Abstract
The prevalence of Salmonella from chicken and pig slaughterhouses in Henan, China and antimicrobial susceptibility of these isolates to antibiotics was determined. From 283 chicken samples and 240 pig samples collected, 128 and 70 Salmonella isolates were recovered with an isolation rate of 45.2 and 29.2% respectively. The predominant serovars in chicken samples were S. enterica serovar Enteritidis, S. enterica serovar Hadar and S. enterica serovar Indiana, while those in pig samples were S. enterica serovar Typhimurium, S. enterica serovar Derby and S. enterica serovar Enteritidis. Resistance to ciprofloxacin was 8.6 and 10.0% for isolates from chickens and pigs respectively, whereas resistance to cefotaxime was 5.5 and 8.6%, respectively. Multidrug resistance (resistance to three or more classes of antimicrobial agent) was markedly higher in pig isolates (57.1%) than in chicken isolates (39.8%). Of particular concern was the detection of ciprofloxacin and cefotaxime co-resistant S. enterica serovar Indiana isolates, which pose risk to public health. All 16 S. enterica serovar Indiana isolates detected were resistant to ciprofloxacin, among which 11 were co-resistant to cefotaxime. The S. enterica serovar Indiana isolates accumulated point mutations in quinolone resistance determination regions of gyrA (S83F/D87G or S83F/D87N) and parC (T57S/S80R). Two plasmid mediated quinolone resistant determinants were found with aac (6')-Ib-cr and oqxAB in 16 and 12 S. enterica serovar Indiana isolates respectively. Cefotaxime-resistance of S. enterica serovar Indiana was associated with the acquisition of a bla CTX-M-65 gene. The potential risk of ciprofloxacin and cefotaxime co-resistant S. enterica serovar Indiana infection is a significant concern due to limited alternative treatment options. Reduction of Salmonella in chicken and pig slaughterhouses, in particular, ciprofloxacin and cefotaxime co-resistant S. enterica serovar Indiana will be an important measure to reduce the public health burden of Salmonella infections.
Introduction
Foodborne salmonellosis is an important public health problem worldwide [1]. There is an estimated 94 million annual cases of non-typhoidal Salmonella infections with 155,000 deaths and 85% of these cases are foodborne [2]. Salmonellosis is also one of the main causes of morbidity in China, accounting for 75% (30 million cases) of the foodborne diseases [3]. Children younger than 5-years were the group most affected by Salmonella-related infection in several cities in China [4,5,6]. Chickens and pigs are the main reservoirs of human foodborne non-typhoidal Salmonella infections. Epidemiological investigations have shown that the contaminated raw or undercooked chicken- and pig-meat was the primary vehicle for transmission to humans [7,8]. Fluoroquinolones and extended-spectrum cephalosporins are the most frequently used antimicrobial agents for treating invasive salmonellosis in humans, especially children and the elderly [1,5]. On the other hand, antimicrobials are also used for animal growth promotion and this application can select for and disseminate antimicrobial-resistant Salmonella. Multidrug-resistant (MDR) Salmonella strains that emerged in animal populations may pass from food animals to humans via the food chain [1]. Recent studies have shown that food-producing animals and retail food products might be important reservoirs of fluoroquinolone and/or extended-spectrum cephalosporin-resistant Salmonella which have already been reported in many countries [5,9]. Salmonella enterica serovar Indiana (S. enterica serovar Indiana) is one of the most frequently isolated Salmonella serovars from many sources [10,11]. And more importantly, the ciprofloxacin and ceftriaxone co-resistant S. enterica serovar Indiana have been isolated from retail chickens in Beijing and sick animals (ducks and pigs) in Gangdong province, China [10,12].
The prevalence of these co-resistant isolates could be amplified through cross-contamination during the slaughtering process [13]. However, limited data are available for the distribution and characteristics of ciprofloxacin and cefotaxime co-resistant Salmonella isolates from chicken and pig slaughterhouses.
In Salmonella, ciprofloxacin resistance is mainly attributed to mutations in gyrA and parC genes [14]. Plasmid-mediated quinolone resistance (PMQR) determinants such as qnrA, qnrB, qnrD, qnrS, and oqxAB may also contribute to ciprofloxacin resistance [12,15]. Resistance to cefotaxime or other extended spectrum beta-lactams is usually due to intracellular production of extended spectrum β-lactamases (ESBLs). The most commonly found ESBL types in Asia are the CTX-M group, which are usually located on transmissible plasmids that tend to disseminate among members of Enterobacteriacea [16]. In this study, Salmonella isolated from chicken and pig slaughterhouses were tested for their susceptibility to antimicrobial compounds commonly used in human medicine. The genetic basis of resistance to ciprofloxacin and cefotaxime resistance and the genetic relationships among these isolates were also determined.
Materials and Methods
Sample collection and Salmonella serotyping
From February to December in 2011, 283 whole chicken carcasses post cooling bath samples from chicken slaughterhouses with processing capacity of 10,000 ~50,000 chickens per day were collected from 2 cities (Hebi and Zhoukou) in Henan province, China. The chicken slaughterhouses were Qixian TIYA and Tangying YUDA in Hebi and Zhoukou TIYA and Shaizai TIYA in Zhoukou. For pig samples, 120 surface swabs of four sites of pig carcasses after evisceration and before chilling and 120 ileocaecal lymph nodes without fat or connective tissue samples from pig slaughterhouses with processing capacity of 2,000~3,000 pigs per day were collected from 2 cities (Hebi and Luohe) in Henan province, China. The pig slaughterhouses were Chengsheng and Xihui in Hebi and Luohe respectively. Both the chicken and pig slaughterhouses were the largest-scale slaughtering and process lines in the regions sampled. No more than five samples were collected from the same slaughterhouse per visit during the study. Each sample was put into an aseptic plastic bag, marked and shipped to the laboratory within 24 h on ice. These chickens and pigs were killed as part of the slaughterhouse routine. The whole chicken carcasses and ileocaecal lymph nodes from pigs were purchased during the project.
For whole chicken carcasses samples, each sample was immediately removed from the bag in which it was transported and then placed into a 3500 stomacher bag (Seward, UK) followed by the addition of 500 mL buffered peptone water (BPW; Becton-Dickinson, USA) per kilogram and thoroughly manually massaged for 3–5 minutes to ensure the surface, internal and external of the chicken carcass were fully in contact with the rinse. Then samples were incubated at 37°C for 22–26 h.
Surface swabs for sampling of the pig carcasses were performed after evisceration and before chilling. A surface of about 100 cm2 per site was swabbed using one single abrasive sponge for 4 sites (hind limb, abdomen, mid-dorsal region, jowl). The swabs were transferred to 100 mL BPW and incubated at 37°C for 18–20 h. For Lymph nodes samples from pigs, lymph nodes from each pig were removed and pooled, surface de-contaminated before analysis by dipping into 75% (v/v) alcohol and drying in air for 3~5 minutes. Twenty-five grams of lymph nodes were placed into a plastic bag with 225 mL BPW added and mechanistically homogenized by banging with a hammer and then incubated at 37°C for 18–20 h.
For all the samples, 0.5- and 0.1-mL of the pre-enrichment culture were transferred to 10 mL tetrathionate broth (TT; Becton-Dickinson, USA) and Rappaport-Vassiliadis (RV; Becton-Dickinson, USA) broth and incubated at 42±1°C with shaking at 100 rpm for 22–24 h. After selective enrichment, a loopful of TT or RV broth culture was streaked onto xylose lysine tergitol 4 (XLT4; Becton-Dickinson, USA) agar, and incubated at 37±1°C overnight. Three presumptive Salmonella colonies on each XLT4 plate were picked and inoculated onto a triple sugar iron slant (Becton-Dickinson, USA) and incubated at 35±1°C for 24 h. Isolates with typical Salmonella morphologies were confirmed by amplification of the invA gene by PCR [17] and the API 20E biochemical identification system (BioMérieux, Beijing, China). For all confirmed Salmonella isolates, serovars were determined by slide agglutination with commercial Salmonella antisera (Statens Serum Institute, Denmark) following the Kauffmann-White scheme.
Antimicrobial susceptibility testing
Antimicrobial susceptibility of all Salmonella isolates was determined by the agar dilution method and interpreted according to the Clinical and Laboratory Standards Institute guidelines (CLSI) with the current MIC breakpoints being used [18]: (compounds are abbreviated in parentheses; resistance breakpoints are given in square brackets): ampicillin (AMP [≥32mg/mL]), cefotaxime (CTX [≥4mg/mL]), ceftazidime (CAZ [≥16mg/mL]), chloramphenicol (CHL [≥32mg/mL]), ciprofloxacin (CIP [≥1mg/mL]), gentamicin (GEN [≥16mg/mL]), imipenem (IMP [≥4mg/mL]), nalidixic acid (NAL [≥32mg/mL]), tetracycline (TET [≥16mg/mL]), tigecycline (TGC [≥4mg/mL]) and trimethoprim-sulfamethoxazole (SXT [≥4/76mg/mL]). Multidrug resistance was defined as resistance to three or more classes of antimicrobial agent. Isolates with an MIC 1 mg/L for ceftriaxone or ceftazidime were further screened for extended-spectrum β-lactamase (ESBL) production by determination of synergy between 0.25 and128 mg/L ceftazidime or cefotaxime and 4 mg/L clavulanate. The tigecycline MIC interpretive breakpoint was recommended by the European Committee on Antimicrobial Susceptibility Testing (EUCAST-2012; www.eucast.org). Escherichia coli ATCC 25922 and Klebsiella pneumoniae ATCC 700603 were used as quality control organisms in antimicrobial susceptibility tests.
PCR amplification and DNA sequencing of S. enterica serovar Indiana isolates
The quinolone resistance determination regions (QRDRs) of gyrA, gyrB, parC and parE in Salmonella isolates were amplified by PCR as described previously [14], sequenced, and then compared to the genes from Salmonella enterica serovar Typhimurium LT2 to identify any mutated positions in the target gene. The presence of one or more PMQR genes qnrA, qnrB, qnrC, qnrD, qnrS, qepA, oqxAB and aac(6’)Ib-cr was determined by PCR and sequencing using primers described previously [12,15]. The genotypes of ESBL-positive isolates were determined by PCR targeting known β-lactamase genes as previously described (bla TEM, bla SHV, bla CMY, bla CTX-M, and bla OXA) [12,19]. All the PCR products were either directly sequenced or cloned into pMD18-T plasmid (Takara Biotechnology Cooperation, Dalian, China) for sequence analysis at Takara Biotechnology Cooperation. The sequences obtained were analyzed by Sequencher 4.6 software (Gene Codes Corporation, Ann Arbor, MI, USA). A search for homologous sequences was performed using the BLASTn program at the U.S. National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST/).
Pulsed-Field Gel Electrophoresis (PFGE) and Multilocus sequence typing (MLST) of S. enterica serovar Indiana isolates
Genomic DNA for PFGE was prepared in accordance with the method described previously [20]. Briefly, the genomic DNA of the test strains were digested with 50 U XbaI for 2 hours and separated on 1% agarose SeaKem Gold gel with the CHEF DR III system (Bio-Rad, Hercules, California). The PFGE patterns were interpreted with Bionumerics software (Applied Math, Belgium) using Unweighted Pair Group Method with Arithmetic Mean (UPGMA). Bands which were smaller than 20.5 kb were not included in the analysis. Patterns indistinguishable by computer and visual inspection were assigned the same pattern designation, position tolerance of 1%. Clusters were defined as DNA patterns sharing ≥80% similarity (A, B, C).
MLST analysis was conducted by sequencing fragments of seven housekeeping genes (thrA, purE, sucA, hisD, aroC, hemD, dnaN) and sequence types (STs) were assigned by comparison to the S. enterica MLST database (http://mlst.warwick.ac.uk/mlst/dbs/Senterica/documents/primersEnterica_html) [21].
Plasmid replicon typing and conjugation experiment of S. enterica serovar Indiana isolates
Plasmids were isolated using Plasmid Plus Midi kit (Qiagen, Germany). Replicon typing of plasmids was performed using a PCR-based method as previously described [22]. Conjugation experiments were performed for all Salmonella isolates using a sodium azide-resistant E. coli J53 as recipient strain as previously described [23]. Transconjugants were selected on LB agar plates containing cefotaxime (2 μg/mL) and sodium azide (100 μg/mL).
Statistical analysis
The Pearson Chi-Square test were used to determine the significant difference (p < 0.05) of prevalence of Salmonella in chicken and pigs with different characteristics and the rates of resistance between ciprofloxacin and cefotaxime co-resistant Salmonella isolates and ciprofloxacin and cefotaxime none-co-resistant Salmonella isolates. Statistical software SPSS for Windows (version 17.0, SPSS, Inc., Chicago, IL) was used for descriptive analysis.
Results
Prevalence and serotypes of Salmonella isolated from chicken and pig samples
Out of the 283 chicken carcasses collected in Hebi (n = 141) and Zhoukou (n = 142), 128 Salmonella isolates were recovered with an isolation rate of 45.2%. The prevalence of Salmonella was found to be similar between Hebi (59/141, 41.8%) and Zhoukou (69/142, 48.6%) (χ2 = 1.300, P = 0.254). Samples collected in the autumn showed the highest contamination rate (77.5%) (Table 1). Serotyping was performed on all isolates. The 128 Salmonella isolates belonged to 7 serovars with 10 isolates being defined as untypable. The top three serovars were S. enterica serovar Enteritidis (n = 76, 59.4%), S. enterica serovar Hadar (n = 15, 11.7%) and S. enterica serovar Indiana (n = 11, 8.6%) (Table 2).
Table 1. Prevalence of Salmonella isolates from chicken slaughterhouses and pig slaughterhouses.
| Chickens | Pigs | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hebi | zhoukou | Total | Hebi | Luohe | Total | |||||||
| Seasons* | No. of samples | No. of positive samples | No. of samples | No. of positive samples | No. of samples | No. of positive samples | No. of samples | No. of positive samples | No. of samples | No. of positive samples | No. of samples | No. of positive samples |
| Spring | 35 | 15(42.9%) | 38 | 9(23.7%) | 73 | 24(32.9%) | 33 | 8(24.2%) | 36 | 14(38.9%) | 69 | 22(31.9%) |
| Summer | 35 | 12(34.3%) | 35 | 19(54.3%) | 70 | 31(44.3%) | 33 | 15(45.5%) | 24 | 9(37.5%) | 57 | 24(42.1%) |
| Autumn | 36 | 25(69.4%) | 35 | 30(85.7%) | 71 | 55(77.5%) | 33 | 7(21.2%) | 33 | 7(21.2%) | 66 | 14(21.2%) |
| Winter | 35 | 7(20.0%) | 34 | 11(32.4%) | 69 | 18(26.1%) | 21 | 9(42.9%) | 27 | 1(3.7%) | 48 | 10(20.8%) |
| Total | 141 | 59(41.8%) | 142 | 69(48.6%) | 283 | 128(45.2%) | 120 | 39(32.5%) | 120 | 31(25.8%) | 240 | 70(29.2%) |
*Definition of seasons: Spring (March, April, May), Summer (June, July, August), Autumn (September, October, November), Winter (December, January, February)
Table 2. Serotyping of Salmonella isolates from chicken slaughterhouses and pig slaughterhouses.
| Chickens | Pigs | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of isolates | No. of isolates | ||||||||||||
| Serovar | Hebi(n = 59) | Zhoukou (n = 69) | Total (n = 128) | Serovar | Hebi(n = 39) | Luohe (n = 31) | Total (n = 70) | ||||||
| Enteritidis | 28 | (47.5%) | 48 | (69.6%) | 76 | (59.4%) | Typhimurium | 15 | (38.5%) | 5 | (16.2%) | 20 | (28.6%) |
| Hadar | 0 | (0.0%) | 15 | (21.7%) | 15 | (11.7%) | Derby | 1 | (2.6%) | 18 | (58.1%) | 19 | (27.1%) |
| Indiana | 11 | (18.6%) | 0 | (0.0%) | 11 | (8.6%) | Enteritidis | 5 | (12.8%) | 1 | (3.2%) | 6 | (8.6%) |
| Typhimurium | 7 | (11.9%) | 0 | (0.0%) | 7 | (5.5%) | Indiana | 5 | (12.8%) | 0 | (0.0%) | 5 | (7.2%) |
| Infantis | 0 | (0.0%) | 5 | (7.2%) | 5 | (3.9%) | Muenster | 5 | (12.8%) | 0 | (0.0%) | 5 | (7.2%) |
| Senftenberg | 3 | (5.1%) | 0 | (0.0%) | 3 | (2.3%) | London | 4 | (10.2%) | 0 | (0.0%) | 4 | (5.7%) |
| Kentucky | 1 | (1.7%) | 0 | (0.0%) | 1 | (0.8%) | Agona | 2 | (5.1%) | 1 | (3.2%) | 3 | (4.3%) |
| Untyped | 9 | (15.2%) | 1 | (1.5%) | 10 | (7.8%) | Aberdeen | 1 | (2.6%) | 0 | (0.0%) | 1 | (1.4%) |
| Meleagridis | 0 | (0.0%) | 1 | (3.2%) | 1 | (1.4%) | |||||||
| Chester | 0 | (0.0%) | 1 | (3.2%) | 1 | (1.4%) | |||||||
| Thompson | 0 | (0.0%) | 1 | (3.2%) | 1 | (1.4%) | |||||||
| Untyped | 1 | (2.6%) | 3 | (9.7%) | 4 | (5.7%) | |||||||
Out of the 240 samples taken from pigs in Hebi (surface swabs = 60, lymph nodes = 60) and Luohe (surface swabs = 60, lymph nodes = 60), 70 Salmonella isolates were recovered with an isolation rate of 29.2%. The prevalence of Salmonella was found to be similar between Hebi (39/120, 32.5%) and Luohe (31/120, 25.8%) (χ2 = 1.291, P = 0.256) (Table 1). However there was a significant difference between surface swabs samples (26/120, 21.7%) and ileocaecal lymph nodes (44/120, 36.7%) (χ2 = 6.534, P = 0.011). Samples collected in the summer period showed the highest contamination rate (42.1%)(Table 1). Of these 70 pig isolates, 11 serovars were found with 4 isolates being untypable. The top three serovars were S. enterica serovar Typhimurium (n = 20, 28.6%), S. enterica serovar Derby (n = 19, 27.1%), and S. enterica serovar Enteritidis (n = 6, 8.6%) (Table 2).
Antimicrobial susceptibility testing
Of the 128 chicken isolates studied, 6 were susceptible to all antimicrobial compounds tested and all isolates were susceptible to imipenem and tigecycline. Resistance to nalidixic acid was common (117/128, 91.4%), followed by ampicillin (84/128, 65.6%), tetracycline (60/128, 46.9%). Eleven isolates (8.6%) were resistant to ciprofloxacin, and were all S. enterica serovar Indiana. Seven isolates (5.5%, 7/128) (S. enterica serovar Indiana = 6 and S. enterica serovar Infantis = 1) were ESBL-producers, of which six (85.7%, 6/7) were also resistant to ciprofloxacin. Twenty antimicrobial resistance profiles were identified among the 128 chicken isolates. Multiple drug resistance accounted for 39.8% (51/128) of the chicken isolates. The top 3 antimicrobial resistance profiles of the chicken isolates were AMP-NAL (n = 33, 27.0%), NAL alone (n = 22, 18.0%), and AMP -NAL-TET (n = 19, 15.6%) (Table 3).
Table 3. The top 10 antimicrobial resistance profiles of Salmonella isolates recovered from chicken slaughterhouses in Henan, China.
| Resistant profiles | No. of resistant isolates | ||||||
|---|---|---|---|---|---|---|---|
| Enteritidis | Hadar | Typhimurium | Indiana | Infantis | Other Serovars | Total | |
| AMP-CAZ-CHL-CIP-CTX-GEN-NAL-SXT-TET* | 0 | 0 | 0 | 6 | 0 | 0 | 6 |
| AMP-CHL-CIP-GEN-NAL-SXT-TET | 0 | 0 | 0 | 5 | 0 | 0 | 5 |
| AMP-CHL-GEN-NAL-TET | 0 | 0 | 6 | 0 | 0 | 0 | 6 |
| AMP-GEN-NAL-TET | 2 | 2 | 0 | 0 | 0 | 0 | 4 |
| AMP-GEN-NAL | 6 | 0 | 0 | 0 | 0 | 0 | 6 |
| AMP-NAL-TET | 17 | 2 | 0 | 0 | 0 | 0 | 19 |
| NAL-TET | 0 | 8 | 1 | 0 | 0 | 2 | 11 |
| AMP-NAL | 32 | 1 | 0 | 0 | 0 | 0 | 33 |
| NAL | 18 | 0 | 0 | 0 | 4 | 0 | 22 |
| TET | 0 | 1 | 0 | 0 | 0 | 1 | 2 |
*AMP, ampicillin; CAZ, ceftazidime; CHL, Chloramphenicol; CIP, ciprofloxacin; CTX, cefotaxime; GEN, gentamicin; SXT, trimethoprim/sulfamethoxazole; TET, tetracycline.
Of the 70 pig isolates, 13 were susceptible to all antimicrobials tested and all isolates were susceptible to imipenem and tigecycline. Resistance to tetracycline was common (47/70, 67.1%) among these isolates, followed by nalidixic acid (45/70, 64.3%), chloramphenicol (42/70, 60.0%). Seven isolates (10.0%, 7/70) of which five and two were S. enterica serovar Indiana and S. enterica serovar Typhimurium respectively, were found to be resistant to ciprofloxacin. Six isolates (8.6%, 6/70) (S. enterica serovar Indiana = 5, S. enterica serovar Enteritidis = 1) were ESBL-producers and 5 (83.3%, 5/6) of these were also resistant to ciprofloxacin. Eighteen antimicrobial resistance profiles were identified among the 70 pig isolates. Multiple drug resistance accounted for 57.1% (40/70) of the pig collection. The top 3 antimicrobial resistance profiles of pig isolates were CHL-NAL-TET (n = 11, 19.3%), AMP-CHL-GEN-NAL-SXT-TET (n = 8, 14.0%), and TET alone (n = 6, 10.5%) (Table 4).
Table 4. The top 10 antimicrobial resistance profiles of Salmonella isolates recovered from pig slaughterhouses in Henan, China.
| Resistant profiles | No. of resistant isolates | ||||||
|---|---|---|---|---|---|---|---|
| Typhimurium | Derby | Indiana | Enteritidis | London | Other Serovars | Total | |
| AMP-CAZ-CHL-CIP-CTX-GEN-NAL-SXT-TET * | 0 | 0 | 5 | 0 | 0 | 0 | 5 |
| AMP-CHL-GEN-NAL-SXT-TET | 8 | 0 | 0 | 0 | 0 | 0 | 8 |
| AMP-CHL-GEN-NAL-TET | 5 | 0 | 0 | 0 | 0 | 0 | 5 |
| AMP-CHL-GEN-SXT-TET | 0 | 0 | 0 | 0 | 3 | 0 | 3 |
| AMP-CHL-NAL-SXT-TET | 1 | 0 | 0 | 0 | 0 | 1 | 2 |
| CHL-NAL-TET | 0 | 11 | 0 | 0 | 0 | 0 | 11 |
| AMP-NAL | 1 | 0 | 0 | 3 | 0 | 1 | 5 |
| CHL-TET | 0 | 2 | 0 | 0 | 0 | 0 | 2 |
| NAL | 0 | 0 | 0 | 2 | 0 | 0 | 2 |
| TET | 0 | 0 | 0 | 0 | 0 | 6 | 6 |
*AMP, ampicillin; CAZ, ceftazidime; CHL, Chloramphenicol; CIP, ciprofloxacin; CTX, cefotaxime; GEN, gentamicin; SXT, trimethoprim/sulfamethoxazole; TET, tetracycline.
In total, 11 S. enterica serovar Indiana isolates were identified as being co-resistant to ciprofloxacin and cefotaxime, with 6 and 5 of these being isolated from chickens and pigs, respectively. These isolates were also resistant to 7 other antimicrobial agents (AMP, CAZ, CHL, GEN, NAL, SXT, TET) (Table 5). In contrast, the ciprofloxacin and cefotaxime -co-resistant Salmonella isolates alone showed high rate of resistance to most of the antibiotics except nalidixic acid (Table 5).
Table 5. Resistance phenotypes of ciprofloxacin and cefotaxime co-resistance Salmonella isolates and none ciprofloxacin and cefotaxime co-resistance Salmonella isolates from chicken and pig slaughterhouses.
| Antimicrobials | No. of resistant isolates | ||||
|---|---|---|---|---|---|
| co-Resistant Isolates (n = 11) | Non co-Resistant Isolates (n = 187) | p-Value* | |||
| Ampicillin | 11 | (100.0%) | 107 | (57.2%) | 0.013 |
| Ceftazidime | 11 | (100.0%) | 2 | (1.1%) | <0.001 |
| Chloramphenicol | 11 | (100.0%) | 51 | (27.3%) | <0.001 |
| Ciprofloxacin | 11 | (100.0%) | 7 | (3.7%) | <0.001 |
| Cefotaxime | 11 | (100.0%) | 2 | (1.1%) | <0.001 |
| Gentamicin | 11 | (100.0%) | 44 | (23.5%) | <0.001 |
| Imipenem | 0 | (0.0%) | 0 | (0.0%) | - |
| Nalidixic acid | 11 | (100.0%) | 151 | (80.7%) | 0.228 |
| Tetracycline | 11 | (100.0%) | 96 | (51.3%) | 0.005 |
| Tigecycline | 0 | (0.0%) | 0 | (0.0%) | - |
| Trimethoprim-sulfamethoxazole | 11 | (100.0%) | 22 | (11.8%) | <0.001 |
* p value were calculated using the chi-square by SPSS version 17.0.
Nine isolates were ciprofloxacin and cefotaxime non-co-resistant with 7 and 2 Salmonella isolates being resistant to ciprofloxacin only and cefotaxime only respectively. The ciprofloxacin resistant only isolates were from two different serovars, and 5 of these were S. enterica serovar Indiana isolates from chickens along with 2 S. enterica serovar Typhimurium isolates from pigs. The cefotaxime resistant only isolates were from two different serovars, a S. enterica serovar Infant isolate from a chicken sample and a S. enterica serovar Enteritidis isolate from a pig sample.
Identification of quinolone resistance determinants of S. enterica serovar Indiana isolates
Sixteen isolates were resistant to ciprofloxacin with minimum inhibitory concentration (MIC) values between 64 and 128 μg /mL. PCR and sequencing was used to identify point mutations in the QRDRs of gyrA, gyrB, parC or parE. in all 16 ciprofloxacin-resistant S. enterica serovar Indiana isolates among which 11 isolates were also resistant to cefotaxime. No mutations were found in gyrB nor parE in any of the isolates. All possessed 4 point mutations in QRDRs of which 8 isolates had gyrA (S83F/D87N), parC (T57S/S80R) and 8 isolates had gyrA (S83F/D87G), parC (T57S/S80R) mutations.
We also determined the carriage of the PMQR determinants and found that all 16 isolates carried aac(6')-Ib-cr, among which 12 isolates also carried oqxAB. None of the isolates carried qnrA, qnrB, qnrC, qnrD, qnrS or qepA.
Occurrence of β-lactamase-encoding genes of S. enterica serovar Indiana isolates
Among the 16 S. enterica serovar Indiana isolates, 11 carried genes encoding CTX-M-type ESBLs and the subtype of bla CTX-M was identified as bla CTX-M-65 for all 11 isolates. The ESBL genotypes, bla OXA-1 and bla TEM-1 were also identified in 16 and 5 isolates respectively.
PFGE and MLST analysis of S. enterica serovar Indiana isolates
To further elucidate the genetic relationships of the 16 ciprofloxacin and/or cefotaxime resistant S. enterica serovar Indiana isolates by MLST and PFGE. All 16 isolates belonged to the same sequence type ST17. By PFGE, the 16 isolates were clustered into 12 pulsotypes. A dendrogram was constructed using these PFGE patterns. Isolates grouped into the same PFGE patterns mostly were recovered from the same regions. The predominant pulsotype was denoted as CN001, which included five isolates from the same pig slaughterhouse in Hebi on the same day (23rd, February 2011). The remaining 11 pulsotypes contained a single isolate (Fig 1). Cluster A consisted of mainly ciprofloxacin and cefotaxime co-resistant Salmonella isolates of which the isolates from chicken and pig slaughterhouses were different. Cluster C contained only ciprofloxacin resistant Salmonella isolates with the exception of isolate 11C28. Those isolates containing point mutations in gyrA (S83F/D87N) and parC (T57S/S80R) were all grouped together in cluster A whilst those with point mutations in gyrA (S83F/D87G) and parC (T57S/S80R) were mostly grouped into clusters B and C.
Fig 1. Dendrogram of 16 S. enterica serovar Indiana isolates constructed based on XbaI PFGE patterns.
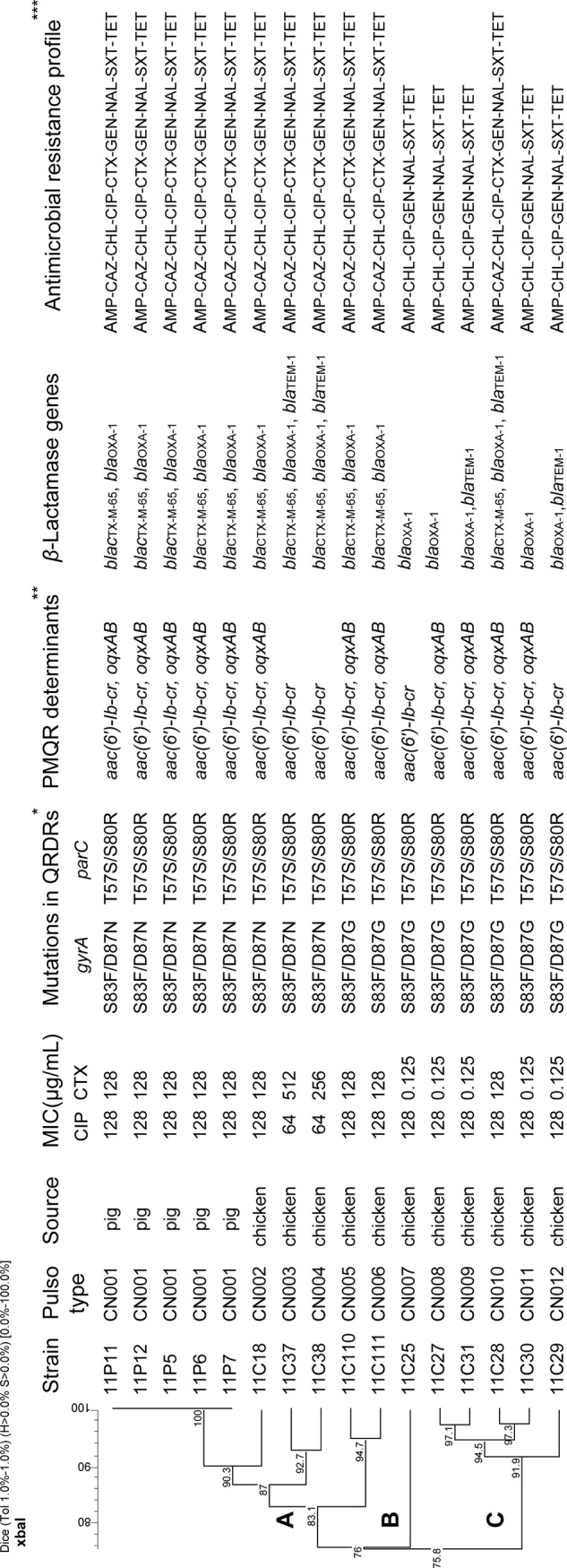
Stains information, pulsotype, mutations in QRDRs, resistance determinants/genes and resistance profiles are shown on the right. * PMQR, Plasmid Mediated Quinolone Resistance. **QRDRs, Quinolone Resistance Determining Regions. ***AMP, ampicillin; CAZ, ceftazidime; CHL, chloramphenicol; CIP, ciprofloxacin; CTX, cefotaxime; GEN, gentamicin; SXT, trimethoprim/sulfamethoxazole; TET, tetracycline.
Plasmid replicon typing and conjugation experiment of ESBL-S. enterica serovar Indiana isolates
Conjugation experiments were conducted on these isolates using E. coli J53 as a recipient strain. However, no transconjugants were obtained. Seven isolates contained plasmids belonging to the IncN group and none of the 18 plasmids was detected in the rest of 4 ESBL-Salmonella isolates.
Discussion
In this study, we screened processed chickens and pigs in slaughterhouses collected from 3 districts of Henan province which was one of the largest food-animal processing provinces in China for the presence of Salmonella, and further characterized the isolates using serotyping and of antimicrobial susceptibility testing. Specifically the genotypic mechanisms of ciprofloxacin-resistant and cefotaxime-resistant isolates were identified.
There had been a few studies on prevalence of Salmonella in poultry in retail markets in China wherein values varied from 38.9 to 65.3% depending on the geographical [24]. Our isolation rate of 45.2% from chickens in slaughterhouses was similar to that of 47.9% reported earlier from retail markets in the same (Henan) province [24], suggesting similar levels of contamination in the entire supply chain. However, our isolation rate was much higher than that (22.1%) from a slaughterhouse in Anhui province reported by Wang et al. [11]. The top serovars found in this study were S. enterica serovar Enteritidis, S. enterica serovar Indiana and S. enterica serovar Typhimurium. Two of the three (S. enterica serovar Enteritidis, S. enterica serovar Indiana) were also counted in the top three Salmonella serovars (S. enterica serovar Enteritidis, S. enterica serovar Hadar and S. enterica serovar Indiana) in chickens in China as reported by other studies [10,25,26]. These serovars were within the top four serovars (S. enterica serovar Typhimurium, S. enterica serovar Enteritidis, S. enterica serovar Derby, S. enterica serovar Indiana) isolated from patients in Henan from 2006 to 2007 [27], consistent with poultry as a major source of Salmonella infection in humans.
The isolation of Salmonella from pigs was much lower and the prevalent serovars were also different. The isolation rate of 29.2% was higher than the isolation rate from slaughterhouses reported by the EFSA (10.3%) [28] and Li et al. (12.0%) [29], but similar to that from pork in retail market (31%) reported by Yang et al. [30]. The differences were likely due to sources or regional difference. The common serovars found in this study (S. enterica serovar Typhimurium, S. enterica serovar Derby and S. enterica serovar Enteritidis) were also reported to be common in pigs by the EFSA [28].
Salmonella is still the leading cause of hospitalization in USA [31] and MDR Salmonella has become a significant public health concern worldwide [32]. Our data showed that nearly half of the isolates (46.0%) from Henan province were multidrug resistant which was the same as that (46.0%) from patients in Guangdong province [4] and a somewhat higher than that (34.9%) in Beijing [10] but was sharply lower than that (93%) in Anhui province [11]. The high occurrence of multidrug resistant Salmonella in slaughterhouses indicates that cross-contamination in the subsequent retail process poses serious risk to public health. With the large-scale use, and often abuse, of antimicrobials over time, fluoroquinolone and/or extended-spectrum cephalosporin resistant Salmonella isolates have been detected from patients in numerous locations with variable prevalence in China, such as 6.7% in Gangdong province [4], 12.2% in Wuhan [5], 17.2% in Shanghai [6] and 26.9% in Henan province [27]. Importantly, 10.1% Salmonella isolates exhibited resistance to ciprofloxacin or third generation cephalosporin in this study. Fluoroquinolone resistance in Salmonella is a unique problem in China and other Asian countries where there is no strict control on its availability [5,33], in contrast to countries with strict antimicrobial controls, such as the United States, where less than 3% of Salmonella isolates were considered resistant to ciprofloxacin [34] and in Belgium, wherein resistance to ciprofloxacxin was undetectable in isolates from both healthy pigs and chickens [35].
In this study, we found 11 Salmonella isolates that were co-resistant to ciprofloxacin and cefotaxime and all of which were S. enterica serovar Indiana. The appearance of ciprofloxacin and cefotaxime co-resistant S. enterica serovar Indiana is alarming. It seems that the co-resistance only developed recently since the study by Xia et al. in 2009 [27] found that S. enterica serovar Indiana from Henan province were susceptible to cefotaxime [27]. The emergence of cefotaxime resistant S. enterica serovar Indiana could either be due to acquisition of cefotaxime resistance genes by local strains or through spread from other regions where ciprofloxacin and cefotaxime co-resistant S. enterica serovar Indiana has been reported such as Beijing [10] or by cross-genus spread from ciprofloxacin and cefotaxime co-resistant E. coli which has been reported in Gangdong province, China [36].
Two PMQR determinants were identified in our S. enterica serovar Indiana isolates withaac(6')-Ib-cr present in all 16 isolates and oqxAB present in 12 isolates. The distribution of the oqxAB-positive S. enterica serovar Indiana isolates on the PFGE dendrogram (Fig 1) suggests that it was acquired independently 3 times. OqxAB, a novel efflux pump, that mediates resistance to olaquindox and also extrudes antibiotics such as chloramphenicol and fluoroquinolones, broadening the resistance phenotype to these latter antibiotics [37]. Olaquindox, a quinoxaline derivative antibiotic, is as a growth promoter that has been in use in animals for decades in China. The latter feature is suggestive of a constant selective pressure for the acquisition and dissemination of oqxAB [12,38]. The co-existence of PMQR genes, aac(6')-Ib-cr and oqxAB, had been detected previously in both human and animal Salmonella isolates in China [12]. Although PMQR confers only low-level quinolone resistance, it can facilitate the emergence of high-level resistance via mutations in a topoisomerase gene gyrA, parC or both [39]. This may be also the case for the 16 S. enterica serovar Indiana isolates in this study, PMQR combined with multiple mutations in QRDRs [gyrA (S83F/D87N) or gyrA (S83F/D87G), parC (T57S/S80R)] resulted in high-level resistance to ciprofloxacin (MIC, ≥ 64μg/mL).
Uniquely in China, our study found that bla CTX-M-65 was carried by the 11 cefotaxime resistant S. enterica serovar Indiana isolates. In contrast bla CTX-M-14 and bla CTX-M-27 have been found in S. enterica serovar Indiana isolates from chickens and pigs in Gangdong [12] and bla CTX-M-24 was found in S. enterica serovar Indiana isolates from chickens in Shandong [40]. These bla CTX-M genes are usually located on conjugative plasmids [16]. The bla CTX-M-24 found in S. enterica serovar Indiana by Lai et al. was shown to be encoded on a unique class 1 integron with multiple resistance genes co-located on a IncHI2 plasmid [40] whilst bla CTX-M-14 and bla CTX-M-27 were found in S. enterica serovar Indiana by Jiang et al. and was shown to be encoded on IncHI2 and/or IncN plasmid [12]. Similarly bla CTX-M-24,bla CTX-M-27 and bla CTX-M-65 have been reported in E. coli from both animals and humans in China [36,41]. It is possible that these bla CTX-M genes were acquired by Salmonella from E. coli.
The genetic relationship of the S. enterica serovar Indiana isolates based on PFGE revealed that all except one ciprofloxacin and cefotaxime co-resistant isolate were grouped together in one cluster, denoted as cluster A, suggesting a single origin. The pig isolates were identical and belonged to this cluster, indicating likely the possible spread from chickens to pigs. The ciprofloxacin and cefotaxime co-resistant isolate located outside the cluster was likely to have acquired the bla CTX-M-65 gene independently. The isolate also carried a different gyrA mutation (S83F/D87G), different from those of the isolates in cluster A, lending further support to its independent origin. Therefore both clonal spread and independent acquisition of cefotaxime resistance may have acted to contribute to the emergence and spread of ciprofloxacin and cefotaxime co-resistant S. enterica serovar Indiana in Henan province.
Multidrug resistant S. enterica serovar Indiana has already been reported in human infections in China [6,27]. All S. enterica serovar Indiana isolates from human infections in Henan were resistant to ciprofloxacin but susceptible to cefotaxime [27]. It is inevitable that the ciprofloxacin and cefotaxime co-resistant S. enterica serovar Indiana emerged in Henan will transmit to humans, posing a significant threat to public health. The co-resistance limits the optimal treatment alternatives of salmonellosis in humans.
Conclusion
The prevalence of Salmonella in the chicken and pig slaughterhouses in Henan was found to be high, and represented by a small number of different serovars such as S. enterica serovar Enteritidis, S. enterica serovar Hadar and S. enterica serovar Indiana in chickens and S. enterica serovar Typhimurium, S. enterica serovar Derby and S. enterica serovar Enteritidis in pigs. This finding highlights the fact that food-producing animals at processing plants are a significant reservoir of antimicrobial-resistant Salmonella. Our data may contribute to the development of interventions at the processing level to interrupt the transmission of Salmonella to humans. Ciprofloxacin and cefotaxime co-resistant S. enterica serovar Indiana possibly emerged only recently through acquisition of bla CTX-M-65. The potential risk of ciprofloxacin and cefotaxime co-resistant S. enterica serovar Indiana infections in humans requires urgent attention. The emergence of ciprofloxacin and cefotaxime co-resistance also underscores the importance of strict control on the use of antibiotics in animal production to prevent the emergence of antibiotic resistance in Salmonella.
Acknowledgments
We thank Professor Séamus Fanning from UCD Centre for Food Safety, School of Public Health, Physiotherapy and Population Science, UCD Centre for Molecular Innovation and Drug Discovery, University College Dublin, Ireland for his valuable suggestions.
Data Availability
All relevant data are within the paper.
Funding Statement
The authors have no support or funding to report.
References
- 1. White DG, Zhao S, Sudler R, Ayers S, Friedman S, Chen S, et al. The isolation of antibiotic-resistant Salmonella from retail ground meats. New England Journal of Medicine. 2001; 345: 1147–1154. [DOI] [PubMed] [Google Scholar]
- 2. Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, et al. The global burden of nontyphoidal Salmonella gastroenteritis. Clinical Infectious Diseases. 2010; 50: 882–889. 10.1086/650733 [DOI] [PubMed] [Google Scholar]
- 3. Wu H, Xia X, Cui Y, Hu Y, Xi M, Wang X, et al. Prevalence of extended-spectrum β-lactamase-producing Salmonella on retail chicken in six provinces and two national cities in the People's Republic of China. Journal of Food Protection. 2013; 76: 2040–2044. 10.4315/0362-028X.JFP-13-224 [DOI] [PubMed] [Google Scholar]
- 4. Liang Z, Ke B, Deng X, Liang J, Ran L, et al. Serotypes, seasonal trends, and antibiotic resistance of non-typhoidal Salmonella from human patients in Guangdong Province, China, 2009–2012. BMC Infect Dis. 2015; 15: 53 10.1186/s12879-015-0784-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cui S, Li J, Sun Z, Hu C, Jin S, Li F, et al. Characterization of Salmonella enterica isolates from infants and toddlers in Wuhan, China. Journal of antimicrobial chemotherapy. 2009; 63: 87–94. 10.1093/jac/dkn452 [DOI] [PubMed] [Google Scholar]
- 6. Li Y, Xie X, Xu X, Wang X, Chang H, et al. Nontyphoidal salmonella infection in children with acute gastroenteritis: prevalence, serotypes, and antimicrobial resistance in Shanghai, China. Foodborne Pathog Dis. 2014; 11: 200–206. 10.1089/fpd.2013.1629 [DOI] [PubMed] [Google Scholar]
- 7. M'ikanatha NM, Sandt CH, Localio AR, Tewari D, Rankin SC, Whichard JM, et al. Multidrug-resistant Salmonella isolates from retail chicken meat compared with human clinical isolates. Foodborne pathogens and disease. 2010; 7: 929–934. 10.1089/fpd.2009.0499 [DOI] [PubMed] [Google Scholar]
- 8. Sandt CH, Fedorka-Cray PJ, Tewari D, Ostroff S, Joyce K, et al. A comparison of Non-Typhoidal Salmonella from humans and food animals using pulsed-field gel electrophoresis and antimicrobial susceptibility patterns. PLoS One. 2013; 8: e77836 10.1371/journal.pone.0077836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lunguya O, Lejon V, Phoba M- F, Bertrand S, Vanhoof R, Glupczynski Y, et al. Antimicrobial resistance in invasive non-typhoid Salmonella from the Democratic Republic of the Congo: Emergence of decreased fluoroquinolone susceptibility and extended-spectrum Beta lactamases. PLoS neglected tropical diseases. 2013; 7: e2103 10.1371/journal.pntd.0002103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang Y, Chen Q, Cui S, Xu X, Zhu J, Luo H, et al. Enumeration and characterization of Salmonella isolates from retail chicken carcasses in Beijing, China. Foodborne pathogens and disease. 2014; 11: 126–132. 10.1089/fpd.2013.1586 [DOI] [PubMed] [Google Scholar]
- 11. Wang H, Ye K, Wei X, Cao J, Xu X, Zhou G. Occurrence, antimicrobial resistance and biofilm formation of Salmonella isolates from a chicken slaughter plant in China. Food Control. 2013; 33: 378–384. [Google Scholar]
- 12. Jiang HX, Song L, Liu J, Zhang XH, Ren YN, Zhang WH, et al. Multiple transmissible genes encoding fluoroquinolone and third-generation cephalosporin resistance co-located in non-typhoidal Salmonella isolated from food-producing animals in China. Int J Antimicrob Agents. 2014; 43: 242–247. 10.1016/j.ijantimicag.2013.12.005 [DOI] [PubMed] [Google Scholar]
- 13. Hue O, Le Bouquin S, Lalande F, Allain V, Rouxel S, Petetin I, et al. Prevalence of Salmonella spp. on broiler chicken carcasses and risk factors at the slaughterhouse in France in 2008. Food Control. 2012; 22: 1158–1164. [Google Scholar]
- 14. Chen S, Cui S, McDermott PF, Zhao S, White DG, Paulsen I, et al. Contribution of target gene mutations and efflux to decreased susceptibility of Salmonella enterica serovar Typhimurium to fluoroquinolones and other antimicrobials. Antimicrobial agents and chemotherapy. 2007; 51: 535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu JH, Deng YT, Zeng ZL, Gao JH, Chen L, Arakawa Y, et al. Coprevalence of plasmid-mediated quinolone resistance determinants QepA, Qnr, and AAC (6’)-Ib-cr among 16S rRNA methylase RmtB-producing Escherichia coli isolates from pigs. Antimicrobial agents and chemotherapy. 2008; 52: 2992–2993. 10.1128/AAC.01686-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Munday C, Xiong J, Li C, Shen D, Hawkey P. Dissemination of CTX-M type β-lactamases in Enterobacteriaceae isolates in the People’s Republic of China. International journal of antimicrobial agents. 2004; 23: 175–180. [DOI] [PubMed] [Google Scholar]
- 17. Malorny B, Hoorfar J, Hugas M, Heuvelink A, Fach P, Ellerbroek L, et al. Interlaboratory diagnostic accuracy of a Salmonella specific PCR-based method. International journal of food microbiology. 2003; 89: 241–249. [DOI] [PubMed] [Google Scholar]
- 18.Performance Standards for Antimicrobial Susceptibility Testing: Seventeenth Informational Supplement M100-S22. CLSI, Wayne, PA, USA. 2012.
- 19. Xu L, Ensor V, Gossain S, Nye K, Hawkey P. Rapid and simple detection of bla CTX− M genes by multiplex PCR assay. Journal of medical microbiology. 2005; 54: 1183–1187. [DOI] [PubMed] [Google Scholar]
- 20. Ribot EM, Fair M, Gautom R, Cameron D, Hunter S, Swaminathan B, et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157: H7, Salmonella, and Shigella for PulseNet. Foodbourne Pathogens & Disease. 2006; 3: 59–67. [DOI] [PubMed] [Google Scholar]
- 21. Achtman M, Wain J, Weill FX, Nair S, Zhou Z, Sangal V, et al. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica . PLoS Pathog. 2012; 8: e1002776 10.1371/journal.ppat.1002776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. Journal of Microbiological Methods. 2005; 63: 219–228. [DOI] [PubMed] [Google Scholar]
- 23. Jacoby GA, Chow N, Waites KB. Prevalence of plasmid-mediated quinolone resistance. Antimicrob Agents Chemother. 2003; 47: 559–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang B, Xi M, Wang X, Cui S, Yue T, Hao H. Prevalence of Salmonella on raw poultry at retail markets in China. Journal of food protection. 2011; 74: 1724–1728. 10.4315/0362-028X.JFP-11-215 [DOI] [PubMed] [Google Scholar]
- 25. Yang B, Qiao L, Zhang X, Cui Y, Xia X, Cui S, et al. Serotyping, antimicrobial susceptibility, pulse field gel electrophoresis analysis of Salmonella isolates from retail foods in Henan Province, China. Food Control. 2012; 32: 228–235. [Google Scholar]
- 26. Lu Y, Wu CM, Wu GJ, Zhao HY, He T, Cao XY, et al. Prevalence of antimicrobial resistance among Salmonella isolates from chicken in China. Foodborne Pathogens and Disease. 2011; 8: 45–53. 10.1089/fpd.2010.0605 [DOI] [PubMed] [Google Scholar]
- 27. Xia S, Hendriksen RS, Xie Z, Huang L, Zhang J, Guo W, et al. Molecular characterization and antimicrobial susceptibility of Salmonella isolates from infections in humans in Henan Province, China. Journal of clinical microbiology. 2009; 47: 401–409. 10.1128/JCM.01099-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Authority EFS. Report of the Task Force on Zoonoses Data collection on the analysis of the baseline survey on the prevalence of Salmonella in slaughter pigs, Part A. The EFSA Journal. 2008; 135: 1–111. [Google Scholar]
- 29. Li R, Lai J, Wang Y, Liu S, Li Y, Liu K, et al. Prevalence and characterization of Salmonella species isolated from pigs, ducks and chickens in Sichuan Province, China. Int J Food Microbiol. 2013; 163: 14–18. 10.1016/j.ijfoodmicro.2013.01.020 [DOI] [PubMed] [Google Scholar]
- 30. Yang B, Qu D, Zhang X, Shen J, Cui S, Shi Y, et al. Prevalence and characterization of Salmonella serovars in retail meats of marketplace in Shaanxi, China. Int J Food Microbiol. 2010; 141: 63–72. 10.1016/j.ijfoodmicro.2010.04.015 [DOI] [PubMed] [Google Scholar]
- 31. Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, et al. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis. 2011; 17: 7–15. 10.3201/eid1701.091101p1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Su LH, Chiu CH, Chu C, Ou JT. Antimicrobial resistance in nontyphoid Salmonella serotypes: a global challenge. Clin Infect Dis. 2004; 39: 546–551. [DOI] [PubMed] [Google Scholar]
- 33. Vlieghe ER, Phe T, De Smet B, Veng CH, Kham C, Bertrand S, et al. Azithromycin and ciprofloxacin resistance in Salmonella bloodstream infections in Cambodian adults. PLoS Negl Trop Dis. 2012; 6: e1933 10.1371/journal.pntd.0001933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Medalla F, Hoekstra RM, Whichard JM, Barzilay EJ, Chiller TM, Joyce K, et al. Increase in resistance to ceftriaxone and nonsusceptibility to ciprofloxacin and decrease in multidrug resistance among Salmonella Strains, United States, 1996–2009. Foodborne pathogens and disease. 2013; 10: 302–309. 10.1089/fpd.2012.1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. de Jong A, Smet A, Ludwig C, Stephan B, De Graef E, Vanrobaeys M, et al. Antimicrobial susceptibility of Salmonella isolates from healthy pigs and chickens (2008–2011). Vet Microbiol. 2014; 171: 298–306. 10.1016/j.vetmic.2014.01.030 [DOI] [PubMed] [Google Scholar]
- 36. Xu X, Cui S, Zhang F, Luo Y, Gu Y, Yang B, et al. Prevalence and characterization of cefotaxime and ciprofloxacin co-resistant Escherichia coli isolates in retail chicken carcasses and ground pork, China. Microbial drug resistance. 2014; 20: 73–81. 10.1089/mdr.2012.0224 [DOI] [PubMed] [Google Scholar]
- 37. Hansen LH, Jensen LB, Sorensen HI, Sorensen SJ. Substrate specificity of the OqxAB multidrug resistance pump in Escherichia coli and selected enteric bacteria. J Antimicrob Chemother. 2007; 60: 145–147. [DOI] [PubMed] [Google Scholar]
- 38. Wong MH, Yan M, Chan EW, Biao K, Chen S. Emergence of clinical Salmonella enterica serovar Typhimurium isolates with concurrent resistance to ciprofloxacin, ceftriaxone, and azithromycin. Antimicrobial agents and chemotherapy. 2014; 58: 3752–3756. 10.1128/AAC.02770-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hernández A, Sánchez MB, Martínez JL. Quinolone resistance: much more than predicted. Frontiers in microbiology. 2011; 2: 22 10.3389/fmicb.2011.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lai J, Wang Y, Shen J, Li R, Han J, Foley SL, et al. Unique class 1 integron and multiple resistance genes co-located on IncHI2 plasmid is associated with the emerging multidrug resistance of Salmonella Indiana isolated from chicken in China. Foodborne pathogens and disease. 2013; 10: 581–588. 10.1089/fpd.2012.1455 [DOI] [PubMed] [Google Scholar]
- 41. Li J, Ma Y, Hu C, Jin S, Zhang Q, Ding H, et al. Dissemination of cefotaxime-M-producing Escherichia coli isolates in poultry farms, but not swine farms, in China. Foodborne pathogens and disease. 2010; 7: 1387–1392. 10.1089/fpd.2010.0581 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.


