Abstract
ARID1A is a subunit of the Switch/Sucrose Non-Fermentable (SWI/SNF) chromatin-remodeling complex that regulates gene expression by controlling gene accessibility. ARID1A shows one of the highest mutation rates across different human cancer types. For example, ARID1A is mutated in ~50% of ovarian clear cell carcinoma (OCCC). There is considerable interest in developing cancer therapeutics that correlate with ARID1A mutational status. A recent study demonstrated a synthetic lethality by targeting EZH2 histone methyltransferase activity in ARID1A-mutated OCCC using a clinically applicable small molecule inhibitor. The observed synthetic lethality correlated with inhibition of PI3K/AKT signaling. In addition, there is evidence indicating that ARID1A-mutated cancer may also be subjected to therapeutic intervention by targeting residual SWI/SNF activity, the PI3K/AKT pathway, the DNA damage response, the tumor immunological microenvironment and stabilizing wild-type p53. In summary, we propose EZH2 inhibitor-based combinatorial strategies for targeting ARID1A-mutated cancers.
1. Introduction
The Switch/Sucrose Non-Fermentable (SWI/SNF) complex regulates gene transcription through its ATP-dependent chromatin-remodeling activity. Consistent with its role in gene transcription, the SWI/SNF complex is involved in essential cellular processes such as transformation, development, DNA damage repair and cell cycle regulation. The SWI/SNF complex has garnered substantial attention because subunits of the complex are collectively mutated in >20% of human cancers 1. Among the SWI/SNF subunits, ARID1A has highest mutation rate in human cancers. ARID1A is mutated in ~50% of ovarian clear cell carcinoma (OCCC) 2, 3. ARID1A mutations are typically nonsense or frame-shift, which cause loss of ARID1A protein expression. Further highlighting the importance of the SWI/SNF complex in human cancer, several studies have found correlations between the mutational and/or expressional status of SWI/SNF complex subunits and tumor progression, prognosis and response to chemotherapy. These findings have raised considerable interest in developing targeted therapies that take advantage of ARID1A mutations.
Here we will highlight advances in identifying therapeutic targets for ARID1A-mutated cancers and discuss additional therapeutic targets based on newly gained mechanistic insights into ARID1A’s role in cancer.
2. Epigenetic synthetic lethality by targeting EZH2 in ARID1A-mutated cancers
A recent study examined epigenetic inhibitors that selectively suppress the growth of ARID1A-deficient compared with proficient OCCC cells. The authors identified an inhibitor of EZH2, a histone methyltransferase, which selectively promotes apoptosis in ARID1A-mutated OCCC cells 4. EZH2, a subunit of the polycomb complex, represses gene transcription. Mechanistically, the observed synthetic lethality is due to the antagonistic roles played by ARID1A and EZH2 in regulate the expression of ARID1A/EZH2 target genes. Similar epigenetic antagonism exists between EZH2 and SNF5, a core subunit of the SWI/SNF complex that is often deleted in childhood rhabdoid tumors 5. Consistently, inhibition of EZH2 causes regression of SNF5-deleted rhabdoid tumors 5. In addition, mutations of SMARCA4, the gene encoding the ATPase SWI/SNF subunit BRG1, in non-small lung cancer increases sensitivity to a combinatorial EZH2 and topoisomerase inhibition 6.
EZH2 inhibitors are in clinical development for hematopoietic malignancies such as diffuse large B cell lymphoma. These recent findings indicate that targeting EZH2 using a clinically applicable EZH2 inhibitor represents a strategy for cancers with mutations in subunits of the SWI/SNF complex. Further studies are warranted to determine whether EZH2 inhibition displays a similar selectivity in cancers with genetic inactivation of other SWI/SNF subunits and whether the selectivity of EZH2 inhibition in ARID1A-mutated cancer cells is tissue- and/or genetic context-dependent.
3. Targeting residual SWI/SNF complex as a specific vulnerability in ARID1A-mutated cancers
The observation that knockdown of SWI/SNF’s catalytic subunit, BRG1, inhibits the growth of SNF5-deficient rhabdoid tumors suggests that the survival of SNF5-deficient tumors depends upon residual SWI/SNF complex activity 7. Residual SWI/SNF complex dependence was further validated by examining mutually exclusive SWI/SNF subunits. Specifically, ARID1A and ARID1B are mutually exclusive in their association with the SWI/SNF complex. The survival of ARID1A-mutated cancer cells depends upon the presence of ARID1B in the residual SWI/SNF complex 8. Likewise, BRG1 and BRM1 (encoded by the SMARCA2 gene) are mutually exclusive subunits of the SWI/SNF complex and survival of SMARCA2-mutated cells depends upon the residual BRG1-containing complex 9. Conversely, knockdown of BRM1 selectively suppresses the growth of BRG1-deficient cells. Collectively, these findings raise the possibility of targeting residual SWI/SNF complex based on mutual exclusivity of different subunits. The challenge of this potential approach lies in the development of a subunit-specific inhibitor given the structure and functional similarities.
4. Targeting mutual exclusivity between TP53 and ARID1A mutation
Genetic profiling of ARID1A-mutated ovarian cancer reveals enrichment of wild-type TP53 in these tumors 10. Functional characterization reveals that ARID1A and p53 function in the same pathway to regulate the expression of p53 target genes 10. It is therefore possible that stabilization of wild-type p53 might be sufficient to overcome the effects of ARID1A loss and reactivate p53 target tumor suppressor genes. Notably, Nutlin 3, a p53 stabilizer, suppresses the growth of ARID1A-mutated A2780 ovarian cancer cells 11. Clinically applicable p53-stabilizers have been developed 12. Although it is unlikely that a p53-stabilizer itself will have substantial clinical activity, it provides a target for combinational therapies.
5. Targeting PI3K/AKT Signaling in ARID1A-mutated cancers
ARID1A mutation often co-exists with genetic alterations that lead to activation of the PI3K/AKT pathway. These include gain-of-function mutations in the PIK3CA oncogene in OCCC or inactivation of the tumor suppressor PTEN in ovarian endometrioid carcinoma (OEC). In an immunohistochemical analysis of OCCC tumors, loss of nuclear ARID1A expression correlated to an increase in AKT phosphorylation 13. Combination of conditional inactivation of ARID1A with activation of PIK3CA or inactivation of PTEN drives the development of OCCC and OEC, respectively 14. PIK3IP1, an inhibitor of PI3K/AKT, plays a major role in the observed synthetic lethality between ARID1A mutation and EZH2 inhibition 4. ARID1A-mutated cells are more sensitive to PI3K/AKT inhibitors compared with ARID1A wild-type cells 4, 15.. Notably, inhibitors of mTOR, the downstream effector activated by PI3K/AKT signaling, such as temsirolimus and everolimus, are now in clinical trials for OCCC. The single-agent inhibition of PI3K/AKT is likely not sufficient to eradicate the disease. Consistently, in an ARID1A/PIK3CA mouse model of OCCC, an inhibitor of PI3K only increased survival by 3.5 weeks 14.
6. Targeting the SWI/SNF-dependent DNA damage response in ARID1A-mutated cancers
In addition to modulating signaling through gene regulation, the SWI/SNF complex is implicated in DNA damage repair. SWI/SNF complexes often localize to sites of DNA double-strand breaks (DSB) and facilitate phosphorylation of histone H2AX via ATM/ATR 16. Thus, SWI/SNF mutated cancers could be sensitive to DNA damage-inducing chemotherapeutics. Counter intuitively, ARID1A-mutated OCCC typically lacks genomic instability, and OCCC tumors are less responsive to DNA damage inducing platinum-based chemotherapy. This suggests that the role of the SWI/SNF complex in DNA damage might be subunit- and/or tissue dependent.
7. Targeting tumor microenvironment in ARID1A-mutated cancers
The link between chronic inflammation and carcinogenesis is well-characterized and is a hallmark of cancer. Chronic inflammation and expression of pro-inflammatory cytokines (e.g. IL6) are important for escape from anti-tumor immune responses. Recent evidence suggests that ARID1A protects against inflammation-driven tumorigenesis 14. In a mouse model, ARID1A loss and PIK3CA mutation cooperate to promote OCCC through sustained IL6 production. Subsequently, IL6 knockdown resulted in significantly smaller tumors, indicating the potential for anti-IL6 therapies in ARID1A-mutated cancers. Given the recent success in targeting immune checkpoints, it will be interesting to evaluate the impact of ARID1A mutation on anti-tumor immunity and whether ARID1A-mutated cancers are sensitive to reactivation of anti-tumor immunity.
8. Conclusion
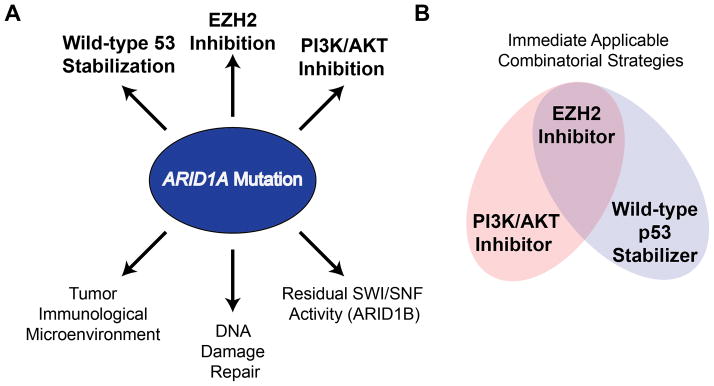
Recent genome-wide sequencing studies have revealed frequent ARID1A mutations in a variety of cancer types. Clinical and pathological studies suggest a great need to develop precision therapy that correlates with ARID1A mutational status. In this review, we discussed literature on therapeutic targets with the potential of specifically and selectively targeting ARID1A-mutated cancers (Figure 1A). They include EZH2, residual SWI/SNF activity, the PI3K/AKT pathway, the DNA damage response, the tumor immunological microenvironment and stabilizing wild-type p53. The synthetic lethality between EZH2 inhibition and ARID1A mutation presents a unique opportunity for developing novel combination therapeutic strategies that correlate with ARID1A mutation, the very definition of precision medicine.
Figure 1. Potential Therapeutic Targets in ARID1A-Mutated Cancer.
ARID1A-containing SWI/SNF chromatin remodeling-complex regulates multiple biological processes related to tumor suppression. A) ARID1A mutation and/or loss of expression leads to atypical signaling and cellular functions. In ARID1A-mutated cancers, the indicated pathways are potential targets that are selective against ARID1A mutation. B) To achieve a sustained clinical response, combinatorial therapies will be necessary. An EZH2 inhibitor-based approach presents a unique opportunity for combinatorial strategies.
9. Expert Opinion
EZH2 inhibition is synthetic lethal with ARID1A mutation and causes the regression of established ARID1A-mutated OCCC in vivo. These findings indicate that the newly discovered synthetic lethality between ARID1A mutation and EZH2 inhibition could be developed as an urgently needed therapeutic for ARID1A-mutated OCCC. It will be important to investigate whether this approach can be extended into other cancers with ARID1A mutation. Since EZH2 inhibition has also been shown to inhibit the growth of SNF5-deficient rhabdoid tumors, it will be interesting to determine whether EZH2 inhibition-based synthetic lethality extends to mutations in other SWI/SNF complex subunits.
Despite the well-described advantages of selectivity and limited toxicity of targeted cancer therapy, clinical trials have extensively demonstrated that targeted therapy, including synthetic lethality-based therapy, often leads to the development of resistance and is not sufficient to eradiate cancer. Combinational therapeutic strategies offer a solution for this major clinical challenge. Clinically applicable drugs that target EZH2, stabilize wild-type p53 or inhibit PI3K/AKT signaling have already been developed. Based on the genetic makeup of ARID1A-mutated cancers such as OCCC, an EZH2 inhibitor in combination with a PI3K/AKT signaling inhibitor or wild-type p53 stabilizer may represent a therapeutic strategy that conveys a sustained clinical response (Figure 1B). Further studies are warranted to investigate potential side effects and pharmacodynamics of these proposed combinatorial approaches. In the long-term, given the recent evidence that ARID1A suppresses tumor-promoting inflammation, it will be interesting to explore EZH2 inhibition in combination with reagents that target the tumor immunological microenvironment.
Acknowledgments
This work was supported by US National Institutes of Health/National Cancer Institute grants (R01CA160331 and R01CA163377 to R.Z.), a US Department of Defense ovarian cancer academy award (OC093420 to R.Z.) and an Ovarian Cancer Research Fund program project (to R.Z.). B.G.B. is supported by an American Cancer Society postdoctoral fellowship (PF-13-058-01-TBE). Support of Core Facilities was provided by Cancer Center Support Grant (CCSG) CA010815 to The Wistar Institute.
Footnotes
Declaration of Interest.
The study sponsors had no role in the design of the study, the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed above.
References
- 1.Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nature genetics. 2013 Jun;45(6):592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones S, Wang TL, Shih Ie M, Mao TL, Nakayama K, Roden R, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010 Oct 8;330(6001):228–31. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3*.Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010 Oct 14;363(16):1532–43. doi: 10.1056/NEJMoa1008433. References 2 and 3 are the first studies showing that ARID1A is mutated in ovarian clear cell carinoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4**.Bitler BG, Aird KM, Garipov A, Li H, Amatangelo M, Kossenkov AV, et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med. 2015 Mar;21(3):231–8. doi: 10.1038/nm.3799. This is the first study showing the synthetic lethality between EZH2 inhibition and ARID1A mutation in ovarian clear cell carcinoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson BG, Wang X, Shen X, McKenna ES, Lemieux ME, Cho YJ, et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell. 2010 Oct 19;18(4):316–28. doi: 10.1016/j.ccr.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fillmore CM, Xu C, Desai PT, Berry JM, Rowbotham SP, Lin YJ, et al. EZH2 inhibition sensitizes BRG1 and EGFR mutant lung tumours to TopoII inhibitors. Nature. 2015 Apr 9;520(7546):239–42. doi: 10.1038/nature14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Sansam CG, Thom CS, Metzger D, Evans JA, Nguyen PT, et al. Oncogenesis caused by loss of the SNF5 tumor suppressor is dependent on activity of BRG1, the ATPase of the SWI/SNF chromatin remodeling complex. Cancer Res. 2009 Oct 15;69(20):8094–101. doi: 10.1158/0008-5472.CAN-09-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helming KC, Wang X, Wilson BG, Vazquez F, Haswell JR, Manchester HE, et al. ARID1B is a specific vulnerability in ARID1A-mutant cancers. Nat Med. 2014 Mar;20(3):251–4. doi: 10.1038/nm.3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oike T, Ogiwara H, Tominaga Y, Ito K, Ando O, Tsuta K, et al. A synthetic lethality-based strategy to treat cancers harboring a genetic deficiency in the chromatin remodeling factor BRG1. Cancer Res. 2013 Sep 1;73(17):5508–18. doi: 10.1158/0008-5472.CAN-12-4593. [DOI] [PubMed] [Google Scholar]
- 10*.Guan B, Wang TL, Shih Ie M. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 2011 Nov 1;71(21):6718–27. doi: 10.1158/0008-5472.CAN-11-1562. This is the first study showing the enrichment of wild-type TP53 in ARID1A-mutated ovarian clear cell carcinoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meijer A, Kruyt FA, van der Zee AG, Hollema H, Le P, ten Hoor KA, et al. Nutlin-3 preferentially sensitises wild-type p53-expressing cancer cells to DR5-selective TRAIL over rhTRAIL. Br J Cancer. 2013 Nov 12;109(10):2685–95. doi: 10.1038/bjc.2013.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004 Feb 6;303(5659):844–8. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 13.Wiegand KC, Hennessy BT, Leung S, Wang Y, Ju Z, McGahren M, et al. A functional proteogenomic analysis of endometrioid and clear cell carcinomas using reverse phase protein array and mutation analysis: protein expression is histotype-specific and loss of ARID1A/BAF250a is associated with AKT phosphorylation. BMC cancer. 2014;14:120. doi: 10.1186/1471-2407-14-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandler RL, Damrauer JS, Raab JR, Schisler JC, Wilkerson MD, Didion JP, et al. Coexistent ARID1A-PIK3CA mutations promote ovarian clear-cell tumorigenesis through pro-tumorigenic inflammatory cytokine signalling. Nature communications. 2015;6:6118. doi: 10.1038/ncomms7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samartzis EP, Gutsche K, Dedes KJ, Fink D, Stucki M, Imesch P. Loss of ARID1A expression sensitizes cancer cells to PI3K- and AKT-inhibition. Oncotarget. 2014 Jul 30;5(14):5295–303. doi: 10.18632/oncotarget.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park JH, Park EJ, Lee HS, Kim SJ, Hur SK, Imbalzano AN, et al. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. EMBO J. 2006 Sep 6;25(17):3986–97. doi: 10.1038/sj.emboj.7601291. [DOI] [PMC free article] [PubMed] [Google Scholar]