Abstract
Mammalian lymphoid immunity is mediated by fast and slow responders to pathogens. Fast innate lymphocytes are active within hours after infections in mucosal tissues. Slow adaptive lymphocytes are conventional T and B cells with clonal antigen receptors that function days after pathogen exposure. A transcription factor (TF) regulatory network guiding early T cell development is at the core of effector function diversification in all innate lymphocytes, and the kinetics of immune responses is set by developmental programming. Operational units within the innate lymphoid system are not classified by the types of pathogen-sensing machineries but rather by discrete effector functions programmed by regulatory TF networks. Based on the evolutionary history of TFs of the regulatory networks, fast effectors likely arose earlier in the evolution of animals to fortify body barriers, and in mammals they often develop in fetal ontogeny prior to the establishment of fully competent adaptive immunity.
Keywords: T cells, transcription factor regulatory network, evolution of immunity, fetal lymphopoiesis, γδ T cells, cytokines
INTRODUCTION
The immune system deploys cytokines to demarcate the tissue areas of hyperstasis precipitated by infections. Cytokines fortify the mucosal barrier and marshal a cadre of activated, specialized cell types that collectively control infectious agents at sites of infection. Tightly controlled duration of action and choice of cytokines tailored to specific damaging events are essential for a rapid restoration of tissue homeostatic set points. Unique combinations of effector cytokines are made by specialized lymphoid effector subsets, which are considered distinct cell lineages. Emergence in the past decade of lymphoid effector subsets that are classified as performing innate function has firmly established that mammalian lymphoid immunity consists of at least three layers of overlapping defense against pathogens (Figure 1). Each lymphoid layer is arrayed at different proximity to the entry points for pathogens, from the mucosal epithelia, where most pathogens first interact with host cells, to the more distal locations in the secondary lymphoid tissues, such as lymph nodes (LNs), where T and B lymphocytes are alerted and mobilized for reaction and deployment against pathogen-derived products. The barrier proximal layer is populated by fast-acting innate lymphoid cells (ILCs) that lack somatically generated clonal antigen receptors (1). Intermixed among them and extending to lymphoid tissues is another group of fast-acting innate effectors that express T or B cell antigen receptors (2, 3). Arrayed behind these fast effector subsets are the conventional T and B cells of adaptive immunity, whose participation in pathogen clearance is kinetically and strategically delayed in animals that have not previously encountered the same pathogen (4, 5). Remarkably, each layer of protection includes cell subsets that can secrete a near identical repertoire of effector cytokines that operationally define them. Mosmann, Coffman, and colleagues first described the lymphoid division of labor within adaptive helper CD4+ αβ T cells by identifying IFN-γ-producing Th1 cells and IL-4-producing Th2 cells that are primarily responsible for bacterial and parasitic clearances, respectively (6). Subsequently, the discovery of IL-17-producing CD4+ T cells, which are termed Th17 cells (7, 8) and are involved in evoking tissue inflammation and neutrophil recruitment, entrenched the triad of effector cytokine classes and their cellular sources, which dominantly dictate pathogen-tailored immunity: type 1 (IFN-γ, TNF), type 2 (IL-4, IL-5, and IL-13), and type 3 (IL-17 and IL-22). The adaptive CD4+ effector T cell subsets are specialized by a cohort of transcription factors (TFs) that are responsible for the cytokine signatures, with T-bet (Tbx21), GATA3, and RORγt (Rorc) segregating with Th1, Th2, and Th17 cells, respectively (9–12). The same division of effector cytokine production and subset-defining TFs is evident among all fast innate lymphoid effector (ILE) cells studied in some depth. Therefore, in nearly all tissues where lymphocytes patrol, critical effector cytokines are produced by multiple fast and slow responders.
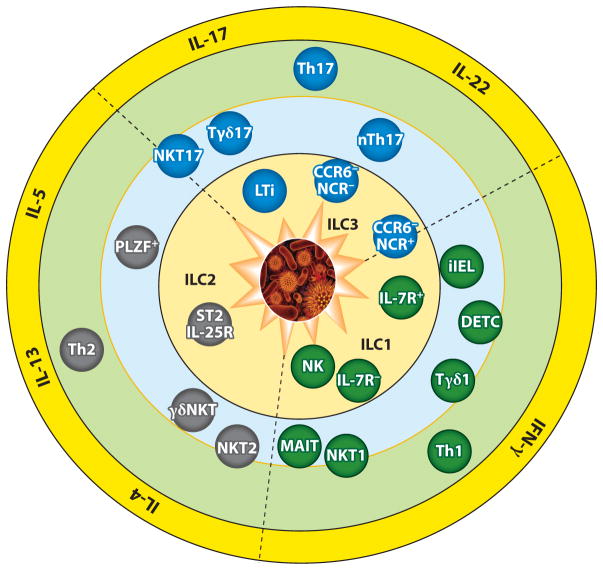
Figure 1.
Three overlapping layers of T cell and T cell–like immunity and representative effector subsets in each layer. A pathogen breach of the body barrier activates each layer in succession, with the fast-acting innate lymphocytes arrayed most proximal to the barrier surfaces. The subsets are described in the text. The effector repertoire of each layer is similar, broadly divided into three wedges united by the type of cytokines each can produce. Heterogeneity of functional units (at the cell lineage level) within the fast inner layers is complex, whereas for the slow outer adaptive ring it is more limited, although inflammations can drive conversions of established Th subsets. Conversely, clonal variations at the outer ring are extremely high because individual cells express distinct antigen receptors, whereas the subsets in the inner rings often express canonical antigen receptors and/or pathogen-associated pattern receptors. The middle ring contains innate effectors that express either αβTCR or γδTCR. For the latter, those found in lymphoid tissues (such as Tγδ1) are mostly naive, express clonal TCRs, and primarily exhibit default IFN-γ production, similar to NK cells. Their numbers in mammals are dwarfed by those of the innate γδT cells in tissues, owing to the vast surface areas of mucosal tissues such as the skin and the gut. Type 1 cytokine–producing cells are marked as IFN-γ enabled, ranging from cNK cells in the inner ring to the adaptive Th1 cells in the outer ring. Cells in the inner rings are mostly tissue tropic, although some (such as cNK and Tγδ1 cells) are circulatory. The number of type 2 cytokine (IL-4/5/13)-producing cell subsets is the smallest, and IL-4 production in peripheral tissues is constrained. Type 3 cytokine–producing fast effector cells exhibit distinct profiles of IL-17 and IL-22 secretion, with the innermost fast effectors biased toward IL-22 production, whereas in the middle ring IL-22 production is tightly regulated, with skin Tγδ17 cells primed for IL-17, but not IL-22, secretion. For simplicity, the classification of cytokine types shown is not comprehensive; some cytokines are not listed (an example is IL-9, which is considered a type 2 cytokine) or detailed (for example, the IL-17 family has six members, some with unique activities). Also, additional Th subsets, such as Bcl6+ T follicular helper (Tfh) cells and PU.1+ Th9 cells, are not depicted because clear corollary populations among ILE cells have not been established. NKT and γδT cells can secrete cytokines made by these Th subsets (IL-9, IL-10, and IL-21) and can promote B cell maturation and antibody production, but it is unclear whether corresponding specialized subsets exist. ST2 (Il1rl1) and IL-25R (IL-17RB) are receptors for two epithelial cytokines, IL-33 and IL-17E (IL-25), respectively, and both are activators of various type 2 cytokine–producing cells. Abbreviations: DETC, dendritic epidermal T cell; iIEL, intestinal intraepithelial lymphocyte; LTi, lymphoid tissue–inducer cell; MAIT, mucosal-associated invariant T cell; NCR, NK cell receptor; PLZF, promyelocytic leukemia zinc finger.
Clearly, what distinguish each layer of lymphoid protection are the apparatuses for sensing damaging pathogens. Together, these provide a built-in redundancy in effector cytokine production, presumably such that evolving pathogens are less likely to evade host sensing altogether. Although the pathogen-sensing mechanisms for some lymphoid subsets remain a mystery, gene regulatory circuits controlling effector subset differentiation in each layer of immunity are coming into sharper focus. In this review we describe TF regulators of murine innate effector subsets and their function and evolutionary origin, in support of the thesis that the fast-acting lymphoid compartment of the mammalian immune system is built using common TF regulatory networks that arose in a stepwise manner during evolution, prior to the emergence of TFs and effector molecules dedicated to fostering the slow, adaptive compartment. Moreover, in mammals, the TF network for fast responses is largely constructed in the fetus, before functional adaptive immunity is established. Thus, ILE cells are built for speed—in evolution, in ontogeny, and in early phases of differentiating cells—exploiting versatile, shared gene regulatory circuits for immediate reactions against invasive microorganisms.
DIVERSITY OF ILE CELLS
Operational Definition of Mammalian Lymphoid Immunity
In a simplified scheme the immune system can be described as an interconnected network of components of the two territories populated by fast-acting and slow-acting lymphocytes. Functional parameters of the former are hardwired, whereas pathogen-driven adaptive response initiated in naive, unprogrammed lymphocytes requires time to alter the chromatin landscape of clonal responders. The cost of exquisite pathogen specificity generated by the adaptive, inducible gene circuit is the potential danger associated with slower kinetics of response. This danger is mitigated by the fast response that, although not necessarily tailored to specific pathogens, does permit the initial confinement of foreign entities to sites of infection (3, 13). In metazoan (animal) evolution, immune responses that are slow are associated with more complex organisms (14, 15), suggesting that the fast innate response is the basic foundation of immunity that has been elaborated further to generate pathogen-adaptive immunity. Thus, it is constructive to deduce molecular properties of fast lymphoid effectors to map basic molecular principles responsible for the division of labor in the immune system. This exploration has been led by the identification and characterization of TFs that impose unique, aggregate molecular features of distinct effector functions. Before we discuss the design principle of fast innate lymphocytes, we describe the spectrum of cell types belonging to this class. Although both fast responder T cells and ILCs are innate lymphocytes, given the nomenclature already established for mucosal ILCs (16), we refer to all fast-acting lymphocytes as ILE cells. Innate B lymphocytes, such as B1 cells, which will not be covered here, are also under thematically similar molecular control (17).
Fast ILE Cells Expressing γδ TCR
Tissue-localized murine γδ T cells are the prototypic innate T cells (18). The existence of γδ T cells was revealed in the early 1980s, when the genes coding for γδ and αβ TCRs were discovered; as such, γδ T cells were classified as adaptive T cells. However, it became apparent soon after that γδ and αβ T cells possess radically distinct kinetics of response: Specific subsets of γδ T cells were identified as the earliest lymphoid responders after infection and injury (18); they are primarily an alternative source of IFN-γ (19), mimicking natural killer (NK) cells, the first ILCs discovered (1, 20). Among these Th1-like cells (Figure 1) are the self-renewing, epidermal-resident T cells expressing the canonical Vγ3-Vδ1TCR [dendritic epidermal T cells (DETCs) (21); the TCRγ nomenclature is based on Reference 22] and Vγ5TCR+ intestinal intraepithelial cells (iIELs) positioned as ordered sentries in the epithelial barrier (23–26). iIELs are necessary for optimal induction of antimicrobial compounds by the epithelia and prevention of deregulated inflammatory αβ T cell responses, and their maintenance is controlled in part by dietary aryl hydrocarbon (Ah) (25, 26).
The segregation of γδ T cells into distinct effector lineages was eventually established with the discovery of a neonatally derived γδ cell subset poised to produce both IFN-γ and IL-4 (27), and the RORγt+, IL-17-producing γδ T cells (Tγδ17), which are the source of IL-17 detected within hours after infection with Mycobacterium tuberculosis in the lung (28, 29). The former express semi-invariant Vδ6.3TCR and the TF promyelocytic leukemia zinc finger (PLZF, Zbtb16), making them similar in properties to αβ T cells coexpressing an invariant TCR and NK cell markers (NKT cells, discussed below), and are referred to as γδ NKT cells (30–32). They are selectively expanded during bacterial infection, in particular by Listeria monocytogenes (33), and likely contribute to protecting the host from infection-induced tissue damage (34). Tγδ17 cells primarily express Vγ2 or Vγ4TCR chains and promote efficient neutrophil recruitment to sites of infection (18, 28). More recently, murine dermal Tγδ17 cells have been identified as primary skin sentinels responsible for initiating acute skin inflammation, emphasizing the barrier-tropic tendencies of all innate γδ T cells (35–37). Notably, Vγ4TCR+ Tγδ17 cells are also essential for long-term anamnestic immunity against oral, but not systemic, L. monocytogenes infection (38), supporting the postulated existence of memory in innate lymphocytes (39). In the lung and gut, Tγδ17 cells produce IL-22 in a nuclear hormone receptor–dependent manner to modulate tissue fibrosis and inflammation, respectively (40, 41). The receptors for the vitamin A metabolite retinoic acid (RAR) and for Ah (Ahr) directly activate Il22 transcription, reinforcing the nutrient-sensing capacity as one shared feature of ILE cells (25, 41, 42).
Importantly, the acquisition of effector function by γδ T cell subsets occurs primarily during intrathymic differentiation (18, 32, 43, 44). The extent of lineage diversification in the γδ T lineage was determined by taking advantage of the observed high degree of correlation between Vγ and/or Vδ gene usage and effector function. Comprehensive gene expression profiling [undertaken by the Immunological Genome Project (www.immgen.org; 45)] of immature γδTCR+ thymocyte subsets separated based on the TCR V gene usage unequivocally demonstrated that the lineage divergence occurs prior to or immediately upon TCR acquisition (32). The next intrathymic maturation step equips γδ T cell subsets with tools required for routing to target tissues by chemokine receptor induction (CCR6, CCR10, CCR9, and CXCR6 for Tγδ17 cells, DETCs, iIELs, and NKT cells, respectively) and cytokine synthesis. Given that the difference in transcriptomes between the γδ subsets is comparable in scope to the extent to which a γδ subset is divergent from αβ thymocytes (32), and dramatically greater than the constrained difference noted between CD4+ helper and CD8+ cytotoxic αβ thymocyte subsets (46), γδTCR+ thymocytes are composed of multiple prewired effector lineages that are presumed to undergo distinct developmental checkpoints prior to deployment to tissues. Thus, γδ T cells localized to the body barriers highlight three principal cellular properties of ILE cells: a division into effector subsets, precocious programming of effector functions in tissues of origin, and prewired body-geography acuity.
Unconventional, Innate-Like αβ TCR+ T Cells
The thymus is the factory for the conventional, naive, adaptive αβ T cells that seed the lymphatic and blood systems. However, intrathymic T cell development is highly versatile and generates a plethora of unconventional T cell subsets that constitute ILE cells. That αβTCR+ T cells are not created equal and also contain fast, innate responders became the mainstream understanding after the discovery of αβ thymocytes that are capable of copious IL-4 and IFN-γ production prior to thymic egress (47). Some of these cells express the NK cell lineage marker NK1.1 and a unique Vα14-Jα28TCR chain and are referred to as invariant NKT (iNKT) cells (48). They recognize the nonclassical MHC class I molecule CD1d (49), which presents phospholipids and glycolipids, the most famous being α-galactosylceramide from marine sponges (50). NKT cells perform myriad functions in pathogen clearance and immune regulation, are localized to nonlymphoid tissues such as the liver, and are amenable for manipulation to treat inflammatory disorders and promote tumor rejection.
iNKT ILE cells act as a rapid, dual source of IL-4 and IFN-γ production, deviating from the conventional Th1 and Th2 dichotomy, by virtue of the entirely different class of effector-specifying TF PLZF (51–53). Recent systematic gene expression studies have revealed that iNKT cells are more closely aligned with innate γδ T, NK, and memory CD8+ T cells than with conventional αβ T cells (54). Moreover, whereas it has been understood that iNKT cell development follows intrathymic stepwise maturational phase transitions after TCR engagement (55), evidence is emerging that the distinct thymic NKT subsets may not represent maturation intermediates but rather different NKT effector subtypes (56). In the classic model, the ability to turn on both IL-4 and IFN-γ was thought to be a property associated with a transitional, semimature state prior to the acquisition of NK1.1 expression, after which iNKT cells are terminally mature and only produce IFN-γ (57). A revised model posits that, like γδ T cell subsets, phenotypically distinct iNKT thymic subsets are functionally distinct lineages and that T-bet, PLZF, and RORγt generate IFN-γ+ NKT1, IL-4/13+ NKT2 (NK1.1−), and IL-17+ NKT17 cells, respectively (Figure 1) (56).
A close relative of the iNKT cell is the PLZF+ mucosal-associated invariant T (MAIT) cell, which also expresses an invariant TCR α-chain [Vα19-Jα33 (58)] and recognizes microbial riboflavin metabolites in association with another nonclassical MHC class I molecule, MR1 (59). MAIT cells require commensals for full maturation and are enriched in the human intestinal lamina propria (LP), other sites in gut-associated lymphoid tissues (GALTs), and blood (60), where they respond to most, but not all, bacteria, presumably by recognizing conserved microbial products (59, 61). MAIT cells are relatively rare in inbred mouse strains unless they are infected by intracellular bacteria and yeasts, such as Francisella tularensis and Candida albicans (60, 62). Activated, functionally competent intestinal PLZF+ MAIT cells are detectable in nonlymphoid tissues, such as the lung and intestine, as early as the second trimester in human fetuses (63). MAIT cells exported from the thymus are phenotypically naive, and they acquire PLZF and RORγt expression soon after arriving in mucosal tissues (58, 63). They produce IFN-γ, IL-17, and IL-22 upon stimulation, but whether MAIT cells are also composed of distinct effector subsets has not been established (61, 63). A recent discovery of a human IFN-γ+ invariant αβ T cell subset restricted to the nonclassical MHC CD1b and recognizing a lipid glucose monomycolate derived from Mycobacterium tuberculosis (64) suggests that more iNKT-like innate αβ T cell subsets with unique TCRs exist in mammals.
Another prominent αβTCR+ ILE subset is the CD8αα+ T cells embedded in the intestinal epithelia that cooperate with γδTCR+ iIELs. They are distinct from pathogen-activated conventional memory cytotoxic T cells found in the intestinal epithelium that express CD8αβ heterodimers. The surveillance density for iIELs is high in the small intestine (1 iIEL per 5–10 epithelial cells) but drops off considerably in the colon. Moreover, the sentinel composition is ordered, with γδ IELs more prevalent in the upper intestine and αβ IELs in the distal regions (65). CD8αα+ αβ iIELs are generated by interactions between TCRs and self-ligands in the thymus (66) in the context of TGF-β signaling (67). CD8αα homodimers can bind to the stress-inducible, mucosal epithelia–biased MHC class Ib TL antigen (68), providing additional inputs in modulating iIEL activation. Testifying to the complementary functions of γδ and αβ iIELs, both are preloaded with abundant granzymes to kill infected epithelial cells (23) and are required for protection against Citrobacter rodentium, a model pathogen for human Escherichia coli infections (26, 41). CD8αα iIELs express CD160, a ligand for the multifunctional epithelial conduit herpes virus entry mediator (HVEM, Tnfrsf14), which is required for normal induction of antimicrobial peptides and the receptor for IL-22, the essential epithelial barrier–fortifying cytokine (26, 69).
Arrayed alongside these αβTCR+ ILE cells in diverse tissues are hematopoietic MHC class II–selected, dual IFN-γ/IL-4-producing T cells (70); natural Th17 cells (71); innate PLZF+ CD4 T cells (72); and Eomesodermin (Eomes, a close relative of Tbx21)-expressing innate CD8+ T cells (31), which are all generated in the thymus and exhibit preprogrammed effector function. These T cell subsets share the features of an alternative intrathymic T cell selection process: First, their selection in the thymus is primarily, but not exclusively, based on nonclassical MHC class Ib molecules, such as CD1, MR1, T10/T22, Qa1, and H2-M3, that are highly conserved and relatively nonpolymorphic compared with classical MHC molecules (73). These MHC platforms, often induced upon infection or damage, accommodate pathogen-specific moieties such as glycolipids, riboflavin metabolites, and formylated methionine–containing peptides. Second, the selection process is often based on interactions with thymocytes, rather than with epithelial and professional antigen-presenting cells (74). This broadens the useful range of TCR-self ligand avidity by channeling conventionally forbidden self-reactivity to generate entirely different effector T cell subsets. Third, the selection entails the expression of innate or memory T cell–biased TFs, such as PLZF, EOMES, and ID2 (31, 75, 76), that make up the core TF regulatory networks of ILE cells, as elaborated below.
ILCs, TCR-Less T Cells?
Mammalian slow adaptive immunity is built upon a substratum of lymphoid surveillance for host integrity that operates independently of clonal antigen receptors. This construct was reinforced by the recent discoveries of ILCs (77–81) that are related to NK cells and lymphoid tissue–inducer (LTi) cells, the former being the prototypic ILCs activated by viruses and the latter identified as facilitators of lymphoid tissue genesis by providing lymphotoxins (LTs) (82). ILCs are closely aligned with innate T cells in development and function, sharing common features such as predominant localization to mucosal tissues, including the gut and lungs; preprogrammed segregation into effector subsets along the same principle as TCR+ ILE cells; and utilization of T cell regulatory gene circuits. Although the new classification of the diverse ILC subsets has stressed commonality to conventional NK (cNK) cells (16), shared regulators of the earliest steps of T cell and ILC subset specification argue for a more integrated lineage relationship with T cells.
Conforming to the division of labor classification of T cells, murine ILCs are segregated into three subsets based on cytokine signatures (Figure 1). ILC1s produce IFN-γ in a T-bet- and/or EOMES-dependent manner and include cNK cells (83–85). ILC2s produce copious amounts of type 2 cytokines IL-5 and IL-13, and some IL-4 upon encounter with diverse pathogens, most notably the parasitic worm Nippostrongylus brasiliensis (80, 81, 86). ILC3s encompass the prototypic ILC subset, LTi cells and others of various cell surface phenotypes that secrete LTα/β [required for lymphoid tissue organogenesis (87–89)], and the type 3 cytokines IL-17 and IL-22 (77–79, 90), which have evolved to protect against enterogenic pathogens such as C. rodentium. Murine ILC3s expressing the NKp46 NK cell receptor can also produce IFN-γ in a T-bet concentration–dependent manner (91, 92), reminiscent of pathogenic Th17 cells that produce both IL-17 and IFN-γ (93). These three ILC subsets are generated under the control of the same TF networks that determine CD4+ T cell subset differentiation: IFN-γ, IL-5/13, and IL-17/22 production in ILC subsets is programmed by T-bet/EOMES (94–97), GATA3 (98–100), and RORγt (88), respectively. However, additional TF nodes that do not conform to the CD4+ T effector differentiation gene circuit distinguish ILCs from CD4+ αβ T cells: Some examples are RORα, which contributes to Th17 development and is necessary for the generation of ILC2s but not ILC3s (12, 101, 102); T-bet, which is required for NKp46+ ILC3 generation but has no role in IL-22 production by T cells (91, 92); and PLZF, which is required to generate ILC2 and ILC3 subsets (103) but which among T cells only governs the functional maturation of NKT cells and their close ILE relatives (104).
TF NETWORKS OVERSEEING INNATE LYMPHOID LINEAGE SPECIFICITY
Organized TF Modules Regulating Lymphopoiesis
Modules of coordinately regulated genes are often shared among distinct hematopoietic cell types, and unique amalgamations of available modules confer cell identity (105–109). Each of these modules contains apex regulatory TFs that specify both positive and negative net activities of the component genes of a given module (45, 110). Effector repertoires of distinct layers of ILE cells are comparable as a consequence of shared core regulatory TF networks that are responsible for specifying effector cell lineage fate. Hence, mapping interconnectivities of TFs across ILE subsets is necessary for tracing evolutionary diversification of lymphoid effector subtypes and identifying limited numbers of distinguishing gene nodes embedded in the common regulatory circuits that ultimately make a cell phenotypically and functionally unique.
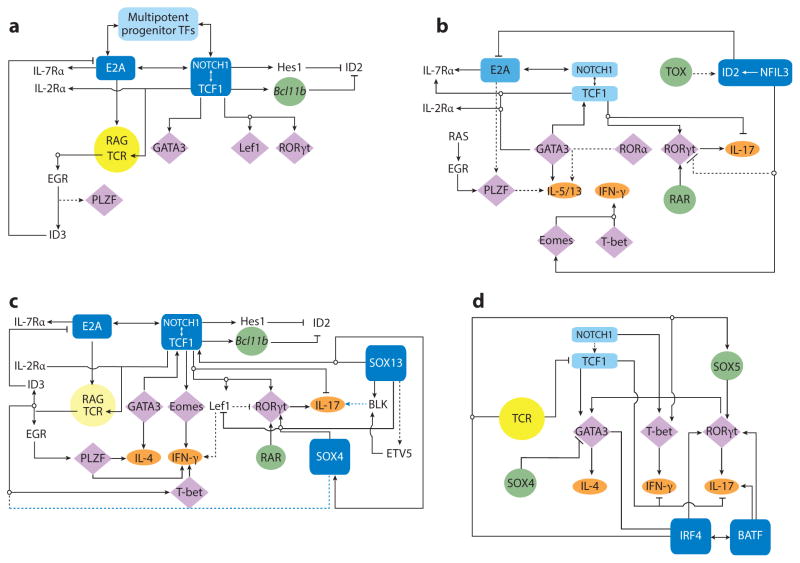
The current hierarchical TF framework for constructing T cells and distinct effector subtypes is robust, and much of the effort in mapping gene circuits in ILE subsets is based on the existing modular gene networks of T cells (111–114). Recent insightful reviews on ordered and coordinated activities of TFs at different phases of T cell development and during innate and adaptive effector subset differentiation have been published in this journal (e.g., 115). Hence, the main focus here is not to catalog all known TFs involved in lymphoid effector lineage specification. Rather, we highlight the basic TF network commonality of ILE cells in a simplified framework. Within the confines of the shared mainframe, select strategies of nodal TF modifiers that have been incorporated to generate specialized cell types are discussed (Figure 2).
Figure 2.
The basic TF network circuitry of T cell development and variations incorporated into it generate lymphoid effector cell types. Minimal components of each circuit are depicted. Cytokine receptor–signaling inputs controlling TF expression are not shown. (a) The foundation regulatory TF circuit of T cell development is the NOTCH-TCF1 hub (rectangles) that intimately integrates with the E2A regulatory domain and induces central genes (circles) necessary for T cell development from ETPs. This process operates in conjunction with TFs already expressed in LMPPs or CLPs that are active at multiple states of T cells (Ikaros, Myb, Runx1, Gfi1, Sox4, and others belong to this cluster). E2A induces the expression of IL-7R and RAG proteins required for the V(D)J recombination of Tcr genes. E2A and TCF1 control chromatin accessibility of the Tcr loci and regulate their ordered gene rearrangements. TCR signaling, along with E2A, can promote PLZF expression. But in adaptive T cells, PLZF is under an autoregulatory feedback loop involving ID3, induced by TCR signaling that in turn inhibits E2A to maintain a naive state of mature thymocytes. Absence of TCF1 in mice leads to a complete loss of T cell–committed DN2 precursor cells, and TCF1 controls the expression of one of the markers of DN2, IL-2R α-chain. TCR signaling, NOTCH, and TCF1 cooperate to turn on the effector lineage controlling TFs GATA3, LEF1, and RORγt (diamonds), but during intrathymic αβT cell development, they do not access the target cytokine gene loci. NOTCH and TCF1 target genes, most notably Bcl11b, inhibit genes active in non–T cell lymphoid lineages, such as ID2, which is required for ILE development. (b) Three sequentially incorporated ILE TFs, NFIL3 (nuclear factor, IL-3 regulated), ID2, and TOX (thymocyte selection–associated high-mobility group box), constitute the ILC hub. The high level of ID2 expression, induced by NFIL3, possibly in concert with TOX, leads to a muted activity of E2A that in turn impedes the cooperative NOTCH-E2A outputs. The consequences are lack of antigen receptor expression and varied dependence on NOTCH signaling for ILC differentiation. NFIL3 can inhibit Rorc expression in Th17 cells (dashed lines represent connections by function only; direct gene-to-gene interactions have not been confirmed in indicated cell types), and conversely, it turns on Eomes in NK cells to regulate IFN-γ (ovals, effector cytokines). In other ILC1s, T-bet, not EOMES, drives IFN-γ production. For ILC2s, TCF1 regulates Gata3 and cytokine receptor genes critical for ILC2 development. RORα is essential for ILC2 differentiation, but its upstream inducers and downstream targets are unmapped. PLZF is transiently induced, perhaps by RAS signaling in conjunction with E2A, and is also required for ILC2s, but its target genes are unmapped in ILCs. As in T cells, TCF1 inhibits IL-17 production and promotes IL-2R α-chain expression in ILC3s. TCF1 may contribute to Rorc transcription in ILCs directly, as in T cells, or indirectly, by promoting IL-7R expression, which in turn enhances RORγt expression (not depicted). RAR and other positive regulators of RORγt further entrench the ILC3 program. (c) Incorporation of two additional HMG TFs, SOX13 and SOX4, institutes the TCR+ ILE program. Activities of TCF1 are modulated by its interacting partners SOX13 and SOX4, which together turn on Rorc, with additional inputs from RAR and ETV5. SOX13 also induces Etv5, Blk, and Il7r and inhibits Lef1 to specify the Tγδ17 cell program. SOX4 regulates iNKT cell development, potentially by calibrating TCR signals, but the molecular mechanism remains to be ascertained. PLZF, ID3, and T-bet induction is initiated by TCR signaling. TCF1 positively regulates Gata3, to generate IL-4-producing cells, and Eomes and Lef1, to control IFN-γ synthesis. (d) A simplified adaptive Th effector circuit illustrates the dominance of TCR signaling as the arbiter of functional specialization. TFs belonging to the IRF-BATF axis, along with others induced by TCR and cytokine receptor signaling, constitute the hub. IRF4 and BATF cooperatively induce RORγt. IRF4 further controls Gata3 expression, whereas BATF amplifies IL-17 synthesis. The ILE HMG TFs still exert modulatory functions as NOTCH enhances expression of effector cell type–specific TFs and promotes cytokine gene expression. Conversely, TCF1 negatively regulates IFN-γ and IL-17 production, but its activity is downmodulated by TCR signaling. SOX4 and SOX5, two additional HMG TFs, modulate Th2 and Th17 cell differentiation, respectively. SOX4 inhibits GATA3 function, whereas SOX5 promotes Rorc expression. Abbreviations: CLP, clonogenic lymphoid precursor; ETP, early thymic progenitor; HMG, high-mobility group; ILC, innate lymphoid cell; ILE, innate lymphoid effector; iNKT cell, invariant NKT cell; LMPP, lymphoid-primed multipotent progenitor; RAG, recombination activating gene; RAR, retinoic acid receptor; TF, transcription factor.
Apex TFs of T Cell Development: NOTCH-TCF1 and E2A
ILE subsets at the mucosal barriers share the regulatory TF network of T cells first identified in the early thymic progenitors (ETPs: cKit+IL-7RloCD44+CCR9+ thymocytes that do not express hematopoietic lineage-specific markers, Lin−). The core apex TFs of T cell development are NOTCH, TCF1, GATA3, and E2A and its regulators, inhibitor of DNA-binding (ID) proteins (116–121). Whereas the T cell lineage specification is initiated by NOTCH1 and orchestrated by its target, TCF1 (mouse Tcf7) (122–124), the earliest steps of mucosal ILC differentiation are governed by ID2, which titrates the class 1 basic helix-loop-helix (bHLH) TFs E2A and HEB, and others (85, 125, 126). In a streamlined scheme (Figure 2a,b), the binary decision of progenitors to generate T cell– or ILC-committed progenies entails two discrete but eventually interlocking events: First, antagonists and modulators of HLH E proteins calibrate the aggregate activities of self-reinforcing TCF1 and E2A, which directly control Tcr gene accessibility and Rag1/2 induction (120, 127); and second, integration of additional TF nodes in committed progenies permits fast effector programming of ILE cells.
The genetic framework of T cells is thus constructed of two connected hubs (Figure 2a): NOTCH-TCF1 and E2A, which interacts with multitudes of TF nodal inputs. Intrathymic T cell development is absolutely dependent on TCF1, a nuclear effector of canonical WNT signaling (128, 129) and a member of a large family of high-mobility group (HMG) box TFs that are essential for programming ILE function, as expanded below. HMG TFs are architectural TFs binding to the minor groove of DNA and providing structural platforms for the formation of multicomplex transcriptionary machines (130). TCF1 operates in conjunction with hematopoietic TFs inherited from blood-borne multipotent progenitors, with Ikaros (Ikzf1), E2A (Tcf3), Myb, Runx1, Gfi1, and Sox4 prominently constructing the lymphoid context in which TCF1 specifies T cell fate (106, 108, 119). NOTCH1 and TCF1 induce Hes1, Gata3, and Bcl11b; together these bring about T cell lineage specification, which involves turning on genes essential for T cell development [HEB (Tcf12), Rorc, Lef1, Gata3, Id3] (121, 122, 124, 131) and suppressing genes of progenitors and alternate cell fates, such as B, NK, and dendritic cells [PU.1 (Spi1), Bcl11a, Hhex, Id2, Lmo2] (132–135). GFI-1-mediated suppression of Id2 (136, 137) in concert with TCF1 (138) and BCL11b may be critical to limiting the development of ID2-dependent thymocytes.
The ILC Network: Variations on the Apex TF Network Centered on ID2
That there exists an extensive overlap in the lineage origins and molecular compositions of T cells and ILCs is underscored by the finding that the historically termed T cell–committed thymic precursors (cKit+IL-7R+CD25+CD44+Lin−, termed DN2) can give rise to thymic NK cells and ILC2s (101). The two key distinguishing features of the regulatory circuit of ILCs originating from clonogenic lymphoid precursors (CLPs: cKit+IL-7R+FLK3+Lin−) are the prominence of ID2 as a decisive switch that dampens E2A activity (80, 125, 139) and modulated dependence on NOTCH signaling (126), which together institute an innate-biased program (Figure 2b). NOTCH-TCF1 is dispensable for cNK cell development but critical for ILC2s (101, 140). Within ILC3 subsets there is further parsing, with adult NKp46+IL-22+ ILC3s being the most reliant (36, 141, 142), whereas for LTi cells, NOTCH is required for the earliest stages of development but sustained signaling prevents maturation (126). A similar need for noncontinuous NOTCH signaling was observed with murine γδ ILE cells (143).
Within the modified NOTCH circuit, ID2 enforces the ILC regulatory network by dampening E2A activities. ID2 was identified as an early marker of innate T cells, preferentially expressed in thymic and bone marrow NK progenitors (144, 145) and other αβTCR+ T cells with preprogrammed effector function such as CD8αα+ T cells (146, 147) and iNKT cells (148). In TCR+ ILE cells, ID2 functions in concert with ID3, but its impact is often constrained to cell maintenance (75, 76, 149). Id2 is required for the development of all ILC subsets from CLPs (80, 125), except thymic NK cells (150). ID2+ ILC progenitors (termed common help-like ILC progenitors, ChILPs) have lost the capacity to differentiate into adaptive T and B cells but possess the potential to generate all ILC subsets, except cNK cells, in vivo (85). Importantly, ID2+ ILC progenitors (IL-7R+α4β7+CD25−Lin−, ChILPs) do not express ILC subset–defining TFs (RORγt, T-bet, EOMES) other than GATA3. This finding suggests that, as in the T cell lineage, distinct functional effector subsets arise after the ILC lineage program is specified in progenitors.
The ID2 hub is connected to NFIL3, TOX, and GATA3 nodes to orchestrate the development of all ILCs (Figure 2b), with variable outputs impinging on NK cells. To illustrate, whereas GATA3 is required for the development of all IL-7R+ ILCs (99, 100), bone marrow–derived cNK cell development is relatively intact in the absence of Gata3, but thymic NK cell development is impaired and Gata3-deficient cNK cells are incapable of IFN-γ production (100, 150). Conversely, NFIL3, a basic leucine zipper TF, is required for the development of EOMES-expressing, mature cNK cells (151) but not mucosal tissue-resident NK (trNK) cells (152). Nfil3 is expressed in CLPs prior to Id2 induction, and NFIL3 binds to cis-acting DNA regulatory elements (CREs) of Id2 and Eomes to promote their expression (153) and may conversely dock onto the Rorc CRE to inhibit its expression (154). ILC subsets other than trNK cells also fail to develop robustly in the absence of Nfil3 (155, 156). Although TCR+ ILE cells have not been analyzed in depth, the development of adaptive T cells is normal in Nfil3−/− mice. Together, these results indicate that NFIL3 is a candidate apex TF for nearly all ILCs.
TOX is another HMG box TF that has pleiotropic effects on multiple lymphocyte subsets. When it is absent in mice, NK, LTi, and NKp46+ ILC3 development is impaired (157). The link between TOX and Id2 in NK cell development is unclear, as the defects in NK cell development cannot be corrected by enforced expression of exogenous Id2 (157), suggesting that TOX may function in a parallel circuit to the ID2 hub, with a possible convergence in target gene regulation. CD4+ αβ T cell development is also severely impaired in Tox−/− mice, in part because TOX regulates the expression of the key CD4 T cell lineage–specifying factor Th-POK (Zbtb7a) (158). So far, TOX has not been reported to be essential for γδTCR+ ILE cells, but its closest relative, TOX2, is exclusively expressed in γδ T cells (www.immgen.org), suggesting that TOX2 may perform some of the functions of TOX in γδ T cell development.
Whereas it was intuitive that ILC progenitors are marked by Id2 expression, a surprising finding was that a subset of ILC progenitors expressed PLZF transiently during ILC subset lineage specification (103). As described, PLZF is the dominant NKT cell programmer. These ID2+PLZF+ precursors are essentially devoid of RORγt but actively transcribe Tcf7, Tox, and Gata3 and display substantial ILC subset generative capacity in vivo, except for cNK and LTi cells. Analysis of developmental potential of Zbtb16-deficient bone marrow stem cells showed that PLZF is required for normal ILC2 and trNK (CD49a+DX5neg) cell development but appears to exert lesser influences over other ILC subsets. These results suggest that PLZF+ ILC precursors lie downstream of ChILPs (85). In aggregate, like conventional T cells, all ILE cells, except some ILC1s, are dependent on IL-7 for development (159), and ILCs require the same set of core TFs (NOTCH-TCF1, GATA3, E2A, and ID2/3) as T cells, although the regulatory domains of these TFs are distinct between T cells and mucosal ILCs.
Variations on the TCF1-Controlled Network Generate TCR+ ILE Cells
An alternative variation on the core T cell regulatory hub incorporates two HMG TFs, SOX13 and SOX4, to generate ILE cells that express TCRs (Figure 2c). Once ETPs become restricted in developmental potential to T cells (cKitint DN2, termed DN2b), TCR and NOTCH signaling delivers the precursors to the CD4+CD8+ (double positive, DP) state from which mature αβ thymocytes of adaptive and ILE lineages arise, depending on the specificity of the TCR (55, 66). γδTCR+ ILE cells diverge at the DN2 stage (160), and for Tγδ17 cells, they cannot be generated from precursors beyond this point (36, 161). This stage is also the pivot point where further maturation potential to become NK cells and ILC2s is lost (101).
Although data are not complete, so far the largest TF family that regulates ILE differentiation is made up of HMG TFs, two of which have already been highlighted, TCF1 and TOX. HMG TFs are classified into three groups based on the specificity of the HMG domain for target gene CREs: TOX; sex-determining region Y (SRY)-related HMG box (SOX); and TCF/LEF-1. Of these, the SOX TF family is the most diverse, with more than 20 members segregated into subgroups based on HMG domain homologies (162). The CREs recognized by diverse HMG TFs are group specific, with some shared characteristics (163). In vivo competitive-binding affinities of different HMG TFs for a given consensus CRE have not been studied in detail. Nevertheless, theoretical permutations in the number of gene regulatory modes of HMG TFs are very high given the inter- and intragroup interactivity as well as the combinatorial association of diverse co-TFs and chromatin regulators. Deployments of multiple closely related HMG TF family members in one cell type entail some functional redundancies as well as plasticity in the gene networks they supervise. A systematic application of this design principle operates to produce γδTCR+ ILE cells, which require several HMG TFs that are expressed from the ETP stage onward, most notably SOX13 and SOX4 (Figure 2c).
In thymocytes, Sox13 is expressed exclusively in immature γδTCR+ thymocytes (164) and during the first phase of Vα14+ iNKT cell development (54). In the fetal ontogeny, SOX13 controls the overall number of γδTCR+ thymocytes produced (164). In adults its major function is to maintain the production of Tγδ17 cells, where its expression is the highest (36). SOX13 is the primary inducer of IL-7R (165) and another γδ T cell–specific gene, Etv5, in Tγδ17 cells (110), both critical for the subset’s differentiation and maintenance. Among the known convergent targets, SOX13 and SOX4 cooperatively control the two requisite Tγδ17 genes, B lymphoid kinase (Blk) and Rorc (Figure 2c) (36). In contrast, SOX4 is not essential for Rorc expression in immature αβ thymocytes or during Th17 cell differentiation. This restricted domain of function may be linked to the possibility of TCF1 displacing SOX4 to control Rorc transcription in αβTCR+ thymocytes (124, 166, 167), and the TCR/IL-6-STAT3 induction of SOX5 [a SOX13 sibling (164)], which operates as an integrator of cytokine receptor and TCR signaling to promote Rorc transcription in Th17 cells (168). Thus, sequential docking of SOX4 (in precursors), TCF1 (thymocytes), and SOX5 (T cells) to the target gene CRE can perpetuate a regulatory gene circuit, with distinct inducers of each TF dictating the operation time frame of the circuit as needed. A similar serial engagement of SOX proteins has been reported to regulate neuronal cell differentiation (169).
SOX4 was initially identified as an essential TF of hematopoiesis and B lymphopoiesis (108, 170). SOX4 is broadly expressed among thymocyte subsets, but expression is particularly high in precursors and TCR+ ILE subsets. SOX4 functions in ILCs have not yet been characterized in depth, but Sox4 is normally repressed by GFI-1 in ILC2s, presumably to inhibit aberrant IL-17 production (171). Testifying to the communal gene network of ILE cells, iNKT cells, but not conventional αβ T cells, are absolutely dependent on Sox4 for differentiation. However, in contrast to its role in Tγδ17 cells, SOX4 is dispensable for Rorc transcription in αβ iNKT cells (N. Malhotra, unpublished observations). This divergence dovetails with the hybrid nature of iNKT cells (54), blending the innate programming and adaptive TCR signaling components that construct an entirely distinct developmental program (55, 172).
The major interacting partner of SOX13 and SOX4 is TCF1 (164, 173), and the global chromatin occupancy pattern of TCF1 in γδ T cells is controlled by SOX13 (J. Kang, unpublished observations). Although γδTCR+ thymocytes are generated in Tcf7−/− mice, their effector subset programming is grossly imbalanced, with an overload to the Il17 inductive state (36), consistent with an inhibitory role of TCF1 in Il17 transcription in αβ T cells (174). TCF1 promotes IFN-γ+ effectors, in part by modulating Lef1 and Eomes, and SOX13, in turn, can inhibit TCF1 function and repress the expression of Lef1 (36). Hence, insertion of one modifying Sox13 node onto the core T cell regulatory gene circuit can radically alter its functional outputs (Figure 3c). Indeed, whereas thymocytes expressing γδTCR or αβTCR are highly divergent in global gene expression profiles [~10% of genes expressed in at least one lineage with ≥2-fold change in relative expression values (32)], only three TFs, SOX13, ETV5, and TOX2, have been classified as γδ T lineage specific (www.immgen.org), indicating that a large-scale alteration in the output of the T cell TF regulatory circuit can be achieved by a limited number of nodal innovations that affect the regulatory hub.
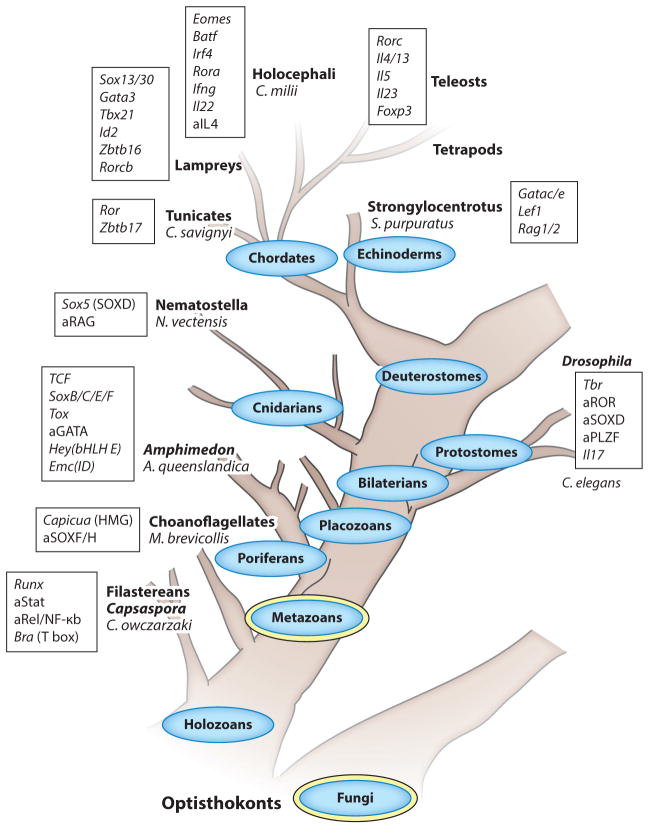
Figure 3.
A simplified phylogenetic tree of TFs and cytokines directing lymphoid effector subset development and function. Depicted are the major phyla (ovals); the kingdoms they belong to (double ovals) and subphyla, classes, or genera they include (bold, referred to in the text); and extant organisms (species) of Opisthokonta (domain Eukaryota, animals and fungi supergroup, no taxonomic rank). Only the holozoan (animals and related unicellular organisms) branch that generated animals (metazoans) is detailed, and the tree is drawn not to a timescale but according to the sequence in which the phyla emerged. Only the clear-cut orthologs of rodent TFs and cytokines (rectangles) are noted in species where they first appear, and for early genes that are not clearly defined as homologs (because a member is missing key functional domains, for instance), they are denoted as ancestral and preceded by the letter a. Lymphocytes only exist in deuterostomes, but it is clear that immune mechanisms, such as the complement cascades and pathogen-associated molecular patterns, predate the emergence of mobile cell types mediating immune responses. Filastereans (Capsaspora owczarzaki), single-cell eukaryotes, and the earliest clade of the holozoan branch are already endowed with intracellular mediators of innate sensors, such as NF-κB, the STAT TF that is activated by cytokine and growth factor receptors; and an ancestral Runx gene linked to the mammalian Runx1, which is essential for definitive hematopoiesis. The closest extant unicellular relatives of animals are choanoflagellates, and they provide evidence for the emergence of sequence-specific HMG TFs, related to Capicua (Cic) and SoxF/H members (Sox17, 18, or 30), ~900 Mya. A significant expansion of the repertoire of immune-related TFs is observed in Porifera (sponges), and a large family of invertebrate IL-17 is found in oysters (Protostomia). A Tbr gene is annotated in the genome of Lottia gigantea, a protostome mollusk, but not in the genome of flies. An ancestral gene for Rag is present in Nematostella vectensis (sea anemone) and further innovated in Strongylocentrotus purpuratus (purple sea urchin), where orthologs of Rag1/2 emerged. Definitive evidence for a discrete ILE subset is first obtained in lampreys, where Il17-expressing SOX13+ lymphocytes that home to mucosal tissues have been discovered. The diversification of mammalian cytokines and central antigen receptor–activated TFs controlling αβT cell effector lineage specification initiates in vertebrates, first in cartilaginous fishes (Callorhinchus milii). The emergence of type 2 cytokines appears to lag behind the emergence of type 1 cytokines, as a single Il4/13 gene and an Il5 homolog are catalogued in teleosts, with only an ancestral Il4 gene in C. milii. Among the central modulators of Th cell specification, the mammalian inducers of type 3 cytokines RORγt and IL-23 appear last in teleosts. Abbreviations: HMG, high-mobility group; ILE, innate lymphoid effector; TF, transcription factor.
A Distinct Combination of HMG TFs Regulates αβTCR+ Effector Subsets
Adaptive CD4+ T cell effector differentiation is ruled by TCR signaling in conjunction with the cytokine milieu created by infections and the host innate immune responses. Cytokine-induced STAT proteins in concert with TFs induced by TCR signaling, most notably IRF and BATF, are principal enforcers of effector lineage fate, distinctly controlling the expression of T-bet, GATA3, and RORγt (Figure 2d) (96, 111, 175–178). The ILE effector lineage TF circuitry is, for the most part, subsumed in naive adaptive αβ T cells, but upon activation of the T cells it becomes critical in at least three major components of adaptive immunity.
First, TCF1, LEF1, SOX5, and ETV6 (most closely related to ETV5, which is operative in Tγδ17 cells) are central modulators of Th17 cell differentiation (111, 167, 168, 174, 179, 180). Second, the NOTCH TF (intracellular N-terminal-truncated NOTCH) enhances the outputs of all regulatory gene circuits of Th subsets (181), and TCF1 promotes Th2 differentiation by inducing GATA3 (182). Conversely, SOX4 has been shown to oppose the development of Th2 cells by inhibiting GATA3 function (183), recalling the contrasting function of TCF1 and SOX4 in Tγδ17 differentiation (36). Third, the ILE regulatory network regains a widespread propagation during the acquisition of an antigen-specific memory state by adaptive T cells. TCF1 is necessary to generate memory CD8+ T cells, accomplished in part by inducing the expression of EOMES (96, 184, 185). In vitro studies suggested that SOX4 can affect key regulatory nodes underpinning the memory state, including Id2, Tcf7, and Eomes, but the exact functional network connectivity in vivo has not been decisively established (109). ID2 and ID3 expression is induced after TCR signaling, and they control the generation of naive αβ T cells (186) and memory CD8+ T cells after pathogen encounter. In the latter process, higher amounts of ID3 favor the memory state, whereas enhanced ID2 expression is associated with the acute effector state (187). The commonality of molecular events occurring after naive αβ T cell activation and during ILE differentiation is clear, as ILE cells are exported from tissues of origin as activated, memory-like, and often long-lived lymphocytes. In aggregate, the ILE TF regulatory networks are fundamental for the generation of diverse cell states after activation, and they are likely the initial generator of lymphocyte division of labor in vertebrates. To focus adaptive lymphocytes for clonal TCR–mediated instructive signals, a developmental phase has been imposed on them whereby a prolonged naive state has been instituted. This phase involves active suppression of the intrinsic ILE TF circuits (124, 186, 188, 189) and rewiring of the network in which TCR signal intermediates become the primary arbiter of adaptive αβ T cell lineage fate.
EARLY EVOLUTIONARY ORIGIN OF ILE REGULATORY CIRCUITS
A Window to the Genesis of ILE Subsets in Animal Evolution
The core design principles of ILE regulatory gene circuits have been established in evolution. Thus, understanding the construct first requires knowledge of the evolutionary timing of the emergence of the regulatory network components. This daunting challenge is now approachable, given the recent explosion of whole-genome sequencing of primitive extant holozoans (all gene annotations are based on Ensembl, unless noted otherwise). TFs controlling T effector cell lineages in mammals did not emerge until vertebrates, but their homologs exist in the earliest animals and fungi. The complexity of the lymphocyte-relevant gene circuits, most notably the number of siblings in each TF family, correlates with the degree of multicellularity and is only evident during metazoan evolution. In particular, the TF family explosion can be traced to the Cambrian period, when bilaterians diverged from the earliest holozoans (190) (Figure 3). Here, the focus is on the effector lineage–specifying TFs, the HMG superfamily, bHLH, GATA, T-bet/EOMES, PLZF, and retinoid nuclear hormone receptors. We merge two datasets to estimate the sequence in which distinct lymphoid effector types originated. The first traces the emergence and diversification of the TFs and the advent of innovative effector molecules during animal evolution in the last ~580 million years (191). The second tracks the TF expression patterns among hematopoietic cell subsets of various species, where available. It is important to note that the complexity of most of these TF families and the upstream factors that regulate them, such as WNT, TGF-β, and NOTCH, most often predates, and does not necessarily correlate with, the evolution of increasingly diversified immune systems, given that they also direct fundamental body plans and multiple tissue generation (192). Thus, caution is needed when drawing conclusions from genome datasets about functional TF interconnections without hematopoietic lineage–specific information.
The Earliest ILE Subset Identified Is Tγδ17 Cells
To date, the most convincing phylogenetic evidence for a vertebrate ILE subset is γδ T cells, discovered in the jawless vertebrates lampreys and hagfishes and characterized by the biased expression of a Sox gene [likely an ortholog of Sox30 or Sox13 by Ensembl, but referred to as Sox13 by Hirano et al. (193)]. These SOX13+ T cells express highly diverse, somatically generated variable lymphocyte antigen receptor C (VLRC) locus proteins containing a leucine-rich repeat (LRR) domain found in other innate sensors of pathogens (15). The VLR generation is activation-induced (cytidine) deaminase (AID) dependent but recombination activating gene (RAG) independent (194). Unlike the lymphoid tissue–resident VLRA+ αβ T cell equivalent, VLRC+ T cells are preferentially localized to the skin, like their mammalian counterparts, and are potentially capable of IL-17 (B/C/D) production, albeit with lower transcript amounts at the population level as compared with VLRA+ T cells (193). This remarkable finding strongly suggests that the different genetic blueprints for mucosa-homing innate T cells and lymphoid tissue–resident T cells with adaptive features predate divergent strategies for equipping T cells with clonal antigen receptors. The absence of Rag genes in lampreys (195) and their precocious emergence in sea urchins, further down the evolutionary tree (196), presage acquisition of different gene clusters dedicated to antigen sensing and tailoring of them to the preexisting gene networks geared for unique effector profiles, rather than the other way around. The same conclusion is inescapable in mammals, as γδTCR or αβTCR expression does not dictate whether the cells are fast innate or slow adaptive effectors. Therefore, the regulatory TF networks take precedence over pathogen-sensing machineries in specifying lymphoid effector cell fate.
Stepwise Emergence of ILE TFs and Cytokines in Holozoan Evolution
Tracing the origin of the effector-specifying gene networks is essential to the pursuit of the design principles of the mammalian immune system. We start by briefly summarizing evolutionary diversification of the effector-defining TFs and cytokines, with an emphasis on those currently known to regulate both innate and adaptive effectors, in the order they evolved in holozoans, and then synthesize a possible timeline of the successive emergence of ILE subsets.
Sequence-specific HMG TFs
One of the earliest TF families in all organisms is the HMG TFs (197–199). Although these regulators are present in the genomes of lower eukaryotes, more complex HMG TFs that recognize CREs and participate in modulating WNT signaling did not emerge until the metazoans. Fungi are equipped with one distantly related TCF gene (ScRox1p), and the closest extant unicellular relatives of animals, the choanoflagellates Monosiga brevicollis, have a homolog of the protostome (including insects, bivalves, and worms) HMG TF Capicua (Cic), which is critical for dorsoventral patterning of the embryo (200). True orthologs of mammalian TCF are acquired in Porifera (sponges), the earliest branch of metazoans, with one Tcf gene in Amphimedon and two in Sycon ciliatum (201, 202).
SOX-like TFs are found in lower holozoans (Figure 3), with a rapid evolutionary diversification in Porifera, where a cluster of Sox genes are acquired and maintained: S. ciliatum has 11 Sox genes belonging to 4 distinct subfamilies, but choanoflagellates only contain one distantly related Sox gene (200, 203, 204). The mammalian SOXB/C (Sox4)/E subfamilies appeared first, along with Tox, in holozoan evolution and are present in sponges (201). The SOXD family members (Sox5/6/13) likely arose last, at the time of the first bilaterians (190), where Sox5 is represented in the genome of cnidarians (the sea anemone Nematostella vectensis and jellyfish, among others) and a Sox13-related gene is present in protostome deer ticks (205). Two SOXD TFs, Sox5 and a Sox13/30-related gene, have been annotated in the lamprey genome (195).
bHLH
These TFs are ancient; the DNA-binding motif is found in fungi, and there are approximately 70 genes in bilaterians (206). E2A and HEB (class 1) construct the lymphoid transcriptome and are essential for T lymphocyte development in mammals (117, 120). The ancestral class 1 bHLH TF Hey is evident in the genome of sponges, placing the origin of positive regulators of hematopoiesis at the root of Metazoa (206). The ID bHLH TF family in mammals consists of four genes, and its ortholog in insects is Emc, homologs of which are present in at least one species of sponges (191) and in the genome of cnidarian Nematostella. An Id2 homolog was first catalogued in sea urchins (Figure 3) (207), and homologs of most vertebrate TFs directing hematopoiesis are operational in adult echinoderm coelomocytes, amalgams of immunocytes that respond to microorganisms (207, 208), supporting the timeline of the emergence of the common hematopoietic gene network at the time of deuterostome divergence, about 560 million years ago (Mya).
GATA
Zinc finger TFs of this type (two zinc fingers, four cysteine residues) are found in multiple branches of the eukaryotic tree. In the holozoan phylogeny they are absent in the genome of choanoflagellates, and they appear first in sponges, where a single Gata gene serves as an endomesoderm differentiation regulator (202). The emergence of two distinct classes of Gata genes is clear in deuterostomes; they eventually reached their zenith in mammals where three genes in each class make up the full complement. Gata1-3 are primarily expressed in hematopoietic cells, and sea urchins express Gatac and Gatae, with the former being a homolog of the mammalian hematopoietic Gata genes. Gatac is preferentially expressed in immature precursor immunocytes and is necessary to generate daughter blastocoel cells that participate in immune responses in the sea urchin larvae (209). The specialization of Gata3 as a lymphoid regulator has presumably occurred in the common ancestor of vertebrates, given that the nearest chordate relatives, tunicates, lack Gata3 (210). In lampreys, the division into T and B cells is further cemented by distinct expression patterns of effector-determining TFs, including GATA3, which is expressed in greater amounts in αβ T cell equivalents relative to SOX13hi γδ–like T cells and not expressed significantly in lamprey B cell equivalents (193).
T-box TFs
EOMES and T-bet drive the transcription of Ifng, and they belong to the T-box Brain (Tbr) subfamily of a large cluster of T-box TFs. Whereas the original T-box TF, Brachyury (Bra), is found in Filasterea (211), some in the holozoan branch, such as choanoflagellates, have not retained any T-box TFs, and Amphimedon (sponges) has lost Bra (190). This discontinuity is also apparent with the Tbr subfamily, which has a sponge ancestor, but was lost in other lower metazoans until the emergence of bilaterians (211, 212). A Tbx21 homolog is found first in lampreys, prior to the existence of the Ifng gene. An Eomes homolog is then found in cartilaginous fishes (Callorhinchus milii, elephant shark, a holocephalan, early cartilaginous fish that diverged from bony vertebrates 450 Mya), where the Ifng gene first emerges (213).
PLZF: Zbtb16
Zbtb16 belongs to a large family of zinc finger and BTB (broad complex, tram-track, bric-a-brac) TFs and is a unique marker of ILE cells responsible for turning on both type 1 and type 2 cytokines, a hallmark of NKT and NKT-like cells. Zbtb16, among its diverse activities, controls spermatogonial stem cell renewal, and the translocation of this gene to the Rara locus is a causative transformation event in acute myeloid leukemia (214). Caenorhabditis elegans EOR-1 (215) and Drosophila CG6792 (216) have been identified as mammalian PLZF orthologs, indicating the likely origin of mammalian PLZF in bilaterians. The two protostome genes are regulated by RAS/ERK signaling during development, paralleling the direct RAS-EGR2 induction of PLZF in iNKT cell development in rodents (104). In chordates, one Zbtb gene has been annotated in the genome of tunicates (217); it is most closely related to Zbtb17 (Miz-1), which has multiple functions, including the regulation of IL-7R expression (218). A Zbtb16 homolog is first found in lampreys (195), but its expression pattern among lymphoid cell subsets has not been reported.
ROR
RORα and RORγ are nuclear hormone receptor TFs critical for fundamental biological processes such as circadian rhythm regulation, cerebellum development, and lymphoid tissue generation (219). RORα/γ may normally function as sensors of lipid homeostasis, given that one of their few candidate natural agonists is 7-oxygenated sterol (220), which is an important intermediary in bile acid metabolism. Ancestral Ror genes Nhr-23 and Hr-46 evolved in protostomes C. elegans and Drosophila, respectively, and are related to Ror (two transcripts) in the genome of tunicates. The Rorb ortholog Rorcb is found in lampreys (Figure 3), and a Rora homolog first emerges in elephant sharks (213). Rorc, however, is an evolutionarily recent innovation: The gene is missing in jawless vertebrates (195) and elephant sharks (213). Rorc is first recognized in teleosts (ray-finned fishes), with basal expression in lymphoid tissues and the gut that can be further induced by bacterial challenges (221). In rodents, RORγt is highly expressed in αβ DP thymocytes, where it prolongs survival and allows the assembly of Tcr genes that rearrange late in DP cells, such as the Vα14 gene segment of iNKT cells. Hence, Rorc−/− mice lack not only ILC3s and Th17 and Tγδ17 cells, but also iNKT cells (222). In the fish thymus, a Rorc paralog is only expressed marginally (223), suggesting that there have been additional modifications in the Rorc CRE and/or incorporations of innovative inducers to allow constitutive expression in thymocytes, thereby facilitating the TCR-mediated selection.
Cytokines
Effector molecules that define CD4+ Th subsets are not found in invertebrates, except for the members of the TNF and IL-17 family (207). IL-17 has structural similarity to cysteine-knot growth factors, such as TGF-β and nerve growth factor (224), distinguishing it from all other mammalian cytokines. The echinoderm purple sea urchin genome contains ~30 genes with similarity to Il17, but no clear evidence for other interleukins before chordates has emerged (207). Nematodes express a protein with some similarity to IL-17C (225), and oysters have retained five invertebrate Il17 genes (226), indicating an ancient function of IL-17 in lower animals that has been preserved and expanded in mammals, despite the emergence of other cytokines with similar activities. One most likely primordial function of IL-17 is the maintenance of mucosal epithelia integrity (226, 227), and the IL-17 expression pattern in lampreys (228) is consistent with this supposition, as the tissue barriers express high amounts of IL-17. Another basic activity of IL-17 is its role as an alarmin. In hemocytes (primitive hematopoietic tissues) of oysters, IL-17 is rapidly induced upon bacterial exposure (225). IL-17A/F can mobilize other innate lymphocytes to the site of infection, such as phagocytes/neutrophils and NK cells during bacterial and fungal infections, respectively, and prime their function (229). From this vantage point, the tissue residency of effector cytokine-producing cells is a fundamental defensive strategy, complemented by the emergence of adaptive immunity and the secondary lymphoid tissue network, which has permitted additional layers of roving defensive units that can be recruited to tissue sites of infections.
Definitive evidence for cytokines specifically required for ILE development and function prior to cartilaginous fishes is also scant. A gene encoding the pan-lymphoid cytokine IL-7 first appeared in the vertebrate cartilaginous fish C. milii, despite the existence of highly complex lymphocytes in the lower jawless vertebrates (193). Orthologs of the lymphoid tissue–inducing factor LTβ, indispensable for secondary lymphoid tissue genesis, have not been uncovered in teleosts that lack LNs; only a distantly related LTα gene has been identified (230). IL-17 may promote the generation of inflammation-induced ectopic lymphoid tissues in species lacking LTβ and an organized lymphatic system (231). IFN-γ is central to defense against pathogens by virtue of its ability to kill infected cells and activate the myeloid cell types mediating the classical innate immunity, which is coordinated by pathogen-associated molecular pattern receptors. Genes with sequence similarities to Ifng have not been uncovered in vertebrates prior to cartilaginous fishes either, suggesting that other inflammatory cytokines that predate IFN-γ, namely, TNF, IL-1, IL-6, and IL-8, (195) are functionally sufficient in lower animals and/or that there are IFN-γ-equivalent cytokines with highly divergent sequences present in chordates. The type 3 cytokine IL-22, essential for barrier and mucosal tissue homeostasis (69), and an ancestral Il4 gene emerged in cartilaginous fishes (213). Clear evidence for IL-5 and IL-23, a mammalian inducer of IL-17, before bony vertebrates (teleosts and beyond) is lacking. These results suggest that cell types capable of production of a limited repertoire of type 2 cytokines against parasites likely existed in the earliest jawed vertebrates, but their identity is unclear, as the existence of CD4+ lymphocytes in the cartilaginous fish C. milii has been questioned (213).
A Model of Sequential Appearance of ILE Subsets in Evolution
The following model of effector lineage diversification integrates the evolutionary emergence of effector-subset-determining TFs with an enhanced granularity in the definition of lymphoid cell subsets and effector molecules of lower vertebrates and can serve as a starting point for further refinements. The first possible ILE subset that arose is IL-17-producing γδ T cells in lampreys, ~550 Mya. As in rodents, the initial regulatory TF circuit candidate for this subset is TCF/SOX and Rorcb, and possibly STAT, in the regulation of Il17 expression, but this has not been experimentally verified. Whether ILC subsets arose in parallel with the first identifiable γδ T cells in lampreys is unknown. This coevolution is possible given that the two defining TFs of ILC progenitors, Id2 and Zbtb16, were operational, and GATA3 and T-bet that control ILC2 and ILC1 development also emerged in jawless vertebrates. However, the cytokine profiles of these putative ILCs would have been quite different from those of mammals, as type 1/2 cytokines did not emerge until cartilaginous fishes, some 50 million years later.
Early IL-17+ effectors were complemented by cytotoxic effectors. The complete perforin-dependent granule-exocytosis pathway originated in jawed vertebrates, at about the same time as true cytotoxic T cells, but perforin and granzyme-like genes exist in more primitive animals, such as sea urchins (207) and cnidarians (232), suggesting that target cell killing may be the earliest form of pathogen eradication. Thus, effectors capable of lysing infected cells (cytotoxic granules and TNF) or activating myeloid cell types using TNF, and those involved in protecting the body barrier by producing IL-17 constitute the earliest triad of ILE cells, which was well entrenched in jawless vertebrates. Given the segregated expression of effector molecules in different lamprey lymphocyte subsets, it is predicted that the basic effector cell lineage specification in lymphocytes is a vertebrate trait.
The next effector cell subsets to arise were type 1 and type 2 cytokine secretors, first in elephant sharks (C. milii), along with the emergence of IFN-γ and its inducer EOMES, providing an alternate mode of Th1-like cell differentiation. At this juncture, type 3 secretors diversified, with Rora and IL-22 available. Critically, with the RAG-based clonal antigen receptor diversity generator in place, the TCR-induced factors that drive the mammalian Th17 cells, such as Batf and Irf4, were potentially now available to generate fast and slow temporally regulated IL-17 responses to infections (Figures 2 and 3) (178). After the transition to bony vertebrates (teleosts), the prototypical Th17 cytokines IL-17A/F emerged, with further innovation of IL-23 and Rorc to complement Rora to fine-tune, and eventually dominate, IL-17 production. The full complement of type 2 cytokine–producing cells was integrated into the chordate immune system at this juncture (Figure 3).
Three general patterns of the faster phylogenetic origin of ILE cells emerge: First, the diversification of central TF families controlling fast effector functions closely parallels increasing complexities in lymphoid effector repertoire and specialization, although the ancestors of each TF family, for the most part, exist as single genes in lower vertebrates and invertebrates. Second, the type of antigen receptors is unlikely to have been the principal discriminator of fast and slow effectors given that fundamentally different design principles of antigen receptor diversifiers were co-opted into preexisting, conserved TF networks that specify effector function. These primary gene networks of fast ILE cells are recursively recruited to generate distinct effector subsets in all T cells as they undergo progressive differentiation. Third, slow effectors that mobilize intracellular TCR signal intermediates to orchestrate effector outputs are relatively late arrivals, in cartilaginous fishes. This would imply a significant time gap between the consolidation of TF circuit design geared for lymphocyte function (at the time of the first vertebrates) and the superimposition of TCR signals as the arbiter of αβ T cell developmental phases and functional states.
FAST, FETAL ORIGIN OF ILE CELLS FOR THE PROTECTION OF NEWBORNS
Fetal Lymphopoiesis
Fast ILE cells likely existed before adaptive T helper effectors during animal evolution, and in mammals they are also fast developmentally, as they arise first in ontogeny, before functional adaptive lymphocytes. The generation of fast lymphoid effectors begins in fetuses, where a developmental clock in stem cells tightly governs the type of effectors that are generated, often in synchronized waves (233, 234). In contrast, murine adaptive T effectors are not observed until near birth and are functionally imbalanced postnatally, with a strong bias to type 2 cytokine–producing cells (235). Normal mouse gestation lasts up to 21 days. The primitive hematopoiesis is in place until embryonic day 8 (E8), geared toward the production of nucleated erythrocytes and primitive myeloid cells, among which are yolk sac (YS)-derived, tissue-resident macrophages in adult mice that originated in embryos, and not from adult bone marrow cells (236). Whereas the definitive hematopoiesis can be observed in several different fetal tissues, lymphopoiesis is thought to originate from the transit of hemangioblasts (hemogenic endothelium) from the aorta, gonads, and mesonephros (AGM) region to the fetal liver (FL) at E10 (237). The expanded FL progenitors then migrate to the fetal thymus at E11. Hematopoiesis soon shifts from the FL to the bone marrow, which becomes the central factory of blood cells.
In E11 FL there are only a handful of long-term repopulating hematopoietic stem cells (HSCs) (238), and data support the existence of E9.5 Tcf7+ progenitors with lymphoid-biased developmental potential prior to the major expansion of HSCs (239, 240). Whether these progenitors, originating from the YS, are the primary source of thymus-settling progenitors (TSPs) at E11 (see below) or fetal-origin ILE cells has not been adequately investigated (241), but in transplantation assays the donor fetal subsets generate T cells that preferentially populate the liver and gut rather than the spleen (239), mimicking the tissue-homing property of ILE cells. Sometime between E9 and E12, CLPs arise in the FL, and among them, α4β7+ CLPs transit to the fetal gut and generate ILC subsets. After E12.5, LTi cells are observed in the fetal gut (42), whereas the first sizeable population of γδTCR+ thymocytes (DETC precursors) is seen at E13.5. Single precursors that can generate NK or T cells (Lin−CD122+NK1.1negDX5neg) are observed at E13.5, and they populate various hematopoietic tissues and generate phenotypically mature, cytotoxic NK cells by E17 (241). These cells may be the presumptive precursors of the fetal-origin, liver-resident NK cells, which are not efficiently generated from adult bone marrow stem cells (97, 242).
Similar regulatory gene network architectures operating within TCR+ ILE cells and ILCs raise the possibility of a shared ontogenic origin. As previously noted, the first TSPs at E11 are already committed to T/NK cell lineage despite the absence of full thymic structural integrity (243, 244). E12 TSPs are closely aligned with the phenotype of CLPs rather than lymphoid-primed multipotent progenitors (LMPPs: cKit+IL-7RloSca1+Lin−) or ETPs of newborns and adult mice. Moreover, the majority of E14 fetal thymic cKit+IL-7R+Lin− precursors express α4β7, and they can give rise to α4β7+RORγt+ ILCs in vitro when NOTCH signaling is absent (126). This first fetal wave of TSPs is geared for production of γδ DETCs and αβTCR+ cells with surprisingly rapid transition to a more mature TCRβhi phenotype, with an attendant dearth of transitional DP thymocytes (234). While the properties of αβ cells arising from E12 TSPs were interpreted as an indication of distinct kinetics of T cell differentiation, an alternative possibility is that these early αβ T cells are innate-like, as suggested by high expression of PLZF, decreased proliferative capacity, and limited TCR diversity due to near absence of the TCR junctional sequence diversifier terminal deoxynucleotidyl transferase (245). Hence, an appealing model is that the early lymphoid progenitors are programmed to generate ILE cells, irrespective of tissues. The notion that distinct, ontogenically ordered progenitors give rise to different lymphocyte lineages was distilled into the layered immune system model by the Herzenbergs in the late 1980s (246).
Fetal-Restricted Generation of ILE Cells
The fetal thymic microenvironment provides as-yet-undefined ingredients for the generation of early preactivated innate T cells that are not produced in adults. The best-characterized fetal ILE cells are T cell subsets that express γδTCRs of a constrained repertoire and appear in an ontogenically ordered manner by the imposition of temporally ordered gene rearrangements on the TCR Vγ gene segments: First, Vγ3 rearranges at E13 (to generate DETCs), then Vγ4 and Vγ2 at E15 and E16, respectively (to generate Tγδ17 cells), followed by Vγ5 (247). Vγ3+ DETC precursors are only found in the fetal thymus, and they promote the development of medullary thymic epithelial cells (mTECs) that express the TF Aire (248). The negative selection of self-reactive αβ T cells is in part mediated by Aire+ mTECs. Thus, innate DETC precursors shape the antigen-sensing repertoire of adaptive T cells in early life, corollary to the LTi function in priming lymphoid tissue architecture for T cell function (248). Tγδ17 cells or their precursors have also been suggested to develop only from fetal or neonatal progenitors, and they may strongly prefer fetal microenvironments for differentiation, although the developmental requirements for Vγ4 and Vγ2 cells differ substantially (36). IL-17 produced by an unidentified thymic cell type(s) has been implicated as an inhibitory factor for Tγδ17 cell differentiation in adults (249). The accumulation of other γδ subsets begins primarily in the neonatal thymus. γδ NKT cells exhibit a restricted developmental window, with most being produced soon after birth, but some can be produced inefficiently from adult bone marrow cells (27).
ILC generation also encompasses a fetal-restricted ontogenic window, but unlike γδ ILE cells, most ILCs can be generated efficiently from adult bone marrow–derived CLPs. By E15, Id2+ ChILP and Id2+PLZF+ ILC precursors are amply evident in FL. Two subsets, trNK cells and LTi cells, are predominantly produced from FL-derived progenitors (82), whereas all others are understood to expand dramatically after birth upon interactions with commensals (1). RORγt expression in fetal LTi cells is dependent on RAR docking onto the Rorc CRE. The agonist that initiates this circuit is retinoic acid that is metabolized from the mother’s dietary vitamin A (42).
Modulators Controlling the Generation of Fetal-Origin ILE Cells
There are currently only three well-established candidate apex regulators of fetal-restricted lymphopoiesis: SOX17 (250), Ikaros (251), and Lin28b (252). However, this number is certain to grow given that a network modeling of regulatory TFs based on the embryonic stem cell transcriptome has identified many more candidate factors that are active only in fetal hematopoiesis, and a significant fraction of those are expressed in lymphocytes. Among them are Id2, Nfil3, Sox13, and Zbtb16 that are expressed predominantly in HSCs from the YS, AGM, and/or FL at E12.5, but not in tissues of older fetuses. This pattern contrasts with the mainstream, which does not exhibit temporal selectivity, as exemplified by Hhex, Ikzf1, Lmo2, Myb, and Runx1, which are expressed in HSCs of all stages (253). This finding offers a tantalizing prospect of ILE-restricted progenitors transiently emerging in a narrow window of time at the beginning of lymphopoiesis.
Among the three known fetal regulators, Lin28b best embodies the predicted features of a conductor of ILE gene circuits. Lin28 is a zinc finger RNA-binding protein that specifically inhibits the processing of Let-7 microRNA (Mirlet7) into the functional isoform (254). Let-7 is a bilaterian innovation to tune regulatory gene networks of multipotential progenitors and their tempo of differentiation. It controls self-renewal and proliferation of adult stem cells by targeted degradations of HMGa2, a general chromatin modifier (belonging to a distinct class from the sequence-specific HMG TFs) that in turn regulates the fetal-biased modulator Igf2bp2 (255). Let-7 also directly dictates Myc and Ras mRNA turnover to control cell proliferation and metabolic states (254). The high self-renewal and robust proliferative capacity of fetal HSCs are linked to low Let-7 expression, which is maintained by the restricted Lin28b expression in fetal hematopoietic progenitors and their progenies. Remarkably, enforced expression of Lin28b in adult HSCs resulted in a skewed developmental propensity toward innate lymphocytes, including γδ NKT and αβ iNKT ILE cells. For the latter, a pronounced enrichment for the thymic subsets that express PLZF and/or RORγt was observed (252). Whether PLZF+ ILC progenitors are also expanded upon Lin28b expression has not been examined. Although the molecular link between Lin28b and Zbtb16 has not been proffered, the elevation of RAS upon Let-7 inhibition is one candidate inducer of Zbtb16 via the ERK activation (254).
A fascinating aspect of the fetal-origin ILE cells is their longevity, the ability to self-renew in the mucosal barrier and in the thymus, in the absence of progenitor contributions (2, 21, 249). Intrathymic T cell progenitors are intrinsically long-lived, and long-term generative capacity and T cell outputs from transplanted thymi can be sustained without inputs from bone marrow–derived progenitors (256, 257). Given that competitions for precursor niches in the adult thymus are the basis for the newly arriving T cell progenitors to completely displace preexisting progenitors, the ability of fetal-derived, effector lineage–committed immature ILE cells to persist into adulthood suggests a superior niche occupancy combined with the imprint of fetal HSCs’ enhanced regenerative capacity (258). Identification of additional fetal lymphopoiesis-restricted regulators of transcription will allow more complete mapping of gene circuits underlying the unipotent progenitor-like properties of ILE subsets. Surveys have yielded a simple design principle: Single gene nodes such as Sox17 and Lin28b are sufficient to impose fetal stem cell–and ILE-like properties, respectively, onto adult HSCs and their progenies (252, 258). These precedents recall the limited gene nodal innovations on the core regulatory TF network of T cell development that specify distinct ILE subsets.
SUMMARY AND PERSPECTIVES
The initial classification of innate immunity encompassing myeloid cells, originating from Elie Mechnikov’s discovery of phagocytes that engulf bacteria, is firmly rooted. However, the increasing recognition of diverse lymphocyte subsets with the ability to kill infected cells and produce effector molecules immediately after pathogen encounter has revealed that most of the quintessential properties used to define functional lymphocyte lineages are imprecise. Central among them are the somatically rearranged antigen receptors and immunological memory as the hallmarks of slow adaptive immunity. Inconsistent with these defining parameters, fast innate lymphocytes include not only ILCs that lack clonal antigen receptors, but also T cell subsets expressing αβTCR or γδTCR. Moreover, NK cells and γδTCR+ ILE cells have been reported to exhibit properties consistent with enhanced recall responses when encountering pathogens previously defended against (39). Hence, an emphasis on the kinetics of immune responses, anatomical distributions, and the type of effector cytokines synthesized may simply, and more accurately, segregate the mammalian immune system. This functional classification of immunity is supported by the utilization of shared TF regulatory networks of T cell development to generate most fast-acting ILE subsets. This network of fast effectors reemerges in adaptive lymphocytes and assumes gene circuitry prominence in distinct functional states after activation, to generate long-lived memory cells that can react like fast innate effectors in nonlymphoid tissues where most ILE cells roam. Thus, upon terminal differentiation, the properties of innate and adaptive lymphocytes converge, except for the critical distinction of focused specificity of adaptive cells for the pathogen that initially elicited the response. Future studies that incorporate genome-wide assessments of the TF networks highlighted here in diverse metazoan species, their modes of function in primitive as well as the most advanced lymphocyte subsets, and detailed gene circuitry maps that integrate highly dynamic external modulators of the networks will further solidify the regulatory TF network–based classification of the mammalian fast and slow immune systems.
FUTURE ISSUES.
System-wide regulatory gene network modeling of ILC subsets in comparison with all hematopoietic cell subsets should determine the extent of lineage relationship between ILCs and TCR-expressing ILE cells.
Do dedicated T cell progenitors for ILE cells exist prior to the onset of cell fate–instructive signals, such as those from TCR?
What are the agonists and intracellular signaling intermediates controlling the regulatory TF networks in ILE subsets?
What is the molecular basis of pathogen-specific memory responses in ILE cells, and how does it compare with that of adaptive immunity?
Do functionally distinct ILE subsets exist in invertebrates and lower animals? If so, what is the sphere of influence of the immune regulatory TF networks in primitive immunity?
Acknowledgments
This work integrates several central issues in immunology spanning decades, and we regret that the space constraint prevents a full accounting of important contributions made by many colleagues. We thank J. Huh, L. Berg, and W. Held for discussions and review of the manuscript and members of the lab, present and past, for discoveries and insights that shaped this review. This work was supported by grants from the National Institutes of Health (CA100382, AI101301, and AI107551).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Spits H, Di Santo JP. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat Immunol. 2011;12:21–27. doi: 10.1038/ni.1962. [DOI] [PubMed] [Google Scholar]
- 2.Sumaria N, Roediger B, Ng LG, Qin J, Pinto R, et al. Cutaneous immunosurveillance by self-renewing dermal γΔT cells. J Exp Med. 2011;208:505–18. doi: 10.1084/jem.20101824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kastenmuller W, Torabi-Parizi P, Subramanian N, Lammermann T, Germain RN. A spatially-organized multicellular innate immune response in lymph nodes limits systemic pathogen spread. Cell. 2012;150:1235–48. doi: 10.1016/j.cell.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheridan BS, Lefrançois L. Regional and mucosal memory T cells. Nat Immunol. 2011;12:485–91. doi: 10.1038/ni.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mueller SN, Gebhardt T, Carbone FR, Heath WR. Memory T cell subsets, migration patterns, and tissue residence. Annu Rev Immunol. 2013;31:137–61. doi: 10.1146/annurev-immunol-032712-095954. [DOI] [PubMed] [Google Scholar]
- 6.Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clones. I Definition according to profiles of lymphokine activities and secreted protein. J Immunol. 1986;136:2348–57. [PubMed] [Google Scholar]
- 7.Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–32. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 8.Park H, Li Z, Yang XO, Chang SH, Nurieva R, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–41. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–96. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 10.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–69. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 11.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, et al. The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–33. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 12.Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang SJ, Liang HE, Reizis B, Locksley RM. Regulation of hierarchical clustering and activation of innate immune cells by dendritic cells. Immunity. 2008;29:819–33. doi: 10.1016/j.immuni.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laird DJ, De Tomaso AW, Cooper MD, Weissman IL. 50 million years of chordate evolution: seeking the origins of adaptive immunity. PNAS. 2000;97:6924–26. doi: 10.1073/pnas.97.13.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boehm T, McCurley N, Sutoh Y, Schorpp M, Kasahara M, Cooper MD. VLR-based adaptive immunity. Annu Rev Immunol. 2012;30:203–20. doi: 10.1146/annurev-immunol-020711-075038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, et al. Innate lymphoid cells—a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–49. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 17.Montecino-Rodriguez E, Dorshkind K. B-1 B cell development in the fetus and adult. Immunity. 2012;36:13–21. doi: 10.1016/j.immuni.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonneville M, O’Brien RL, Born WK. γΔ T cell effector functions: a blend of innate programming and acquired plasticity. Nat Rev Immunol. 2010;10:467–78. doi: 10.1038/nri2781. [DOI] [PubMed] [Google Scholar]
- 19.Chen L, He W, Kim ST, Tao J, Gao Y, et al. Epigenetic and transcriptional programs lead to default IFN-γ production by γΔ T cells. J Immunol. 2007;178:2730–36. doi: 10.4049/jimmunol.178.5.2730. [DOI] [PubMed] [Google Scholar]
- 20.Vosshenrich CA, Di Santo JP. Developmental programming of natural killer and innate lymphoid cells. Curr Opin Immunol. 2013;25:130–38. doi: 10.1016/j.coi.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 21.Jameson J, Havran WL. Skin γΔ T-cell functions in homeostasis and wound healing. Immunol Rev. 2007;215:114–22. doi: 10.1111/j.1600-065X.2006.00483.x. [DOI] [PubMed] [Google Scholar]
- 22.Garman RD, Doherty PJ, Raulet DH. Diversity, rearrangement and expression of murine T cell gamma genes. Cell. 1986;45:733–42. doi: 10.1016/0092-8674(86)90787-7. [DOI] [PubMed] [Google Scholar]
- 23.Shires J, Theodoridis E, Hayday AC. Biological insights into TCRγΔ+ and TCRαβ+ intraepithelial lymphocytes provided by serial analysis of gene expression (SAGE) Immunity. 2001;15:419–34. doi: 10.1016/s1074-7613(01)00192-3. [DOI] [PubMed] [Google Scholar]
- 24.Ismail AS, Behrendt CL, Hooper LV. Reciprocal interactions between commensal bacteria and gamma delta intraepithelial lymphocytes during mucosal injury. J Immunol. 2009;182:3047–54. doi: 10.4049/jimmunol.0802705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Innocentin S, Withers DR, Roberts NA, Gallagher AR, et al. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell. 2011;147:629–40. doi: 10.1016/j.cell.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 26.Shui JW, Larange A, Kim G, Vela JL, Zahner S, et al. HVEM signalling at mucosal barriers provides host defence against pathogenic bacteria. Nature. 2012;488:222–25. doi: 10.1038/nature11242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grigoriadou K, Boucontet L, Pereira P. Most IL-4-producing gamma delta thymocytes of adult mice originate from fetal precursors. J Immunol. 2003;171:2413–20. doi: 10.4049/jimmunol.171.5.2413. [DOI] [PubMed] [Google Scholar]
- 28.Stark MA, Huo Y, Burcin TL, Morris MA, Olson TS, Ley K. Phagocytosis of apoptotic neutrophils regulates granulopoiesis via IL-23 and IL-17. Immunity. 2005;22:285–94. doi: 10.1016/j.immuni.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 29.Lockhart E, Green AM, Flynn JL. IL-17 production is dominated by γΔ T cells rather than CD4 T cells during Mycobacterium tuberculosis infection. J Immunol. 2006;177:4662–69. doi: 10.4049/jimmunol.177.7.4662. [DOI] [PubMed] [Google Scholar]
- 30.Azuara V, Levraud JP, Lembezat MP, Pereira P. A novel subset of adult gamma delta thymocytes that secretes a distinct pattern of cytokines and expresses a very restricted T cell receptor repertoire. Eur J Immunol. 1997;27:544–53. doi: 10.1002/eji.1830270228. [DOI] [PubMed] [Google Scholar]
- 31.Lee YJ, Jameson SC, Hogquist KA. Alternative memory in the CD8 T cell lineage. Trends Immunol. 2011;32:50–56. doi: 10.1016/j.it.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narayan K, Sylvia KE, Malhotra N, Yin CC, Martens G, et al. Intrathymic programming of effector fates in three molecularly distinct γΔ T cell subtypes. Nat Immunol. 2012;13:511–18. doi: 10.1038/ni.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuzaki G, Hiromatsu K, Yoshikai Y, Muramori K, Nomoto K. Characterization of T-cell receptor gamma delta T cells appearing at the early phase of murine Listeria monocytogenes infection. Immunology. 1993;78:22–27. [PMC free article] [PubMed] [Google Scholar]
- 34.Mombaerts P, Arnoldi J, Russ F, Tonegawa S, Kaufmann S. Different roles of αβ and γΔ T cells in immunity against an intracellular bacterial pathogen. Nature. 1993;365:53–56. doi: 10.1038/365053a0. [DOI] [PubMed] [Google Scholar]
- 35.Cai Y, Shen X, Ding C, Qi C, Li K, et al. Pivotal role of dermal IL-17-producing γΔ T cells in skin inflammation. Immunity. 2011;35:596–610. doi: 10.1016/j.immuni.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malhotra N, Narayan K, Cho OH, Sylvia KE, Yin C, et al. A network of high-mobility group box transcription factors programs innate interleukin-17 production. Immunity. 2013;38:681–93. doi: 10.1016/j.immuni.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gray EE, Ramirez-Valle F, Xu Y, Wu S, Wu Z, et al. Deficiency in IL-17-committed Vγ4+ γΔ T cells in a spontaneous Sox13-mutant CD45.1+ congenic mouse substrain provides protection from dermatitis. Nat Immunol. 2013;14:584–92. doi: 10.1038/ni.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheridan BS, Romagnoli PA, Pham QM, Fu HH, Alonzo F, 3rd, et al. γΔ T cells exhibit multi-functional and protective memory in intestinal tissues. Immunity. 2013;39:184–95. doi: 10.1016/j.immuni.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun JC, Ugolini S, Vivier E. Immunological memory within the innate immune system. EMBO J. 2014;33:1295–303. doi: 10.1002/embj.201387651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simonian PL, Wehrmann F, Roark CL, Born WK, O’Brien RL, Fontenot AP. γΔ T cells protect against lung fibrosis via IL-22. J Exp Med. 2010;207:2239–53. doi: 10.1084/jem.20100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mielke LA, Jones SA, Raverdeau M, Higgs R, Stefanska A, et al. Retinoic acid expression associates with enhanced IL-22 production by γΔ T cells and innate lymphoid cells and attenuation of intestinal inflammation. J Exp Med. 2013;210:1117–24. doi: 10.1084/jem.20121588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van de Pavert SA, Ferreira M, Domingues RG, Ribeiro H, Molenaar R, et al. Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature. 2014;508:123–27. doi: 10.1038/nature13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jensen KD, Su X, Shin S, Li L, Youssef S, et al. Thymic selection determines γΔ T cell effector fate: Antigen-naive cells make interleukin-17 and antigen-experienced cells make interferon γ. Immunity. 2008;29:90–100. doi: 10.1016/j.immuni.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ribot JC, deBarros A, Pang DJ, Neves JF, Peperzak V, et al. CD27 is a thymic determinant of the balance between interferon-γ- and interleukin 17-producing γΔ T cell subsets. Nat Immunol. 2009;10:427–36. doi: 10.1038/ni.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shay T, Kang J. Immunological Genome Project and systems immunology. Trends Immunol. 2013;34:602–9. doi: 10.1016/j.it.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mingueneau M, Kreslavsky T, Gray D, Heng T, Cruse R, et al. The transcriptional landscape of αβ T cell differentiation. Nat Immunol. 2013;14:619–32. doi: 10.1038/ni.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bendelac A, Schwartz RH. CD4+ and CD8+ T cells acquire specific lymphokine secretion potentials during thymic maturation. Nature. 1991;353:68–71. doi: 10.1038/353068a0. [DOI] [PubMed] [Google Scholar]
- 48.Makino Y, Kanno R, Ito T, Higashino K, Taniguchi M. Predominant expression of invariant Vα14+ TCR α chain in NK1.1+ T cell populations. Int Immunol. 1995;7:1157–61. doi: 10.1093/intimm/7.7.1157. [DOI] [PubMed] [Google Scholar]
- 49.Bendelac A, Lantz O, Quimby ME, Yewdell JW, Bennink JR, Brutkiewicz RR. CD1 recognition by mouse NK1+ T lymphocytes. Science. 1995;268:863–65. doi: 10.1126/science.7538697. [DOI] [PubMed] [Google Scholar]
- 50.Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, et al. CD1d-restricted and TCR-mediated activation of Vα14 NKT cells by glycosylceramides. Science. 1997;278:1626–29. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 51.Savage AK, Constantinides MG, Han J, Picard D, Martin E, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–64. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doulatov S, Notta F, Rice KL, Howell L, Zelent A, et al. PLZF is a regulator of homeostatic and cytokine-induced myeloid development. Genes Dev. 2009;23:2076–87. doi: 10.1101/gad.1788109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cohen NR, Brennan PJ, Shay T, Watts GF, Brigl M, et al. Shared and distinct transcriptional programs underlie the hybrid nature of iNKT cells. Nat Immunol. 2013;14:90–99. doi: 10.1038/ni.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Godfrey DI, Stankovic S, Baxter AG. Raising the NKT cell family. Nat Immunol. 2010;11:197–206. doi: 10.1038/ni.1841. [DOI] [PubMed] [Google Scholar]
- 56.Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol. 2013;14:1146–54. doi: 10.1038/ni.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benlagha K, Kyin T, Beavis A, Teyton L, Bendelac A. A thymic precursor to the NK T cell lineage. Science. 2002;296:553–55. doi: 10.1126/science.1069017. [DOI] [PubMed] [Google Scholar]
- 58.Treiner E, Duban L, Bahram S, Radosavljevic M, Wanner V, et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. 2003;422:164–69. doi: 10.1038/nature01433. [DOI] [PubMed] [Google Scholar]
- 59.Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012;491:717–23. doi: 10.1038/nature11605. [DOI] [PubMed] [Google Scholar]
- 60.Le Bourhis L, Martin E, Peguillet I, Guihot A, Froux N, et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol. 2010;11:701–8. doi: 10.1038/ni.1890. [DOI] [PubMed] [Google Scholar]
- 61.Dusseaux M, Martin E, Serriari N, Peguillet I, Premel V, et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17–secreting T cells. Blood. 2011;117:1250–59. doi: 10.1182/blood-2010-08-303339. [DOI] [PubMed] [Google Scholar]
- 62.Meierovics A, Yankelevich WJ, Cowley SC. MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. PNAS. 2013;110:E3119–28. doi: 10.1073/pnas.1302799110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leeansyah E, Loh L, Nixon DF, Sandberg JK. Acquisition of innate-like microbial reactivity in mucosal tissues during human fetal MAIT-cell development. Nat Commun. 2014;5:3143. doi: 10.1038/ncomms4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van Rhijn I, Kasmar A, de Jong A, Gras S, Bhati M, et al. A conserved human T cell population targets mycobacterial antigens presented by CD1b. Nat Immunol. 2013;14:706–13. doi: 10.1038/ni.2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beagley KW, Fujihashi K, Lagoo AS, Lagoo-Deenadaylan S, Black CA, et al. Differences in intraepithelial lymphocyte T cell subsets isolated from murine small versus large intestine. J Immunol. 1995;154:5611–19. [PubMed] [Google Scholar]
- 66.Yamagata T, Mathis D, Benoist C. Self-reactivity in thymic double-positive cells commits cells to a CD8αα lineage with characteristics of innate immune cells. Nat Immunol. 2004;5:597–605. doi: 10.1038/ni1070. [DOI] [PubMed] [Google Scholar]
- 67.Konkel JE, Maruyama T, Carpenter AC, Xiong Y, Zamarron BF, et al. Control of the development of CD8αα+ intestinal intraepithelial lymphocytes by TGF-β. Nat Immunol. 2011;12:312–19. doi: 10.1038/ni.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leishman AJ, Naidenko OV, Attinger A, Koning F, Lena CJ, et al. T cell responses modulated through interaction between CD8αα and the nonclassical MHC class I molecule, TL. Science. 2001;294:1936–39. doi: 10.1126/science.1063564. [DOI] [PubMed] [Google Scholar]
- 69.Rutz S, Eidenschenk C, Ouyang W. IL-22, not simply a Th17 cytokine. Immunol Rev. 2013;252:116–32. doi: 10.1111/imr.12027. [DOI] [PubMed] [Google Scholar]
- 70.Li W, Sofi MH, Yeh N, Sehra S, McCarthy BP, et al. Thymic selection pathway regulates the effector function of CD4 T cells. J Exp Med. 2007;204:2145–57. doi: 10.1084/jem.20070321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marks BR, Nowyhed HN, Choi JY, Poholek AC, Odegard JM, et al. Thymic self-reactivity selects natural interleukin 17-producing T cells that can regulate peripheral inflammation. Nat Immunol. 2009;10:1125–32. doi: 10.1038/ni.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Prince AL, Watkin LB, Yin CC, Selin LK, Kang J, et al. Innate PLZF+CD4+ αβ T cells develop and expand in the absence of Itk. J Immunol. 2014;193:673–87. doi: 10.4049/jimmunol.1302058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodgers JR, Cook RG. MHC class Ib molecules bridge innate and acquired immunity. Nat Rev Immunol. 2005;5:459–71. doi: 10.1038/nri1635. [DOI] [PubMed] [Google Scholar]
- 74.Bix M, Coles M, Raulet D. Positive selection of Vβ8+CD4−CD8− thymocytes by class I molecules expressed by hematopoietic cells. J Exp Med. 1993;178:901–8. doi: 10.1084/jem.178.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li J, Wu D, Jiang N, Zhuang Y. Combined deletion of Id2 and Id3 genes reveals multiple roles for E proteins in invariant NKT cell development and expansion. J Immunol. 2013;191:5052–64. doi: 10.4049/jimmunol.1301252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Verykokakis M, Krishnamoorthy V, Iavarone A, Lasorella A, Sigvardsson M, Kee BL. Essential functions for ID proteins at multiple checkpoints in invariant NKT cell development. J Immunol. 2013;191:5973–83. doi: 10.4049/jimmunol.1301521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–70. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 78.Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, et al. RORγt and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Luci C, Reynders A, Ivanov II, Cognet C, Chiche L, et al. Influence of the transcription factor RORγt on the development of NKp46+ cell populations in gut and skin. Nat Immunol. 2009;10:75–82. doi: 10.1038/ni.1681. [DOI] [PubMed] [Google Scholar]
- 80.Moro K, Yamada T, Tanabe M, Takeuchi T, Ikawa T, et al. Innate production of TH2 cytokines by adipose tissue-associated c-Kit+Sca-1+ lymphoid cells. Nature. 2010;463:540–44. doi: 10.1038/nature08636. [DOI] [PubMed] [Google Scholar]
- 81.Neill DR, Wong SH, Bellosi A, Flynn RJ, Daly M, et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature. 2010;464:1367–70. doi: 10.1038/nature08900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cherrier M, Eberl G. The development of LTi cells. Curr Opin Immunol. 2012;24:178–83. doi: 10.1016/j.coi.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 83.Bernink JH, Peters CP, Munneke M, te Velde AA, Meijer SL, et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat Immunol. 2013;14:221–29. doi: 10.1038/ni.2534. [DOI] [PubMed] [Google Scholar]
- 84.Fuchs A, Vermi W, Lee JS, Lonardi S, Gilfillan S, et al. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-gamma-producing cells. Immunity. 2013;38:769–81. doi: 10.1016/j.immuni.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Klose CS, Flach M, Mohle L, Rogell L, Hoyler T, et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 2014;157:340–56. doi: 10.1016/j.cell.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 86.Nussbaum JC, Van Dyken SJ, von Moltke J, Cheng LE, Mohapatra A, et al. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature. 2013;502:245–48. doi: 10.1038/nature12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sonnenberg GF, Monticelli LA, Elloso MM, Fouser LA, Artis D. CD4+ lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity. 2011;34:122–34. doi: 10.1016/j.immuni.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sawa S, Cherrier M, Lochner M, Satoh-Takayama N, Fehling HJ, et al. Lineage relationship analysis of RORγt+ innate lymphoid cells. Science. 2010;330:665–69. doi: 10.1126/science.1194597. [DOI] [PubMed] [Google Scholar]
- 89.Kruglov AA, Grivennikov SI, Kuprash DV, Winsauer C, Prepens S, et al. Nonredundant function of soluble LTα3 produced by innate lymphoid cells in intestinal homeostasis. Science. 2013;342:1243–46. doi: 10.1126/science.1243364. [DOI] [PubMed] [Google Scholar]
- 90.Takatori H, Kanno Y, Watford WT, Tato CM, Weiss G, et al. Lymphoid tissue inducer-like cells are an innate source of IL-17 and IL-22. J Exp Med. 2009;206:35–41. doi: 10.1084/jem.20072713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Klose CS, Kiss EA, Schwierzeck V, Ebert K, Hoyler T, et al. A T-bet gradient controls the fate and function of CCR6-RORγt+ innate lymphoid cells. Nature. 2013;494:261–65. doi: 10.1038/nature11813. [DOI] [PubMed] [Google Scholar]
- 92.Rankin LC, Groom JR, Chopin M, Herold MJ, Walker JA, et al. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat Immunol. 2013;14:389–95. doi: 10.1038/ni.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ghoreschi K, Laurence A, Yang XP, Tato CM, McGeachy MJ, et al. Generation of pathogenic TH17 cells in the absence of TGF-beta signalling. Nature. 2010;467:967–71. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Townsend MJ, Weinmann AS, Matsuda JL, Salomon R, Farnham PJ, et al. T-bet regulates the terminal maturation and homeostasis of NK and Vα14i NKT cells. Immunity. 2004;20:477–94. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 95.Gordon SM, Chaix J, Rupp LJ, Wu J, Madera S, et al. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 2012;36:55–67. doi: 10.1016/j.immuni.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lazarevic V, Glimcher LH, Lord GM. T-bet: a bridge between innate and adaptive immunity. Nat Rev Immunol. 2013;13:777–89. doi: 10.1038/nri3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Daussy C, Faure F, Mayol K, Viel S, Gasteiger G, et al. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J Exp Med. 2014;211:563–77. doi: 10.1084/jem.20131560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mjosberg J, Bernink J, Golebski K, Karrich JJ, Peters CP, et al. The transcription factor GATA3 is essential for the function of human type 2 innate lymphoid cells. Immunity. 2012;37:649–59. doi: 10.1016/j.immuni.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 99.Hoyler T, Klose CS, Souabni A, Turqueti-Neves A, Pfeifer D, et al. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 2012;37:634–48. doi: 10.1016/j.immuni.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yagi R, Zhong C, Northrup DL, Yu F, Bouladoux N, et al. The transcription factor GATA3 is critical for the development of all IL-7Rα-expressing innate lymphoid cells. Immunity. 2014;40:378–88. doi: 10.1016/j.immuni.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wong SH, Walker JA, Jolin HE, Drynan LF, Hams E, et al. Transcription factor RORα is critical for nuocyte development. Nat Immunol. 2012;13:229–36. doi: 10.1038/ni.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Halim TY, MacLaren A, Romanish MT, Gold MJ, McNagny KM, Takei F. Retinoic-acid-receptor-related orphan nuclear receptor alpha is required for natural helper cell development and allergic inflammation. Immunity. 2012;37:463–74. doi: 10.1016/j.immuni.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 103.Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014;508:397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Constantinides MG, Bendelac A. Transcriptional regulation of the NKT cell lineage. Curr Opin Immunol. 2013;25:161–67. doi: 10.1016/j.coi.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Segal E, Shapira M, Regev A, Pe’er D, Botstein D, et al. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet. 2003;34:166–76. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 106.Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yosef N, Regev A. Impulse control: temporal dynamics in gene transcription. Cell. 2011;144:886–96. doi: 10.1016/j.cell.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Laurenti E, Doulatov S, Zandi S, Plumb I, Chen J, et al. The transcriptional architecture of early human hematopoiesis identifies multilevel control of lymphoid commitment. Nat Immunol. 2013;14:756–63. doi: 10.1038/ni.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hu G, Chen J. A genome-wide regulatory network identifies key transcription factors for memory CD8+ T-cell development. Nat Commun. 2013;4:2830. doi: 10.1038/ncomms3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jojic V, Shay T, Sylvia K, Zuk O, Sun X, et al. Identification of transcriptional regulators in the mouse immune system. Nat Immunol. 2013;14:633–43. doi: 10.1038/ni.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ciofani M, Madar A, Galan C, Sellars M, Mace K, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rothenberg EV. The chromatin landscape and transcription factors in T cell programming. Trends Immunol. 2014;35:195–204. doi: 10.1016/j.it.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yui MA, Rothenberg EV. Developmental gene networks: a triathlon on the course to T cell identity. Nat Rev Immunol. 2014;14:529–45. doi: 10.1038/nri3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yosef N, Shalek AK, Gaublomme JT, Jin H, Lee Y, et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 2013;496:461–68. doi: 10.1038/nature11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.De Obaldia ME, Bhandoola A. Transcriptional regulation of innate and adaptive lymphocyte lineages. Annu Rev Immunol. 2015;33:607–42. doi: 10.1146/annurev-immunol-032414-112032. [DOI] [PubMed] [Google Scholar]
- 116.Washburn T, Schweighoffer E, Gridley T, Chang D, Fowlkes BJ, et al. Notch activity influences the αβ versus γΔ T cell lineage decision. Cell. 1997;88:833–43. doi: 10.1016/s0092-8674(00)81929-7. [DOI] [PubMed] [Google Scholar]
- 117.Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–40. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Maillard I, Fang T, Pear WS. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu Rev Immunol. 2005;23:945–74. doi: 10.1146/annurev.immunol.23.021704.115747. [DOI] [PubMed] [Google Scholar]
- 119.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.de Pooter RF, Kee BL. E proteins and the regulation of early lymphocyte development. Immunol Rev. 2010;238:93–109. doi: 10.1111/j.1600-065X.2010.00957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 2012;149:467–82. doi: 10.1016/j.cell.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Weber BN, Chi AW, Chavez A, Yashiro-Ohtani Y, Yang Q, et al. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476:63–68. doi: 10.1038/nature10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Germar K, Dose M, Konstantinou T, Zhang J, Wang H, et al. T-cell factor 1 is a gatekeeper for T-cell specification in response to Notch signaling. PNAS. 2011;108:20060–65. doi: 10.1073/pnas.1110230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dose M, Emmanuel AO, Chaumeil J, Zhang J, Sun T, et al. β-Catenin induces T-cell transformation by promoting genomic instability. PNAS. 2014;111:391–96. doi: 10.1073/pnas.1315752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, et al. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature. 1999;397:702–6. doi: 10.1038/17812. [DOI] [PubMed] [Google Scholar]
- 126.Cherrier M, Sawa S, Eberl G. Notch, Id2, and RORγt sequentially orchestrate the fetal development of lymphoid tissue inducer cells. J Exp Med. 2012;209:729–40. doi: 10.1084/jem.20111594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ikawa T, Kawamoto H, Goldrath AW, Murre C. E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment. J Exp Med. 2006;203:1329–42. doi: 10.1084/jem.20060268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, et al. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature. 1995;374:70–74. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- 129.Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–80. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 130.Giese K, Cox J, Grosschedl R. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell. 1992;69:185–95. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- 131.Li L, Zhang JA, Dose M, Kueh HY, Mosadeghi R, et al. A far downstream enhancer for murine Bcl11b controls its T-cell specific expression. Blood. 2013;122:902–11. doi: 10.1182/blood-2012-08-447839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jogi A, Persson P, Grynfeld A, Pahlman S, Axelson H. Modulation of basic helix-loop-helix transcription complex formation by Id proteins during neuronal differentiation. J Biol Chem. 2002;277:9118–26. doi: 10.1074/jbc.M107713200. [DOI] [PubMed] [Google Scholar]
- 133.Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 2010;329:89–93. doi: 10.1126/science.1188989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li P, Burke S, Wang J, Chen X, Ortiz M, et al. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 2010;329:85–89. doi: 10.1126/science.1188063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Garcia-Ojeda ME, Klein Wolterink RGJ, Lemaitre F, Richard-Le Goff O, Hasan M, et al. GATA-3 promotes T-cell specification by repressing B-cell potential in pro-T cells in mice. Blood. 2013;121:1749–59. doi: 10.1182/blood-2012-06-440065. [DOI] [PubMed] [Google Scholar]
- 136.Yucel R, Karsunky H, Klein-Hitpass L, Moroy T. The transcriptional repressor Gfi1 affects development of early, uncommitted c-Kit+ T cell progenitors and CD4/CD8 lineage decision in the thymus. J Exp Med. 2003;197:831–44. doi: 10.1084/jem.20021417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li H, Ji M, Klarmann KD, Keller JR. Repression of Id2 expression by Gfi-1 is required for B-cell and myeloid development. Blood. 2010;116:1060–69. doi: 10.1182/blood-2009-11-255075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yu S, Zhou X, Steinke FC, Liu C, Chen SC, et al. The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity. 2012;37:813–26. doi: 10.1016/j.immuni.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Satoh-Takayama N, Lesjean-Pottier S, Vieira P, Sawa S, Eberl G, et al. IL-7 and IL-15 independently program the differentiation of intestinal CD3-NKp46+ cell subsets from Id2-dependent precursors. J Exp Med. 2010;207:273–80. doi: 10.1084/jem.20092029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yang Q, Monticelli LA, Saenz SA, Chi AW, Sonnenberg GF, et al. T cell factor 1 is required for group 2 innate lymphoid cell generation. Immunity. 2013;38:694–704. doi: 10.1016/j.immuni.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Possot C, Schmutz S, Chea S, Boucontet L, Louise A, et al. Notch signaling is necessary for adult, but not fetal, development of RORγt+ innate lymphoid cells. Nat Immunol. 2011;12:949–58. doi: 10.1038/ni.2105. [DOI] [PubMed] [Google Scholar]
- 142.Lee JS, Cella M, McDonald KG, Garlanda C, Kennedy GD, et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol. 2012;13:144–51. doi: 10.1038/ni.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Garbe AI, Krueger A, Gounari F, Zuniga-Pflucker JC, von Boehmer H. Differential synergy of Notch and T cell receptor signaling determines αβ versus γΔ lineage fate. J Exp Med. 2006;203:1579–90. doi: 10.1084/jem.20060474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ikawa T, Fujimoto S, Kawamoto H, Katsura Y, Yokota Y. Commitment to natural killer cells requires the helix-loop-helix inhibitor Id2. PNAS. 2001;98:5164–69. doi: 10.1073/pnas.091537598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Carotta S, Pang SH, Nutt SL, Belz GT. Identification of the earliest NK-cell precursor in the mouse BM. Blood. 2011;117:5449–52. doi: 10.1182/blood-2010-11-318956. [DOI] [PubMed] [Google Scholar]
- 146.Kim JK, Takeuchi M, Yokota Y. Impairment of intestinal intraepithelial lymphocytes in Id2 deficient mice. Gut. 2004;53:480–86. doi: 10.1136/gut.2003.022293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yamagata T, Benoist C, Mathis D. A shared gene-expression signature in innate-like lymphocytes. Immunol Rev. 2006;210:52–66. doi: 10.1111/j.0105-2896.2006.00371.x. [DOI] [PubMed] [Google Scholar]
- 148.Monticelli LA, Yang Y, Knell J, D’Cruz LM, Cannarile MA, et al. Transcriptional regulator Id2 controls survival of hepatic NKT cells. PNAS. 2009;106:19461–66. doi: 10.1073/pnas.0908249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang B, Lin YY, Dai M, Zhuang Y. Id3 and Id2 act as a dual safety mechanism in regulating the development and population size of innate-like γΔ T cells. J Immunol. 2014;192:1055–63. doi: 10.4049/jimmunol.1302694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Vosshenrich CA, Garcia-Ojeda ME, Samson-Villeger SI, Pasqualetto V, Enault L, et al. A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nat Immunol. 2006;7:1217–24. doi: 10.1038/ni1395. [DOI] [PubMed] [Google Scholar]
- 151.Gascoyne DM, Long E, Veiga-Fernandes H, de Boer J, Williams O, et al. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol. 2009;10:1118–24. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- 152.Seillet C, Huntington ND, Gangatirkar P, Axelsson E, Minnich M, et al. Differential requirement for Nfil3 during NK cell development. J Immunol. 2014;192:2667–76. doi: 10.4049/jimmunol.1302605. [DOI] [PubMed] [Google Scholar]
- 153.Male V, Nisoli I, Kostrzewski T, Allan DS, Carlyle JR, et al. The transcription factor E4bp4/Nfil3 controls commitment to the NK lineage and directly regulates Eomes and Id2 expression. J Exp Med. 2014;211:635–42. doi: 10.1084/jem.20132398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yu X, Rollins D, Ruhn KA, Stubblefield JJ, Green CB, et al. TH17 cell differentiation is regulated by the circadian clock. Science. 2013;342:727–30. doi: 10.1126/science.1243884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Seillet C, Rankin LC, Groom JR, Mielke LA, Tellier J, et al. Nfil3 is required for the development of all innate lymphoid cell subsets. J Exp Med. 2014;211:1733–40. doi: 10.1084/jem.20140145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Geiger TL, Abt MC, Gasteiger G, Firth MA, O’Connor MH, et al. Nfil3 is crucial for development of innate lymphoid cells and host protection against intestinal pathogens. J Exp Med. 2014;211:1723–31. doi: 10.1084/jem.20140212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Aliahmad P, de la Torre B, Kaye J. Shared dependence on the DNA-binding factor TOX for the development of lymphoid tissue-inducer cell and NK cell lineages. Nat Immunol. 2010;11:945–52. doi: 10.1038/ni.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Aliahmad P, Kadavallore A, de la Torre B, Kappes D, Kaye J. TOX is required for development of the CD4 T cell lineage gene program. J Immunol. 2011;187:5931–40. doi: 10.4049/jimmunol.1101474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kang J, Coles M. IL-7: the global builder of the innate lymphoid network and beyond, one niche at a time. Semin Immunol. 2012;24:190–97. doi: 10.1016/j.smim.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Narayan K, Kang J. Disorderly conduct in γΔ versus αβ T cell lineage commitment. Semin Immunol. 2010;22:222–27. doi: 10.1016/j.smim.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Shibata K, Yamada H, Nakamura M, Hatano S, Katsuragi Y, et al. IFN-γ-producing and IL-17-producing γΔ T cells differentiate at distinct developmental stages in murine fetal thymus. J Immunol. 2014;192:2210–18. doi: 10.4049/jimmunol.1302145. [DOI] [PubMed] [Google Scholar]
- 162.Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol. 2000;227:239–55. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- 163.Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–23. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Melichar HJ, Narayan K, Der SD, Hiraoka Y, Gardiol N, et al. Regulation of γΔ versus αβ T lymphocyte differentiation by the transcription factor SOX13. Science. 2007;315:230–33. doi: 10.1126/science.1135344. [DOI] [PubMed] [Google Scholar]
- 165.Turchinovich G, Hayday AC. Skint-1 identifies a common molecular mechanism for the development of interferon-γ-secreting versus interleukin-17-secreting γΔ T cells. Immunity. 2011;35:59–68. doi: 10.1016/j.immuni.2011.04.018. [DOI] [PubMed] [Google Scholar]
- 166.Wang R, Xie H, Huang Z, Ma J, Fang X, et al. T cell factor 1 regulates thymocyte survival via a RORγt-dependent pathway. J Immunol. 2011;187:5964–73. doi: 10.4049/jimmunol.1101205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Keerthivasan S, Aghajani K, Dose M, Molinero L, Khan MW, et al. β-Catenin promotes colitis and colon cancer through imprinting of proinflammatory properties in T cells. Sci Transl Med. 2014;6:225ra28. doi: 10.1126/scitranslmed.3007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Tanaka S, Suto A, Iwamoto T, Kashiwakuma D, Kagami S, et al. Sox5 and c-Maf cooperatively induce Th17 cell differentiation via RORγt induction as downstream targets of Stat3. J Exp Med. 2014;211:1857–74. doi: 10.1084/jem.20130791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Shim S, Kwan KY, Li M, Lefebvre V, Sestan N. Cis-regulatory control of corticospinal system development and evolution. Nature. 2012;486:74–79. doi: 10.1038/nature11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Schilham MW, Oosterwegel MA, Moerer P, Ya J, de Boer PA, et al. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380:711–14. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- 171.Spooner CJ, Lesch J, Yan D, Khan AA, Abbas A, et al. Specification of type 2 innate lymphocytes by the transcriptional determinant Gfi1. Nat Immunol. 2013;14:1229–36. doi: 10.1038/ni.2743. [DOI] [PubMed] [Google Scholar]
- 172.Cohen NR, Brennan PJ, Shay T, Watts GF, Brigl M, et al. Shared and distinct transcriptional programs underlie the hybrid nature of iNKT cells. Nat Immunol. 2013;14:90–99. doi: 10.1038/ni.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sinner D, Kordich JJ, Spence JR, Opoka R, Rankin S, et al. Sox17 and Sox4 differentially regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Mol Cell Biol. 2007;27:7802–15. doi: 10.1128/MCB.02179-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yu Q, Sharma A, Ghosh A, Sen JM. T cell factor-1 negatively regulates expression of IL-17 family of cytokines and protects mice from experimental autoimmune encephalomyelitis. J Immunol. 2011;186:3946–52. doi: 10.4049/jimmunol.1003497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wei G, Abraham BJ, Yagi R, Jothi R, Cui K, et al. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity. 2011;35:299–311. doi: 10.1016/j.immuni.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Vahedi G, Takahashi H, Nakayamada S, Sun HW, Sartorelli V, et al. STATs shape the active enhancer landscape of T cell populations. Cell. 2012;151:981–93. doi: 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhu J, Jankovic D, Oler AJ, Wei G, Sharma S, et al. The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity. 2012;37:660–73. doi: 10.1016/j.immuni.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Murphy TL, Tussiwand R, Murphy KM. Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat Rev Immunol. 2013;13:499–509. doi: 10.1038/nri3470. [DOI] [PubMed] [Google Scholar]
- 179.Esser C, Rannug A, Stockinger B. The aryl hydrocarbon receptor in immunity. Trends Immunol. 2009;30:447–54. doi: 10.1016/j.it.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 180.Muranski P, Borman ZA, Kerkar SP, Klebanoff CA, Ji Y, et al. Th17 cells are long lived and retain a stem cell-like molecular signature. Immunity. 2011;35:972–85. doi: 10.1016/j.immuni.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Bailis W, Yashiro-Ohtani Y, Fang TC, Hatton RD, Weaver CT, et al. Notch simultaneously orchestrates multiple helper T cell programs independently of cytokine signals. Immunity. 2013;39:148–59. doi: 10.1016/j.immuni.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yu Q, Sharma A, Oh SY, Moon HG, Hossain MZ, et al. T cell factor 1 initiates the T helper type 2 fate by inducing the transcription factor GATA-3 and repressing interferon-gamma. Nat Immunol. 2009;10:992–99. doi: 10.1038/ni.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kuwahara M, Yamashita M, Shinoda K, Tofukuji S, Onodera A, et al. The transcription factor Sox4 is a downstream target of signaling by the cytokine TGF-beta and suppresses TH2 differentiation. Nat Immunol. 2012;13:778–86. doi: 10.1038/ni.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Jeannet G, Boudousquie C, Gardiol N, Kang J, Huelsken J, Held W. Essential role of the Wnt pathway effector Tcf-1 for the establishment of functional CD8 T cell memory. PNAS. 2010;107:9777–82. doi: 10.1073/pnas.0914127107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhou X, Yu S, Zhao DM, Harty JT, Badovinac VP, Xue HH. Differentiation and persistence of memory CD8+ T cells depend on T cell factor 1. Immunity. 2010;33:229–40. doi: 10.1016/j.immuni.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Miyazaki M, Rivera RR, Miyazaki K, Lin YC, Agata Y, Murre C. The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. Nat Immunol. 2011;12:992–1001. doi: 10.1038/ni.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Yang CY, Best JA, Knell J, Yang E, Sheridan AD, et al. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat Immunol. 2011;12:1221–29. doi: 10.1038/ni.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Yu Q, Erman B, Park JH, Feigenbaum L, Singer A. IL-7 receptor signals inhibit expression of transcription factors TCF-1, LEF-1, and RORγt: impact on thymocyte development. J Exp Med. 2004;200:797–803. doi: 10.1084/jem.20032183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Rawlings JS, Gatzka M, Thomas PG, Ihle JN. Chromatin condensation via the condensin II complex is required for peripheral T-cell quiescence. EMBO J. 2011;30:263–76. doi: 10.1038/emboj.2010.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Larroux C, Luke GN, Koopman P, Rokhsar DS, Shimeld SM, Degnan BM. Genesis and expansion of metazoan transcription factor gene classes. Mol Biol Evol. 2008;25:980–96. doi: 10.1093/molbev/msn047. [DOI] [PubMed] [Google Scholar]
- 191.Nichols SA, Dirks W, Pearse JS, King N. Early evolution of animal cell signaling and adhesion genes. PNAS. 2006;103:12451–56. doi: 10.1073/pnas.0604065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kortschak RD, Samuel G, Saint R, Miller DJ. EST analysis of the cnidarian Acropora millepora reveals extensive gene loss and rapid sequence divergence in the model invertebrates. Curr Biol. 2003;13:2190–95. doi: 10.1016/j.cub.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 193.Hirano M, Guo P, McCurley N, Schorpp M, Das S, et al. Evolutionary implications of a third lymphocyte lineage in lampreys. Nature. 2013;501:435–38. doi: 10.1038/nature12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Rogozin IB, Iyer LM, Liang L, Glazko GV, Liston VG, et al. Evolution and diversification of lamprey antigen receptors: evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat Immunol. 2007;8:647–56. doi: 10.1038/ni1463. [DOI] [PubMed] [Google Scholar]
- 195.Smith JJ, Kuraku S, Holt C, Sauka-Spengler T, Jiang N, et al. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet. 2013;45:415–21. doi: 10.1038/ng.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Fugmann SD, Messier C, Novack LA, Cameron RA, Rast JP. An ancient evolutionary origin of the Rag1/2 gene locus. PNAS. 2006;103:3728–33. doi: 10.1073/pnas.0509720103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Sebe-Pedros A, de Mendoza A, Lang BF, Degnan BM, Ruiz-Trillo I. Unexpected repertoire of metazoan transcription factors in the unicellular holozoan Capsaspora owczarzaki. Mol Biol Evol. 2011;28:1241–54. doi: 10.1093/molbev/msq309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ryan JF, Baxevanis AD. Hox, Wnt, and the evolution of the primary body axis: insights from the early-divergent phyla. Biol Direct. 2007;2:37. doi: 10.1186/1745-6150-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Suga H, Chen Z, de Mendoza A, Sebe-Pedros A, Brown MW, et al. The Capsaspora genome reveals a complex unicellular prehistory of animals. Nat Commun. 2013;4:2325. doi: 10.1038/ncomms3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.King N, Westbrook MJ, Young SL, Kuo A, Abedin M, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–88. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier ME, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–26. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Leininger S, Adamski M, Bergum B, Guder C, Liu J, et al. Developmental gene expression provides clues to relationships between sponge and eumetazoan body plans. Nat Commun. 2014;5:3905. doi: 10.1038/ncomms4905. [DOI] [PubMed] [Google Scholar]
- 203.Jager M, Queinnec E, Houliston E, Manuel M. Expansion of the SOX gene family predated the emergence of the Bilateria. Mol Phylogenet Evol. 2006;39:468–77. doi: 10.1016/j.ympev.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 204.Fortunato S, Adamski M, Bergum B, Guder C, Jordal S, et al. Genome-wide analysis of the sox family in the calcareous sponge Sycon ciliatum: multiple genes with unique expression patterns. EvoDevo. 2012;3:14. doi: 10.1186/2041-9139-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Hoy MA, Yu F, Meyer JM, Tarazona OA, Jeyaprakash A, Wu K. Transcriptome sequencing and annotation of the predatory mite Metaseiulus occidentalis (Acari: Phytoseiidae): a cautionary tale about possible contamination by prey sequences. Exp Appl Acarol. 2013;59:283–96. doi: 10.1007/s10493-012-9603-4. [DOI] [PubMed] [Google Scholar]
- 206.Simionato E, Ledent V, Richards G, Thomas-Chollier M, Kerner P, et al. Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol Biol. 2007;7:33. doi: 10.1186/1471-2148-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hibino T, Loza-Coll M, Messier C, Majeske AJ, Cohen AH, et al. The immune gene repertoire encoded in the purple sea urchin genome. Dev Biol. 2006;300:349–65. [Google Scholar]
- 208.Rast JP, Smith LC, Loza-Coll M, Hibino T, Litman GW. Genomic insights into the immune system of the sea urchin. Science. 2006;314:952–56. doi: 10.1126/science.1134301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Solek CM, Oliveri P, Loza-Coll M, Schrankel CS, Ho EC, et al. An ancient role for Gata-1/2/3 and Scl transcription factor homologs in the development of immunocytes. Dev Biol. 2013;382:280–92. doi: 10.1016/j.ydbio.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 210.Yamada L, Kobayashi K, Degnan B, Satoh N, Satou Y. A genomewide survey of developmentally relevant genes in Ciona intestinalisIV Genes for HMG transcriptional regulators, bZip and GATA/Gli/Zic/Snail. Dev Genes Evol. 2003;213:245–53. doi: 10.1007/s00427-003-0316-x. [DOI] [PubMed] [Google Scholar]
- 211.Sebe-Pedros A, Ariza-Cosano A, Weirauch MT, Leininger S, Yang A, et al. Early evolution of the T-box transcription factor family. PNAS. 2013;110:16050–55. doi: 10.1073/pnas.1309748110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Horton AC, Gibson-Brown JJ. Evolution of developmental functions by the Eomesodermin, T-brain-1, Tbx21 subfamily of T-box genes: insights from amphioxus. J Exp Zool. 2002;294:112–21. doi: 10.1002/jez.10151. [DOI] [PubMed] [Google Scholar]
- 213.Venkatesh B, Lee AP, Ravi V, Maurya AK, Lian MM, et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature. 2014;505:174–79. doi: 10.1038/nature12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Zhang T, Xiong H, Kan LX, Zhang CK, Jiao XF, et al. Genomic sequence, structural organization, molecular evolution, and aberrant rearrangement of promyelocytic leukemia zinc finger gene. PNAS. 1999;96:11422–27. doi: 10.1073/pnas.96.20.11422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Howard RM, Sundaram MV. C. elegans EOR-1/PLZF and EOR-2 positively regulate Ras and Wnt signaling and function redundantly with LIN-25 and the SUR-2 Mediator component. Genes Dev. 2002;16:1815–27. doi: 10.1101/gad.998402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Maeng O, Son W, Chung J, Lee KS, Lee YH, et al. The BTB/POZ-ZF transcription factor dPLZF is involved in Ras/ERK signaling during Drosophila wing development. Mol Cells. 2012;33:457–63. doi: 10.1007/s10059-012-2179-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Imai KS, Hino K, Yagi K, Satoh N, Satou Y. Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: towards a comprehensive understanding of gene networks. Development. 2004;131:4047–58. doi: 10.1242/dev.01270. [DOI] [PubMed] [Google Scholar]
- 218.Kosan C, Saba I, Godmann M, Herold S, Herkert B, et al. Transcription factor miz-1 is required to regulate interleukin-7 receptor signaling at early commitment stages of B cell differentiation. Immunity. 2010;33:917–28. doi: 10.1016/j.immuni.2010.11.028. [DOI] [PubMed] [Google Scholar]
- 219.Jetten AM. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl Recept Signal. 2009;7:e003. doi: 10.1621/nrs.07003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Wang Y, Kumar N, Solt LA, Richardson TI, Helvering LM, et al. Modulation of retinoic acid receptor-related orphan receptor alpha and gamma activity by 7-oxygenated sterol ligands. J Biol Chem. 2010;285:5013–25. doi: 10.1074/jbc.M109.080614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Monte MM, Wang T, Costa MM, Harun NO, Secombes CJ. Cloning and expression analysis of two ROR-γ homologues (ROR-γa1 and ROR-γa2) in rainbow trout Oncorhynchus mykiss. Fish Shellfish Immunol. 2012;33:365–74. doi: 10.1016/j.fsi.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 222.Egawa T, Eberl G, Taniuchi I, Benlagha K, Geissmann F, et al. Genetic evidence supporting selection of the Vα14i NKT cell lineage from double-positive thymocyte precursors. Immunity. 2005;22:705–16. doi: 10.1016/j.immuni.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 223.Du L, Yang X, Yang L, Wang X, Zhang A, Zhou H. Molecular evidence for the involvement of RORα and RORγ in immune response in teleost. Fish Shellfish Immunol. 2012;33:418–26. doi: 10.1016/j.fsi.2012.05.033. [DOI] [PubMed] [Google Scholar]
- 224.Hymowitz SG, Filvaroff EH, Yin JP, Lee J, Cai L, et al. IL-17s adopt a cystine knot fold: structure and activity of a novel cytokine, IL-17F, and implications for receptor binding. EMBO J. 2001;20:5332–41. doi: 10.1093/emboj/20.19.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Roberts S, Gueguen Y, de Lorgeril J, Goetz F. Rapid accumulation of an interleukin 17 homolog transcript in Crassostrea gigas hemocytes following bacterial exposure. Dev Comp Immunol. 2008;32:1099–104. doi: 10.1016/j.dci.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 226.Li J, Zhang Y, Zhang Y, Xiang Z, Tong Y, et al. Genomic characterization and expression analysis of five novel IL-17 genes in the Pacific oyster, Crassostrea gigas. Fish Shellfish Immunol. 2014;40:455–65. doi: 10.1016/j.fsi.2014.07.026. [DOI] [PubMed] [Google Scholar]
- 227.Ramirez-Carrozzi V, Sambandam A, Luis E, Lin Z, Jeet S, et al. IL-17C regulates the innate immune function of epithelial cells in an autocrine manner. Nat Immunol. 2011;12:1159–66. doi: 10.1038/ni.2156. [DOI] [PubMed] [Google Scholar]
- 228.Tsutsui S, Nakamura O, Watanabe T. Lamprey (Lethenteron japonicum) IL-17 upregulated by LPS-stimulation in the skin cells. Immunogenetics. 2007;59:873–82. doi: 10.1007/s00251-007-0254-2. [DOI] [PubMed] [Google Scholar]
- 229.Bar E, Whitney PG, Moor K, Reis e Sousa C, LeibundGut-Landmann S. IL-17 regulates systemic fungal immunity by controlling the functional competence of NK cells. Immunity. 2014;40:117–27. doi: 10.1016/j.immuni.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 230.Goetz FW, Planas JV, MacKenzie S. Tumor necrosis factors. Dev Comp Immunol. 2004;28:487–97. doi: 10.1016/j.dci.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 231.Rangel-Moreno J, Carragher DM, de la Luz Garcia-Hernandez M, Hwang JY, Kusser K, et al. The development of inducible bronchus-associated lymphoid tissue depends on IL-17. Nat Immunol. 2011;12:639–46. doi: 10.1038/ni.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Miller DJ, Hemmrich G, Ball EE, Hayward DC, Khalturin K, et al. The innate immune repertoire in Cnidaria—ancestral complexity and stochastic gene loss. Genome Biol. 2007;8:R59. doi: 10.1186/gb-2007-8-4-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Ikuta K, Kina T, MacNeil I, Uchida N, Peault B, et al. A developmental switch in thymic lymphocyte maturation potential occurs at the level of hematopoietic stem cells. Cell. 1990;62:863–74. doi: 10.1016/0092-8674(90)90262-d. [DOI] [PubMed] [Google Scholar]
- 234.Ramond C, Berthault C, Burlen-Defranoux O, de Sousa AP, Guy-Grand D, et al. Two waves of distinct hematopoietic progenitor cells colonize the fetal thymus. Nat Immunol. 2014;15:27–35. doi: 10.1038/ni.2782. [DOI] [PubMed] [Google Scholar]
- 235.Rose S, Lichtenheld M, Foote MR, Adkins B. Murine neonatal CD4+ cells are poised for rapid Th2 effector-like function. J Immunol. 2007;178:2667–78. doi: 10.4049/jimmunol.178.5.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Sieweke MH, Allen JE. Beyond stem cells: self-renewal of differentiated macrophages. Science. 2013;342:1242974. doi: 10.1126/science.1242974. [DOI] [PubMed] [Google Scholar]
- 237.Medvinsky A, Rybtsov S, Taoudi S. Embryonic origin of the adult hematopoietic system: advances and questions. Development. 2011;138:1017–31. doi: 10.1242/dev.040998. [DOI] [PubMed] [Google Scholar]
- 238.Kumaravelu P, Hook L, Morrison AM, Ure J, Zhao S, et al. Quantitative developmental anatomy of definitive haematopoietic stem cells/long-term repopulating units (HSC/RUs): role of the aorta-gonad-mesonephros (AGM) region and the yolk sac in colonisation of the mouse embryonic liver. Development. 2002;129:4891–99. doi: 10.1242/dev.129.21.4891. [DOI] [PubMed] [Google Scholar]
- 239.Yoshimoto M, Porayette P, Glosson NL, Conway SJ, Carlesso N, et al. Autonomous murine T-cell progenitor production in the extra-embryonic yolk sac before HSC emergence. Blood. 2012;119:5706–14. doi: 10.1182/blood-2011-12-397489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Boiers C, Carrelha J, Lutteropp M, Luc S, Green JC, et al. Lymphomyeloid contribution of an immune-restricted progenitor emerging prior to definitive hematopoietic stem cells. Cell Stem Cell. 2013;13:535–48. doi: 10.1016/j.stem.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 241.Tang Y, Peitzsch C, Charoudeh HN, Cheng M, Chaves P, et al. Emergence of NK-cell progenitors and functionally competent NK-cell lineage subsets in the early mouse embryo. Blood. 2012;120:63–75. doi: 10.1182/blood-2011-02-337980. [DOI] [PubMed] [Google Scholar]
- 242.Sojka DK, Plougastel-Douglas B, Yang L, Pak-Wittel MA, Artyomov MN, et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife. 2014;3:e01659. doi: 10.7554/eLife.01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Michie AM, Carlyle JR, Schmitt TM, Ljutic B, Cho SK, et al. Clonal characterization of a bipotent T cell and NK cell progenitor in the mouse fetal thymus. J Immunol. 2000;164:1730–33. doi: 10.4049/jimmunol.164.4.1730. [DOI] [PubMed] [Google Scholar]
- 244.Douagi I, Colucci F, Di Santo JP, Cumano A. Identification of the earliest prethymic bipotent T/NK progenitor in murine fetal liver. Blood. 2002;99:463–71. doi: 10.1182/blood.v99.2.463. [DOI] [PubMed] [Google Scholar]
- 245.Gilfillan S, Benoist C, Mathis D. Mice lacking terminal deoxynucleotidyl transferase: adult mice with a fetal antigen receptor repertoire. Immunol Rev. 1995;148:201–19. doi: 10.1111/j.1600-065x.1995.tb00099.x. [DOI] [PubMed] [Google Scholar]
- 246.Herzenberg LA, Herzenberg LA. Toward a layered immune system. Cell. 1989;59:953–54. doi: 10.1016/0092-8674(89)90748-4. [DOI] [PubMed] [Google Scholar]
- 247.Xiong N, Kang C, Raulet DH. Redundant and unique roles of two enhancer elements in the TCRγ locus in gene regulation and γΔ T cell development. Immunity. 2002;16:453–63. doi: 10.1016/s1074-7613(02)00285-6. [DOI] [PubMed] [Google Scholar]
- 248.Roberts NA, White AJ, Jenkinson WE, Turchinovich G, Nakamura K, et al. Rank signaling links the development of invariant γΔ T cell progenitors and Aire+ medullary epithelium. Immunity. 2012;36:427–37. doi: 10.1016/j.immuni.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Haas JD, Ravens S, Duber S, Sandrock I, Oberdorfer L, et al. Development of interleukin-17-producing γΔ T cells is restricted to a functional embryonic wave. Immunity. 2012;37:48–59. doi: 10.1016/j.immuni.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 250.Kim I, Saunders TL, Morrison SJ. Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell. 2007;130:470–83. doi: 10.1016/j.cell.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Wang JH, Nichogiannopoulou A, Wu L, Sun L, Sharpe AH, et al. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–49. doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- 252.Yuan J, Nguyen CK, Liu X, Kanellopoulou C, Muljo SA. Lin28b reprograms adult bone marrow hematopoietic progenitors to mediate fetal-like lymphopoiesis. Science. 2012;335:1195–200. doi: 10.1126/science.1216557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.McKinney-Freeman S, Cahan P, Li H, Lacadie SA, Huang HT, et al. The transcriptional landscape of hematopoietic stem cell ontogeny. Cell Stem Cell. 2012;11:701–14. doi: 10.1016/j.stem.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Shyh-Chang N, Daley GQ. Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell. 2013;12:395–406. doi: 10.1016/j.stem.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Copley MR, Babovic S, Benz C, Knapp DJ, Beer PA, et al. The Lin28b-let-7-Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat Cell Biol. 2013;15:916–25. doi: 10.1038/ncb2783. [DOI] [PubMed] [Google Scholar]
- 256.Martins VC, Ruggiero E, Schlenner SM, Madan V, Schmidt M, et al. Thymus-autonomous T cell development in the absence of progenitor import. J Exp Med. 2012;209:1409–17. doi: 10.1084/jem.20120846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Peaudecerf L, Lemos S, Galgano A, Krenn G, Vasseur F, et al. Thymocytes may persist and differentiate without any input from bone marrow progenitors. J Exp Med. 2012;209:1401–8. doi: 10.1084/jem.20120845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.He S, Kim I, Lim MS, Morrison SJ. Sox17 expression confers self-renewal potential and fetal stem cell characteristics upon adult hematopoietic progenitors. Genes Dev. 2011;25:1613–27. doi: 10.1101/gad.2052911. [DOI] [PMC free article] [PubMed] [Google Scholar]