Abstract
Understanding how the gut microbiota is affected by diarrhea episodes may help explain alterations in intestinal function among children in low-income settings. This study examined the composition of the gut microbiome of Nicaraguan children both during diarrhea episodes and while free of diarrhea for at least 2 months. Relative abundances of bacterial taxa, phylogenetic diversity, and species richness were determined by 16S amplicon sequencing and compared between paired diarrhea and recovery samples. A total of 66 stools were provided by 25 children enrolled in a 1-year cohort study of diarrhea etiologies. Children in our cohort had a mean age of 21.9 months; 64% were breast-fed, and 10% had received an antibiotic during the diarrhea episode. Overall, phylogenetic diversity and species richness did not differ significantly between diarrhea and recovery stools. However, of children who had a bacterial enteropathogen detected in any diarrhea stool, none experienced an increase in phylogenetic diversity in recovery, whereas of those in whom no bacterial enteropathogens were detected in their diarrhea stool(s), 59% experienced an increase in phylogenetic diversity in recovery (P = 0.008). This preliminary study suggests that recovery of the gut microbiota after a diarrhea episode may take longer time than previously thought and may be pathogen specific.
Introduction
The gut microbiota serves important functions in the human host, including enteropathogen displacement by colonization competition, metabolism of energy substrates, and development of enteric immunity.1 The gut microbiota undergoes a dynamic process of colonization and development during the first years of life. Soon after birth, the aerobic gut environment encourages the growth of aerobes and facultative anaerobes (such as Enterococcus, Streptococcus, and Lactobacillus).2,3 Decreasing oxygen levels favor the growth of obligate anaerobes (such as Bifidobacterium, Clostridium, and Bacteroides).4,5 Weaning and introduction of solid foods further alter the gut microbiota community structure, until the composition resembles that of an adult later in childhood.4,6,7
Besides changes related to age, several other factors have been reported to alter gut microbiota community structure, including diet, birth route (vaginal versus Cesarean delivery), exposure to antibiotics, nutritional status, genetic factors, and diarrhea episodes.2,6,8 Diarrhea episodes have been shown to be associated with an overall decrease in phylogenetic diversity.9–12 Although modifications in the relative abundance of taxa differ by study site, studies have shown increases in the relative abundance of the phylum Proteobacteria and decreases in Bifidobacterium and Lactobacillus during diarrhea episodes.10–12 These alterations in the gut microbiota during diarrhea episodes in the early years of life may have a substantial impact on the subsequent development of the gut microbiota and its related functions. These effects may be especially important in developing world settings, where children commonly experience several diarrhea episodes per year.13
Although several studies have reported on changes in the gut microbiota during diarrhea episodes in developing world settings, such as Nicaragua, less is known about the composition and diversity of the gut microbiota during recovery after diarrhea episodes. Understanding whether and how the gut microbiota changes after a diarrhea episode may inform our understanding of the potential impact of diarrhea episodes on the functions of the gut microbiota. The goal of this study is to compare features of the gut microbiota, including composition and diversity, during diarrhea episodes and after resolution of symptoms in young Nicaraguan children.
Materials and Methods
Study design.
Twenty-five children from a larger population-based study of infectious diarrhea etiologies in León, Nicaragua,14 were selected to participate in this study. For the larger study, children were visited in their households every 14 days between January 25, 2010 and January 24, 2011 by field workers who assessed diarrhea episodes and collected sociodemographic information. Children were weighed, and weight-for-age percentiles were calculated using World Health Organization standards. Diarrhea was defined as an increase in stool frequency to at least three loose stools per 24-hour period or as a substantial change in stool consistency (bloody, very loose, or watery) after at least 3 diarrhea-free days. Stools were collected from children who developed diarrhea during the diarrhea episode and from enrolled children without diarrhea in the preceding 2 months who served as diarrhea-free controls.
Study population.
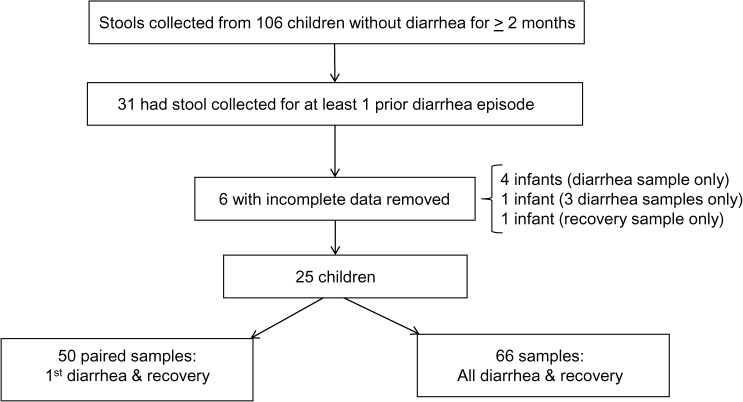
Among the 826 children in the larger study, 106 enrolled children without diarrhea in the preceding 2 months served as age-, sex-, and neighborhood-matched controls for children who developed diarrhea during the study. Among these 106 children, 31 had previously provided at least one stool during a diarrhea episode in the 1-year study and were therefore eligible for this analysis. Six of the 31 children had at least one stool that was not evaluable and so they were removed from the analysis, which resulted in 25 children selected for this study (Figure 1 ). Each child provided at least one stool during diarrhea and one stool during recovery. Nine children provided stool samples from multiple diarrhea episodes and 16 children provided stool samples from a single diarrhea episode, such that a total of 66 diarrhea and 25 recovery stools were included. The study was approved by the Institutional Review Boards of the National Autonomous University of Nicaragua, León (UNAN-León) and the University of North Carolina at Chapel Hill.
Figure 1.
Selection of 25 child participants from larger population-based study.
Stool collection.
Stool specimens were obtained in a sterile plastic container or from the child's soiled diaper and were transported within 2 hours from the household to the Microbiology Laboratory of UNAN-León at 4°C. Separate aliquots were stored in 10% (w/v) suspension of stool with phosphate-buffered saline (pH = 7.2) and stored at −20°C for microbiome analysis.
Detection of enteropathogens.
Detection of enteropathogens in stool samples was described in detail previously.14 In brief, stools were analyzed by enzyme immunoassays to detect rotavirus and adenovirus, and by real-time polymerase chain reaction (PCR) for norovirus and sapovirus. Conventional culturing techniques were used to detect the bacterial enteropathogens, including Shigella spp., Salmonella spp., Campylobacter spp., and Escherichia coli. Positive E. coli cultures were assayed by multiplex PCR for the following pathotypes: enteropathogenic E. coli, enterotoxigenic E. coli, enteroaggregative E. coli, enteroinvasive E. coli, and enterohemorrhagic E. coli, as described by Vilchez.15 Direct microscopy was used for detection of the parasitic enteropathogens, with concentration and acid-fast staining for the detection of Cryptosporidium spp., as described by Garcia.16
16S rDNA amplicon sequencing and sequencing data analysis.
DNA from stool samples was extracted using the QIAmp DNA Stool kit (Qiagen, Hilden, Germany). Modifications were performed in the disruption step to ensure optimal bacterial lysis, especially for gram-positive bacteria. In brief, 200 mg of sample was transferred to a microtube containing 0.2 g of autoclaved 11-μm-diameter glass beads (Sigma, St. Louis, MO) and 1.4 mL of ASL buffer (Qiagen). Samples were then homogenized in a TissueLyser II instrument (Qiagen) at 25 Hz for 2 minutes according to the manufacturer's protocol.
For amplicon library preparation, the V1-V2 hypervariable region of the 16S rDNA was amplified from total bacterial DNA isolated from each sample using a forward primer composed of the Ion Torrent adapter A (5′-CCATCTCATCCCTGCGTGTCTCCGACTCAG-3′), a 10-bp Ion Xpress™ barcode, unique to each sample (Life Technologies, Grand Island, NY) and the universal bacterial primer 8F (5′-AGAGTTTGATCCTGGCTCAG-3′). The reverse primer consisted of the Ion Torrent trP1 adapter (5′-CCTCTCTATGGGCAGTCGGTGAT-3′) (Life Technologies) followed by the reverse bacterial primer 338R (5′-GCTGCCTCCCGTAGGAGT-3′).
PCR reactions contained 5–10 ng of DNA template, 2.5 units of HotStar Hi-fidelity DNA polymerase (Qiagen), 1 × HotStar Hi-Fidelity PCR buffer containing dNTPs (Qiagen), and 0.6 μM of each primer. Reaction conditions consisted of an initial denaturation for 5 minutes at 94°C followed by 35 cycles of denaturing at 94°C for 60 seconds, annealing at 57°C for 60 seconds, and extension at 72°C for 60 seconds, and a final extension of 72°C for 10 minutes. No-template negative controls were amplified for all barcode primer sets. PCR products were gel purified individually using the E-Gel Electrophoresis System (Life Technologies) and standardized before pooling. Purified samples were pooled at equimolar concentrations and sequenced on an Ion Torrent PGM (Life Technologies) using the Ion PGM 400 Sequencing Kit (Life Technologies) and the Ion 318 Chip Kit (Life Technologies) in the Microbiome Core Facility at the University of North Carolina at Chapel Hill.
Bioinformatic analysis of bacterial 16S amplicon data was conducted using the Quantitative Insights Into Microbial Ecology (QIIME) software pipeline17 as previously described.7,18 Sequencing data were demultiplexed and filtered for quality control. Sequences were then aligned and clustered into operational taxonomic units (OTU) using the QIIME implementation of UCLUST.19 ChimeraSlayer was used for chimera removal.20,21 OTUs were assigned to taxonomic categories at a 97% similarity level to approximate species level. After taxonomic assignment of OTUs, sequences were aligned and phylogenetic trees were built with FastTree 2.1.3 (freely available at http://www.microbesonline.org/fasttree).22 Rarefaction analysis was done using a random selection of 3,418 sequences from each sample to ensure an even sampling depth and was used to determine phylogenetic diversity and species richness. Beta diversity estimates were calculated within QIIME using weighted and unweighted UniFrac distances23 between samples at a depth of 3,418 sequences per sample.
Statistical analyses.
Characteristics of children and their diarrhea episodes were summarized by frequencies for categorical variables and sample means for continuous variables. The Wilcoxon signed-rank test was used to assess differences in the relative abundances of bacterial taxa between paired diarrhea and recovery samples. This analysis included only the stool from the first diarrhea episode for children who experienced multiple diarrhea episodes, to allow the longest interval between diarrhea and recovery. Statistical significance was assessed using a false discovery rate (FDR) correction to account for multiple hypothesis tests. Paired t tests were used to compare phylogenetic diversity and species richness between diarrhea and recovery stools. This analysis was stratified by age group, as phylogenetic diversity and species richness are known to increase with age.24,25 As above, we included only the stool from the first diarrhea episode for children who experienced multiple diarrhea episodes. Spearman correlation coefficients were calculated to estimate the correlations between phylogenetic diversity or species richness and age, and between phylogenetic diversity or species richness and time since the last diarrhea episode. Fisher exact test was used to compare changes in phylogenetic diversity and species richness between diarrhea episodes and recovery by enteropathogen category. Fisher exact test was also used to compare changes in phylogenetic diversity and species richness between diarrhea episodes and recovery by category of enteropathogen detected in recovery stools. In these analyses, changes in phylogenetic diversity and species richness were categorized as either increased or decreased. We estimated associations between age, sex, time between diarrhea and recovery stool collection, antibiotic receipt, breast-feeding, and bottle-feeding with phylogenetic diversity and species richness using linear regression models with generalized estimating equations to account for clustering on the level of the individual. These analyses were performed using SAS versions 9.3 and 9.4 (Cary, NC).
Results
Characteristics of children and diarrhea episodes.
The 25 children included in the study had a mean age of 21.9 months, 64% were receiving breast milk, and 4% had a weight-for-age percentile below the 5th percentile. Among the 41 diarrhea episodes experienced by the children, 10% were treated with an antibiotic and 5% resulted in hospitalization (Table 1). Recovery stool samples were collected on average 189 days (standard deviation [SD] = 77 days, range = 62–335 days) after the most recent diarrhea episode.
Table 1.
Characteristics of children and diarrhea episodes
| Characteristics of children (N = 25) | |
|---|---|
| Mean age in months* | 21.9 (SD = 12.4) |
| Gender, % female | 48% (12/25) |
| Receiving breast milk* | 64% (16/25) |
| Weight-for-age percentile†, % > 5th percentile | 96% (24/25) |
| Mother completed any secondary education | 76% (19/25) |
| Poverty index, % with basic needs met | 88% (22/25) |
| Indoor toilet | 84% (21/25) |
| Non-dirt floor | 96% (24/25) |
| Mean diarrhea episodes per year (stools collected) | 2.6 (SD = 0.8) |
| Characteristics of diarrhea episodes (N = 41) | |
| Maximum number of stools per 24 hours, mean (SD) | 3.6 (SD = 1.2) |
| Fever | 27% (11/41) |
| Vomiting | 24% (10/41) |
| Bloody diarrhea | 0% (0/41) |
| Hospitalized | 5% (2/41) |
| Received antibiotic for diarrhea | 10% (4/41) |
| Enteropathogen detected | 61% (25/41) |
| Viral enteropathogen detected | 32% (13/41) |
| Bacterial enteropathogen detected | 15% (6/41) |
| Parasitic enteropathogen detected | 2% (1/41) |
| Mixed category infection detected | 12% (5/41) |
SD = standard deviation.
At the time of first diarrhea episode.
World Health Organization weight-for-age percentile on study entry.
Enteropathogens detected among stool samples.
At least one enteropathogen was detected in 61% of diarrhea stools. The proportions of the 41 diarrhea stools in which viral, bacterial, parasitic, or mixed category enteropathogens were detected were 32%, 15%, 2%, and 12%, respectively (Table 1). Viral enteropathogens identified in diarrhea stool samples included norovirus (n = 13), sapovirus (n = 5), and rotavirus (n = 1). Bacterial enteropathogens identified in diarrheal stool samples included enteropathogenic E. coli (n = 4), enterotoxigenic E. coli (n = 3), enterohemorrhagic E. coli (n = 1), Shigella flexneri (n = 1), and Campylobacter spp. (n = 1). Parasitic enteropathogens identified in diarrheal stool samples included Giardia lamblia (n = 1), Cryptosporidium spp. (n = 1), and Entamoeba histolytica/dispar (n = 3).
Enteropathogens were also detected in 44% of the recovery stools. The proportions of the 25 recovery stools in which viral, bacterial, parasitic, or mixed category enteropathogens were detected were 12%, 20%, 4%, and 8%, respectively. Enteropathogens identified in recovery stool samples included norovirus (n = 3), adenovirus (n = 1), enteropathogenic E. coli (n = 3), enterotoxigenic E. coli (n = 3), enteroaggregative E. coli (n = 1), G. lamblia (n = 1), Cryptosporidium spp. (n = 1) and E. histolytica/dispar (n = 1).
Microbiome characterization of diarrhea and recovery stools.
After filtering for quality and length, a total of 541,186 sequences (7,313 reads ± 4,598 per stool sample) were assigned to 3,211 OTUs at ≥ 97% similarity, clustering into 242 genera, 116 families, 53 orders, 27 classes, and 13 phyla. The microbial composition of both diarrhea and recovery stools was dominated by the phylum Firmicutes, followed by Proteobacteria, Cyanobacteria, Actinobacteria, Bacteroidetes, and Verrucomicrobia (Figure 2 ). Members of the phyla TM7, Fusobacteria, Tenericutes, Chloroflexi, Thermi, Spirochetes, and Synergistetes were detected at levels below 1.0%.
Figure 2.
Relative abundances of phyla and families detected in the first diarrhea stool vs. recovery stool.
In further analysis, we included only the first diarrhea stool for children who experienced multiple diarrhea episodes. At the phylum level, there was an overrepresentation of the phylum Fusobacteria in diarrhea (relative abundance = 0.26%, SD = 0.47%) compared with recovery stools (0.06%, SD = 0.15%) (Wilcoxon signed-rank test, unadjusted P = 0.0036; FDR adjusted P = 0.05). Likewise, the phylum Bacteroidetes was more abundant in diarrhea (4.5%, SD = 4.2%) compared with recovery stools (2.7%, SD = 3.1%) (Wilcoxon signed-rank test, unadjusted P = 0.0316; FDR adjusted P = 0.16). Figure 2 shows the relative abundances of phyla and families identified in the first diarrhea stool versus recovery stool.
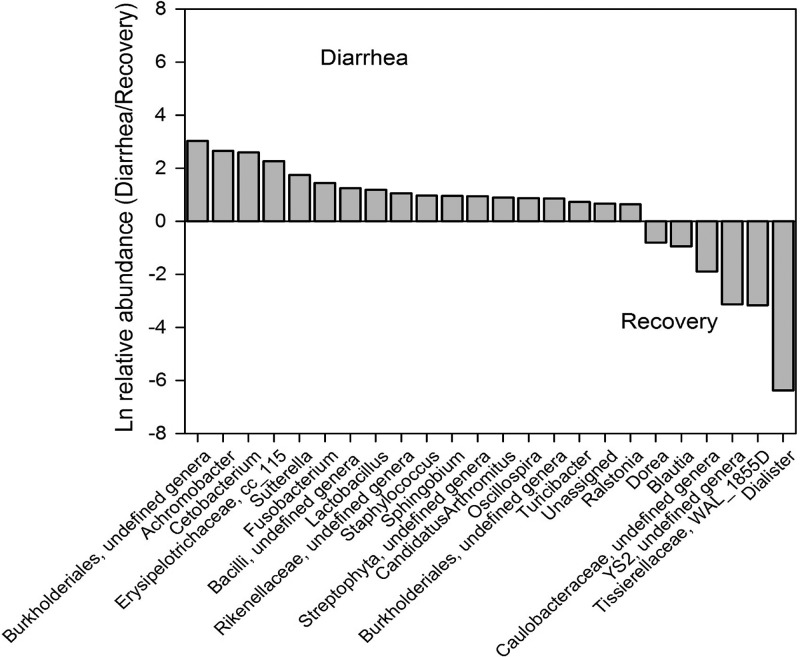
At the genus level, unclassified genera in the family Rickenellaceae within the Bacteroidetes phylum, two genera within the Cyanobacteria phylum, 11 genera within the Firmicutes phylum, two genera within the Fusobacteria phylum, and seven genera within the Proteobacteria phylum were over- or underrepresented in the diarrhea and recovery samples (Wilcoxon signed-rank test, unadjusted P < 0.05) (Figure 3 ). However, only one genus, Cetobacterium, remained statistically significant after controlling for multiple comparisons (FDR adjusted P = 0.0217). Cetobacterium and Fusobacterium in the phylum Fusobacteria were overrepresented in the diarrhea group: 13.5- and 4.2-fold (e2.60 and e1.43), respectively (Figure 3). Achromobacter, a Betaproteobacteria of the Burkholderiales order, and Lactobacillus, a Firmicutes genus that could be considered a marker of a healthy gut microbiota, were 14.3- and 3.3-fold (e2.66 and e1.19) overrepresented in the diarrhea group. Conversely, four out of six genera overrepresented in the recovery group (Dorea, Blautia, Dialister, and Sporobacterium WAL_1855D) were of the order Clostridiales in the phylum Firmicutes.
Figure 3.
Ratio of the natural log of relative abundances of genera that differed* between the first diarrhea stool vs. recovery stool. *P < 0.05 unadjusted Wilcoxon signed-rank test. Using Wilcoxon signed-rank test adjusted by false discovery rate (FDR), only the difference in the relative abundance of Cetobacterium was statistically significant (P = 0.0217).
Phylogenetic diversity and species richness.
For children under 2 years of age, the mean phylogenetic diversity and species richness were lower in recovery stools as compared with diarrhea stools, but the detected differences did not reach statistical significance. Among children aged 2–5 years, mean phylogenetic diversity and species richness values were significantly lower in recovery stools as compared with diarrhea stools (Figure 4 ). Phylogenetic diversity increased with age (Spearman correlation coefficient [ρ] = 0.27, P = 0.0253); similarly, species richness increased with age (ρ = 0.27, P = 0.0273). Neither phylogenetic diversity nor species richness in recovery increased with time since the most recent diarrhea episode (ρ = −0.12, P = 0.3258; ρ = −0.17, P = 0.1662, respectively).
Figure 4.
Phylogenetic diversity and species richness in diarrhea and recovery stools by age group (whiskers indicate the ranges of phylogenetic diversity or species richness values; *P < 0.01 paired t test).
Among the eight children who experienced any diarrhea episode in which a bacterial enteropathogen was detected, none experienced an increase in phylogenetic diversity in recovery; conversely, among the children who experienced diarrhea episode(s) in which no bacterial enteropathogens were detected, 59% (10/17) experienced an increase in phylogenetic diversity in recovery (P = 0.008). We did not find an association between the change in phylogenetic diversity in diarrhea and recovery and the detection in any diarrhea stool of either viral (P = 1.0), parasitic (P = 1.0) or mixed-category enteropathogens (P = 1.0). For species richness, we observed the same relationships as above for phylogenetic diversity. Among the children who experienced any diarrhea episode in which a bacterial enteropathogen was detected, 13% (1/8) experienced an increase in species richness in recovery, as compared with 59% (10/17) in children who did not experience any diarrhea episodes in which a bacterial enteropathogen was detected (P = 0.042). Also, we did not find an association between the change in species richness in diarrhea and recovery and the detection of either viral (P = 1.0), parasitic (P = 0.34), or mixed category enteropathogens (P = 0.18) in any diarrhea stool. Further, in recovery, there was no association between the detection of any category of enteropathogen in recovery stools and change in either phylogenetic diversity (P = 0.66 for bacterial; P = 1.0 for viral; P = 1.0 for parasitic; and P = 0.50 for mixed) or species richness (P = 0.41 for bacterial; P = 0.57 for viral; P = 1.0 for parasitic; and P = 0.49 for mixed).
In linear regression modeling, while increasing age was associated with an increase in phylogenetic diversity and species richness, there were no associations between sex, time between diarrhea and recovery stool collection, antibiotic receipt, and breast-feeding or bottle-feeding and these outcomes (results not shown).
Discussion
A growing body of evidence shows that the gut microbiota, which develops during early life, has functions important to human health. Although prior studies have examined changes in gut microbiota during diarrhea episodes, less is known about changes during recovery. In our study, we did not find an increase in phylogenetic diversity or species richness of the gut microbiome in children after at least 2 months after resolution of symptoms from a diarrhea episode. These findings suggest that diarrhea episodes may be more disruptive to the gut microbiota than previously thought. As has been reported previously,7,24,25 we found that phylogenetic diversity trended upward with age. In this study, we found that children without bacterial enteropathogens detected in their diarrheal stool were more likely to have a higher phylogenetic diversity in recovery. It is possible that the action of bacterial enteropathogens continues undetected after resolution of symptoms from diarrhea episodes, prolonging the time needed for a complete recovery of the gut microbiota. Since diversity was lower in recovery in all cases in which a bacterial enteropathogen was detected, the mechanisms by which these pathogens impact microbial diversity are likely different and may include toxin production and invasion of tissues through different mechanisms.
Further, we found that microbiome composition differed during diarrhea episodes and recovery. Recovery stools had a lesser relative abundance of Lactobacillus, thought to be a marker of a healthy microbiome.26,27 While this finding was unexpected, a previous study of Indian children found that the abundance of Lactobacillus species increased at the end of diarrhea episodes and then decreased after 3 months of recovery.28 Also, as compared with diarrhea stools, recovery stools had a greater relative abundance of genera of the order Clostridiales (Dorea, Blautia, Dialister, and Sporobacterium WAL_1855D). Some of these Clostridiales genera are known to produce the short-chain fatty acid, butyrate, which promotes the growth of the intestinal epithelium. While Clostridium difficile was not found to be more abundant in recovery stools, it is possible that the same gut environment in recovery that promotes the colonization of the other Clostridiales genera may also support colonization by C. difficile. A previous study followed microbiome composition in children treated for cholera until 28 days after the onset of diarrhea and also found an increase in Clostridiales.29 Overall, the decrease in Lactobacillus, a facultative anaerobe, and the increase in anaerobic Clostridiales genera may reflect decreasing oxygen availability in the gut during recovery as compared with a diarrhea episode.
A limitation of this study is that it was performed with 25 children; our findings should be confirmed with a larger study. Also, we acknowledge that 16S sequencing may underestimate bifidobacterial taxa30; our approach could have been strengthened by the addition of primers specific to known members of this genus. Further, we acknowledge that recent evidence shows that detection of bacterial enteropathogens, particularly, Shigella spp., may be increased through the use of molecular diagnostic techniques over conventional bacterial culture.31 Therefore, we may have underdetected bacterial enteropathogens in our samples. Finally, because we did not collect a sample before the first diarrhea episode, we were unable to determine whether the microbiome composition returned to a “pre-diarrhea” state or attained a “new normal” composition.
Overall, this study suggests that diarrhea episodes may have a longer effect on changes in gut microbiota than previously thought, and these effects may be pathogen specific. Future studies may identify the functional implications of these changes and potential interventions to limit the burden of diarrheal disease.
ACKNOWLEDGMENTS
We would like to acknowledge Ley Killeya-Jones for her assistance with figures and editing of the manuscript.
Footnotes
Financial support: Financial support for this study was provided from the University Research Council at the University of North Carolina at Chapel Hill. Microbiome analysis was performed at the University of North Carolina's Microbiome Core Facility (NIH/NIDDK Grant P30 DK34987).
Authors' addresses: Sylvia Becker-Dreps, Department of Family Medicine, UNC Chapel Hill, Chapel Hill, NC, E-mail: sbd@unc.edu. Imane Allali, Laboratory of Biochemistry and Immunology, Mohammed V University, Rabat, Morocco, E-mail: imane_allali@med.unc.edu. Andrea Monteagudo, Microbiome Core Facility, UNC Chapel Hill, Chapel Hill, NC, E-mail: amonm@email.unc.edu. Samuel Vilchez, Department of Microbiology and Parasitology, National Autonomous University of Nicaragua, Leon, Nicaragua, E-mail: samuelvilchez@gmail.com. Michael G. Hudgens, Department of Biostatistics, UNC Gillings School of Global Public Health, Chapel Hill, NC, E-mail: mhudgens@email.unc.edu. Elizabeth T. Rogawski, Department of Epidemiology, UNC Gillings School of Global Public Health, Chapel Hill, NC, E-mail: rogawski@unc.edu. Ian M. Carroll, Department of Medicine, UNC Chapel Hill, Chapel Hill, NC, E-mail: ian_carroll@med.unc.edu. Luis Enrique Zambrana, Center for Demographic and Health Research (CIDS), National Autonomous University of Nicaragua, Leon, Nicaragua, E-mail: lamarcoleta@gmail.com. Felix Espinoza, Department of Microbiology and Parasitology, National Autonomous University of Nicaragua, Leon, Nicaragua, E-mail: espinozafelix08@gmail.com. M. Andrea Azcarate-Peril, Microbiome Core Facility, UNC Chapel Hill, Chapel Hill, NC, E-mail: andrea_azcarate-peril@med.unc.edu.
References
- 1.Buccigrossi V, Nicastro E, Guarino A. Functions of intestinal microflora in children. Curr Opin Gastroenterol. 2013;29:31–38. doi: 10.1097/MOG.0b013e32835a3500. [DOI] [PubMed] [Google Scholar]
- 2.Johnson CL, Versalovic J. The human microbiome and its potential importance to pediatrics. Pediatrics. 2012;129:950–960. doi: 10.1542/peds.2011-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin R, Nauta AJ, Ben Amor K, Knippels LMJ, Knol J, Garssen J. Early life: gut microbiota and immune development in infancy. Benef Microbes. 2010;1:367–382. doi: 10.3920/BM2010.0027. [DOI] [PubMed] [Google Scholar]
- 4.Matamoros S, Gras-Leguen C, Le Vacon F, Potel G, de La Cochetiere M-F. Development of intestinal microbiota in infants and its impact on health. Trends Microbiol. 2013;21:167–173. doi: 10.1016/j.tim.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Fouhy F, Ross RP, Fitzgerald GF, Stanton C, Cotter PD. Composition of the early intestinal microbiota: knowledge, knowledge gaps and the use of high-throughput sequencing to address these gaps. Gut Microbes. 2012;3:203–220. doi: 10.4161/gmic.20169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hopkins MJ, Sharp R, Macfarlane GT. Variation in human intestinal microbiota with age. Dig Liver Dis. 2002;34((Suppl 2)):S12–S18. doi: 10.1016/s1590-8658(02)80157-8. [DOI] [PubMed] [Google Scholar]
- 7.Thompson AL, Monteagudo-Mera A, Cadenas MB, Lampl ML, Azcarate-Peril MA. Milk- and solid-feeding practices and daycare attendance are associated with differences in bacterial diversity, predominant communities, and metabolic and immune function of the infant gut microbiome. Front Cell Infect Microbiol. 2015;5:3. doi: 10.3389/fcimb.2015.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Huërou-Luron I, Blat S, Boudry G. Breast- v. formula-feeding: impacts on the digestive tract and immediate and long-term health effects. Nutr Res Rev. 2010;23:23–36. doi: 10.1017/S0954422410000065. [DOI] [PubMed] [Google Scholar]
- 9.Pop M, Walker AW, Paulson J, Lindsay B, Antonio M, Hossain MA, Oundo J, Tamboura B, Mai V, Astrovskaya I, Corrada Bravo H, Rance R, Stares M, Levine MM, Panchalingam S, Kotloff K, Ikumapayi UN, Ebruke C, Adeyemi M, Ahmed D, Ahmed F, Alam MT, Amin R, Siddiqui S, Ochieng JB, Ouma E, Juma J, Mailu E, Omore R, Morris JG, Breiman RF, Saha D, Parkhill J, Nataro JP, Stine OC. Diarrhea in young children from low-income countries leads to large-scale alterations in intestinal microbiota composition. Genome Biol. 2014;15:R76. doi: 10.1186/gb-2014-15-6-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monira S, Shabnam SA, Alam NH, Endtz HP, Cravioto A, Alam M. 16S rRNA gene-targeted TTGE in determining diversity of gut microbiota during acute diarrhoea and convalescence. J Health Popul Nutr. 2012;30:250–256. doi: 10.3329/jhpn.v30i3.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma C, Wu X, Nawaz M, Li J, Yu P, Moore JE, Xu J. Molecular characterization of fecal microbiota in patients with viral diarrhea. Curr Microbiol. 2011;63:259–266. doi: 10.1007/s00284-011-9972-7. [DOI] [PubMed] [Google Scholar]
- 12.Solano-Aguilar G, Fernandez KP, Ets H, Molokin A, Vinyard B, Urban JF, Gutierrez MF. Characterization of fecal microbiota of children with diarrhea in 2 locations in Colombia. J Pediatr Gastroenterol Nutr. 2013;56:503–511. doi: 10.1097/MPG.0b013e318282aa12. [DOI] [PubMed] [Google Scholar]
- 13.Kosek M, Bern C, Guerrant RL. The global burden of diarrhoeal disease, as estimated from studies published between 1992 and 2000. Bull World Health Organ. 2003;81:197–204. [PMC free article] [PubMed] [Google Scholar]
- 14.Becker-Dreps S, Bucardo F, Vilchez S, Zambrana LE, Weber DJ, Peña R, Hudgens MG, Morgan DR, Svennson L, Nordgren J, Espinoza F, Paniagua M. Etiology of childhood diarrhea following rotavirus vaccine introduction: a prospective, community-based study in Nicaragua. Pediatr Infect Dis J. 2014;33:1156–1163. doi: 10.1097/INF.0000000000000427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vilchez S, Reyes D, Paniagua M, Bucardo F, Möllby R, Weintraub A. Prevalence of diarrhoeagenic Escherichia coli in children from León, Nicaragua. J Med Microbiol. 2009;58:630–637. doi: 10.1099/jmm.0.007369-0. [DOI] [PubMed] [Google Scholar]
- 16.Garcia LS, Bruckner DA, Brewer TC, Shimizu RY. Techniques for the recovery and identification of Cryptosporidium oocysts from stool specimens. J Clin Microbiol. 1983;18:185–190. doi: 10.1128/jcm.18.1.185-190.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devine AA, Gonzalez A, Speck KE, Knight R, Helmrath M, Lund PK, Azcarate-Peril MA. Impact of ileocecal resection and concomitant antibiotics on the microbiome of the murine jejunum and colon. PLoS One. 2013;8:e73140. doi: 10.1371/journal.pone.0073140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 20.Edgar RC, Haas BJ, Clemente JC, Quince C, Knigh R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methe B, DeSantis TZ. Human Microbiome Consortium Petrosino JF, Knight R, Birren BW. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lozupone C, Hamady M, Knight R. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2011;108((Suppl 1)):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murgas Torrazza R, Neu J. The developing intestinal microbiome and its relationship to health and disease in the neonate. J Perinatol. 2011;31:S29–S34. doi: 10.1038/jp.2010.172. [DOI] [PubMed] [Google Scholar]
- 27.Gutierrez-Castrellon P, Lopez-Velazquez G, Diaz-Garcia L, Jimenez-Gutierrez C, Mancilla-Ramirez J, Estevez-Jimenez J, Parra M. Diarrhea in preschool children and Lactobacillus reuteri: a randomized controlled trial. Pediatrics. 2014;133:e904–e909. doi: 10.1542/peds.2013-0652. [DOI] [PubMed] [Google Scholar]
- 28.Balamurugan R, Janardhan HP, George S, Raghava MV, Muliyil J, Ramakrishna BS. Molecular studies of fecal anaerobic commensal bacteria in acute diarrhea in children. J Pediatr Gastroenterol Nutr. 2008;46:514–519. doi: 10.1097/MPG.0b013e31815ce599. [DOI] [PubMed] [Google Scholar]
- 29.Monira S, Nakamura S, Gotoh K, Izutsu K, Watanabe H, Alam NH, Nakaya T, Horii T, Ali SI, Iida T, Alam M. Metagenomic profile of gut microbiota in children during cholera and recovery. Gut Pathog. 2013;5:1. doi: 10.1186/1757-4749-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milani C, Lugli GA, Turroni F, Mancabelli L, Duranti S, Viappiani A, Mangifesta M, Segata N, van Sinderen D, Ventura M. Evaluation of bifidobacterial community composition in the human gut by means of a targeted amplicon sequencing (ITS) protocol. FEMS Microbiol Ecol. 2014;90:493–503. doi: 10.1111/1574-6941.12410. [DOI] [PubMed] [Google Scholar]
- 31.Liu J, Kabir F, Manneh J, Lertsethtakarn P, Begum S, Gratz J, Becker SM, Operario DJ, Taniuchi M, Janaki L, Platts-Mills JA, Haverstick DM, Kabir M, Sobuz SU, Nakjarung K, Sakpaisal P, Silapong S, Bodhidatta L, Qureshi S, Kalam A, Saidi Q, Swai N, Mujaga B, Maro A, Kwambana B, Dione M, Antonio M, Kibiki G, Mason CJ, Haque R, Iqbal N, Zaidi AK, Houpt ER. Development and assessment of molecular diagnostic tests for 15 enteropathogens causing childhood diarrhoea: a multicentre study. Lancet Infect Dis. 2014;14:716–724. doi: 10.1016/S1473-3099(14)70808-4. [DOI] [PubMed] [Google Scholar]