Abstract
IMPORTANCE
Stroke is the second leading cause of death and the third leading cause of years of life lost. Genetic factors contribute to stroke prevalence, and candidate gene and genome-wide association studies (GWAS) have identified variants associated with ischemic stroke risk. These variants often have small effects without obvious biological significance. Exome sequencing may discover predicted protein-altering variants with a potentially large effect on ischemic stroke risk.
OBJECTIVE
To investigate the contribution of rare and common genetic variants to ischemic stroke risk by targeting the protein-coding regions of the human genome.
DESIGN, SETTING, AND PARTICIPANTS
The National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (ESP) analyzed approximately 6000 participants from numerous cohorts of European and African ancestry. For discovery, 365 cases of ischemic stroke (small-vessel and large-vessel subtypes) and 809 European ancestry controls were sequenced; for replication, 47 affected sibpairs concordant for stroke subtype and an African American case-control series were sequenced, with 1672 cases and 4509 European ancestry controls genotyped. The ESP's exome sequencing and genotyping started on January 1, 2010, and continued through June 30, 2012. Analyses were conducted on the full data set between July 12, 2012, and July 13, 2013.
MAIN OUTCOMES AND MEASURES
Discovery of new variants or genes contributing to ischemic stroke risk and subtype (primary analysis) and determination of support for protein-coding variants contributing to risk in previously published candidate genes (secondary analysis).
RESULTS
We identified 2 novel genes associated with an increased risk of ischemic stroke: a protein-coding variant in PDE4DIP (rs1778155; odds ratio, 2.15; P = 2.63 × 10−8) with an intracellular signal transduction mechanism and in ACOT4 (rs35724886; odds ratio, 2.04; P = 1.24 × 10−7) with a fatty acid metabolism; confirmation of PDE4DIP was observed in affected sibpair families with large-vessel stroke subtype and in African Americans. Replication of protein-coding variants in candidate genes was observed for 2 previously reported GWAS associations: ZFHX3 (cardioembolic stroke) and ABCA1 (large-vessel stroke).
CONCLUSIONS AND RELEVANCE
Exome sequencing discovered 2 novel genes and mechanisms, PDE4DIP and ACOT4, associated with increased risk for ischemic stroke. In addition, ZFHX3 and ABCA1 were discovered to have protein-coding variants associated with ischemic stroke. These results suggest that genetic variation in novel pathways contributes to ischemic stroke risk and serves as a target for prediction, prevention, and therapy.
According to the 2010 Global Burden of Disease study, stroke is the second leading cause of death and the third leading cause of years of life lost.1,2 Ischemic stroke is the overt symptomatic manifestation of brain infarction, but the burden of cerebrovascular disease is much greater, with many more covert infarctions not resulting in the diagnosis of clinical stroke. More than 7% of asymptomatic adults in the general population have radiographic evidence of brain infarction,3 with substantially higher rates in the elderly population.4 Although many effective treatments for stroke exist, novel strategies for stroke prediction, prevention, and therapy need to be found.
Epidemiologic and family studies5,6 support an inherited component to stroke risk. Family-based linkage studies, including Cerebral Autosomal Dominant Arteriopathy With Sub-cortical Infarcts and Leukoencephalopathy and Cerebral Autosomal Recessive Arteriopathy With Subcortical Infarcts and Leukoencephalopathy,7 identified rare forms of stroke. Ischemic stroke studies include large candidate gene association meta-analyses,8,9 mendelian randomization,10 studies of affected sibpair families,11 and genome-wide association studies (GWAS).12–14 The joint International Stroke Genetics Consortium/Wellcome Trust Case Control Consortium 2 (WTCCC2) effort14 identified HDAC9 (7p21.1; GenBank NM_058176) and confirmed associations of ischemic stroke with variants in PITX2 (GenBank KJ891816) and ZFHX3 (GenBank NM_006885) (cardioembolic stroke) and in 9p21 (large-vessel/atherosclerotic stroke). The Australian Stroke Genetics Collaborative, the WTCCC2, and the International Stroke Genetics Consortium have identified and replicated the chromosome 6p21.1 large-artery susceptibility locus.13 Discovering the missing heritability for ischemic stroke could provide critical insights into the cause of the disease, novel pathways, and therapeutic targets.15
Rare, protein-coding variation could contribute a burden of risk for ischemic stroke, attributable to multiple variants in a gene rather than any individual variant.16,17 Exome sequencing in ischemic stroke has been performed only as a pilot study.18 Herein, we present results from what we believe to be the first large-scale study of protein-coding region variants in ischemic stroke from the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (ESP). We describe analyses used to identify novel stroke susceptibility genes, determine genes and variants that are specific to stroke subtypes, and establish replication of previous GWAS and candidate gene associations.
Methods
Participating Studies and Design
The NHLBI ESP ascertained samples of European American and African American ancestry. Data were obtained on NHLBI cohorts (Atherosclerosis Risk in Communities, Coronary Artery Risk Development in Young Adults, Cardiovascular Health Study, Framingham Heart Study, Jackson Heart Study, the Multi-Ethnic Study of Atherosclerosis, and the Women's Health Initiative [WHI]) and 2 National Institute of Neurological Disorders and Stroke studies (Siblings With Ischemic Stroke Study [SWISS] and Ischemic Stroke Genetic Study [ISGS)]). Characteristics of the participants analyzed are presented in Table 1.
Table 1.
Characteristics of Samples Used in the Discovery and Replication Analyses
| Cases |
Controls |
|||||||
|---|---|---|---|---|---|---|---|---|
| Female, No./Total No. (%) | Type of Stroke, No. (%) |
Female, No./Total No. (%) | ||||||
| Sample | Age Mean (SD), y | Small Vessel | Large Vessel | Cardioembolic | Undetermined | Age, Mean (SD), y | ||
| EA ESP | 312/365 (85.5) | 62.4 (11.6) | 249 (68.2) | 114 (31.2) | 0 | 2 (0.5) | 460/809 (56.9) | 58.5 (11.7) |
|
| ||||||||
| AA ESP | 68/83 (81.9) | 57.0 (9.2) | 57 (68.7) | 23 (27.7) | 0 | 3 (3.6) | 215/317 (67.8) | 59.3 (8.3) |
|
| ||||||||
| SWISS | 36/94 (38.3) | 67.4 (11.5) | 56 (59.6) | 38 (40.4) | 0 | 0 | NA | NA |
|
| ||||||||
| WHI ExomeChip | 1672/1672 (100) | 68.7 (5.8) | 224 (13.4) | 107 (6.4) | 605 (36.2) | 736 (44.0) | 4509/4509 (100) | 66.9 (6.4) |
Abbreviations: AA, African ancestry; EA, European ancestry; ESP, Exome Sequencing Project; NA, not available; SWISS, Siblings With Ischemic Stroke Study; WHI, Women's Health Initiative.
Discovery analysis by exome sequencing focused on small-vessel and large-vessel subtypes in unrelated persons of European ancestry. Replication was conducted in small- and large-vessel subtypes in SWISS and ESP African American individuals, as well as ExomeChip data from European American participants in the WHI that included cardioembolic sub-types. Exome sequencing was performed on 365 unrelated cases (327 cases from NHLBI cohorts and 38 cases from ISGS: 114 with large-vessel subtype, 249 with small-vessel subtype, and 2 undetermined), 47 affected sibpairs from SWISS (94 individuals, used in replication: 28 pairs concordant for small-vessel subtype and 19 pairs concordant for large-vessel sub-type), and African Americans (83 cases and 317 controls, used in replication). ExomeChip genotyping was performed on 1672 unrelated WHI ischemic stroke cases: 107 with large-vessel sub-type, 224 with small-vessel subtype, 605 with cardioembolic subtype, and 736 of undetermined subtype. Cases classified as undetermined were included in the overall case-control analysis but not the subtype-specific analyses. Controls (n = 809) were identified as participants with exome sequence data who had not experienced a stroke or myocardial infarction at baseline or on subsequent follow-up and as individuals who were originally ascertained on the basis of low blood pressure or decreased low-density lipoprotein cholesterol levels. Controls for ExomeChip analysis were 4509 WHI participants who self-reported having no history of stroke and myocardial infarction and no family history of stroke.
Ischemic Stroke Classification
Ischemic stroke was defined as a typical clinical syndrome with radiologic confirmation of brain infarction or the absence of an alternative diagnosis for the clinical syndrome. Stroke subtyping was based on the Trial of Org 10172 in Acute Stroke Treatment classification system.19 Brain imaging by computed tomographic scanning, magnetic resonance imaging, or autopsy confirmation was available to classify all cases. Relevant institutional review boards approved participating studies, and all participants provided written informed consent for participating in the original study as well as for genetic research. There was no financial compensation for use of existing data and samples from the contributing cohorts as part of this study.
Genetic Data
Exome sequencing was performed at the Broad Institute and the University of Washington (eAppendix 2 in the Supplement). ExomeChip genotyping was performed at the Broad Institute (eAppendix 3 in the Supplement). The ESP exome sequencing and ExomeChip genotyping started on January 1, 2010, and continued through June 30, 2012. Analyses were conducted on the full data set between July 12, 2012, and July 13, 2013. A total of 225 239 protein-coding variants were common to both ESP exome sequencing and ExomeChip geno-typing, of which 119 963 were polymorphic in the full ESP sample (n = 7355). The self-reported ancestry and genetic ancestry distributions by principal components analysis are shown in eFigure 1 in the Supplement.
Statistical Analysis
Single-variant tests and 2 gene-based tests were used to determine the association with ischemic stroke, conditional on the minor allele frequency (MAF) of individual variants (eAppendix 4 in the Supplement). Variants with MAF of 0.5% or more and at least 100 observations with a nonmissing genotype were analyzed using a single-variant test. For gene-based tests, only missense, splice, or nonsense variants were considered. Variants with MAF of 5% or less were analyzed with the sequence kernel association test (SKAT),20 or variants with MAF of 1% or less analyzed with the combined multivariate and collapsing method, using variants with MAF of less than 1% (CMC/T1 test).21 For SKAT and CMC/T1, only genes with cumulative MAF of 0.5% or more were assessed. Analyses were conducted separately for exome sequence (combining data from all cohorts and ISGS) and ExomeChip (WHI) data; a meta-analysis was conducted using seqMeta software (http://cran.r-project.org/web/packages/seqMeta/).22 Covariate adjustment included participant's age at baseline and sex, as appropriate. For autosomal variants, a log-additive genetic model was used; for sex chromosome variants, a dominance model was used.
Ischemic stroke and subtype analyses used 2037 cases and 5318 controls with ESP exome sequencing or ExomeChip data. Association of ischemic stroke with any single variant used P < 4.17 × 10−7 (0.05/119 963) as the threshold for exomewide significance. Restriction to variants with MAF of 0.005 or more and at least 100 observations with a nonmissing genotype yielded 25 467 variants, with a significance threshold at P < 1.96 × 10−6. Gene-level associations used a variety of statistical tests and MAF cutoffs23; CMC/T1 is most powerful when all variants in a gene influence risk in the same direction; SKAT is more powerful when the variants in a gene influence risk in opposite directions. For CMC/T1, genes were included for individual variants with MAF of 0.01 or less and cumulative MAF of 0.005 or more, resulting in 6173 informative genes with a threshold of P < 8.10 × 10−6. For SKAT, there were 8361 genes with MAF of 0.05 or less and cumulative MAF of 0.005 or more, with a threshold of P < 5.98 × 10−6. In the discovery phase, these analyses were conducted for all ischemic stroke, small-vessel subtype, and large-vessel subtype. Affected sibpairs from SWISS were analyzed using identity-by-descent methods (eAppendix 4 in the Supplement). Regions of excess identity by descent were examined, focusing on missense, nonsense, stop-gain/loss functional variants. Single-variant and aggregate identity-by-descent probabilities of each gene across all available sibpairs were calculated to generate logarithm-of-the-odds scores for excess identity-by-descent sharing, adjusting for local recombination rates based on European ancestry estimates (HapMap phase 3), repeated for sibpairs concordant for small-vessel and large-vessel subtypes. The distribution of scores from the identity-by-descent analyses is shown in eFigure 2 in the Supplement.
Targeted secondary analysis of 59 candidate genes previously associated with ischemic stroke, stroke risk factors, and mendelian syndromes yielded 134 variants. Most of these variants are not located in protein-coding regions; however, proxies were identified with strong linkage disequilibrium with ESP-defined protein-coding variants. For single-variant tests, significance was defined as P < 1.86 × 10−4. For gene-based tests, 33 genes contained polymorphic variants for analysis and significance was defined as P < 7.58 × 10−4 since 2 gene-based tests (CMC/T1 and SKAT) were used.
Results
Exome Variants Associated With Ischemic Stroke
Single-variant tests identified 2 protein-coding variants significantly associated with ischemic stroke (Table 2). A common (MAF, 0.316) missense variant, rs1778155, in PDE4DIP (phosphodiesterase 4D-interacting protein; 1q21.1; GenBank NM_014644) was associated with ischemic stroke (odds ratio [OR], 2.15; P = 2.63 × 10−8). An infrequent (MAF, 0.017) missense variant, rs35724886, in ACOT4 (acyl-coenzyme A thioesterase 4; 14q24.3; GenBank NM_152331) was associated with ischemic stroke (OR, 2.04; P = 1.2 × 10−7). Although not reaching exomewide significance, a common (MAF, 0.379) variant, rs5001076, in PRIM2 (primase, DNA, polypeptide 2 [58 kilodaltons (kDa)]); 6p11.2; GenBank NM_000947) showed evidence of increased risk with ischemic stroke (OR, 2.11; P = 2.96 × 10−6) (Table 2).
Table 2.
Association Results for Coding Region Variants and Risk of Ischemic Stroke With a Meta-analysis P<5×10−6 Displayed
| OR |
P Value |
No. of SNVs in SKAT | SKAT Gene-Level P Value |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Location | Gene Size, kb | Associated SNV | MAF | Discovery | Replication | Meta-analysis | Discovery | Replication | Meta-analysis | Discovery | Replication | Meta-analysis | |
| PDE4DIP | 1q21.1 | 240.030 | rs1778155 | 0.316 | 2.147 | NA | 2.147 | 2.63 × 10−8 | NA | 2.63 × 10−8 | 42 | .97 | .24 | .45 |
|
| ||||||||||||||
| ACOT4 | 14q24.3 | 4.791 | rs35724886 | 0.017 | 2.044 | NA | 2.044 | 1.24 × 10−7 | NA | 1.24 × 10−7 | 1 | 1.36 × 10−7 | NA | 1.36 × 10−7 |
|
| ||||||||||||||
| PRIM2 | 6p11.2 | 333.773 | rs5001076 | 0.379 | 2.105 | NA | 2.105 | 2.96 × 10−6 | NA | 2.96 × 10−6 | 4 | 7.2 × 10−5 | NA | 7.2 × 10−5 |
Abbreviations: kb, kilobase; MAF, minor allele frequency; OR, odds ratio; SNV, single-nucleotide variant; SKAT, sequence kernel association test.
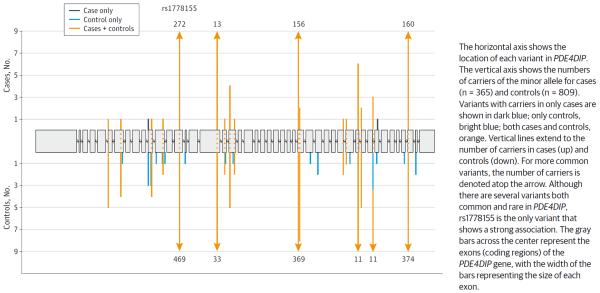
Rare and infrequent protein-coding variants in PDE4DIP from exome sequencing in 365 cases and 809 controls and their estimated effect on ischemic stroke risk are shown in the Figure. A large complex gene (240.030 kilobases [kb]; 2346 amino acids, 265 kDa), PDE4DIP has 86 distinct introns, 64 different messenger RNAs, 49 alternatively spliced variants, and at least 10 transcription factor binding sites in its promoter. In contrast, ACOT4 is a small gene (4.791 kb; 421 amino acids, 46 kDa) with 2 distinct introns and 1 spliced messenger RNA. Both genes are expressed in the brain and in other tissues.
Figure.
Associations With Ischemic Stroke in PDE4DIP
The results of no single-variant or gene-based test attained statistical significance for the small-vessel (lacunar) stroke subtype (Table 3). PDE4DIP rs1778155 had direction and size of effect on risk consistent in the all-stroke and small-vessel stroke categories (OR, 2.03; meta-analysis P = 7.96 × 10−6). ACOT4 rs35724886 also had the same direction and similar size of effect on risk (OR, 1.95; meta-analysis P = 2.43 × 10−5). One novel gene, CEP164 (centrosomal protein 164 kDa; 11q23.3; GenBank NM_014956) with 24 rare variants (cumulative MAF, 0.01) approached statistical significance (SKAT meta-analysis P = 7.93 × 10−6).
Table 3.
Association Results for Coding Region Variants and Risk of Small-Vessel Ischemic Stroke With Meta-analysis P<5×10−4 Displayed
| Test Results |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR |
P Value |
|||||||||
| Gene | Location | Gene Size, kb | SNV | MAF | ESP Sequence | Exome-Chip | Meta-analysis | ESP Sequence | Exome-Chip | Meta-analysis |
| Single-Variant Test Used | ||||||||||
|
| ||||||||||
| PDE4DIP | 1q21.1 | 240.030 | Best, rs1778155 | 0.308 | 2.029 | NA | 2.029 | 7.96 × 10−6 | NA | 7.96 × 10−6 |
|
| ||||||||||
| ACOT4 | 14q24.3 | 4.791 | Best, rs35724886 | 0.018 | 1.952 | NA | 1.952 | 2.43 × 10−5 | NA | 2.43 × 10−5 |
|
| ||||||||||
| Gene-Based Test Used (SKAT) | ||||||||||
|
| ||||||||||
| CEP164 | 11q23.3 | 98.712 | n = 24 | Cumulative, 0.010 | NA | NA | NA | .15 | 4.9 × 10−6 | 7.93 × 10−6 |
Abbreviations: kb, kilobase; MAF minor allele frequency; NA, not applicable; OR, odds ratio; SKAT, sequence kernel association test; SNV, single-nucleotide variant.
Within large-vessel (atherosclerotic) stroke, no single-variant or gene-based test attained statistical significance (Table 4). PDE4DIP rs1778155 had a strong effect and consistent direction (OR, 2.40; P = 2.18 × 10−5). Single-variant tests did not support ACOT4 rs35724886, but gene-based tests of many infrequent or rare variants provided suggestive evidence for association (eg, SPSB3 [GenBank KJ899913], SLC22A5 [GenBank KJ897580], PGAP1 [GenBank NM_024989], KIF21A [GenBank NM_001173464], and MX1 [GenBank NM_001144925]) with large-vessel subtype.
Table 4.
Association Results for Coding Region Variants and Risk of Large-Vessel Ischemic Stroke With Meta-analysis P<5×10−4 Displayed
| Test Results |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR |
P Value |
|||||||||
| Gene | Location | Gene Size, kb | SNV | MAF | ESP-Sequence | Exome-Chip | Meta-analysis | ESP-Sequence | Exome-Chip | Meta-analysis |
| Single-Variant Test Used | ||||||||||
|
| ||||||||||
| SPSB3 | 16p13.3 | 16.989 | Best, rs147735377 | 0.006 | NA | 58.225 | 58.225 | NA | 9.62 × 10−6 | 9.62 × 10−6 |
|
| ||||||||||
| PDE4DIP | 1q21.1 | 240.030 | Best, rs1778155 | 0.302 | 2.40 | NA | 2.403 | 2.18 × 10−5 | NA | 2.18 × 10−5 |
|
| ||||||||||
| DNAH14 | 1q42.12 | 503.033 | Best, rs41267347 | 0.080 | 2.33 | 2.03 | 2.181 | .0016 | .0058 | 3.06 × 10−5 |
|
| ||||||||||
| MPDZ | 9p23 | 173.887 | Best, rs188840960 | 0.009 | 2.51 | 30.35 | 8.497 | .2056 | 4.45 × 10−6 | 3.90 × 10−5 |
|
| ||||||||||
| ZNF778 | 16q24.3 | 11.855 | Best, rs117690401 | 0.040 | 4.380 | NA | 4.380 | 4.88 × 10−5 | NA | 4.88 × 10−5 |
|
| ||||||||||
| SLC22A5 | 5q31.1 | 25.906 | Best, rs28383481 | 0.005 | 0.9289 | 172.89 | 16.853 | .9439 | 4.25 × 10−8 | 5.48 × 10−5 |
|
| ||||||||||
| PGAP1 | 2q33.1 | 94.793 | Best, rs62185645 | 0.009 | 23.482 | NA | 23.482 | 6.01 × 10−5 | NA | 6.01 × 10−5 |
|
| ||||||||||
| KIF21A | 12q12 | 150.163 | Best, rs78616703 | 0.005 | 2.44 | 84.71 | 16.290 | 0.3811 | 3.07 × 10−6 | 6.06 × 10−5 |
|
| ||||||||||
| MX1 | 21q22.3 | 38.911 | Best, rs140022520 | 0.005 | 51.953 | NA | 51.953 | 9.51 × 10−5 | NA | 9.51 × 10−5 |
|
| ||||||||||
| ACOT4 | 14q24.3 | 4.791 | Best, rs35724886 | 0.015 | 2.384 | NA | 2.384 | 1.07 × 10−4 | NA | 1.07 × 10−4 |
|
| ||||||||||
| Gene-Based Test Used (SKAT) | ||||||||||
|
| ||||||||||
| SPSB3 | 16p13.3 | 16.989 | n = 3 | Cumulative, 0.009 | NA | NA | NA | .3169 | 1.05 × 10−5 | 1.33 × 10−5 |
|
| ||||||||||
| FAM151A | 1p32.3 | 14.380 | n = 11 | Cumulative, 0.053 | NA | NA | NA | 5.12 × 10−4 | .3479 | 4.20 × 10−5 |
|
| ||||||||||
| MX1 | 21q22.3 | 38.911 | n = 8 | Cumulative, 0.007 | NA | NA | NA | 5.20 × 10−5 | .0696 | 5.97 × 10−5 |
|
| ||||||||||
| SLC22A5 | 5q31.1 | 25.906 | n = 11 | Cumulative, 0.011 | NA | NA | NA | .9600 | 6.05 × 10−8 | 7.04 × 10−5 |
|
| ||||||||||
| PGAP1 | 2q33.1 | 94.793 | n = 7 | Cumulative, 0.013 | NA | NA | NA | 5.85 × 10−5 | .9556 | 8.96 × 10−5 |
|
| ||||||||||
| KIF21A | 12q12 | 150.163 | n = 15 | Cumulative, 0.074 | NA | NA | NA | .3302 | 1.05 × 10−6 | 1.08 × 10−4 |
Abbreviations: ESP, Exome Sequencing Project; kb, kilobase; MAF, minor allele frequency; NA, not applicable; OR, odds ratio; SNV, single-nucleotide variant; SKAT, sequence kernel association test.
Cases of cardioembolic ischemic stroke were not selected for ESP exome sequencing; these participants were sequenced only if they were selected for other (nonstroke) phenotypes or were included later for ExomeChip genotyping. No statistically significant associations with cardioembolic stroke were found by either single-variant or gene-based tests. The most significant single-variant association was with PPIP5K2 rs35671301 (GenBank NM_001276277) (diphosphoinositol pentakisphosphate kinase 2; 5q21.1; OR, 3.59; MAF, 0.011; meta-analysis P = 1.01 × 10−5). The SKAT test also provided support for PPIP5K2 on risk in the cardioembolic stroke (5 variants with cumulative MAF, 0.104; meta-analysis P = 1.51 × 10−5).
Replication of Exome Variants Associated With Ischemic Stroke
Single-nucleotide polymorphisms in PDE4DIP and ACOT4 were analyzed in an independent set of affected sibpairs from SWISS and unrelated ESP African American participants. The PDE4DIP rs1778155 variant had significantly increased allele sharing identity by descent (P = 1.16 × 10−4) and was observed in 16 of 19 large-vessel concordant pairs with a logarithm of the odds of 6.07 (OR, 2.41; MAF, 0.302; P = 2.18 × 10−5) and a false discovery rate of P = 6.39 × 10−3. ACOT4 rs35724886 was not polymorphic in either the SWISS or WHI ExomeChip data. A significant association with ischemic stroke was observed for PDE4DIP rs1778155 in the ESP African American samples (MAF, 0.191; OR, 2.34, P = 7.50 × 10−4). This variant failed genotype quality control metrics on the ExomeChip and was not analyzed. In African Americans, no significant association was detected for ACOT4 rs35724886 with ischemic stroke although the effect size was similar (OR, 1.86; MAF, 0.180; P = .31).
Analysis of Previously Identified Candidate Genes and Variants
We identified 59 published candidate genes to be analyzed with the single-variant and gene-based tests. A single variant in ABCA1 (adenosine triphosphate-binding cassette, subfamily A [ABC1], member 1; 9q31.1; GenBank NM_005502) was associated with all ischemic stroke (meta-analysis P = 1.94 × 10−4) and in both subtypes (small-vessel meta-analysis P = .008; large-vessel meta-analysis P = .02). Variants in ZFHX3 (zinc finger homeobox 3; 16q22.3), a candidate gene for atrial fibrillation, were nominally associated with all ischemic stroke using the CMC/T1 test (P = .001), primarily in the cardioembolic subtype (P = .04). Results for all candidate genes are presented in eTables 1 through 4 in the Supplement.
Discussion
We report the results of ESP exome sequencing and ExomeChip genotyping of 2037 cases of ischemic stroke and 5318 controls, with replication in an African American population and a collection of affected sibpairs. We present evidence supporting the role of protein-coding variants in 2 novel genes, PDE4IP and ACOT4, which are associated with an increased risk of ischemic stroke. We evaluated protein-coding variants in previously reported genes associated with ischemic stroke, and support the role of ABCA1 in large-vessel stroke and ZFHX3 in cardioembolic stroke.
A primary interacting partner of PDE4DIP is PDE4D (phosphodiesterase 4D, cAMP-specific; GenBank NM_001104631), originally identified as linked to ischemic stroke in Icelandic families.24 The PDE4D variants have been extensively studied25–28 for association with ischemic stroke risk in diverse populations, with equivocal results. A meta-analysis29 of ischemic stroke was conducted on 7 studies, using approximately 12 000 cases and 15 000 controls that included 6 single-nucleotide polymorphisms in PDE4D. In the meta-analysis, PDE4D rs702553 had a significant association with ischemic stroke. The PDE4D result, coupled with PDE4DIP identified in the present study,30,31 supports a role of this pathway in ischemic stroke.
The ACOT4 gene encodes an enzyme that catalyzes the hydrolysis of acyl-coenzyme A (CoA) to the free fatty acid and coenzyme A, regulating intracellular levels of acyl-CoA and fatty acids. Selected fatty acids are associated with adverse outcomes including dyslipidemia, inflammation, myocardial infarction, and other cardiovascular mortality.32–36 Human ACOT4 hydrolyzes succinyl-CoA, glutaryl-CoA, and long-chain acyl-CoA reactions catalyzed by multiple enzymes in other species. Expression of ACOT4 is regulated by peroxisome proliferator-activated receptor-α,37,38 a nuclear receptor that has been implicated in vascular and cardiac disease and represents a therapeutic target in diabetes mellitus.39 ACOT4 links risk for ischemic stroke with fatty acid metabolism.
This study, which we believe to be the largest on exome sequencing in stroke, has limitations. The replication of PDE4DIP rs1778155 association with ischemic stroke succeeded in both affected sibpair families and a small case-control cohort of African ancestry; however, it was not replicated in ExomeChip analyses. Furthermore, the ACOT4 rs35724886 infrequent variant was not informative in the replication samples and, therefore, was not replicated. Because these are protein-coding variants, their coverage in GWAS reports (eg, METASTROKE)40 is often incomplete. PDE4DIP rs1778155 has no surrogate within r2 value greater than 0.4 in the 1000 Genomes Project data; ACOT4 rs35724886 has no surrogate within r2 value greater than 0.8 in the ACOT2/ACOT4/ACOT6 cluster. In the joint ImmunoChip/WTCCC2/METASTROKE data,40 the PDE4DIP rs1778155 variant is 20.5 Mb proximal to the SELP (selectin P [granule membrane protein 140kDa, antigen CD62; 1q24.2]; GenBank NM_003005) rs3917792 variant (intronic, associated with all ischemic stroke: OR, 1.14; P = 1.76 × 10−7). However, ACOT4 rs35724886 is only 1.4 Mb from the METASTROKE-identified RGS6 (regulator of G-protein signaling 6; 14q24.2; GenBank NM_001204416) rs2238238 variant (intronic, associated with all ischemic stroke: OR, 1.18; P = 1.65 × 10−6). Thus, METASTROKE provides indirect support for the ACOT4 contribution to ischemic stroke. Several candidate genes and loci were not analyzed, since few protein-coding variants were identified through ESP exome sequencing or included on the ExomeChip (eg, HDAC9, PDE4D). The 9p21.3 region, previously associated with large-vessel ischemic stroke, contains few protein-coding genes and, therefore, few coding-region variants. Only 3 variants in CDKN2A (cyclin-dependent kinase inhibitor 2A; GenBank NG_007485) and 1 variant in CDKN2B-AS1 (CDKIN2B-antisense RNA 1; GenBank HG975381) were analyzed, with no variant associated with overall ischemic stroke or stroke subtype. Finally, our discovery sample focused on small- and large-vessel ischemic stroke subtypes to increase statistical power to detect novel genetic loci and reduce heterogeneity. Subsequent epidemiologically robust sampling approaches, consistent with the underlying distribution of subtypes in the population, should be conducted. Such studies would enable estimation of risk attributable to these genes (eg, PDE4DIP and ACOT4) and pathways to small-and large-vessel stroke and the effect of these variants in individual risk profiles for ischemic stroke.
The NHLBI ESP is the first large-scale exome sequencing project to focus on complex human phenotypes rather than mendelian disorders. Although data from both European Americans and African Americans were sequenced in the ESP, only European Americans had sufficiently large numbers of ischemic stroke to permit robust genetic analyses. The motivation for exploring the exome was that the protein-coding region variants would have greater likelihood of functional impact and, therefore, larger effects on disease risk, although examples of this remain few and require large sample sizes, similar to those of GWAS. Nevertheless, exome sequencing has, as exhibited in the present study, provided novel gene targets and biological pathways for examination on their role in ischemic stroke.
Conclusions
Our study has led to the identification of 2 novel genes (PDE4DIP and ACOT4) and the replication of 2 previously reported candidate genes (ABCA1 and ZFXH3) as containing coding region variants associated with ischemic stroke. These new results suggest that2 pathways, involving cell migration and growth (PDE4DIP) and long-chain fatty acid metabolism (ACOT4), could provide insights into the cause of ischemic stroke and as targets for pharmacologic interventions and therapies.
Supplementary Material
Acknowledgments
Funding/Support: Funding for GO ESP was provided by NHLBI grants RC2 HL-103010 (HeartGO), RC2 HL-102923 (LungGO), and RC2 HL-102924 (Women's Health Initiative Sequencing Project). The exome sequencing was performed through NHLBI grants RC2 HL-102925 (BroadGO) and RC2 HL-102926 (SeattleGO). HeartGO components and their support include Atherosclerosis Risk in Communities (NHLBI contracts N01 HC-55015, N01 HC-55016, N01HC-55017, N01 HC-55018, N01 HC-55019, N01 HC-55020, and N01 HC-55021); Cardiovascular Health Study (NHLBI contracts HHSN268201200036C, HHSN268200800007C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, and N01HC85086); and NHLBI grants U01HL080295, R01HL087652, R01HL105756, R01HL103612, and R01HL120393, with additional contribution from the National Institute of Neurological Disorders and Stroke. Additional support was provided through R01AG023629 from the National Institute on Aging. A full list of principal Cardiovascular Health Study investigators and institutions can be found at CHS-NHLBI.org; Coronary Artery Risk Development in Young Adults (NHLBI contracts N01-HC95095, N01-HC48047, N01-HC48048, N01-HC48049, and N01-HC48050); Framingham Heart Study (NHLBI contract N01-HC-25195 and grants NS17950, AG08122, and AG033193); Jackson Heart Study (NHLBI contracts N01 HC-95170, N01 HC-95171, and N01 HC-95172); Multi-Ethnic Study of Atherosclerosis (NHLBI contracts N01-HC-95159 through N01-HC-95169 and grant 024156). The Siblings with Ischemic Stroke Study (grant R01-NS39987) and the Ischemic Stroke Genetics Study (grant R01-NS42733) contributed phenotypic data and DNA samples. Dr Nalls' participation in this research was supported, in part, by the Intramural Research Program of the National Institute on Aging (grant Z01-AG000954-7). Portions of this study used the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health (NIH) (http://biowulf.nih.gov). The Women's Health Initiative program is funded by the NHLBI through contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C, and HHSN271201100004C, and grant RC2-HL102924 for the Women's Health Initiative Sequencing Project. LungGO components and their support include Cystic Fibrosis Foundation (Cystic Fibrosis Foundation grants GIBSON07K0, KNOWLE00A0, OBSERV04K0, and RDP R026), and NIH (grants R01-HL068890, R02-HL095396, UL1-RR025014, and 5R00-HG004316); Chronic Obstructive Pulmonary Disease (COPDGene; NIH grants U01-HL089897 and U01-HL089856), and the COPD Foundation through contributions made to an industry advisory board comprising AstraZeneca, Boehringer Ingelheim, Novartis, Pfizer, and Sunovian; Acute Lung Injury (NIH grant RC2-HL101779); Lung Health Study (NIH grants RC2-HL066583, R01-HG004738, and HR-46002); Pulmonary Arterial Hypertension (NIH grants P50-HL084946, K23-AR52742, and F32-HL083714); and Asthma (NIH grants RC2-HL101651, HL077916, HL-69197, HL-76285, and M01-RR07122).
Role of the Funder/Sponsor: The funding organization (NIH/NHLBI) had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Author Contributions: Drs Peters and Rich contributed equally to the manuscript. Dr Rich had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Kooperberg, Peters, Rich.
Acquisition, analysis, or interpretation of data: Auer, Nalls, Meschia, Worrall, Longstreth, Seshadri, Burger, Carlson, Carty, Chen, Cupples, DeStefano, Fornage, Hardy, Hsu, Jackson Jarvik, Kim, Lakshminarayan, Lange, Manichaikul, Quinlan, Singleton, Thornton, Nickerson, Rich.
Drafting of the manuscript: Rich.
Critical revision of the manuscript for important intellectual content: All authors.
Statistical analysis: Auer, Nalls, Kooperberg.
Obtained funding: Jackson, Nickerson, Rich.
Administrative, technical, or material support: Kooperberg, Nickerson, Peters Rich.
Study supervision: Nickerson, Peters, Rich.
Conflict of Interest Disclosures: None reported.
Group Information: NHLBI Exome Sequencing Project members are listed in eAppendix 1 in the Supplement.
Additional Contributions: The support of the research institutions, study investigators, field staff, and study participants assisted in creating this resource for biomedical research.
REFERENCES
- 1.Roger VL, Go AS, Lloyd-Jones DM, et al. American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Executive summary: heart disease and stroke statistics–2012 update: a report from the American Heart Association. Circulation. 2012;125(1):188–197. doi: 10.1161/CIR.0b013e3182456d46. [DOI] [PubMed] [Google Scholar]
- 2.Kim AS, Johnston SC. Temporal and geographic trends in the global stroke epidemic. Stroke. 2013;44(6 suppl 1):S123–S125. doi: 10.1161/STROKEAHA.111.000067. [DOI] [PubMed] [Google Scholar]
- 3.Vernooij MW, Ikram MA, Tanghe HL, et al. Incidental findings on brain MRI in the general population. N Engl J Med. 2007;357(18):1821–1828. doi: 10.1056/NEJMoa070972. [DOI] [PubMed] [Google Scholar]
- 4.Vermeer SE, Longstreth WT, Jr, Koudstaal PJ. Silent brain infarcts: a systematic review. Lancet Neurol. 2007;6(7):611–619. doi: 10.1016/S1474-4422(07)70170-9. [DOI] [PubMed] [Google Scholar]
- 5.Kiely DK, Wolf PA, Cupples LA, Beiser AS, Myers RH. Familial aggregation of stroke: the Framingham Study. Stroke. 1993;24(9):1366–1371. doi: 10.1161/01.str.24.9.1366. [DOI] [PubMed] [Google Scholar]
- 6.Schulz UG, Flossmann E, Rothwell PM. Heritability of ischemic stroke in relation to age, vascular risk factors, and subtypes of incident stroke in population-based studies. Stroke. 2004;35(4):819–824. doi: 10.1161/01.STR.0000121646.23955.0f. [DOI] [PubMed] [Google Scholar]
- 7.Meschia JF, Worrall BB, Rich SS. Genetic susceptibility to ischemic stroke. Nat Rev Neurol. 2011;7(7):369–378. doi: 10.1038/nrneurol.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ariyaratnam R, Casas JP, Whittaker J, Smeeth L, Hingorani AD, Sharma P. Genetics of ischaemic stroke among persons of non-European descent: a meta-analysis of eight genes involving approximately 32,500 individuals. PLoS Med. 2007;4(4):e131. doi: 10.1371/journal.pmed.0040131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casas JP, Hingorani AD, Bautista LE, Sharma P. Meta-analysis of genetic studies in ischemic stroke: thirty-two genes involving approximately 18,000 cases and 58,000 controls. Arch Neurol. 2004;61(11):1652–1661. doi: 10.1001/archneur.61.11.1652. [DOI] [PubMed] [Google Scholar]
- 10.Holmes MV, Newcombe P, Hubacek JA, et al. Effect modification by population dietary folate on the association between MTHFR genotype, homocysteine, and stroke risk: a meta-analysis of genetic studies and randomised trials. Lancet. 2011;378(9791):584–594. doi: 10.1016/S0140-6736(11)60872-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meschia JF, Nalls M, Matarin M, et al. Siblings With Ischemic Stroke Study Investigators. Siblings With Ischemic Stroke Study: results of a genome-wide scan for stroke loci. Stroke. 2011;42(10):2726–2732. doi: 10.1161/STROKEAHA.111.620484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Traylor M, Farrall M, Holliday EG, et al. Australian Stroke Genetics Collaborative, Wellcome Trust Case Control Consortium 2 (WTCCC2); International Stroke Genetics Consortium. Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration): a meta-analysis of genome-wide association studies. Lancet Neurol. 2012;11(11):951–962. doi: 10.1016/S1474-4422(12)70234-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holliday EG, Maguire JM, Evans TJ, et al. Australian Stroke Genetics Collaborative; International Stroke Genetics Consortium; Wellcome Trust Case Control Consortium 2. Common variants at 6p21.1 are associated with large artery atherosclerotic stroke. Nat Genet. 2012;44(10):1147–1151. doi: 10.1038/ng.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bellenguez C, Bevan S, Gschwendtner A, et al. International Stroke Genetics Consortium (ISGC); Wellcome Trust Case Control Consortium 2 (WTCCC2). Genome-wide association study identifies a variant in HDAC9 associated with large vessel ischemic stroke. Nat Genet. 2012;44(3):328–333. doi: 10.1038/ng.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marth GT, Yu F, Indap AR, et al. 1000 Genomes Project. The functional spectrum of low-frequency coding variation. Genome Biol. 2011;12(9):R84. doi: 10.1186/gb-2011-12-9-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tennessen JA, Bigham AW, O'Connor TD, et al. Broad GO; Seattle GO; NHLBI Exome Sequencing Project. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337(6090):64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cole JW, Stine OC, Liu X, et al. Rare variants in ischemic stroke: an exome pilot study. PLoS One. 2012;7(4):e35591. doi: 10.1371/journal.pone.0035591. doi:10.1371/journal.pone.0035591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams HP, Jr, Bendixen BH, Kappelle LJ, et al. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke. 1993;24(1):35–41. doi: 10.1161/01.str.24.1.35. [DOI] [PubMed] [Google Scholar]
- 20.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89(1):82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83(3):311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.R Core Team . R: a language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: [Accessed June 1, 2012]. 2012. http://www.R-project.org/ [Google Scholar]
- 23.Lange LA, Hu Y, Zhang H, et al. NHLBI Grand Opportunity Exome Sequencing Project. Whole-exome sequencing identifies rare and low-frequency coding variants associated with LDL cholesterol. Am J Hum Genet. 2014;94(2):233–245. doi: 10.1016/j.ajhg.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gretarsdottir S, Thorleifsson G, Reynisdottir ST, et al. The gene encoding phosphodiesterase 4D confers risk of ischemic stroke. Nat Genet. 2003;35(2):131–138. doi: 10.1038/ng1245. [DOI] [PubMed] [Google Scholar]
- 25.Meschia JF, Brott TG, Brown RD, Jr, et al. SWISS Study Group; ISGS Study Group; MSGD Study Group. Phosphodiesterase 4D and 5-lipoxygenase activating protein in ischemic stroke. Ann Neurol. 2005;58(3):351–361. doi: 10.1002/ana.20585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bevan S, Porteous L, Sitzer M, Markus HS. Phosphodiesterase 4D gene, ischemic stroke, and asymptomatic carotid atherosclerosis. Stroke. 2005;36(5):949–953. doi: 10.1161/01.STR.0000162713.06519.41. [DOI] [PubMed] [Google Scholar]
- 27.Staton JM, Sayer MS, Hankey GJ, et al. Association between phosphodiesterase 4D gene and ischaemic stroke. J Neurol Neurosurg Psychiatry. 2006;77(9):1067–1069. doi: 10.1136/jnnp.2006.092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Worrall BB, Mychaleckyj JC. PDE4D and stroke: a real advance or a case of the emperor's new clothes? Stroke. 2006;37(8):1955–1957. doi: 10.1161/01.STR.0000234048.04053.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon D, Park SK, Kang D, Park T, Park JW. Meta-analysis of homogeneous subgroups reveals association between PDE4D gene variants and ischemic stroke. Neuroepidemiology. 2011;36(4):213–222. doi: 10.1159/000327915. [DOI] [PubMed] [Google Scholar]
- 30.Bamshad MJ, Shendure JA, Valle D, et al. Centers for Mendelian Genomics. The Centers for Mendelian Genomics: a new large-scale initiative to identify the genes underlying rare mendelian conditions. Am J Med Genet A. 2012;158A(7):1523–1525. doi: 10.1002/ajmg.a.35470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee Y, Gamazon ER, Rebman E, et al. Variants affecting exon skipping contribute to complex traits. PLoS Genet. 2012;8(10):e1002998. doi: 10.1371/journal.pgen.1002998. doi:10.1371/journal.pgen.1002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willett WC, Ascherio A. Trans fatty acids: are the effects only marginal? Am J Public Health. 1994;84(5):722–724. doi: 10.2105/ajph.84.5.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smit LA, Katan MB, Wanders AJ, Basu S, Brouwer IA. A high intake of trans fatty acids has little effect on markers of inflammation and oxidative stress in humans. J Nutr. 2011;141(9):1673–1678. doi: 10.3945/jn.110.134668. [DOI] [PubMed] [Google Scholar]
- 34.Mozaffarian D, Katan MB, Ascherio A, Stampfer MJ, Willett WC. Trans fatty acids and cardiovascular disease. N Engl J Med. 2006;354(15):1601–1613. doi: 10.1056/NEJMra054035. [DOI] [PubMed] [Google Scholar]
- 35.Lopez-Garcia E, Schulze MB, Meigs JB, et al. Consumption of trans fatty acids is related to plasma biomarkers of inflammation and endothelial dysfunction. J Nutr. 2005;135(3):562–566. doi: 10.1093/jn/135.3.562. [DOI] [PubMed] [Google Scholar]
- 36.Kromhout D, Menotti A, Bloemberg B, et al. Dietary saturated and trans fatty acids and cholesterol and 25-year mortality from coronary heart disease: the Seven Countries Study. Prev Med. 1995;24(3):308–315. doi: 10.1006/pmed.1995.1049. [DOI] [PubMed] [Google Scholar]
- 37.Hunt MC, Rautanen A, Westin MA, Svensson LT, Alexson SE. Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional evolution results in a reduced number of human peroxisomal ACOTs. FASEB J. 2006;20(11):1855–1864. doi: 10.1096/fj.06-6042com. [DOI] [PubMed] [Google Scholar]
- 38.Westin MA, Hunt MC, Alexson SE. The identification of a succinyl-CoA thioesterase suggests a novel pathway for succinate production in peroxisomes. J Biol Chem. 2005;280(46):38125–38132. doi: 10.1074/jbc.M508479200. [DOI] [PubMed] [Google Scholar]
- 39.van Bilsen M, van Nieuwenhoven FA. PPARs as therapeutic targets in cardiovascular disease. Expert Opin Ther Targets. 2010;14(10):1029–1045. doi: 10.1517/14728222.2010.512917. [DOI] [PubMed] [Google Scholar]
- 40.Kilarski LL, Achterberg S, Devan WJ, et al. GARNET Collaborative Research Group, Wellcome Trust Case Control Consortium 2, Australian Stroke Genetic Collaborative, the METASTROKE Consortium, and the International Stroke Genetics Consortium. Meta-analysis in more than 17,900 cases of ischemic stroke reveals a novel association at 12q24.12. Neurology. 2014;83(8):678–685. doi: 10.1212/WNL.0000000000000707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.