Abstract
Spontaneous deamidation of asparagine is a non-enzymatic post-translational modification of proteins. Residue Asn 321 is the main site of deamidation of the Drosophila melanogaster Hox transcription factor Sex Combs Reduced (Scr). Formation of iso-aspartate, the major deamidation product, is detected by HNCACB triple-resonance NMR spectroscopy. The rate of deamidation is quantified by fitting the decay of Asn NH2 side-chain signals in a time-series of 15N-1H HSQC NMR spectra. The deamidated form of Scr binds to specific DNA target sequences with reduced affinity as determined by an electrophoretic mobility shift assay.
Keywords: asparagine deamidation, dissociation constant, DNA binding, Hox transcription factor, Sex Combs Reduced, NMR spectroscopy
Introduction
Asparagine (Asn) residues in proteins are prone to deamidation, isomerization, racemization, and peptide bond cleavage, under physiological conditions in the absence of enzymatic catalysis [1; 2]. Deamidation to form aspartate or iso-aspartate (isoAsp) is the most common Asn modification. Conversion of Asn to Asp or isoAsp introduces one additional negative charge into the protein. In addition, iso-aspartate forms a non-native β-peptide backbone linkage through the γ-carboxylate, rather than the α-carboxylate [3]. At pH > 5, deprotonation of the backbone amide nitrogen (NH) carboxyl to an Asn residue generates a nucleophile that attacks the side chain carbonyl group to yield a cyclic tetrahedral intermediate. Subsequent elimination of ammonia yields a succinimide intermediate. Hydrolysis of this intermediate on either side of the cyclic imide nitrogen yields isoAsp or Asp with an isoAsp:Asp ratio of 3:1 [1]. The deamidation reaction rate is sensitive primarily to the identity of the amino acid residue that is C-terminal to the Asn residue, Asn-Xxx, in which Xxx is any residue but proline, with the fastest rate observed for Xxx = Gly, because the amino acid side chain of other residues sterically hinders encounter between the Asn side chain and the backbone amide nitrogen of residue Xxx [4; 5]. Deamidation also is fastest for Asn in labile regions of proteins and interactions that hinder mobility, such as tertiary structure, backbone rigidity, and hydrogen bonding, further reduce the reaction rate. Glutamine (Gln) residues are also susceptible to deamidation; however, the reaction usually is much slower and proceeds through a slightly different mechanism than for Asn [6]. Deamidation may have effects on protein structure and function by altering the protein backbone conformation through introduction of the β-peptide linkage, with an additional rotatable dihedral angle, and introduction of a negative charge. Deamidation has been associated with fibril formation [7; 8], age-related changes in crystalline proteins [9; 10], and targeted protein degradation [11].
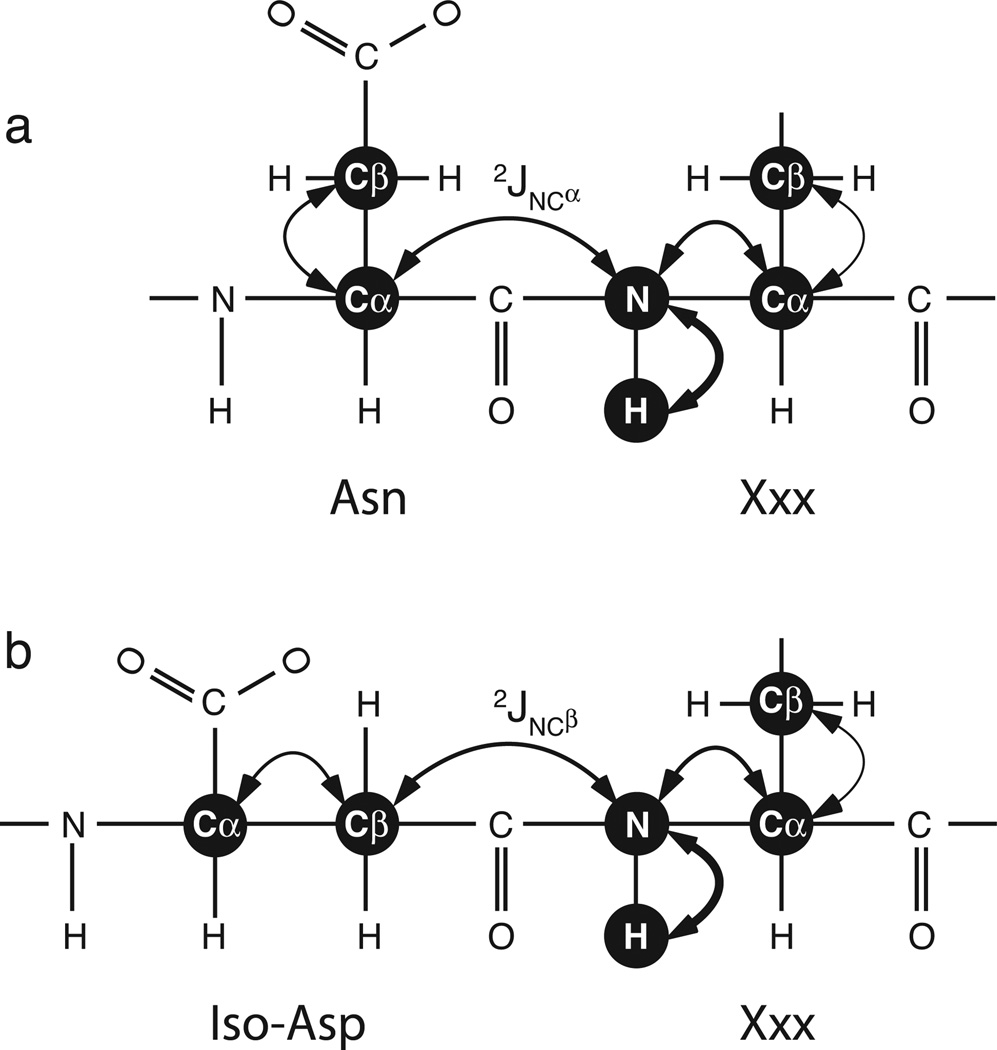
Chazin and coworkers used NMR spectroscopy to identify isoAsp as a result of deamidation of Asn56 in calbindin D9k by means of 1H-1H nuclear Overhauser effects (NOE) [12]. This approach requires careful assessment of the intensities of NOESY crosspeaks characteristic of normal α-linked and variant β-linked peptide bonds. More recently, Kay and coworkers [13] demonstrated that the HNCACB experiment [14] allows facile identification of isoAsp-Xxx peptide bonds, without the difficulties posed by quantifying NOESY cross peaks, because the signs of the inter-residue Cαi−1 and Cβi−1 resonance peaks are inverted relative to the intra-residue Cαi and Cβi resonance peaks in the N-H plane for the residue carboxyl to the site of deamidation. The coherence transfer pathways in the HNCACB experiment for normal α-linked and variant β-linked peptide bonds are shown in Figure 1.
Figure 1.
Coherence transfer for the HNCACB pulse sequence for (a) Asn-Xxx and (b) isoAsp-Xxx dipeptide moieties. The out-and-back coherence transfer steps are shown as double-ended arrows. The sequential transfers “out” from the HN spin are indicated by arrows of decreasing thickness and vice versa. The critical difference between the Asn-Xxx and isoAsp-Xxx results from transfer through the (a) 2JNCα or (b) 2JNCβ scalar coupling interactions, respectively.
Herein, the HNCACB experiment is used to detect deamidation of an Asn residue at position 323 in the Drosophila melanogaster Hox transcription factor, Sex Combs Reduced (Scr; Fig. 2). The rate of deamidation is quantified by monitoring the decay of NH2 resonances in a time series of fHSQC spectra [15]. The approach definitively identifies isoAsp linkages, and other β-peptide moieties, and extends the application of NMR spectroscopy to the quantification of deamidation rates in proteins. As shown by an electrophoretic mobility shift assay, deamidation of Scr reduces its affinity for target DNA sequences in the presence of the cofactors Extradenticle and the HM isoform of Homothorax (Exd-HM).
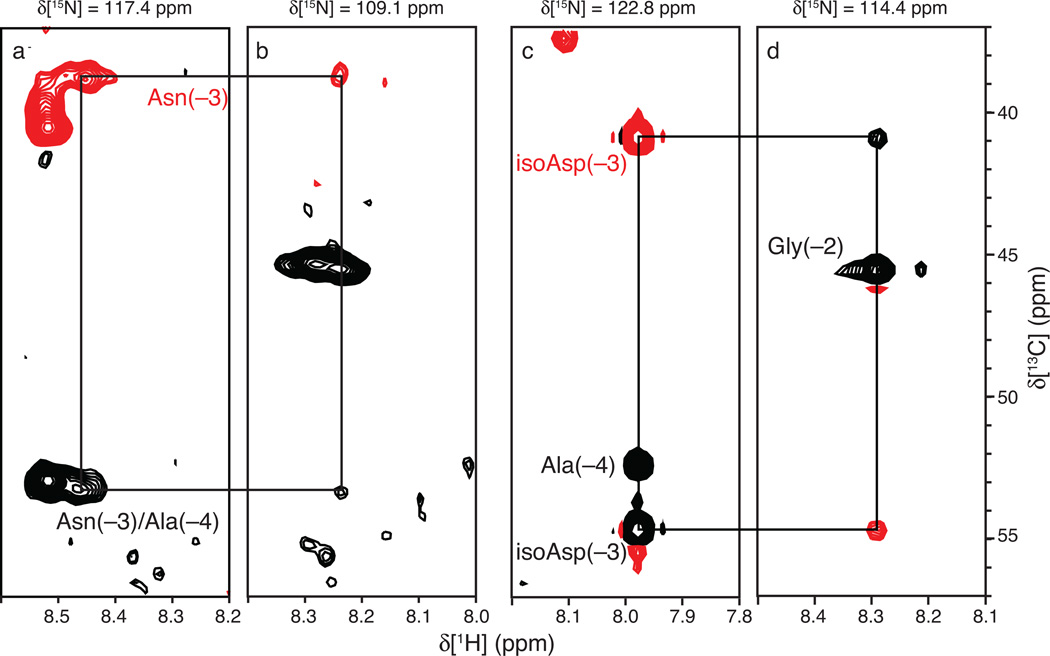
Figure 2.
HNCACB spectra of (a, b) fresh and (c, d) aged Scr. Strip plots are shown for the indicated backbone N-H planes of (a) Asn(−3) and (b) Gly(−2) for a fresh Scr sample and of (c) isoAsp(−3) and (d) Gly(−2) for an aged sample. Positive (negative) cross-peaks are shown as black (red) contours. Black (red) labels denote 13Cδ (13Cδ) resonances. The 13Cδ resonances are degenerate for Asn(−3) and Asn(−4) in the fresh Scr sample. The signs of the inter-residue isoAsp(−3) Cδ and Cδ cross peaks in the aged sample are inverted in the Gly(−2) N-H plane when compared to the signs of the corresponding cross peaks in the fresh sample. This phenomenon arises from the β-linked peptide backbone that is the characteristic feature of isoAsp.
Materials and Methods
Sample preparation
The protein used for NMR spectroscopy consisted of the Scr N-terminal extension and homeodomain, residues 298–385 and incorporated a Cys362Ser mutation. This construct is numbered from −26 to 62 so that residue +1 corresponds to the initial residue of the Scr homeodomain, Thr 324 (in this numbering, Asp 321 is denoted Asp (−3) and Glu 327 is denoted Glu 4). The mutant Asn (−3) to Asp mutant Scr was made by QuikChange mutagenesis (Stratagene). Scr was expressed from a pET-15b plasmid in E. coli BL21 (DE3) pLysS cells and purified by column chromatography, as described elsewhere [16], except that in some preparations, all purification steps were performed at T = 277 K and pH 7.0 to reduce the rate of deamidation. The His-tag was cleaved from the expressed protein using thrombin. [U-13C, U-15N] and [U-15N] Scr was prepared in M9 minimal media prepared with 13C6 glucose and 15NH4Cl as necessary. NMR samples were 1 mM Scr in 10 mM HEPES buffer (pH 7.0) and 50 mM NaCl. Samples were analyzed by mass spectrometry to verify molecular weight and purity. Aged samples were left at room temperature for three weeks prior to assay.
NMR spectroscopy
All experiments were performed at T = 300 K on a Bruker DRX500 and DRX600 NMR spectrometers equipped with a cryogenically cooled triple-resonance probe. All spectra were processed in NMRpipe [17] and analyzed in Sparky [18]. Standard gradient-enhanced HNCACB spectra [14; 19] were acquired with 128 × 36 × 1024 complex points (13C × 15N × 1H) on the DRX500 spectrometer for the fresh (non-deamidated) Scr sample and with 100 × 32 × 512 complex points (13C × 15N × 1H) on the DRX600 spectrometer for the aged (deamidated) protein sample. An HNCACB spectrum optimized for correlations to NH2 spin systems [20] was acquired on the DRX600 spectrometer with 128 × 36 × 1024 complex points (13C × 15N × 1H) for the assignment of Asn and Gln side chain amide moieties. Constant time 15N interferograms were extended by mirror-image linear prediction prior to apodization and Fourier transformation. Lorentzian-to-Gaussian apodization was used for 1H and Kaiser apodizations were used for 15N and 13C dimensions. Data were zero-filled twice in all dimensions. To measure the deamidation rate, fHSQC spectra [15] were collected on the DRX600 spectrometer at 5 h intervals for 5 days using 128 × 512 complex points for the 15N × 1H dimensions. Lorentzian-to-Gaussian apodization was used for 1H and Kaiser apodization was used for 15N dimensions. Data were zero-filled twice in all dimensions prior to Fourier transformation. Intensity decays were fit to single exponential functions to determine rate constants for deamidation using CurveFit (http://www.palmer.hs.columbia.edu/software/curvefit.html). Uncertainties in rate constants were estimated from jackknife simulations.
DNA affinity
Binding affinities of fresh wild-type, aged wild-type, and mutant Scr were measured by electrophoretic mobility shift assay (EMSA). Reaction conditions used 86.5 pM DNA target sequence and 210 nM Exd-HM. The target sequence was the 40 base pair Fkh[250Con] (5’- GATCTCAATGTCAAGATTTATGGCCAGCTGTGGGACGAGG-3’) target site. Binding was carried out in 10 mM Tris pH 7.5, 50 mM NaCl, 1 mM MgCl2, 4% glycerol, 5 mM DTT, 0.5 mM EDTA, 0.2 mg/mL BSA and 50 mM poly(dI-dC) at room temperature for 20 min. The extent of binding was assayed by gel shift on a 4% non-denaturing polyacrylamide gel run at a constant voltage of 100 V for approximately 1 hour. Gels imaged using a Typhoon Phosphorimager (Amersham).
Results
Strip plots from the HNCACB spectrum for the N-H plane of isoAsp(−3) and Gly(−2) are shown in Figure 2. As predicted by the coherence transfer pathways in Fig. 1, the isoAsp(−3) Cαi−1 and Cβi−1 crosspeaks are inverted in the Gly(−2) N-H plane. With the exception of the sign inversion, backbone resonance assignments were established in the usual approach using tripleresonance NMR spectra (not shown); most assignments are transferable from the unmodified species, because the reaction typically only perturbs the chemical shifts of residues in the vicinity of the affected Asn [12].
Although sequence specific assignments of Scr were obtained herein, the HNCACB spectrum can be used to detect deamidation without resonance assignments. In such cases, the probable sites of deamidation can be inferred by estimating the reaction half-time from primary sequence and 3D structure, if available, using the approach of Robinson et al. [4; 5]. For Scr, the Asn(−3)-Gly(−2) motif has a half-time predicted from sequence to be t1/2 = 1.05 days, while the next most labile site, Asn(−5)-Ala(−4), has a predicted t1/2 = 27.5 days at pH 7.4 and T = 310 K (Table 1). Thus, consistent with the resonance assignments, the predicted rates would suggest that Asn −3 is the likely site of deamidation. The difference between predicted and observed rates, presumably reflecting local structural restrictions in Scr, emphasizes that predictions in the absence of sequence assignments should be confirmed by site-directed mutagenesis.
Table 1.
Deamidation rate for side chain NH2 groups in Scr
| Residue | Rate (day−1) | t1/2 (day) | Sequence | Predicted t1/2 (day) |
|---|---|---|---|---|
| Asn −5 | (2.24 ± 1.44) × 10−3 | 310 ± 200 | VNA | 27.5 |
| Asn −3 | (2.68 ± 0.14) × 10−2 | 26 ± 1 | ANG | 1.05 |
| Gln 4 | (6.80 ± 1.20) × 10−3 | 102 ± 18 | RQR | 2300 |
| Gln 12 | (1.44 ± 0.83) × 10−3 | 483 ± 279 | YQT | 5600 |
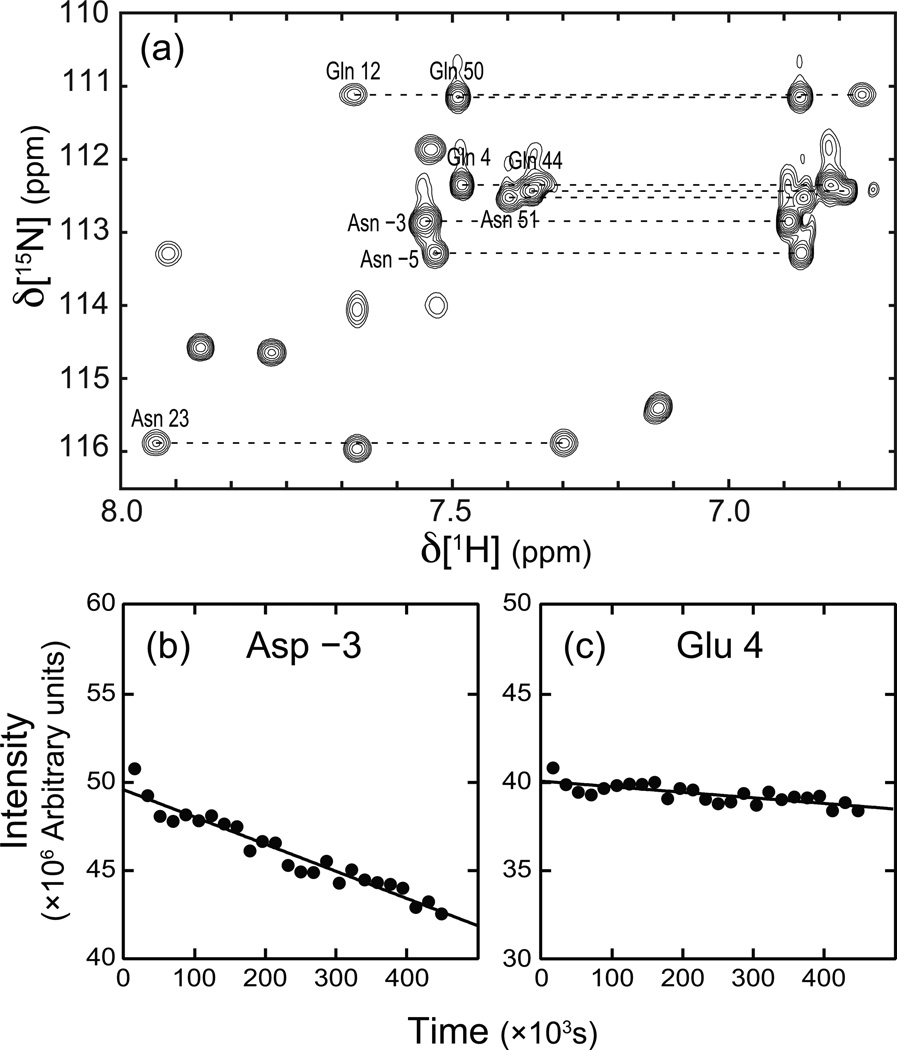
The deamidation rate is calculated for the sum of the intensities of the two side chain NH2 peaks for each residue in Fig. 4a. The predicted t1/2 for the flanking tripeptide sequence (in single letter amino acid code) is derived from Robinson et al. [4; 5]. Fitted deamidation rates for Asn 23, Gln 44, Gln 50, and Asn 51 are < 1.25 × 10−3day−1 and are not significantly different from zero (not shown).
The rate of deamidation in Scr was determined experimentally by monitoring the intensities of the NH2 resonance signals in a time-series of 1H-15N HSQC spectra recorded over a period of five days, using a freshly prepared 15N-labeled sample. Resonance assignments of the NH2 moieties were obtained using an NH2-HNCACB NMR experiment tuned for coherence transfer in secondary amide groups [20] together with backbone assignments derived from the conventional HNCACB spectrum. The resonance assignments for the resolvable resonances are shown in Figure 3. The signal intensities for selected side chain NH2 resonances are plotted as a function of time in Figure 4. The fitted decay rate constants are given in Table 1. Resonance intensities for only Asn −3 and Gln 4 NH2 side chains exhibit the largest decays over the course of the experiment. The significantly different values of t1/2 obtained experimentally, compared with the prediction from the amino acid sequence, reflects tertiary structural effects on the deamidation reaction in Scr.
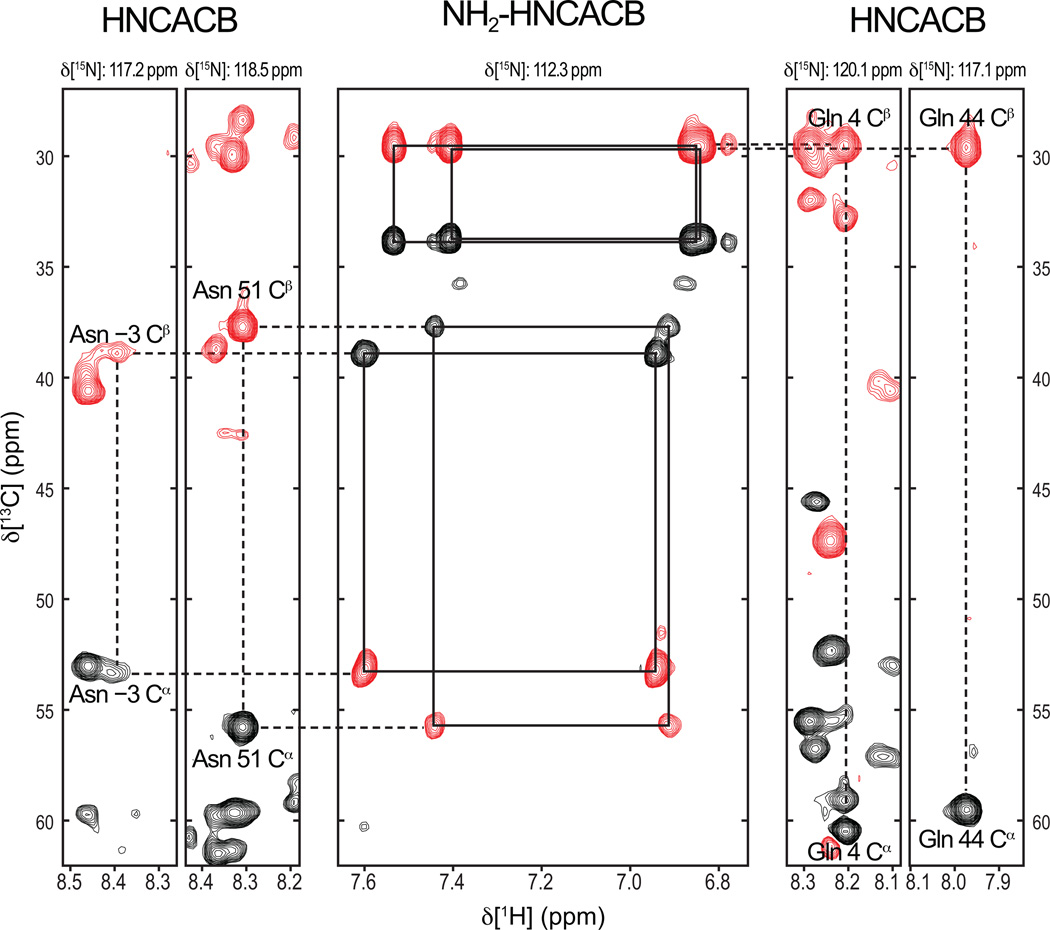
Figure 3.
Assignments of Asn and Gln residues in Scr. Resonance assignments were obtained from conventional HNCACB spectra (left and right panels) and an NH2-HCACB spectrum with coherence transfer delays tuned for NH2 rather than NH moieties. Positive (negative) cross-peaks are shown as black (red) contours. Planes are taken at the indicated 15N resonance positions. The center panel is at a 15N frequency that is midpoint between the 15Nδ of Asn −3 and 15Nδ of Gln 4
Figure 4.
Rate of deamidation Scr. (a) 15NH2 region of the initial 15N-1H HSQC spectrum acquired for Scr in the time course monitoring deamidation. (b-c) Peak intensity, given as the sum of the two 1H-15N amide resonance signals (connected by dashed lines), is plotted as a function of time for two of the eight resolvable side chain resonances. The fitted rate constants for deamidation are (b) (2.68 ± 0.14) × 10−2 day−1 for Asn −3 and (c) (6.80 ± 1.20) × 10−3 day−1 for Gln 4.
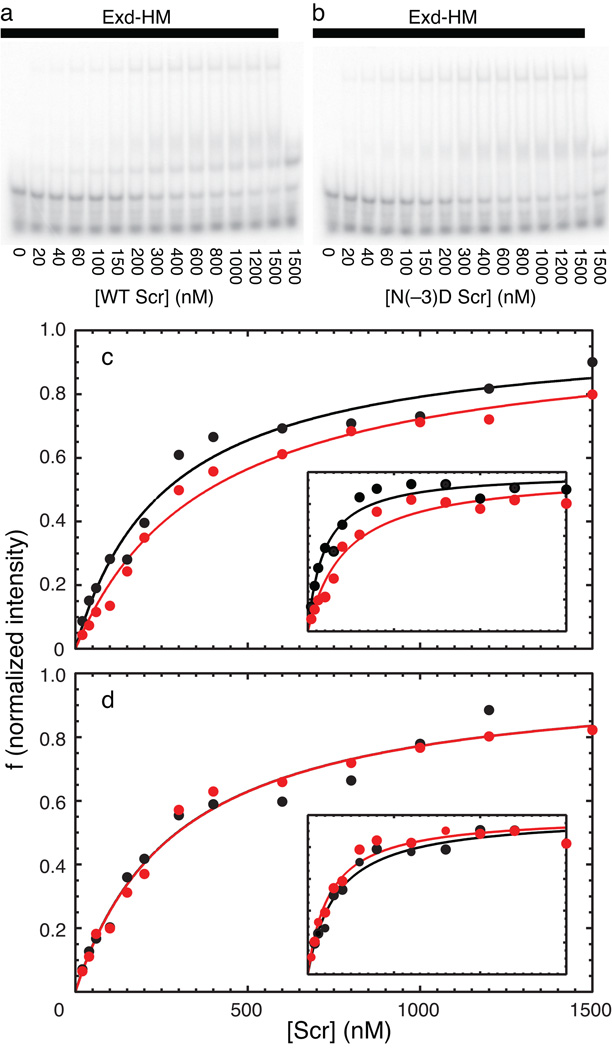
Asn(−3) is three residues N-terminal to the start of the Scr homeodomain (HD). This region of the protein lies between the sequence YPWM, which interacts with the Hox cofactor Extradenticle (Exd), and the start of the homeodomain. Although this portion of the protein is disordered in the two currently available X-ray crystal structures, residues on either side of Asn(−3), including His(−12) and Arg(+3) interact with the minor groove of the DNA (Joshi et al). Therefore, because of its proximity to the DNA, deamidation of Scr at Asn(−3) has the potential to influence DNA binding by alteration of the protein backbone conformation and introduction of a negative charge in the vicinity of the DNA phosphate backbone. The affinities of fresh (unmodified) and aged (partially deamidated) Scr for DNA in the presence and absence of the cofactors Exd-HM were measured by an electrophoretic mobility shift assay (EMSA). Examples of the EMSA analyses and graphs of the measured intensities of the shifted bands are plotted in Figure 5. The derived dissociation constants are given Table 2. Scr binds weakly to DNA in the absence of the Exd-HM cofactor, with values of the dissociation constant near the detection limit of the EMSA experiment. The differences between fresh and aged samples are not significant, consistent with absence of intermolecular interactions of the N-terminal arm with DNA in the absence of the cofactor Exd-HM. Scr binds approximately 3-fold more tightly to DNA in the presence of Exd-HM under these experimental conditions. The aged sample bound more weakly than the fresh sample by a factor of 1.4–2.3, depending upon the data analysis method, with greater inhibition of the most supershifted band. A mutant protein incapable of deamination was prepared by mutating position −3 from Asn to Asp, to mimic one of the reaction products. This sample has a slightly reduced affinity compared to wild-type Scr, but as expected is unaffected by aging.
Figure 5.
Binding isotherms for unmodified and modified Scr. Representative EMSAs for aged (a) wild-type and (b) Asn(−3) → Asp mutant Scr binding to DNA; EMSAs for fresh samples are not shown for brevity. Concentrations of Scr are indicated (nM); Exd-HM is present in all but last lane of each gel (solid bar). Integrated intensities of supershifted bands from EMSAs of (black) fresh or (red) aged (c) wild-type and (d) Asn(−3) → Asp mutant Scr binding to DNA/Exd-HM. The main graphs show results for the sum intensity of all supershifted bands and the insets show results only for the most supershifted (Scr-Exd-HM-DNA) band. Binding of Scr to DNA also was measured in the absence of Exd-HM (not shown). Fitted dissociation constants are given in Table 2.
Table 2.
Binding affinities of wild-type and mutant Scr
Discussion
HNCACB experiment has the advantage that the presence of the major reaction product, isoAsp, of deamidation of Asn residues is readily detected by the sign inversion of the interresidue resonance peaks for the amino acid residue C-terminal to the site of deamidation. The HNCACB experiment, in conjunction with a set of standard triple-resonance experiments, also provides sequence specific identification of the deamidated residue. The rate of deamidation is measured straightforwardly using a time series of 1H-15N correlation spectra to monitor signal loss from side chain NH2 resonances.
These approaches have been applied to deamidation at Asn 321 (and to a lesser extent Glu 327) in the linker region of the D. melanogaster transcription factor Scr. Deamidation of this residue results in weaker binding to target DNA sequences. Scr has a role in mediating segment identity in embryogenesis (day 0–1); additionally, the protein is involved in imaginal disk patterning during larval development (day 1–10) [21]. Whether the deamidation reaction contributes to physiological function of Scr or is a concern only for in vitro studies over long time periods, remains to be determined in future investigations. Interestingly, a closely related D. melanogaster homeodomain protein, Ubx, also has an Asn-Gly motif at the same position as the Asn(−3)-Gly(−2) dipeptide in Scr [22].
Highlights.
Drosophila melanogasterHox transcription factor Sex Combs Reduced (Scr) is spontaneously deamidated at residue Asn 321.
NMR spectroscopy is used to detect and quantify deamidation
Deamidation reduces the affinity of Scr for target DNA.
Acknowledgements
We acknowledge financial support from National Institutes of Health grants T32GM008281 (N.E.O), R01GM054510 (R.S.M), and GM50291 (A.G.P.). We thank Dr. Liping Sun for assistance with EMSA experiments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
RSM and AGP conceived and supervised the study; RSM and AGP designed experiments; NEO, and KL performed experiments; NEO, KL, RSM and AGP analyzed data; NEO, KL, RSM and AGP wrote the manuscript.
References
- 1.Geiger T, Clarke S. Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. Succinimide-linked reactions that contribute to protein degradation. J. Biol. Chem. 1987;262:785–794. [PubMed] [Google Scholar]
- 2.Wright HT, Urry DW. Nonenzymatic deamidation of asparaginyl and glutaminyl residues in proteins. Crit. Rev. Biochem. Mol. Biol. 1991;26:1–52. doi: 10.3109/10409239109081719. [DOI] [PubMed] [Google Scholar]
- 3.Capasso S, Di Donato A, Esposito L, Sica F, Sorrentino G, Vitagliano L, Zagari A, Mazzarella L. Deamidation in proteins: The crystal structure of bovine pancreatic ribonuclease with an isoaspartyl residue at position 67. J. Mol. Biol. 1996;257:492–496. doi: 10.1006/jmbi.1996.0179. [DOI] [PubMed] [Google Scholar]
- 4.Robinson NE, Robinson AB. Molecular clocks. Proc. Natl. Acad. Sci. U. S. A. 2001;98:944–949. doi: 10.1073/pnas.98.3.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson NE, Robinson AB. Prediction of protein deamidation rates from primary and three-dimensional structure. Proc. Natl. Acad. Sci. U. S. A. 2001;98:4367–4372. doi: 10.1073/pnas.071066498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson NE, Robinson ZW, Robinson BR, Robinson AL, Robinson JA, Robinson ML, Robinson AB. Structure-dependent nonenzymatic deamidation of glutaminyl and asparaginyl pentapeptides. J. Pept. Res. 2004;63:426–436. doi: 10.1111/j.1399-3011.2004.00151.x. [DOI] [PubMed] [Google Scholar]
- 7.Dunkelberger EB, Buchanan LE, Marek P, Cao P, Raleigh DP, Zanni MT. Deamidation accelerates amyloid formation and alters amylin fiber structure. J. Am. Chem. Soc. 2012;134:12658–12667. doi: 10.1021/ja3039486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwasa H, Meshitsuka S, Hongo K, Mizobata T, Kawata Y. Covalent structural changes in unfolded GroES that lead to amyloid fibril formation detected by NMR: insight into intrinsically disordered proteins. J. Biol. Chem. 2011;286:21796–21805. doi: 10.1074/jbc.M111.228445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hooi MY, Raftery MJ, Truscott RJ. Age-dependent deamidation of glutamine residues in human gammaS crystallin: deamidation and unstructured regions. Protein Sci. 2012;21:1074–1079. doi: 10.1002/pro.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moreau KL, King JA. Protein misfolding and aggregation in cataract disease and prospects for prevention. Trends Mol. Med. 2012;18:273–282. doi: 10.1016/j.molmed.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dho SH, Deverman BE, Lapid C, Manson SR, Gan L, Riehm JJ, Aurora R, Kwon KS, Weintraub SJ. Control of cellular Bcl-xL levels by deamidation-regulated degradation. PLoS Biol. 2013;11:e1001588. doi: 10.1371/journal.pbio.1001588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chazin WJ, Koerdel J, Thulin E, Hofmann T, Drakenberg T, Forsen S. Identification of an isoaspartyl linkage formed upon deamidation of bovine calbindin D9k and structural characterization by 2D proton NMR. Biochemistry. 1989;28:8646–8653. doi: 10.1021/bi00447a055. [DOI] [PubMed] [Google Scholar]
- 13.Tugarinov V, Muhandiram R, Ayed A, Kay LE. Four-dimensional NMR spectroscopy of a 723-residue protein: Chemical shift assignments and secondary structure of malate synthase G. J. Am. Chem. Soc. 2002;124:10025–10035. doi: 10.1021/ja0205636. [DOI] [PubMed] [Google Scholar]
- 14.Wittekind M, Mueller L. HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the alpha-carbon and beta-carbon resonances in proteins. J. Magn. Reson. Ser. B. 1993;101:201–205. [Google Scholar]
- 15.Mori S, Abeygunawardana C, Johnson MO, Vanzijl PCM. Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J. Magn. Reson. Ser. B. 1995;108:94–98. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- 16.Joshi R, Passner JM, Rohs R, Jain R, Sosinsky A, Crickmore MA, Jacob V, Aggarwal AK, Honig B, Mann RS. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007;131:530–543. doi: 10.1016/j.cell.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 18.Goddard TD, Kneeler DG. Sparky 3. San Francisco: University of California; 1999. [Google Scholar]
- 19.Cavanagh J, Fairbrother WJ, Palmer AG, Skelton NJ, Rance M. Protein NMR Spectroscopy: Principles and Practice. 1 edit. San Diego, CA: Academic Press; 2007. [Google Scholar]
- 20.Farmer BT, Venters RA. Assignment of aliphatic side-chain 1HN/15N resonances in perdeuterated proteins. J. Biomol. NMR. 1996;7:59–71. doi: 10.1007/BF00190457. [DOI] [PubMed] [Google Scholar]
- 21.Martinez-Arias A, Ingham P, Scott M, Akam M. The spatial and temporal deployment of Dfd and Scr transcripts throughout development of Drosophila. Development. 1987;100:673–683. doi: 10.1242/dev.100.4.673. [DOI] [PubMed] [Google Scholar]
- 22.Passner JM, Ryoo HD, Shen L, Mann RS, Aggarwal AK. Structure of DNA-bound Ultrabithorax-Extradenticle homeodomain complex. Nature. 1999;397:714. doi: 10.1038/17833. [DOI] [PubMed] [Google Scholar]