a stronger 3’UTR distribution bias than non-enriched mRNAs, while for Upf1 mutants, both sets of mRNAs were indistinguishable in their enhanced 3’UTR bias (Figure 6B, right graph, compare dashed and solid lines). These findings suggest that Upf1 ATPase activity limits Upf1 accumulation in 3’UTRs to an extent that is greater for non-target than target mRNAs.
Figure 6. Upf1 ATPase mutants accumulate in 3’UTRs with enhanced binding downstream of termination codons and near 3′ ends.
(A) Mean read density for Upf1 WT and mutant CLIP assays across the metagene, normalized to the total number of reads per gene.
(B) Cumulative fraction of genes with 3’UTR read abundance represented as a fraction of total reads in the gene, normalized to nucleotide length, shown for all mRNAs on the left, and for WT enriched and non-WT enriched mRNAs, as defined in Figure 2, on the right. Differences between WT and mutant curves were statistically significant (p-value < 0.001; KS statistic 0.30 for both DEAA and KA compared to WT for non-WT Enriched and 0.20 and 0.19 for DEAA and KA, respectively, compared to WT for WT Enriched)
(C) Mean read densities, shown as percentages of total reads mapped in the region depicted per mRNA, around the first nucleotide of the stop codon, shown on the left, and the 3′ end of annotated transcripts, shown on the right. Solid lines represent regions where differences were found to be significant with a P-value <0.05 (Bonferroni corrected).
Examination of CLIP-seq read density at nucleotide resolution around translation termination codons and at mRNA 3′ ends revealed two peaks that are stronger for Upf1 DEAA and KA than WT Upf1. The first centers around 45 nucleotides downstream of the termination codon (Figure 6C, left), while the second peak occurs upstream of transcript 3′ ends (Figure 6C, right). This difference between mutant and WT Upf1 in binding site distribution proximal to the termination codon and the poly(A) tail was seen for both Upf1-enriched and non-enriched mRNAs (Figure S6F). These findings suggest that Upf1 ATPase activity is needed to limit Upf1 accumulation at these specific 3’UTR sites.
Upf1 dissociates faster from non-target mRNA than target mRNA in an ATPase- and translation-dependent manner in vitro
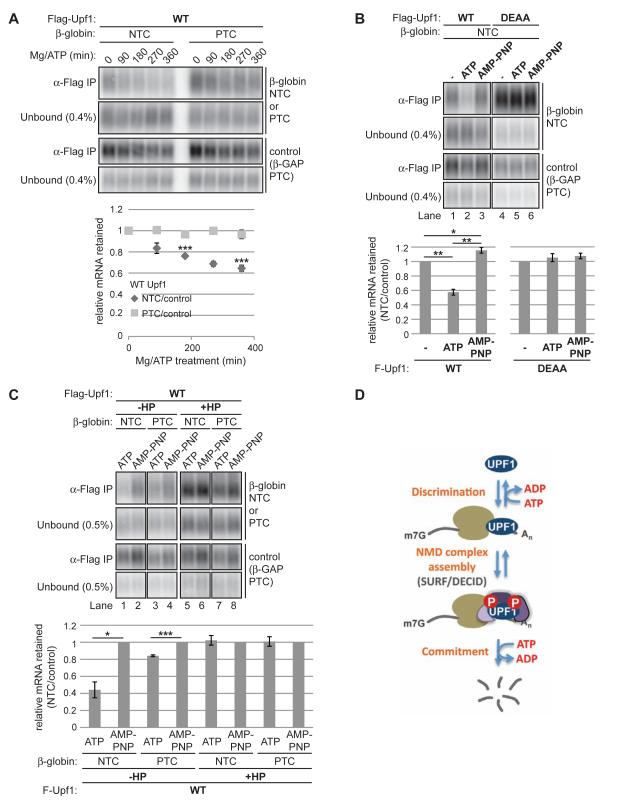
The findings described above are consistent with a mechanism by which Upf1 target binding specificity depends on Upf1 ATPase activity to limit accumulation of Upf1 and assembly of NMD complexes on non-target mRNAs, particularly in 3’UTRs. One possible mechanism accounting for this Upf1 ATPase-dependence is that Upf1 ATP hydrolysis is needed for preferential dissociation of Upf1 from non-targets. Indeed, Upf1 binding to synthetic RNAs is disrupted upon incubation with ATP but not ADP or nonhydrolyzable ATP analogs (Kurosaki et al., 2014; Weng et al., 1998). To test if Upf1 release from non-target mRNAs differs from target mRNAs, we developed in vitro RNA release assays that measure the relative release kinetics of Upf1 from reporter mRNAs. RIPs for Flag-Upf1 were performed from cell extracts with a low level of EDTA to repress the Upf1 ATPase and subsequently supplemented at regular intervals during IPs with Mg2+ and ATP to stimulate Upf1 ATPase activity.
As seen in Figure 7A, Upf1 dissociates from both target and non-target mRNAs over time in the presence of Mg-ATP, despite even IP recovery of Upf1 (Figure S7A) and unchanged overall mRNA levels (Figure S7B). Intriguingly, quantification of the Upf1-mRNA dissociation rate after normalization to the PTC-containing internal control revealed that release of NTC mRNA was faster than its PTC counterpart (Figure 7A, graph below). To avoid significant RNA degradation (SRL, unpublished observations), the release assays were performed at a low temperature (4°C). While we note that the rate of release in these assays is slow, ATP hydrolysis by Upf1 under the same conditions occurred at a comparably slow rate (Figure S7C).
Figure 7. Upf1 ATP hydrolysis is required for faster release of Upf1 from non-target over target mRNA.
(A) Northerns for β-globin and control reporter mRNAs in unbound fractions or coprecipitated with Flag-Upf1 WT from cell lysates treated with MgCl2/ATP for the number of minutes indicated above lanes. Graph under Northerns shows the ratio of βglobin mRNA recovery in IPs to the internal control β-GAP PTC mRNA after subtraction of background from negative control Flag IPs as a function of MgCl2/ATP-treatment duration, normalized to values at time 0. Data are represented as the normalized mean ratio +/− SEM for two to three biological replicates. P-value calculations were restricted to time points with triplicate measurements.
(B) Northerns of β-globin and control mRNAs in unbound fractions or coprecipitated with Flag-Upf1 from untreated lysates (-) or lysates treated with MgCl2/ATP or MgCl2/AMP PNP for 360 minutes. Graphs under Northerns represent mean ratios +/− SEM for triplicate biological repeats calculated as in panel A, normalized to values for untreated lysate samples.
(C) Similar to panel B comparing release of β-globin +/− HP mRNAs used in Figure 4B. Graphs under Northerns are from triplicate biological repeats normalized to values for AMP-PNP-treated samples.
Asterisks denote P-values: *≤0.1, **≤0.05, ***≤0.01 (paired student t-test, two-tailed). See also Figure S7
(D) Model depicting the ATPase-dependent mRNA discrimination step by Upf1 preceding NMD complex assembly. A second mRNA-selective commitment step might occur prior to mRNA degradation. See text for detail.
The preferential Mg-ATP-dependent release of NTC-mRNA was abolished for ATPase-deficient Upf1 DEAA (Figure S7D) and upon replacement of ATP with a nonhydrolysable ATP analog, AMP-PNP (Figure 7B). In fact, addition of AMP-PNP yielded a small but significant increase in NTC mRNA retention by WT Upf1 relative to the PTC internal control, perhaps due to AMP-PNP competition with cellular ATP in the extract. These findings suggest that Upf1 ATP hydrolysis supports faster Upf1 release from non-target mRNAs than from targets.
Since translation limits Upf1 accumulation on non-target mRNA (Figure 4), we next tested the possibility that preferential release of non-target mRNAs by Upf1 is dependent on translation. As shown in Figure 7C, mRNAs translationally repressed by a hairpin structure were blocked in ATP hydrolysis-dependent release in contrast to their unrepressed counterparts. It is important to note that this result cannot distinguish whether mRNA release from Upf1 is affected by translation occurring in the extract or instead by translation-dependent mRNP remodeling in cells. Regardless, these results suggest that Upf1 release of non-target mRNAs is faster than targets and requires Upf1 ATP hydrolysis and translation.
DISCUSSION
The Upf1 ATPase cycle is required for NMD target discrimination
In this study, we present multiple lines of evidence that target mRNA specificity in the NMD pathway is critically dependent on the ATPase cycle of the central NMD factor Upf1 (Figure 7D). First, Upf1 mutations that interfere with distinct aspects of the Upf1 ATPase cycle render Upf1 defective in its preferential association with NMD target mRNAs. This is evidenced by the loss of mRNA specificity for mutant Upf1 blocked in ATP binding or ATP hydrolysis steps of the ATPase cycle (Figures 1, 2) and the failure of ATPase-hyperactive Upf1 to accumulate on mRNA (Figure 1). Consistent with our findings, a recent study also reported loss of selective target mRNA association for Upf1 G495A/G497E which also lacks ATPase activity (Kurosaki et al., 2014). Second, processes central to target discrimination by the NMD pathway affect Upf1 specificity in an ATPase-dependent manner, as both translation and termination-proximal PABPC1 require Upf1 ATPase activity to prevent Upf1 accumulation on non-target mRNAs (Figures 4 and 5). Third, wild-type Upf1 is preferentially retained on target over non-target mRNA in vitro in a manner dependent on ATP hydrolysis (Figure 7A-C). Thus, our findings from mutational interference with three distinct aspects of the Upf1 ATPase cycle as well as the effect of a non-hydrolyzable ATP analog on wild-type Upf1 support the conclusion that the Upf1 ATPase cycle is critical for Upf1 target discrimination.
Upf1 ATPase activity prevents accumulation of NMD complexes on non-target mRNAs
How does Upf1 ATPase activity contribute to Upf1 target specificity? Surprisingly, the principal contribution appears to be in preventing accumulation of Upf1 and NMD complexes on non-target mRNAs. This is evidenced by the observation that Upf1 mutants blocked in the ATPase cycle accumulate in a phosphorylated form along with downstream Smg5-7 NMD factors on non-target mRNAs (Figures 1 and 3). Moreover, Upf1 ATPase activity was required for translation- and PABPC1-dependent inhibition of Upf1 accumulation on non-target mRNAs (Figures 4 and 5). Notably, wild-type Upf1 also associates with non-target mRNAs, but to a lesser degree than Upf1 mutants blocked in the ATPase cycle (Figure 1), suggesting that these Upf1 mutants are stalled at a normal step in the mRNA binding dynamics of Upf1. Despite the increased accumulation of NMD complexes on non-target mRNAs in the presence of Upf1 ATP binding or ATP hydrolysis mutants, we did not observe evidence for endonucleolytic cleavage or accelerated decay of these mRNAs (SRL and JL-A, unpublished observations), consistent with previous observations for ATPase-deficient yeast Upf1 (Sheth and Parker, 2006). Thus, either the large number of mRNPs with which these mutant Upf1 proteins associate renders the availability of Upf1 or Upf1-interacting factors limiting, or Upf1 ATP hydrolysis may have additional roles in downstream steps of the NMD pathway after NMD complex formation, consistent with our previous observations for target mRNAs (Franks et al., 2010, Figure 7D). Alternatively, as proposed previously by others (Hogg and Goff, 2010; Sheth and Parker, 2006), a second discrimination step based on NMD target features may occur after NMD complex assembly and be required for degradation (commitment step in Figure 7D). In metazoans, this could include Smg6-mediated endonucleolytic cleavage stimulated by 3’UTR-associated EJCs (Boehm et al., 2014), which are absent from the non-target mRNAs tested in this study. Regardless, our observations suggest that the Upf1 ATPase cycle plays a key role in ensuring NMD complex assembly specifically on NMD substrates, thereby preventing NMD complexes from accumulating on non-target mRNAs and titrating out cellular NMD machinery.
Translation and PABP as modulators of Upf1 ATPase-dependent mRNA specificity
Manipulating translation revealed that translation promotes Upf1 accumulation on target mRNAs, but inhibits accumulation on non-targets (Figure 4). The latter observation is consistent with previous observations for another non-target mRNA reporter (Zünd et al., 2013); our findings indicate that this effect is dependent on Upf1 ATPase activity (Figure 4). How translation affects Upf1 accumulation on mRNAs may reflect both direct interactions between Upf1 and translational machinery and translation-dependent impacts on mRNP composition. For example, direct contacts of Upf1 with the 40S ribosomal subunit (Min et al., 2013) and/or ribosome release factors (Czaplinski et al., 1998; Ivanov et al., 2008; Kashima et al., 2006; Singh et al., 2008) may contribute to the accumulation of Upf1 on target mRNAs. Conversely, the process of translational elongation may, as previously proposed (Gregersen et al., 2014; Hurt et al., 2013; Zünd et al., 2013), interfere with accumulation of Upf1 on translated regions of mRNAs. Additionally, our observation that termination-proximal PABP inhibits Upf1-mRNA accumulation in an ATPase-dependent manner (Figure 5) raises the possibility that coupling between translation termination and PABP, or PABP-associated factors, prevents Upf1-mRNA association in a manner dependent on the Upf1 ATPase. Indeed, interactions between PABP and eRF3 are well established (Cosson and Berkova, 2002; Hoshino et al., 1999; Kozlov and Gehring, 2010) and NMD targets are thought to undergo aberrant termination at least in part because they lack termination-proximal PABP and/or PABP-associated factors (Amrani et al., 2004; Fatscher et al., 2014; Joncourt et al., 2014; Kervestin and Jacobson, 2012; Peixeiro et al., 2012). The relative contributions of translation elongation and normal termination in limiting Upf1 accumulation on non-target mRNAs in an ATPase-dependent manner, and the mechanism by which PABP acts, remain important issues for future study.
How does Upf1 ATPase activity impact Upf1-mRNA selectivity?
While it is possible that Upf1 ATPase activity affects its recruitment to NMD targets, our observations point instead to a target discrimination mechanism by which Upf1 ATPase activity promotes Upf1 release preferentially from non-target mRNAs (Figure 7D). This is evidenced by the dramatic increase in non-target mRNA accumulation for Upf1 mutants blocked in the ATPase cycle (Figure 1) and the ATPase-dependent release of wild-type Upf1 that occurs preferentially from non-target over target mRNA in vitro (Figure 7). At this point, it remains to be resolved whether the faster dissociation of Upf1 from non-target mRNA reflects direct release or involves translocation along the mRNA. A striking observation from our CLIP assays is that Upf1 mutants blocked in the ATPase cycle exhibit greater binding in mRNA 3’UTRs compared to wild-type Upf1, particularly in regions just downstream of translation termination sites and at mRNA 3′ ends (Figure 6). This suggests that Upf1 ATPase activity is particularly important for release from these sites. Taken together, our observations suggest a mechanism by which Upf1 is initially recruited to both target and non-target mRNAs, but is preferentially released from non-targets by a mechanism that requires activation of the Upf1 ATPase and is dependent on translation. Differential release of an RNA binding regulator for target discrimination may be particularly advantageous in RNA quality control pathways, as it could permit rapid inspection of an entire RNA population under surveillance.
Deeper insight into the Upf1 ATPase-dependence of Upf1 target specificity is likely to come through future study of the specific mechanisms by which target and non-target mRNP components differentially impact Upf1-mRNA accumulation and the Upf1 ATPase. Interestingly, S. cerevisiae translation release factors eRF1 and eRF3 have been found to inhibit the Upf1 ATPase in vitro (Czaplinski et al., 1998), an observation we were able to reproduce with human eRF1 (Supplemental Figure S7E and S7F). This raises the possibility that Upf1 ATPase repression by release factors could cause retention of Upf1 on mRNA by a mechanism influenced by the efficiency of translation termination, which is thought to differ on NMD target and non-target mRNAs, as discussed above. Another factor reported to influence Upf1 ATPase activity in vitro is Upf2, which stimulates Upf1 ATPase activity by a mechanism inhibited by the Upf1 C-terminus (Chakrabarti et al., 2011; Chamieh et al., 2008; Fiorini et al., 2012), a finding we confirmed with full-length human Upf2 (Figure S7G). Future studies should reveal whether Upf2 contributes to the Upf1 ATPase-dependent step required for target discrimination described in this study or if Upf2 promotes a step in the NMD pathway downstream of Upf1-target mRNA binding, as has been previously implicated in yeast NMD (Sheth and Parker, 2006). Collectively, our findings suggest that a key determinant for NMD is a delay in the activation of Upf1 ATPase specifically on target mRNAs, which allows NMD complexes to form and initiate degradation prior to Upf1 ATPase-mediated complex disassembly and complete mRNA degradation (Figure 7D).
EXPERIMENTAL PROCEDURES
For RNA-immunoprecipitation (RIP) and release assays, human Flp-In 293-T-Rex cells (with or without stably expressed Upf1 variants as indicated in figures) were transfected with CMV promoter-driven reporter mRNA constructs, as well as protein expression constructs and, if used, siRNAs for protein depletion, 48-72 hrs before cell harvest. At 20-24 and 3-3.5 hrs before harvest, Flag-Upf1 expression in stable cell lines was induced at close to endogenous levels with tetracycline and cells used in Figure 3 were treated with okadaic acid, respectively. Cell extracts prepared in isotonic lysis buffer were subjected to immunoprecipitation with indicated antibodies (see Extended Experimental Procedures for details). Phosphatase Arrest I (G Biosciences) was included in all extracts prepared for Figure 3 except those pre-treated with λ protein phosphatase. Protein and Trizol (Invitrogen)-extracted RNA samples prepared from input and IP samples were subjected to Western and Northern or RNA-seq analysis, respectively. RIP-seq libraries were prepared using an Illumina TruSeq Stranded Total RNA HT Sample Prep Kit with Ribo-Zero Gold for ribosomal RNA depletion. CLIP-seq libraries were prepared as described previously (Yeo et al., 2009), with slight modifications (see Figure S6A and Extended Experimental Procedures for details).
Supplementary Material
Highlights.
Upf1 association with mRNA inversely correlates with Upf1 ATPase activity.
Upf1 mutants deficient in ATP binding and hydrolysis lose NMD target specificity.
Translation and PABP require Upf1 ATPase to modulate Upf1-mRNA association.
Upf1 ATPase activity is required for its preferential release from non-target mRNA.
ACKNOWLEDGEMENTS
We thank Toby Franks for Myc-Upf1 constructs, Sébastien Durand for the Flag-Smg6 construct derived from a gift of HA-Smg6 from Oliver Muhlemann, Herve Le Hir for the Upf1 ΔNC bacterial expression construct, and Søren Lykke-Andersen for Smg5-3xFlag and Smg7-3xFlag constructs. Thanks to J. Robert Hogg, Dan Pollard, members of the Lykke-Andersen and Yeo labs for feedback on this work. This study was supported by NIH grant R01 GM099717 to JL-A, NIH Grants NS075449, HG004659, HG007005 to GWY, Helen Hay Whitney Postdoctoral Fellowship to SRL and NSF Predoctoral Fellowship to GP. F.M. is supported in part by a UCSD Genetics training grant from the National Institute of General Medical Sciences, T32 GM008666. GWY is an Alfred P Sloan Research Fellow.
Footnotes
AUTHOR CONTRIBUTIONS
SRL and JL-A conceived of the study and wrote the manuscript, with input from GWY, GP, and FM. GWY, GP and FM carried out the RIP-seq and CLIP-seq and JL-A performed in vitro ATPase assays. GWY and GP conducted the bioinformatics analyses. SRL executed all other experiments.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
All sequencing data is accessible through the Gene Expression Omnibus (GEO) accession number GSE69586.
REFERENCES
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- Ankö M-L, Neugebauer KM. RNA-protein interactions in vivo: global gets specific. Trends Biochem. Sci. 2012;37:255–262. doi: 10.1016/j.tibs.2012.02.005. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J. 2007;26:1591–1601. doi: 10.1038/sj.emboj.7601588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgrader P, Cheng J, Maquat LE. Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear level of human triosephosphate isomerase mRNA. Proc. Natl. Acad. Sci. U. S. A. 1993;90:482–486. doi: 10.1073/pnas.90.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Czaplinski K, Trifillis P. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA. 2000;6:1226–1235. doi: 10.1017/s1355838200000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm V, Haberman N, Ottens F, Ule J, Gehring NH. 3′ UTR Length and Messenger Ribonucleoprotein Composition Determine Endocleavage Efficiencies at Termination Codons. Cell Rep. 2014;9:555–568. doi: 10.1016/j.celrep.2014.09.012. [DOI] [PubMed] [Google Scholar]
- Chakrabarti S, Jayachandran U, Bonneau F, Fiorini F, Basquin C, Domcke S, Le Hir H, Conti E. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol. Cell. 2011;41:693–703. doi: 10.1016/j.molcel.2011.02.010. [DOI] [PubMed] [Google Scholar]
- Chakrabarti S, Bonneau F, Schüssler S, Eppinger E, Conti E. Phospho-dependent and phospho-independent interactions of the helicase UPF1 with the NMD factors SMG5-SMG7 and SMG6. Nucleic Acids Res. 2014;42:9447–9460. doi: 10.1093/nar/gku578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat. Struct. Mol. Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Muhlrad D, Lim MK, Parker R, Song H. Structural and functional insights into the human Upf1 helicase core. EMBO J. 2007;26:253–264. doi: 10.1038/sj.emboj.7601464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Han S, Choe J, Park SG, Choi SS, Kim YK. SMG5-PNRC2 is functionally dominant compared with SMG5-SMG7 in mammalian nonsense-mediated mRNA decay. Nucleic Acids Res. 2013;41:1319–1328. doi: 10.1093/nar/gks1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson B, Berkova N. Poly(A)-binding protein and eRF3 are associated in vivo in human and Xenopus cells. Biol. Cell. 2002;94:205–216. doi: 10.1016/s0248-4900(02)01194-2. [DOI] [PubMed] [Google Scholar]
- Cosson B, Couturier A. Poly(A)-Binding Protein Acts in Translation Termination via Eukaryotic Release Factor 3 Interaction and Does Not Influence [PSI+] Propagation. Mol. Cell. Biol. 2002;22:3301–3315. doi: 10.1128/MCB.22.10.3301-3315.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle AB, Stalder L, Mathys H, Orozco RZ, Mühlemann O. Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol. 2008;6:e92. doi: 10.1371/journal.pbio.0060092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle AB, Lykke-Andersen S, Mühlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat. Struct. Mol. Biol. 2009;16:49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- Fatscher T, Boehm V, Weiche B, Gehring NH. The interaction of cytoplasmic poly(A)-binding protein with eukaryotic initiation factor 4G suppresses nonsense-mediated mRNA decay. RNA. 2014:2014. doi: 10.1261/rna.044933.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorini F, Boudvillain M, Le Hir H. Tight intramolecular regulation of the human Upf1 helicase by its N- and C-terminal domains. Nucleic Acids Res. 2012;41:2404–2415. doi: 10.1093/nar/gks1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks T, Lykke-Andersen J. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev. 2007;719:735. doi: 10.1101/gad.1494707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks T, Singh G, Lykke-Andersen J. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense-mediated mRNA decay. Cell. 2010;143:938–950. doi: 10.1016/j.cell.2010.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen LH, Schueler M, Munschauer M, Mastrobuoni G, Chen W, Kempa S, Dieterich C, Landthaler M. MOV10 Is a 5′ to 3′ RNA Helicase Contributing to UPF1 mRNA Target Degradation by Translocation along 3’UTRs. Mol. Cell. 2014:1–13. doi: 10.1016/j.molcel.2014.03.017. [DOI] [PubMed] [Google Scholar]
- Hogg JR, Goff SP. Upf1 senses 3’UTR length to potentiate mRNA decay. Cell. 2010;143:379–389. doi: 10.1016/j.cell.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoof A, Wagner EJ. A brief survey of mRNA surveillance. Trends Biochem. Sci. 2011;36:585–592. doi: 10.1016/j.tibs.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino S. -i., Imai M, Kobayashi T, Uchida N, Katada T. The Eukaryotic Polypeptide Chain Releasing Factor (eRF3/GSPT) Carrying the Translation Termination Signal to the 3’-Poly(A) Tail of mRNA. J. Biol. Chem. 1999;274:16677–16680. doi: 10.1074/jbc.274.24.16677. [DOI] [PubMed] [Google Scholar]
- Hurt JA, Robertson AD, Burge CB. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res. 2013;23:1636–1650. doi: 10.1101/gr.157354.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J, Sato H, Tang Y, Matsuda D, Maquat LE. UPF1 association with the cap-binding protein, CBP80, promotes nonsense-mediated mRNA decay at two distinct steps. Mol. Cell. 2010;39:396–409. doi: 10.1016/j.molcel.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 2008;27:736–747. doi: 10.1038/emboj.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MJO, He F, Spatrick P, Li C, Jacobson A. Association of yeast Upf1p with direct substrates of the NMD pathway. Proc. Natl. Acad. Sci. U. S. A. 2007;104:20872–20877. doi: 10.1073/pnas.0709257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns L, Grimson A, Kuchma SL, Newman CL, Anderson P. Caenorhabditis elegans SMG-2 selectively marks mRNAs containing premature translation termination codons. Mol. Cell. Biol. 2007;27:5630–5638. doi: 10.1128/MCB.00410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joncourt R, Eberle AB, Rufener SC, Mühlemann O. Eukaryotic Initiation Factor 4G Suppresses Nonsense-Mediated mRNA Decay by Two Genetically Separable Mechanisms. PLoS One. 2014;9:e104391. doi: 10.1371/journal.pone.0104391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kervestin S, Jacobson A. NMD: a multifaceted response to premature translational termination. Nat. Rev. Mol. Cell Biol. 2012;13:700–712. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol. Cell. Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov G, Gehring K. Molecular basis of eRF3 recognition by the MLLE domain of poly(A)-binding protein. PLoS One. 2010;5:e10169. doi: 10.1371/journal.pone.0010169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T, Maquat LE. Rules that govern UPF1 binding to mRNA 3’UTRs. Proc. Natl. Acad. Sci. U. S. A. 2013;110:3357–3362. doi: 10.1073/pnas.1219908110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T, Li W, Hoque M, Popp MW-L, Ermolenko DN, Tian B, Maquat LE. A post-translational regulatory switch on UPF1 controls targeted mRNA degradation. Genes Dev. 2014;28:1900–1916. doi: 10.1101/gad.245506.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- Loh B, Jonas S, Izaurralde E. The SMG5-SMG7 heterodimer directly recruits the CCR4-NOT deadenylase complex to mRNAs containing nonsense codons via interaction with POP2. Genes Dev. 2013;27:2125–2138. doi: 10.1101/gad.226951.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losson R, Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc. Natl. Acad. Sci. U. S. A. 1979;76:5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE, Kinniburgh AJ, Rachmilewitz EA, Ross J. Unstable β-Globin mRNA in mRNA-Deficient β° Thalassemia. Cell. 1981;27:543–553. doi: 10.1016/0092-8674(81)90396-2. [DOI] [PubMed] [Google Scholar]
- Mili S, Steitz J. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min EE, Roy B, Amrani N, He F, Jacobson A. Yeast Upf1 CH domain interacts with Rps26 of the 40S ribosomal subunit. Rna. 2013:1105–1115. doi: 10.1261/rna.039396.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Nicholson P, Josi C, Kurosawa H, Yamashita A, Mühlemann O. A novel phosphorylation-independent interaction between SMG6 and UPF1 is essential for human NMD. Nucleic Acids Res. 2014;42:9217–9235. doi: 10.1093/nar/gku645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada-Katsuhata Y, Yamashita A, Kutsuzawa K, Izumi N, Hirahara F, Ohno S. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res. 2012;40:1251–1266. doi: 10.1093/nar/gkr791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixeiro I, Inácio Â, Barbosa C, Silva AL, Liebhaber SA, Romão L. Interaction of PABPC1 with the translation initiation complex is critical to the NMD resistance of AUG-proximal nonsense mutations. Nucleic Acids Res. 2012;40:1160–1173. doi: 10.1093/nar/gkr820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrua O, Libri D. RNA quality control in the nucleus: the Angels’ share of RNA. Biochim. Biophys. Acta. 2013;1829:604–611. doi: 10.1016/j.bbagrm.2013.02.012. [DOI] [PubMed] [Google Scholar]
- Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- Schweingruber C, Rufener SC, Zünd D, Yamashita A, Mühlemann O. Nonsense-mediated mRNA decay - mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim. Biophys. Acta. 2013;1829:612–623. doi: 10.1016/j.bbagrm.2013.02.005. [DOI] [PubMed] [Google Scholar]
- Sheth U, Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A, Ribeiro P, Inácio Â, Liebhaber S, Romão L. Proximity of the poly (A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay. RNA. 2008;14:563–576. doi: 10.1261/rna.815108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Rebbapragada I, Lykke-Andersen J. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 2008;6:e111. doi: 10.1371/journal.pbio.0060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Hoshino S-I, Imataka H, Sonenberg N, Katada T. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in Cap/Poly(A)-dependent translation. J. Biol. Chem. 2002;277:50286–50292. doi: 10.1074/jbc.M203029200. [DOI] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW. ATP is a cofactor of the Upf1 protein that modulates its translation termination and RNA binding activities. RNA. 1998;4:205–214. [PMC free article] [PubMed] [Google Scholar]
- Yamashita a, Ohnishi T, Kashima I, Taya Y, Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001;15:2215–2228. doi: 10.1101/gad.913001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu X-D, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zünd D, Gruber A, Zavolan M, Muhlemann O. Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in 3′ UTRs. Nat. Struct. Mol. Biol. 2013;20:936–943. doi: 10.1038/nsmb.2635. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.