Abstract
Ticks and other arthropods often are hosts to nutrient providing bacterial endosymbionts, which contribute to their host’s fitness by supplying nutrients such as vitamins and amino acids. It has been detected, in our lab, that Ixodes pacificus is host to Rickettsia species phylotype G021. This endosymbiont is predominantly present, and 100% maternally transmitted in I. pacificus. To study roles of phylotype G021 in I. pacificus, bioinformatic and molecular approaches were carried out. MUMmer genome alignments of whole genome sequence of I. scapularis, a close relative to I. pacificus, against completely sequenced genomes of R. bellii OSU85-389, R. conorii, and R. felis, identified 8,190 unique sequences that are homologous to Rickettsia sequences in the NCBI Trace Archive. MetaCyc metabolic reconstructions revealed that all folate gene orthologues (folA, folC, folE, folKP, ptpS) required for de novo folate biosynthesis are present in the genome of Rickettsia buchneri in I. scapularis. To examine the metabolic capability of phylotype G021 in I. pacificus, genes of the folate biosynthesis pathway of the bacterium were PCR amplified using degenerate primers. BLAST searches identified that nucleotide sequences of the folA, folC, folE, folKP, and ptpS genes possess 98.6%, 98.8%, 98.9%, 98.5% and 99.0% identity respectively to the corresponding genes of Rickettsia buchneri. Phylogenetic tree constructions show that the folate genes of phylotype G021 and homologous genes from various Rickettsia species are monophyletic. This study has shown that all folate genes exist in the genome of Rickettsia species phylotype G021 and that this bacterium has the genetic capability for de novo folate synthesis.
Introduction
Symbiotic relationships with microorganisms are widespread across arthropods. It has been reported that all insects, and even a majority of arthropods, carry symbiotic microorganisms [1, 2]. Microbial symbionts vary greatly in the effects on their hosts, ranging from provision of essential nutrients [3–5] and symbiont-mediated protection from pathogens and parasites [6–8], to altering the host's reproductive system or sex determination systems [9, 10]. Recently, it was found that a symbiont-mediated stimulation of host immune system may contribute to host defense against pathogens and natural enemies [2, 11].
Most symbiotic relationships have a biochemical basis [12, 13]. For eukaryotic hosts with limited metabolic capabilities, some of the most discernable benefits of microbial symbionts in a mutualistic relationship involve the provision of essential amino acids [14, 15], vitamins and cofactors [16, 17], and recycling and storage of nitrogenous wastes from hosts [18, 19]. The exchange of these nutritional constituents appears to be a primary driving force for eukaryotic cell evolution [20].
Arthropods that feed primarily or entirely on blood are a breeding ground for forming nutritional interactions with microbial symbionts. For instance, vertebrate blood rarely contains sufficient quantities of essential B-vitamins, which are eight life-essential enzymes and cofactors as well as their derivatives, including biotin and folate [16]. Since B-vitamin synthetic pathways cannot be independently produced by animals [21], the diet of blood feeding arthropods is complemented with bacterially synthesized vitamins [16, 22, 23]. The western black-legged tick, Ixodes pacificus, is a common human biter in the Pacific Coast states of the U.S. [24]. I. scapularis is a closely related tick species of I. pacificus and is found on the east coast of North America. Both I. pacificus and I. scapularis are major vectors of tick-borne diseases, including Lyme borreliosis and anaplasmosis [25]. With limited feeding events during the life cycle, ticks’ sole diet is blood from vertebrates. However, the sources of B-vitamins for I. pacificus ticks are unknown. Due to their exclusive nutrient-poor vertebrate blood diets, ticks must obtain many vitamins through non-dietary means.
It has been detected, in our lab, that I. pacificus is host to a rickettsial endosymbiont, Rickettsia species phylotype G021 [26]. This endosymbiont is predominantly present, and 100% maternally transmitted [27, 28]. The function of the Rickettsia species phylotype G021 in I. pacificus is unknown, but the phylotype does not appear to have an effect on embryogenesis, oviposition and egg hatching of I. pacificus [29].
The acquisition of DNA sequences by the advent of rapid genome sequencing techniques has enabled substantial progress in identification of the biosynthetic capabilities of symbionts in arthropods [16, 23, 30, 31]. Since many bacterial symbionts are obligate intracellular organisms that reside exclusively inside host cells, analysis of many symbionts’ genomes is impeded by the inability to separate bacterial nucleotide sequences away from host’s genomes. One method of discovering genomic sequences of symbionts beyond the arthropods’ genomes that were originally sequenced is by MUMmer, a genome-wide alignment software [32, 33]. In this report, the genome-sequencing project of I. scapularis was used to reveal genomic sequences of the tick’s bacterial partner, the symbiotic Rickettsia species phylotype G021, by MUMmer alignment, and to demonstrate that the bacterium has all the genes for synthesizing folate in the tick host, I. pacificus.
Materials and Methods
Bioinformatics identification of folate biosynthetic genes
The Ixodes scapularis Genome Project is a partnership between the J. Craig Venter Institute, the National Institute Health, and the Broad Institute [34, 35]. In 2008, 570,640 contigs and 369,492 scaffolds of the I. scapularis Genome Project were deposited in the NCBI Trace Archive.
To test if there are Rickettsia sequences in I. scapularis’s Trace Archive database, MUMmer (mummer.sourceforge.net), which is a suffix-tree algorithm, was used to rapidly align the contigs and scaffolds of the I. scapularis genome against three reference Rickettsia genomes. Specifically, NUCMER from MUMmer was used to align nucleotide sequences between the I. scapularis genome and three completely sequenced Rickettsia genomes: R. bellii RML 369-C from Rickettsia bellii group Rickettsia [36], R. conorii subsp. israelensis [37], and R. felis str. LSU-Lb [38] from spotted fever group Rickettsia [39]. Nucleotide sequences of the I. scapularis genome with identities that were greater than 70% with either of the three reference Rickettsia genomes were retrieved by NUCMER.
Several java scripts were developed to manipulate data obtained from the MUMmer searches. The first program, SequenceSearch, takes call numbers from the MUMmer search and returns the matching DNA sequences as output files. A second program, called NucleotideReader, features the ability to read the input nucleotide file, communicate with the NCBI Basic Local Alignment Search Tool BLASTX website that aligns translated DNA with proteins from a non-redundant protein database, and identify the top protein match on the result output HTML of BLASTX and return that protein name. NucleotideReader works for large-batch searches (>5000) and is very useful when analyzing large data sets (S1 File and S2 File). Finally vitamin biosynthesis pathways were created manually from the genome of Rickettsia species by MetaCyc (http://metacyc.org/META/NEW-IMAGE?object=Vitamin-Biosynthesis), a manual curated database containing 2,260 metabolic pathways from 2,600 organisms. The names of the proteins in each vitamin biosynthesis pathway were matched against those in the BLASTX output files. The names and EC numbers of the matched proteins from MetaCyc were recorded.
Tick collection and Identification
I. pacificus adult ticks were collected from rural areas of Humboldt County in Northern California (UTM coordinates: Northing 4530184; Easting 567363; Zone/Sector 51T) by flagging or sweeping grass blades and shrubs with a 1-m2 white cotton cloth. Ticks were classified to the genus and species level based on morphological characteristics [40].
DNA Extraction Method
Ticks were washed in 95% ethanol three times and DNA from the ticks was extracted using the ammonium hydroxide DNA extraction method [41]. Briefly, whole I. pacificus adult ticks were submerged in liquid nitrogen and homogenized to a fine powder. Cells were lysed by 0.7 M ammonium hydroxide and boiled at 100°C for 15 minutes, cooled on ice for 30 seconds and the remaining ammonium was allowed to evaporate from the open tube for 15 minutes. DNA concentrations were determined using a Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Hampton, NH) and DNA was stored at -20°C.
Polymerase Chain Reaction (PCR) primer design and PCR
To amplify whole open reading frames (ORFs) of the folate genes of Rickettsia species phylotype G021, conserved regions that are upstream and downstream of each folate gene among at least nine spotted fever group rickettsiae were identified by BLAST using folate gene sequences of R. buchneri in I. scapularis from the Trace Archive and aligned using alignment software, Codon Code Aligner 3.7.1.2 (http://www.codoncode.com/aligner/) and ClustalX version 2.0. The identified conserved regions were then used as templates for the primer design by Primer3 (www.simgene.com/Primer3) (Table 1).
Table 1. Oligonucleotide primers used in the study.
Folate oligos with annealing temperatures and product sizes were indicated.
| Gene | Primers sequences (5'-3') | Melting temperature (°C) | PCR fragment size (bp) |
|---|---|---|---|
| External primers | |||
| folA2-F | CTACATTTGGAATGTATTGCACT | 51 | 913 |
| folA2-R | ATGCTCTATAACATTATGCACTCG | 53 | |
| folC2-F | TATGATTCCATATTTGGGCTGCG | 55 | 2237 |
| folC2-R | CGGKTTACGAATATTATTCGGTCC | 54 | |
| folE2-F | TACCTCCTAACMTTCGGGCAG | 57 | 1280 |
| folE2-R | GATTCCATCCATAAATGTGCSGG | 56 | |
| folKP2-F | ACCTATCACCCCTTGTGGGTCACA | 62 | 1400 |
| folKP2-R | ATCACCGGTTTCAGGAGTGCATGG | 61 | |
| ptpS2-F | ATCSTCAATTTGAATGCAGGCAG | 56 | 1385 |
| ptpS2-R | CGTGTTCGTATGTCACGCTT | 56 | |
| Internal primers | |||
| bioC-F | TTCTTATTYGGTGAAGCAAT | 49 | 578 |
| bioC-R | TATCGRAGTGCYRAWATTAC | 48 | |
| folA-F | CAGATGCCTTGGTCTTATAC | 50 | 367 |
| folA-R | GCCATTCTGCTAAAAGATTA | 48 | |
| folC-F | RCTCTTTCTGCCTTCCAAAT | 53 | 677 |
| folC-R | GAATTATGGCARGTTTGTGA | 50 | |
| folE-F | ATTCATTGGTGAAGATCCWA | 49 | 409 |
| folE-R | ACGCATHGCATACARCTAT | 51 | |
| folKP-F | TTTCTATARGCAGTAATTTAGGY | 49 | 662 |
| folKP-R | ATAATCKGCAATTAATCGTA | 45 | |
| ptpS-F | TAAACCTCAACAAAACAA | 44 | 416 |
| ptpS-R | ATGATMAAATGTACTCGTCGY | 51 | |
Amplification of the five folate genes of Rickettsia species phylotype G021 in I. pacificus was performed using a gradient PCR protocol: initial denaturation at 95°C for 5 minutes, followed by 40 cycles of denaturation at 95°C for 30 seconds, annealing at the temperatures between 50°C and 60°C for 30 seconds, and extension at 72°C for 1 minute and 30 seconds. Each folate gene was amplified by PCR using two sets of primers: external primers and internal primers (Table 1); only external primers amplified the whole open reading frames of each folate gene. A gradient PCR thermocycler, MJ Research PTC-200 (MJ Research, St. Bruno, Canada), was used to establish optimal annealing temperature for amplification of the genes. Each PCR reaction is composed of 2 μL 50 ng/μL I. pacificus genomic DNA, 1 μL 5 μM forward and reverse primers, 10 μL GoTaq® Green Master Mix (Promega, Madison, WI) and 6 μL PCR grade water. Two positive controls were used in PCR: Rickettsia montanensis DNA, provided by Dr. David H. Walker at the University of Texas Medical Branch, and I. scapularis DNA, extracted from I. scapularis ticks obtained the National Tick Research and Education Resource at Oklahoma State University. Amplification was confirmed by gel electrophoresis using 1% agarose. DNA bands corresponding to the expected gene sizes were used for cloning.
Cloning and plasmid DNA purification
Resulting amplicons of the folate genes from PCR were ligated into StrataClone™ PCR Cloning vector Escherichia coli Psc-A-amp/kan plasmid (Agilent Technologies, La Jolla, CA). A blue-white screen of the clones containing the insert was performed to detect successful ligation. White colonies on LB agar plates containing ampicillin and X-gal were re-streaked and then grown in LB broth with ampicillin overnight at 37°C. Plasmid DNA was extracted from the cells using the Wizard Plus SV Miniprep kit (Promega, Madison, WI). Eco RI restriction enzyme digest and gel electrophoresis were used to confirm the sizes of the clone inserts.
Sequencing and sequence analyses
The extracted plasmid DNA was sequenced through Elim Biopharmaceuticals (https://www.elimbio.com/) using M13 primer and sequence-specific primers. Ten clones for each gene were sequenced. The GenBank accession numbers for the folate genes of phylotype G021 are KT225568 for folA gene, KT225569 for folC gene, KT225570 for folE gene, KT225571 for folKP gene, and KT225572 for ptpS gene. The folate genes of phylotype G021 were used as the query sequences to perform homology searches using BLASTN. Kyoto Encyclopedia of Genes and Genomes (KEGG) was used as a reference knowledge base to interpret the completed Rickettsia genomes for the presence of the folate biosynthetic pathway.
Physical gene maps were generated in order to determine where on the genome the folate genes are located and if the folate genes of phylotype G021 and other Rickettsia species are organized in an operon. Genes from the species with closest identity to those from phylotype G021 were used to select the organisms to be represented in the gene map. NCBI’s GenBank was used to locate the gene loci within each organism’s complete genome. Arrows were used to represent ORFs. Each gene adjacent to the folate genes (upstream and downstream) was mapped the same way to show the physical clustering of gene loci. The directionality of each gene was based on the orientation displayed on GenBank.
Genes from the organisms other than the genus Rickettsia with close identity to the folA gene of phylotype G021 were also chosen to be represented on the gene map. Using BLASTX with the exclusion of the Rickettsiaceae family, the organisms Wolbachia endosymbiont of Drosophila melanogaster and Chlamydophila felis were chosen as members of two of the genera with a total BLAST score of at least 115 (E value less than 2e-28), when compared to folA of phylotype G021. Chlamydia trachomatis was selected due its total BLAST score of 92.8 (E value of 9e-20), when compared to folA of phylotype G021, and its previous characterization as a folate synthesizing endosymbiont [42]. Mycobacterium avium was also selected due to its known role as a folate synthesizing endosymbiont [43].
Phylogenetic tree constructions
To study evolutionary relationship of phylotype G021 to validated Rickettsia species, the database nucleotide collection (nr/nt) from BLAST was used to find homologies to our folate gene sequences. Sequences of the folate genes from phylotype G021, along with homologous folate gene sequences from closely related bacteria, were aligned with ClustalX version 2.0, edited manually using Geneious 8.0 (http://www.geneious.com/), and subjected to phylogenetic analyses by PAUP* 4.0 that is implemented in Geneious 8.0. Jukes and Cantor model was used to determine evolutionary distance values; the values were then used to construct phylograms by a neighbor-joining method [26]. The maximal parsimony method implemented in Geneious 8.0 was also used to confirm the results of the neighbor-joining method.
Results
Bioinformatics approaches were used to gain insight into biosynthesis of vitamins and cofactors of R. buchneri in I. scapularis. Based on MUMmer alignments against complete genomes of R. bellii, R. conorii, and R. felis, 2,903, 7,496, and 7,203 Trace Archive sequences from the I. scapularis genome project with a minimal nucleotide identity of 70% were identified to be homologous to the three reference Rickettsia genome sequences. Overall, MUMmer identified 8,190 unique sequences that are homologous to Rickettsia sequences in the Trace Archive (S1 Table). Among the sequence matches, two vitamin biosynthetic pathways were manually annotated by MetaCyc metabolic reconstructions in the genome of R. buchneri in I. scapularis, including the vitamin B7 (biotin) and B9 (folate) biosynthetic pathways.
MUMmer alignments and MetaCyc metabolic reconstructions revealed that all folate gene orthologues required for de novo folate biosynthesis are present in the genome of R. buchneri in I. scapularis. The five folate genes are folA, coding for dihydrofolate reductase (FolA), folC, coding for dihydrofolate synthase (or folylpolyglutamate synthase, FolC), folE, coding for GTP-Cyclohydrolase I (FolE), the folKP gene encoding a bifunctional protein 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase (FolK) and dihydropteroate synthase (FolP), and ptpS, coding for 6-pyruvoyltetrahydropterin synthase (PtpS-III).
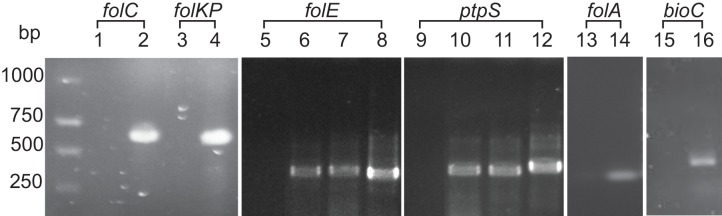
To test if Rickettsia species phylotype G021 has the genetic capacities of biotin and folate biosynthesis, PCR was carried out using degenerate primers designed based on conserved upstream and downstream regions as well as conserved internal sequences of the biotin and folate genes in order to ensure amplifications of complete ORFs of the folate genes of phylotype G021 in I. pacificus (Table 1). All five genes (folA, folC, folE, folKP, and ptpS) of the folate biosynthetic pathway were successfully amplified using gene-specific primers. The sizes of the PCR amplicons match the expected sizes of the folate genes (Fig 1). In contrast, only the bioC gene (encoding malonyl-CoA O-methyltransferase) in the biotin biosynthetic pathway was amplified by the degenerate primers in this study (Fig 1).
Fig 1. Detection of genes from the folate biosynthetic pathway of Rickettsia species phylotype G021 in Ixodes pacificus by PCR.
I. pacificus tick genomic DNA was used as the template for PCR amplification of the genes using internal primers (Table 1). The PCR products were electrophoresed and observed by staining with ethidium bromide. Lane 1 and 2: folC gene amplification without or with I. pacificus DNA, respectively; Lane 3 and 4: folKP gene amplification without or with I. pacificus DNA, respectively; Lane 5 and 6: folE gene amplification without or with I. pacificus DNA, respectively; Lane 7 and 8: folE gene amplification using I. scapularis DNA or Rickettsia montanensis DNA, respectively; Lane 9 and 10: ptpS gene amplification without or with I. pacificus DNA, respectively; Lane 11 and 12: ptpS gene amplification using I. scapularis DNA and Rickettsia montanensis DNA, respectively; Lane 13 and 14: folA gene amplification without or with I. pacificus DNA, respectively. Lane 15 and 16: bioC gene amplification without or with I. pacificus DNA, respectively. bp, base pair.
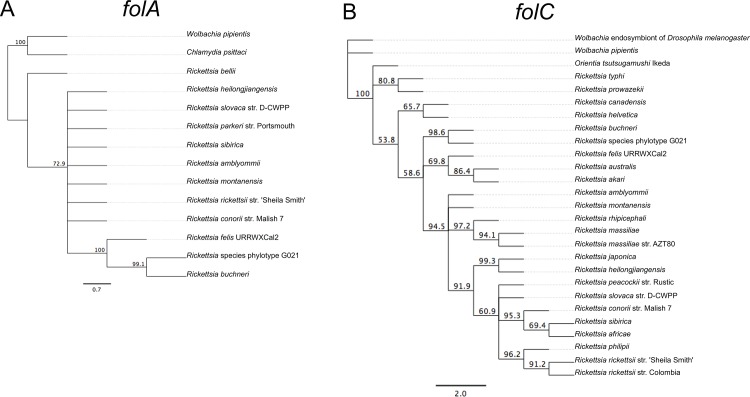
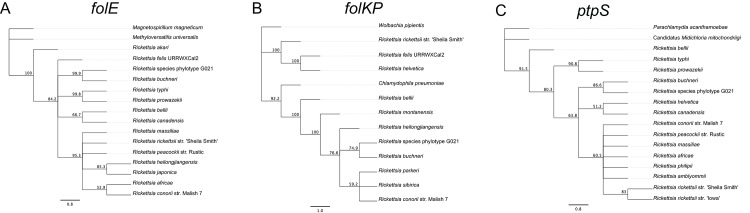
Sequencing of ten clones for each folate gene of Rickettsia species phylotype G021 showed that the folate genes have bacterial origin and no significant homology with any Ixodes genes. BLAST searches revealed that the folA, folC, folE, folKP, and ptpS genes of phylotype G021 possess 98.6%, 98.8%, 98.9%, 98.5% and 99.0% nucleotide sequence identity, respective to the corresponding genes of R. buchneri in I. scapularis [44]. Phylogenetic tree constructions analyses showed that the folate genes of phylotype G021 in I. pacificus and homologous genes from various Rickettsia species from the spotted fever group are monophyletic (Figs 2 and 3). The phylogenetic trees based on the sequences of the folA and folC genes showed that phylotype G021 in I. pacificus is clustered with R. buchneri in I. scapularis and other spotted fever group rickettsiae as well as R. bellii for both folA and folC genes, although the folC gene is also clustered with the folC gene from the typhus group rickettsiae. However, the folA gene from phylotype G021 is on a branch that is distinct from the folA gene from other genera of bacteria, such as Wolbachia species and Chlamydophila felis. Likewise, the folC gene from phylotype G021 falls in a clade that is separated from other genera of bacteria, such as Orientia, Neorickettsia, Wolbachia, and others (Fig 2). In the phylogenetic trees based on the folE, folKP, and ptpS nucleotide sequences, phylotype G021 was again in a cluster of Rickettsia endosymbionts in Ixodes ticks with strong bootstrap support. Phylotype G021 in I. pacificus and R. buchneri in I. scapularis are placed on the same branch with other spotted fever group rickettsiae (Fig 3).
Fig 2. Phylogenetic analyses of the folA and folC genes of Rickettsia species phylotype G021 in Ixodes pacificus.
(A) Phylogram of the folA gene; (B) folC gene. The phylogenetic trees were constructed using the Jukes and Cantor model and the neighbor-joining method included in PAUP*4.0 software. Bootstrap values (based on 1,000 replicates) that are greater than 50% are indicated at the nodes.
Fig 3. Phylogenetic analyses of the folE, folKP, and ptpS genes of Rickettsia species phylotype G021 in Ixodes pacificus.
(A) Phylogram of the folE gene; (B) folKP gene; (C) ptpS gene. The phylogenetic trees were constructed using the Jukes and Cantor model and the neighbor-joining method included in PAUP*4.0 software. Bootstrap values (based on 1,000 replicates) that are greater than 50% are indicated at the nodes.
Subsequent bioinformatic analyses based on sequences of the clone DNA confirmed that the folate biosynthetic pathway-coding genes exist in the genome of Rickettsia species phylotype G021. Specifically, the folA, folC, folE, folKP, and ptpS genes from phylotype G021 have 642, 1,281, 573, 1,329, and 417 nucleotide ORFs, respectively, that encode 213, 426, 190, 442, and 138 amino acids, respectively, with predicted protein molecular masses of 24.65, 47.4, 21.81, 50.56, and 16.13 kDa, respectively. The upstream region of each folate gene from phylotype G021 was studied with the aim of identifying cis-acting control elements. We have identified a putative ribosome-binding site (identical or one nucleotide mismatch to the prokaryotic consensus sequences AGGAGG or AGGA of the ribosome-binding site) around 8 nucleotides immediately preceding each folate gene. Using Softberry BPROM, we have identified a putative E. coli σ 70-like promoter that is located from 31 to 51 nucleotides upstream of the translational start codon of folE gene. However, no putative E. coli σ70-like promoters were identified within 67, 54, 60, and 77 sequenced nucleotides that are upstream of the translation start site of the folA, folC, folKP, and ptpS genes of the sequenced clones, respectively.
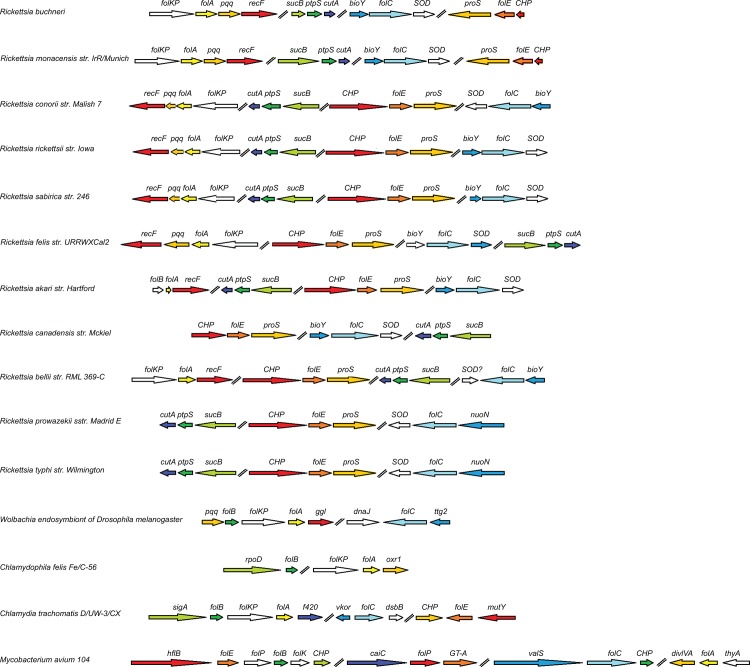
In contrast to a monocistronic organization of the other folate genes, the folA and folKP gene loci are polycistronic; the mRNA is made of folKP-folA-pqq-recF genes (the pqq gene encodes for pyrroloquinoline synthesis protein and recF encodes for a DNA replication and repair protein). Although the polycistronic folKP-folA gene organization is found in many bacteria [45, 46], BLAST searches showed that the folKP-folA-pqq-recF polycistronic organization is only present in the Rickettsia genus. Wolbachia, a closely related genus, have a different genetic organization of the folKP-folA genes, indicating that the two genera employed different evolutionary strategies for acquiring the folate genes (Fig 4) [47].
Fig 4. Physical clustering of folate biosynthetic genes across Rickettsia genus and related bacterial species.
ORFs and coding strand are indicated by arrows. The lengths of the arrows represent the lengths of the genes, with a scale of 0.1 inch = 100 bp. Adjacent arrows represent proximal loci, while the double lines between some arrows represent non-adjacency between loci. The names above the arrows represent the gene name; CHP gene encodes for conserved hypothetical protein with no known function; Abbreviations not described in the text: proS, prolyl-tRNA synthetase; sod, superoxide dismutase; bioY, biotin synthesis BioY protein; cutA, divalent cation tolerance protein; sucB, dihydrolipoamide succinyltransferase; nuoN, NADH dehydrogenase subunit N; ggl, gamma-glutamate ligase; dnaJ, molecular chaperone DnaJ; ttg2, toluene tolerance protein; rpoD, RNA polymerase sigma factor; oxr1, oxidoreductase 1; thyA, thymidylate synthetase; mutY, adenine DNA glycosylase; dsbB, disulfide bond formation protein; vkor, vitamin K epoxide; f420, coenzyme f420 hydrogenase; sigA, RNA polymerase sigma factor RpoD; hflB, ATP-dependent metalloprotease; caiC, crotonobetaine/carnitine-CoA ligase; GT-A, beta-1,4-N-acetyl-galactosaminyl transferase 1; valS, valyl-tRNA synthetase; divlVA, cell division protein DivlVA.
The KEGG pathway database was used as a reference knowledge base to see if the folate biosynthetic pathway is present in other Rickettsia species. Among 44 annotated Rickettsia genomes, other than R. buchneri in I. scapularis and Rickettsia species phylotype G021 in I. pacificus, fifteen Rickettsia species have five folate genes for de novo folate biosynthesis (Fig 4). Specifically, two Rickettsia species (R. bellii RML 369 and R. bellii BSU8S-389) in the Rickettsia belli group and thirteen Rickettsia species (R. conorii subsp. Israelensis, R. felis str. LSU-Lb, R. rickettsii Shelia Smith, R. rickettsii Arizona, R. rickettsia Colombia, R. rickettsii Hauke, R. rickettsii Brazil, R. rickettsia Hino, R. rickettsii Morgan, R. rickettsii R, R. heilongjiangensis, R. montanensis, and R. parkeri) in the spotted fever group have all five folate genes that are required for de novo folate biosynthesis. In contrast, genome sequences from other Rickettsia species revealed that the folKP gene is lost and only some of the five genes of the folate biosynthetic pathway are present. Specifically, some Rickettsia species possess folC, folE, and ptpS genes only, whereas some other species possess only folA, folC, folE, and ptpS genes. The genetic capacity for de novo folate biosynthesis is not related to bacterial pathogenicity since both pathogens (such as R. rickettsii) and endosymbionts (such as Rickettsia species phylotype G021 and R. buchneri) have all five folate genes. Interestingly, all typhus group rickettsiae sequenced so far (two R. typhi genotypes, ten R. prowazekii genotypes) have folC, folE, and ptpS genes, but folA, folK, and folP genes are not present in their genomes (Fig 4).
Discussion
Our findings that R. buchneri in I. scapularis has all five genes required for the synthesis of folate were based on MUMmer alignments and MetaCyc metabolic reconstructions using the NCBI Trace Archive data from the I. scapularis genome sequencing project [34, 35]. The bioinformatics analyses were carried out before the complete annotation of the genome of I. scapularis was released in 2012 (https://www.vectorbase.org/news/ixodes-scapularis-gene-set-iscaw12-released). Our study demonstrated that Rickettsia species phylotype G021 in I. pacificus has the genetic capacity to de novo synthesize tetrahydrofolate. In contrast, only the folA, folC, and folE genes were correctly annotated by the I. scapularis genome sequencing project (http://www.ncbi.nlm.nih.gov/genome/1908). So far, the genome of R. buchneri ISO7T strain [48] has been sequenced but has not been annotated completely (http://www.ncbi.nlm.nih.gov/genome/38153).
Tetrahydrofolate and other folate derivatives are the most essential components for cell growth [49]. Folate is vitamin B9 and is needed by all eukaryotic and prokaryotic organisms for the one-carbon metabolism in nucleotide synthesis and in the methylation of DNA, RNA, proteins, and phospholipids [50–52]. Folate insufficiency causes megaloblastic anemia and neural tube defects, such as spina bifida, in humans [53, 54]. Larvae of Drosophila melanogaster showed prolonged development when fed with a diet containing antibiotics and near-zero folate [47]. Deletions of the genes of the folate biosynthetic pathway in E. coli lead to the production of non-viable phenotypes [55, 56]. Animals cannot synthesize folate, and must therefore obtain it either through diet or mutualistic associations with microbes [16, 47]. The tick genome project revealed that ticks lack genetic capacity for de novo folate synthesis [34, 35]. Interestingly, ticks have the ability to survive as long as a year until the next life stage. Since ticks feed exclusively on nutrient poor (especially vitamin B9 and vitamin B7) vertebrate blood [16], they must obtain folate through non-dietary means.
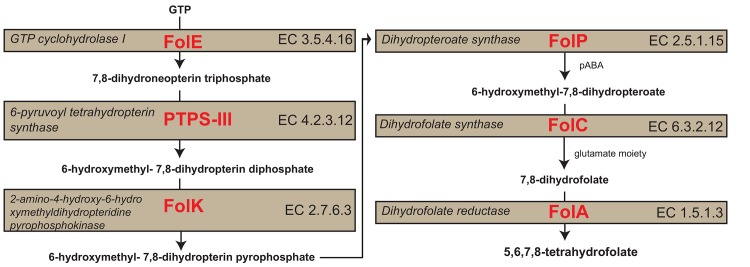
We propose a model for de novo folate biosynthesis by phylotype G021. Guanosine-5'-triphosphate (GTP), from tick hosts or bacterial sources, is catalyzed by a series of enzymatic actions of the FolA, FolC, FolE, FolKP, and PtpS-III proteins from phylotype G021 to synthesize tetrahydrofolate (Fig 5). First, the FolE protein (GTP-Cyclohydrolase I) synthesizes 7,8-dihydroneopterin triphosphate, a pterin-ring molecule, from GTP [57–59]. Then the PTPS-III protein (6-pyruvoyltetrahydropterin synthase) cleaves the side chain of 7,8-dihydroneopterin triphosphate to form 6-hydroxymethyl-7,8-dihydropterin diphosphate [60, 61]. Then the FolKP bifunctional enzyme (2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase and dihydropteroate synthase) joins the pterin ring to para-aminobenzoic acid (pABA) to synthesize 6-hydroxymethyl-7,8-dihydropteroate [62]. A glutamate moiety is then added to 6-hydroxymethyl-7,8-dihydropteroate by FolC (dihydrofolate synthase) to synthesize 7,8-dihydrofolate [63], which is then utilized by the FolA protein (dihydrofolate reductase) to synthesize tetrahydrofolate [43].
Fig 5. The proposed pathway of folate biosynthesis by Rickettsia species phylotype G021 in Ixodes pacificus.
The diagram shows the sequence of intermediates produced from the enzymatic reactions that constitute the pathway for synthesizing folate. The precursor, GTP, is transformed and combined with pABA and glutamate, which eventually leads to the reduced cofactor tetrahydrofolate.
Many bacteria and plants as well as lower eukaryotes have a genetic capacity for de novo folate biosynthesis that requires the presence of a folB gene. The folB gene encodes dihydroneopterin aldolase, an enzyme that catalyzes the substrate 2-amino-4-hydroxy-6-(D-erythro-1,2,3-trihydroxypropyl)-7,8-dihydropteridine to 2-amino-4-hydroxy-6-hydroxymethyl-7,8-dihydropteridine and glycolaldehyde [60, 61]. However, our MUMmer alignments didn’t identify a folB orthologue in the genome of R. buchneri of I. scapularis. Likewise, the folB gene is not present in genomes of many bacteria, including the phyla Acidobacteria, Chloroflexi, Firmicutes, Planctomycetes, and Spirochaetes [61]. Although non-orthologous enzymes could carry out the function of dihydroneopterin aldolase, or a folB gene with a low nucleotide identity (less than the minimum alignment identity of 70% of MUMmer) could be present in the genome, we identified a PtpS orthologue from the Trace Archive sequences of R. buchneri of I. scapularis and amplified the ptpS gene from phylotype G021. Since both FolB and PtpS-III proteins produce the product 6-hydroxymethyl-7,8 dihydropterin, which is required as a substrate for the FolKP protein, PtpS-III protein is considered as a functional alternative of the FolB protein in the folate biosynthesis pathway of many bacteria and Plasmodium species [60] as well as phylotype G021.
Due to the essential functions of folate in cell growth and cell division [50–52], it is not surprising that both pathogenic and endosymbiotic rickettsiae can carry out de novo folate biosynthesis. Having this genetic capacity is important since folate is absolutely required during the period of rapid cell growth and cell division inside hosts, including the engorgement stage after blood meals in arthropod hosts. A readily available source of folate synthesized by phylotype G021 promotes rapid cell proliferation not only for the bacterium but also for I. pacificus tick. This nutritional interaction between endosymbiotic phylotype G021 and I. pacificus may explain the predominant presence of the bacterium in I. pacificus [27, 28]. It should also be noted at this time that many other bacteria belonging to the tick microbiome could also provide folate in addition to phylotype G021. However, the interaction between I. pacificus and phylotype G021 is an important first step in the study of the folate biosynthesis and provides a framework for identifying other potential bacterial folate producers in the tick microbiome.
Although many Rickettsia species from the spotted fever group and Rickettsia belli group have the genetic capacity to synthesize folate, none of the twelve sequenced Rickettsia species or genotypes from the typhus group possess the folA, folK, and folP genes in their genomes (Fig 4). The missing genes indicate that those Rickettsia species lack the genetic capacity for de novo biosynthesis of folate. The major invertebrate hosts for the typhus group Rickettsia are lice and fleas [64]. In contract, hard ticks are the main hosts for the spotted fever and Rickettsia belli group of Rickettsia. It could be true that lice and fleas have other enzymes from either arthropods or other microbial symbionts to convert 6-hydroxymethyl-7,8-dihydropterin to tetrahydrofolate.
Little is known about the nutritional basis for the symbiotic relationship of endosymbionts in ticks, though newly developed systems are beginning to shed light on novel interactions. Kurtti et. al, recently isolated and characterized Rickettsia buchneri from tick embryonic cell lines IRE11 and ISE6 [48]. This approach provides a basis to further testing symbiotic interactions of other Rickettsia species. Employing a similar methodology, the culturing of Rickettsia species phylotype G021 using a tick embryonic cell line could be feasible. Understanding the nutritional interactions between Rickettsia species in ticks may help us to eliminate Lyme disease and other tick-borne diseases by suppressing tick population.
Supporting Information
(JAVA)
(JAVA)
Column 1 and 2 are call numbers. Column 3 is the GenBank accession numbers of homologous proteins. Column 4 is the names of the homologous proteins. Column 5 is the names of the bacterial species. Column 6 and 7 are the percentage identity to top BLASTX results. Column 8 is the AT% for the DNA. Column 9 is the sequences obtained by the MUMmer alignment.
(XLS)
Acknowledgments
We thank Dr. David H. Walker at the University of Texas Medical Branch for providing Rickettsia montanensis DNA.
Data Availability
Data are available for download from Dryad: doi:10.5061/dryad.2jc88.
Funding Statement
This work was supported by National Institute of Health AREA grant 1 R15 AI099902-01. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Duron O, Hurst GD. Arthropods and inherited bacteria: from counting the symbionts to understanding how symbionts count. BMC Biol. 2013;11: 45 10.1186/1741-7007-11-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Douglas AE. The molecular basis of bacterial-insect symbiosis. J Mol Biol. 2014;426: 3830–3837. 10.1016/j.jmb.2014.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Douglas AE. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera . Annu Rev Entomol. 1998;43: 17–37. [DOI] [PubMed] [Google Scholar]
- 4. Zientz E, Dandekar T, Gross R. Metabolic interdependence of obligate intracellular bacteria and their insect hosts. Microbiol Mol Biol Rev. 2004;68: 745–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baumann P. Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol. 2005;59: 155–189. [DOI] [PubMed] [Google Scholar]
- 6. Currie CR, Stuart AE. Weeding and grooming of pathogens in agriculture by ants. Proc Biol Sci. 2001;268: 1033–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Oliver KM, Moran NA, Hunter MS. Variation in resistance to parasitism in aphids is due to symbionts not host genotype. P Natl Acad Sci USA. 2005;102: 12795–12800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Piel J. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. P Natl Acad Sci USA. 2002;99: 14002–14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Breeuwer JA, Jacobs G. Wolbachia: intracellular manipulators of mite reproduction. Exp Appl Acarol. 1996;20: 421–434. [DOI] [PubMed] [Google Scholar]
- 10. Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6: 741–751. 10.1038/nrmicro1969 [DOI] [PubMed] [Google Scholar]
- 11. Gross R, Vavre F, Heddi A, Hurst GD, Zchori-Fein E, Bourtzis K. Immunity and symbiosis. Mol Microbiol. 2009;73: 751–759. 10.1111/j.1365-2958.2009.06820.x [DOI] [PubMed] [Google Scholar]
- 12. Moya A, Pereto J, Gil R, Latorre A. Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat Rev Genet. 2008;9: 218–229. 10.1038/nrg2319 [DOI] [PubMed] [Google Scholar]
- 13. Martin MM. The biochemical basis of the fungus-attine ant symbiosis. Science. 1970;169: 16–20. [DOI] [PubMed] [Google Scholar]
- 14. Baumann P, Baumann L, Lai CY, Rouhbakhsh D, Moran NA, Clark MA. Genetics, physiology, and evolutionary relationships of the genus Buchnera: intracellular symbionts of aphids. Annu Rev Microbiol. 1995;49: 55–94. [DOI] [PubMed] [Google Scholar]
- 15. Akman L, Aksoy S. A novel application of gene arrays: Escherichia coli array provides insight into the biology of the obligate endosymbiont of tsetse flies. P Natl Acad Sci USA. 2001;98: 7546–7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Akman L, Yamashita A, Watanabe H, Oshima K, Shiba T, Hattori M, et al. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia . Nat Genet. 2002;32: 402–407. [DOI] [PubMed] [Google Scholar]
- 17. McCutcheon JP, McDonald BR, Moran NA. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. P Natl Acad Sci USA. 2009;106: 15394–15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Defossez E, Djieto-Lordon C, McKey D, Selosse MA, Blatrix R. Plant-ants feed their host plant, but above all a fungal symbiont to recycle nitrogen. Proc Biol Sci. 2011;278: 1419–1426. 10.1098/rspb.2010.1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Neef A, Latorre A, Pereto J, Silva FJ, Pignatelli M, Moya A. Genome economization in the endosymbiont of the wood roach Cryptocercus punctulatus due to drastic loss of amino acid synthesis capabilities. Genome Biol Evol. 2011;3: 1437–1448. 10.1093/gbe/evr118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vigneron A, Masson F, Vallier A, Balmand S, Rey M, Vincent-Monegat C, et al. Insects recycle endosymbionts when the benefit is over. Curr Biol. 2014;24: 2267–2273. 10.1016/j.cub.2014.07.065 [DOI] [PubMed] [Google Scholar]
- 21. Gerdes S, Lerma-Ortiz C, Frelin O, Seaver SM, Henry CS, de Crecy-Lagard V, et al. Plant B vitamin pathways and their compartmentation: a guide for the perplexed. J Exp Bot. 2012;63: 5379–5395. 10.1093/jxb/ers208 [DOI] [PubMed] [Google Scholar]
- 22. Beard CB, Dotson EM, Pennington PM, Eichler S, Cordon-Rosales C, Durvasula RV. Bacterial symbiosis and paratransgenic control of vector-borne Chagas disease. Int J Parasitol. 2001;31: 621–627. [DOI] [PubMed] [Google Scholar]
- 23. Wu D, Daugherty SC, Van Aken SE, Pai GH, Watkins KL, Khouri H, et al. Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biol. 2006;4: e188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Westrom DR, Lane RS, Anderson JR. Ixodes pacificus (Acari: Ixodidae): population dynamics and distribution on Columbian black-tailed deer (Odocoileus hemionus columbianus). J Med Entomol. 1985;22: 507–511. [DOI] [PubMed] [Google Scholar]
- 25. Burgdorfer W, Lane RS, Barbour AG, Gresbrink RA, Anderson JR. The western black-legged tick, Ixodes pacificus: a vector of Borrelia burgdorferi . Am J Trop Med Hyg. 1985;34: 925–930. [DOI] [PubMed] [Google Scholar]
- 26. Phan JN, Lu CR, Bender WG, Smoak RM 3rd, Zhong J. Molecular detection and identification of Rickettsia species in Ixodes pacificus in California. Vector Borne Zoonotic Dis. 2011;11: 957–961. 10.1089/vbz.2010.0077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cheng D, Vigil K, Schanes P, Brown RN, Zhong J. Prevalence and burden of two rickettsial phylotypes (G021 and G022) in Ixodes pacificus from California by real-time quantitative PCR. Ticks Tick Borne Dis. 2013;4: 280–287. 10.1016/j.ttbdis.2012.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cheng D, Lane RS, Moore BD, Zhong J. Host blood meal-dependent growth ensures transovarial transmission and transstadial passage of Rickettsia sp. phylotype G021 in the western black-legged tick (Ixodes pacificus). Ticks Tick Borne Dis. 2013;4: 421–426. 10.1016/j.ttbdis.2013.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kurlovs AH, Li J, Cheng D, Zhong J. Ixodes pacificus ticks maintain embryogenesis and egg hatching after antibiotic treatment of Rickettsia endosymbiont. PloS one. 2014;9: e104815 10.1371/journal.pone.0104815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kambhampati S, Alleman A, Park Y. Complete genome sequence of the endosymbiont Blattabacterium from the cockroach Nauphoeta cinerea (Blattodea: Blaberidae). Genomics. 2013;102: 479–483. 10.1016/j.ygeno.2013.09.003 [DOI] [PubMed] [Google Scholar]
- 31. Hansen AK, Moran NA. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. P Natl Acad Sci USA. 2011;108: 2849–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Delcher AL, Phillippy A, Carlton J, Salzberg SL. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30: 2478–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Salzberg SL, Dunning Hotopp JC, Delcher AL, Pop M, Smith DR, Eisen MB, et al. Serendipitous discovery of Wolbachia genomes in multiple Drosophila species. Genome Biol. 2005;6: R23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hill CA, Wikel SK. The Ixodes scapularis Genome Project: an opportunity for advancing tick research. Trends Parasitol. 2005;21: 151–153. [DOI] [PubMed] [Google Scholar]
- 35. Pagel Van Zee J, Geraci NS, Guerrero FD, Wikel SK, Stuart JJ, Nene VM, et al. Tick genomics: the Ixodes genome project and beyond. Int J Parasitol. 2007;37: 1297–1305. [DOI] [PubMed] [Google Scholar]
- 36. Ogata H, La Scola B, Audic S, Renesto P, Blanc G, Robert C, et al. Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLOS Genet. 2006;2: e76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sentausa E, El Karkouri K, Robert C, Raoult D, Fournier PE. Genome sequence of Rickettsia conorii subsp. israelensis, the agent of Israeli spotted fever. J Bacteriol. 2012;194: 5130–5131. 10.1128/JB.01118-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gillespie JJ, Driscoll TP, Verhoeve VI, Utsuki T, Husseneder C, Chouljenko VN, et al. Genomic diversification in strains of Rickettsia felis Isolated from different arthropods. Genome Biol Evol. 2015;7: 35–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Merhej V, Angelakis E, Socolovschi C, Raoult D. Genotyping, evolution and epidemiological findings of Rickettsia species. Infect Genet Evol. 2014;25: 122–137. 10.1016/j.meegid.2014.03.014 [DOI] [PubMed] [Google Scholar]
- 40. Furman DP, Loomis EC. The ticks of California (Acari:Ixodida) Berkeley: University of California Press; 1984. viii, 239. [Google Scholar]
- 41. Moran-Cadenas F, Schneider H, Lommano E, Burri C, Moret J, Gern L. A comparison of two DNA extraction approaches in the detection of Borrelia burgdorferi sensu lato from live Ixodes ricinus ticks by PCR and reverse line blotting. Vector Borne Zoonotic Dis. 2007;7: 555–561. [DOI] [PubMed] [Google Scholar]
- 42. Fan H, Brunham RC, McClarty G. Acquisition and synthesis of folates by obligate intracellular bacteria of the genus Chlamydia . J Clin Invest. 1992;90: 1803–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zywno-van Ginkel S, Dooley TP, Suling WJ, Barrow WW. Identification and cloning of the Mycobacterium avium folA gene, required for dihydrofolate reductase activity. FEMS Microbiol Lett. 1997;156: 69–78. [DOI] [PubMed] [Google Scholar]
- 44. Gillespie JJ, Joardar V, Williams KP, Driscoll T, Hostetler JB, Nordberg E, et al. A Rickettsia genome overrun by mobile genetic elements provides insight into the acquisition of genes characteristic of an obligate intracellular lifestyle. J Bacteriol. 2012;194: 376–394. 10.1128/JB.06244-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Adams NE, Thiaville JJ, Proestos J, Juarez-Vazquez AL, McCoy AJ, Barona-Gomez F, et al. Promiscuous and adaptable enzymes fill "holes" in the tetrahydrofolate pathway in Chlamydia species. mBio. 2014;5: e01378–14. 10.1128/mBio.01378-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sybesma W, Starrenburg M, Kleerebezem M, Mierau I, de Vos WM, Hugenholtz J. Increased production of folate by metabolic engineering of Lactococcus lactis . Appl Environ Microb. 2003;69: 3069–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Blatch SA, Meyer KW, Harrison JF. Effects of dietary folic acid level and symbiotic folate production on fitness and development in the fruit fly Drosophila melanogaster . Fly. 2010;4: 312–319. 10.4161/fly.4.4.13258 [DOI] [PubMed] [Google Scholar]
- 48. Kurtti TJ, Felsheim RF, Burkhardt NY, Oliver JD, Heu CC, Munderloh UG. Rickettsia buchneri sp. nov., a rickettsial endosymbiont of the blacklegged tick Ixodes scapularis . Int J Syst Evol Micr. 2015;65: 965–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bermingham A, Derrick JP. The folic acid biosynthesis pathway in bacteria: evaluation of potential for antibacterial drug discovery. Bioessays. 2002;24: 637–648. [DOI] [PubMed] [Google Scholar]
- 50. Sybesma W, Burgess C, Starrenburg M, van Sinderen D, Hugenholtz J. Multivitamin production in Lactococcus lactis using metabolic engineering. Metab Eng. 2004;6: 109–115. [DOI] [PubMed] [Google Scholar]
- 51. Wheeler PR. One-carbon requirements of Mycobacterium leprae: need for the folate pathway. Int J Lepr Other Mycobact Dis. 1992;60: 445–450. [PubMed] [Google Scholar]
- 52. Fenech M. Folate (vitamin B9) and vitamin B12 and their function in the maintenance of nuclear and mitochondrial genome integrity. Mutat Res. 2012;733: 21–33. 10.1016/j.mrfmmm.2011.11.003 [DOI] [PubMed] [Google Scholar]
- 53. Sifakis S, Pharmakides G. Anemia in pregnancy. Ann NY Acad Sci. 2000;900: 125–136. [DOI] [PubMed] [Google Scholar]
- 54. Darnton-Hill I, Mkparu UC. Micronutrients in pregnancy in low- and middle-income countries. Nutrients. 2015;7: 1744–1768. 10.3390/nu7031744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pyne C, Bognar AL. Replacement of the folC gene, encoding folylpolyglutamate synthetase-dihydrofolate synthetase in Escherichia coli, with genes mutagenized in vitro. J Bacteriol. 1992;174: 1750–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fermer C, Swedberg G. Adaptation to sulfonamide resistance in Neisseria meningitidis may have required compensatory changes to retain enzyme function: kinetic analysis of dihydropteroate synthases from N. meningitidis expressed in a knockout mutant of Escherichia coli . J Bacteriol. 1997;179: 831–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hossain T, Rosenberg I, Selhub J, Kishore G, Beachy R, Schubert K. Enhancement of folates in plants through metabolic engineering. P Natl Acad Sci USA. 2004;101: 5158–5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. El Yacoubi B, Bonnett S, Anderson JN, Swairjo MA, Iwata-Reuyl D, de Crecy-Lagard V. Discovery of a new prokaryotic type I GTP cyclohydrolase family. J Biol Chem. 2006;281: 37586–37593. [DOI] [PubMed] [Google Scholar]
- 59. Grochowski LL, Xu H, Leung K, White RH. Characterization of an Fe(2+)-dependent archaeal-specific GTP cyclohydrolase, MptA, from Methanocaldococcus jannaschii . Biochemistry. 2007;46: 6658–6667. [DOI] [PubMed] [Google Scholar]
- 60. Dittrich S, Mitchell SL, Blagborough AM, Wang Q, Wang P, Sims PF, et al. An atypical orthologue of 6-pyruvoyltetrahydropterin synthase can provide the missing link in the folate biosynthesis pathway of malaria parasites. Mol Microbiol. 2008;67: 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pribat A, Jeanguenin L, Lara-Nunez A, Ziemak MJ, Hyde JE, de Crecy-Lagard V, et al. 6-pyruvoyltetrahydropterin synthase paralogs replace the folate synthesis enzyme dihydroneopterin aldolase in diverse bacteria. J Bacteriol. 2009;191: 4158–4165. 10.1128/JB.00416-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gengenbacher M, Xu T, Niyomrattanakit P, Spraggon G, Dick T. Biochemical and structural characterization of the putative dihydropteroate synthase ortholog Rv1207 of Mycobacterium tuberculosis . FEMS Microbiol Lett. 2008;287: 128–135. 10.1111/j.1574-6968.2008.01302.x [DOI] [PubMed] [Google Scholar]
- 63. Wang P, Wang Q, Yang Y, Coward JK, Nzila A, Sims PF, et al. Characterisation of the bifunctional dihydrofolate synthase-folylpolyglutamate synthase from Plasmodium falciparum; a potential novel target for antimalarial antifolate inhibition. Mol Biochem Parasit. 2010;172: 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Azad AF, Beard CB. Rickettsial pathogens and their arthropod vectors. Emerg Infect Dis. 1998;4: 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(JAVA)
(JAVA)
Column 1 and 2 are call numbers. Column 3 is the GenBank accession numbers of homologous proteins. Column 4 is the names of the homologous proteins. Column 5 is the names of the bacterial species. Column 6 and 7 are the percentage identity to top BLASTX results. Column 8 is the AT% for the DNA. Column 9 is the sequences obtained by the MUMmer alignment.
(XLS)
Data Availability Statement
Data are available for download from Dryad: doi:10.5061/dryad.2jc88.