Abstract
This study was performed to evaluate nested polymerase chain reaction (PCR) and real-time PCR methods for detection of Strongyloides stercoralis in fecal samples compared with parasitological methods. A total of 466 stool samples were examined by conventional parasitological methods (formalin ether concentration [FEC] and agar plate culture [APC]). DNA was extracted using an in-house method, and mitochondrial cytochrome c oxidase subunit 1 and 18S ribosomal genes were amplified by nested PCR and real-time PCR, respectively. Among 466 samples, 12.7% and 18.2% were found infected with S. stercoralis by FEC and APC, respectively. DNA of S. stercoralis was detected in 18.9% and 25.1% of samples by real-time PCR and nested PCR, respectively. Considering parasitological methods as the diagnostic gold standard, the sensitivity and specificity of nested PCR were 100% and 91.6%, respectively, and that of real-time PCR were 84.7% and 95.8%, respectively. However, considering sequence analyzes of the selected nested PCR products, the specificity of nested PCR is increased. In general, molecular methods were superior to parasitological methods. They were more sensitive and more reliable in detection of S. stercoralis in comparison with parasitological methods. Between the two molecular methods, the sensitivity of nested PCR was higher than real-time PCR.
Introduction
Strongyloidiasis is estimated to infect 30–100 million people worldwide. This parasitic disease is caused by the intestinal nematode Strongyloides stercoralis.1,2 Most patients with strongyloidiasis are asymptomatic while some have a variety of gastrointestinal, cutaneous, or pulmonary symptoms. Chronic strongyloidiasis may lead to hyperinfection syndrome or life-threatening disseminated infections in immunocompromised patients, such as transplant recipients and those receiving corticosteroid treatment.3,4 As the number of immunocompromised patients is dramatically increasing, severe complicated strongyloidiasis could pose a major problem to such patients.1 Therefore, a precise and reliable diagnosis method is essential for rapid detection of the parasite in at-risk patients to decrease the mortality and morbidity rate of the infection.
Currently, there is no definitive gold standard test for diagnosis of S. stercoralis infection.5 Many parasitological methods have been used for detection of larvae in stool samples, including formalin–ethyl acetate concentration, the Baermann method, the Harada-Mori culture method, and nutrient agar plate culture (APC).2 Parasitological methods are often insensitive, because the parasitic load is usually low and the larval output is minimal.1 To increase the test sensitivity, multiple stool sampling over consecutive days is required.6 Since the introduction of APC as a method for detection of S. stercoralis several studies confirmed its superiority to other parasitological methods.7,8 However, this method is difficult, time consuming (requiring 2–3 days), labor intensive, and also requires freshly collected stool and expert microscopists.2,8
Various serodiagnostic assays including enzyme-linked immunosorbent assay, indirect immunofluorescence assay, and western blot have been reported with variable sensitivity and specificity depending on antigen preparation and immunoglobulin isotopes. Immunodiagnostic methods have higher sensitivity than conventional microscopic methods. They are useful for screening, but have lower specificity because of cross-reactivity with other helminths commonly coinfecting populations in endemic areas.5,9
Among molecular methods, real-time PCR targeting ribosomal RNA (rRNA) genes for specific detection of S. stercoralis DNA in fecal samples has been evaluated in a few studies with variable results.10–15 Accordingly, it appears that the method of DNA extraction is a key factor influencing the results of the evaluated tests. So, to increase the sensitivity of molecular methods, utilization of an appropriate DNA extraction technique is necessary. In addition, comparative efficacy evaluations of different molecular methods will pave the way for identifying the most reliable method.
In this study, two molecular methods (nested PCR and real-time PCR) were evaluated for the diagnosis of S. stercoralis in fecal samples in comparison with two most common parasitological methods (formalin ether concentration [FEC] and APC). This study was performed in an S. stercoralis-endemic area of Iran16 on a large sample size of indigenous people with a wide range of clinical symptoms and parasitic load.
Materials and Methods
Sample collection.
During 2010–2013, a total of 466 fresh stool samples were collected from areas endemic of strongyloidosis in Iran, including Mazandaran, Guilan, Khuzestan, and Hormozgan provinces, and, also, from patients referred to Helminthological Laboratory of School of Public Health, Tehran University of Medical Sciences. Presence of any gastrointestinal, cutaneous, and pulmonary symptoms at the time of sampling was registered.
Parasitological methods.
All fecal samples were examined using FEC and microscopic examination. The samples were also subjected to APC as described previously.7 In brief, 3–4 g of stool was placed on nutrient agar dish. After incubation at 28–30°C for 2–3 days, the dishes were evaluated by light microscopy with low magnification for detection of any larva and adults or their tracks.7,8 For collection of larvae, the surface of the positive agar plate was washed out by lukewarm phosphate buffer saline (PBS) solution.8 Differentiation of S. stercoralis from other probable nematode larvae such as Trichostrongylus spp., hookworm, and also Rhabditis spp. was carried out based on morphological characteristics.17
Stool samples were categorized in three groups according to parasitic load as follows: low infection—FEC-negative and 1–4 larvae counted on agar plate surface; moderate infection—FEC-positive and 5–10 larvae counted on agar plate surface; high infection—FEC-positive and more than 10 larvae counted on agar plate surface.
Molecular methods.
DNA was extracted from all 466 fecal samples and subjected to nested PCR and real-time PCR.
DNA isolation.
Part of each fecal sample was preserved in 70% ethanol at room temperature for DNA isolation. DNA was extracted using an in-house (IH) method described by Repetto and others18 with the following modifications: prior to DNA extraction, stool samples were washed twice with sterile distilled water followed by centrifugation at 2000 × g for 5 minutes to remove the ethanol. Approximately 1 g stool, diluted in 10 mL PBS, was subjected to 5 cycles of freezing (liquid nitrogen) and thawing (in boiling water). About 500 μL PBS-diluted stool was incubated overnight with 500 μL GTES buffer (100 mM glycine, 0.05% sodium dodecyl sulfate [SDS], 100 mM Tris/Cl, and 1 mM ethylenediaminetetraacetic acid [EDTA]) at 37°C,18,19 followed by three cycles of freeze thawing. Samples were mixed with 200 mg of glass beads (0.5 mm in diameter) and shaken vigorously for 5 minutes. The suspension was incubated for 12 hours in nematode lysis buffer (100 mM EDTA, 100 mM NaCl, 100 mM Tris pH 7.5, 0.05% SDS, proteinase K 100 μg/mL) at 37°C.18 Next, the samples were extracted twice with one volume of phenol–chloroform–isoamyl alcohol (25:24:1), and DNA was precipitated with an equal volume of isopropanol and 1 mL of 100% ethanol, respectively. The pellet was washed with 300 μL of 70% ethanol, dried, and eluted in 100 μL of TE buffer (10 mM Tris, 1 mM EDTA pH 8) and stored at −20°C until use.
Nested PCR.
On the basis of alignment of different sequences related to S. stercoralis, deposited in GenBank (accession nos.: AB526297, AB526298, AB526299, AB526300), two sets of primer pairs were designed for nested PCR using DNASIS software (Hitachi, Tokyo, Japan) to enable amplification of a 509-bp target in the first PCR round and a 261-bp target in the second PCR round in the mitochondrial cytochrome c oxidase subunit 1 (cox1) gene. PCR reactions were performed in 20 μL volumes containing 2 × red PCR premix (Ampliqon, Odense, Denmark), 10 pmol of each primer and 4 μL of DNA template for the first amplification round, and 1 μL of 1/40 diluted first-round PCR product for the second amplification round. For the first amplification round, the primers cox F (5′TGG TTT GGG TAC TAG TTG-3′) and cox R (5′-GAT GAG CTC AAA CTA CAC A-3′) were used and for the second amplification round, the primers CNF (5′-TTC TAG TGT TGA TTT GGC T-3′) and CNR (5′-TTA CCA CCA AAA CTA GGA TC-3′) were used. Three negative controls (distilled water) and one positive control (DNA extracted from filariform larvae of S. stercoralis) were included in each round. The cycling conditions comprised an initial denaturation step at 95°C for 6 minutes, 35 cycles of denaturation at 95°C for 45 seconds, annealing at 55°C for 60 seconds, and extension at 72°C for 60 seconds, followed by a final extension at 72°C for 6 minutes. PCR conditions for the second round of amplification consisted of initial denaturation at 95°C for 2 minutes, 25 cycles of 94°C for 15 seconds, 60°C for 30 seconds, and 70°C for 30 minutes, followed by a final extension at 72°C for 6 minutes. A 5-μL aliquot of nested PCR product was electrophoresed on a 1.5% agarose gel and visualized using ultraviolet light after staining with 0.5 μg/mL ethidium bromide.
Sequencing.
To confirm the results of nested PCR, six positive products including two parasitologically positive and four parasitologically negative stool samples were selected randomly and prepared for sequencing. These nested PCR products were purified using a commercial purification kit (Bioneer, Daejeon, South Korea), and sequenced unidirectionally using the forward primer used in the second-round PCR. Sequence results were edited and analyzed by BioEdit version 7.2 (http://www.mbio.ncsu.edu/bioedit/bioedit.html), and consensus sequences were compared with GenBank reference sequences using the BLAST query facility (http://www.ncbi.nlm.nih.gov/).
Real-time PCR.
Real-time PCR was performed using S. stercoralis-specific primers and probe sequences targeting a 101-bp small subunit rDNA region (18S) of S. stercoralis as described previously in a study.10 Forward and reverse primers and probe sequences were as follows: Stro18S-1530F, 5′-GAA TTC CAA GTA AAC GTA AGT CAT TAG C-3′, Stro18S-1630R, 5′-TGC CTC TGG ATA TTG CTC AGT TC-3′, and Stro18S-1586T, FAM-5′-ACA CAC CGG CCG TCG CTG C-3′-TAMRA. PCR reactions were performed using the following reaction mixture: 4 μL of 5 × FIREPol master mix (Solis BioDyne, Tartu, Estonia), 0.2 μM of each S. stercoralis primer, 0.1 μM of probe, 4 μL of DNA template, and 11 μL high pure water in a final volume of 20 μL.
Amplification and detection were performed using an ABI Step One real-time PCR machine (Applied Biosystems, Foster City, CA). The amplification program consisted of a denaturation step at 95°C for 10 minutes, followed by 40 cycles of 15 seconds at 95°C, and a combined annealing/extension step at 60°C for 1 minute. Fluorescence was measured at the end of each extension step.
Analytical sensitivity and specificity of nested PCR and real-time PCR.
To determine the analytical sensitivity of the nested PCR and real-time PCR, genomic DNA was extracted from 10 filariform larvae of S. stercoralis using a DNA extraction kit from Bioneer according to the manufacturer's protocol; five 10-fold serial dilutions were prepared (1 through 10−5). Nested PCR and real-time PCR of these dilutions were carried out in several runs as mentioned above.
PCR specificity was evaluated using extracted DNA from adult Ascaris lumbricoides, Taenia saginata, Enterobius vermicularis, and Rhabditis axei. Moreover, genomic DNA from stool samples positive for Fasciola hepatica, Dicrocoelium denriticum, Hymenolepis nana, Trichostrongylus sp., Entamoeba histolytica, Entamoeba coli, Giardia lamblia, Cryptosporidium sp., Enterocytozoon bieneusi, Blastocystis, and Cystoisospora belli was used for specificity testing, along with DNA from cultured Candida albicans.
Ethical approval.
This study was reviewed and approved by the Ethics Committees of Tehran University of Medical Sciences, Iran.
Data analysis.
Data were analyzed using SPSS software (version 18; SPSS Inc., Chicago, IL) to identify statistically significant differences between observations using the χ2 test and to calculate diagnostic sensitivity and specificity of nested PCR and real-time PCR assays.
Results
Clinical presentation of strongyloidiasis.
Overall, 117 people were diagnosed with S. stercoralis by at least one of the methods. These patients exhibited a variable range of clinical manifestations. Hence, 60 patients were symptomatic, three of whom had hyperinfection/disseminated infection and two of whom had fatal strongyloidiasis; 27 cases exhibited no symptoms; and in 30 cases, symptom status remained unknown because of patients not being able to reliably express the nature of any potential symptoms (because of, e.g., mental retardation).
Sensitivity and specificity.
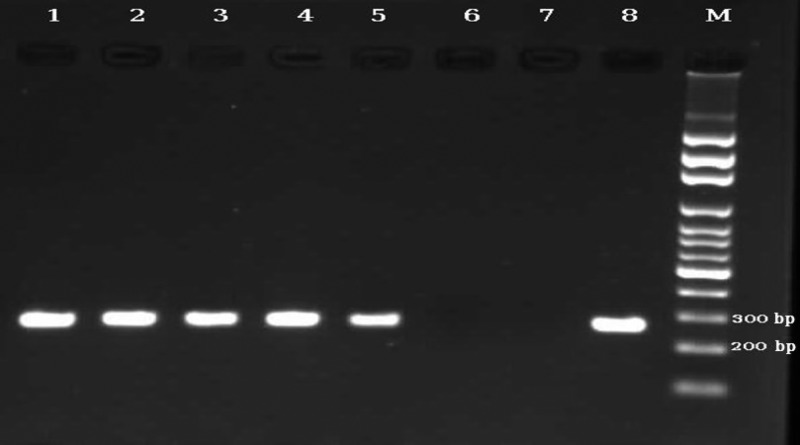
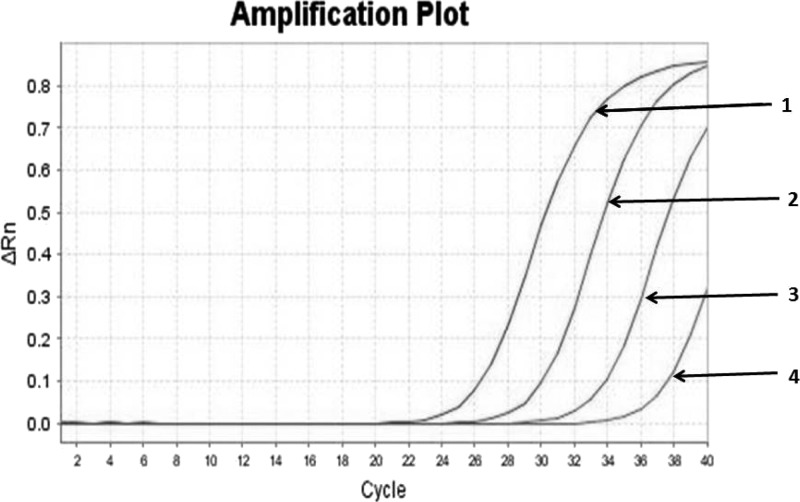
The detection limit for nested PCR and real-time PCR corresponded to 10−4 and 10−3 dilution of DNA of one S. stercoralis filariform larva, respectively (Figures 1 and 2 ). As, except for S. stercoralis, no amplification was detected by nested PCR and real-time PCR from DNAs extracted from all the above mentioned microorganisms, we considered both methods highly specific for S. stercoralis.
Figure 1.
Agarose gel electrophoresis showing the analytical sensitivity of nested polymerase chain reaction (PCR). The PCR products are as follows: lane 1, DNA from one Strongyloides stercoralis filariform larva; lane 2, 10−1 dilution of DNA from one S. stercoralis filariform larva; lane 3, 10−2 dilution of DNA from one S. stercoralis filariform larva; lane 4, 10−3 dilution of DNA from one S. stercoralis filariform larva; lane 5, 10−4 dilution of DNA from one S. stercoralis filariform larva; lane 6, 10−5 dilution of DNA from one S. stercoralis filariform larva; lane 7, negative control; and lane 8, positive control (S. stercoralis). M = 100-bp DNA marker.
Figure 2.
Amplification curves of analytical sensitivity of real-time polymerase chain reaction (PCR). Curve 1: DNA from one Strongyloides stercoralis filariform larva; curve 2: 10−1 dilution of DNA from one S. stercoralis filariform larva; curve 3: 10−2 dilution of DNA from one S. stercoralis filariform larva; curve 4: 10−3 dilution of DNA from one S. stercoralis filariform larva.
Comparison of parasitological methods with nested PCR and real-time PCR.
Among the 466 individuals examined, 85 people (18.2%) were found infected with S. stercoralis by combined parasitological methods (Table 1). The detection rate for APC and FEC alone was 85 (18.2%) and 59 (12.7%), respectively. APC detected all cases identified as positive by FEC in addition to 26 more cases. Therefore, APC was able to detect 1.44 times more positive cases than FEC.
Table 1.
Comparison of the results obtained by parasitological and molecular methods for detection of Strongyloides stercoralis in 466 stool samples
| Methods | FEC | APC | Combination of FEC and APC | ||||
|---|---|---|---|---|---|---|---|
| Positive (59) | Negative (407) | Positive (85) | Negative (381) | Positive (85) | Negative (381) | ||
| Nested PCR | Positive (117) | 59 | 58 | 85 | 32 | 85 | 32 |
| Negative (349) | 0 | 349 | 0 | 349 | 0 | 349 | |
| Real-time PCR | Positive (88) | 57 | 31 | 72 | 16 | 72 | 16 |
| Negative (378) | 2 | 376 | 13 | 365 | 13 | 365 | |
APC = agar plate culture; FEC = formalin ether concentration; PCR = polymerase chain reaction.
Among 85 parasitologically positive cases, low, moderate, and high infection rates were found in 26 (30.6%), 22 (25.9%), and 37 (43.5%) cases, respectively.
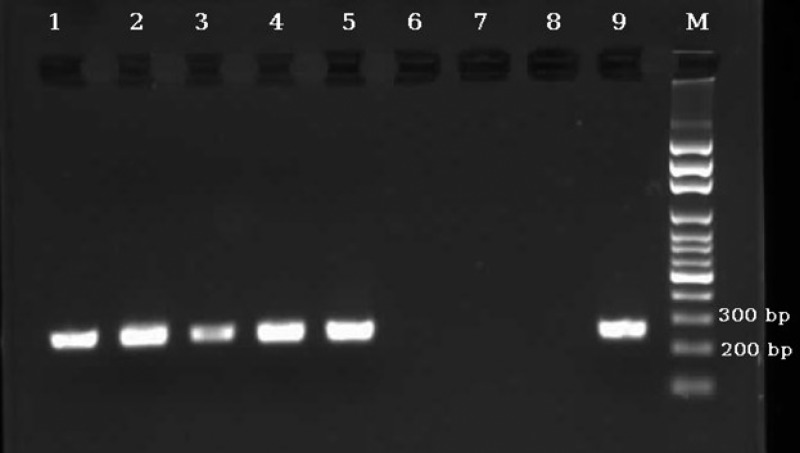
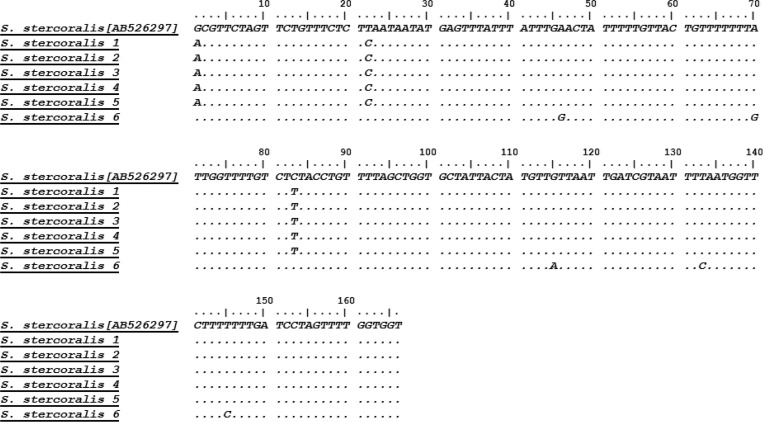
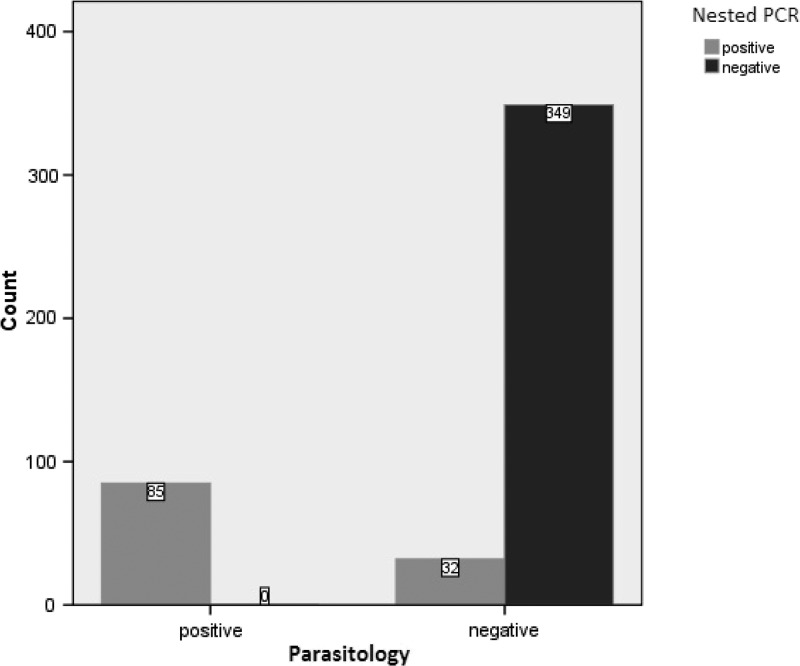
Using nested PCR, 117/466 samples (25.1%) were found positive for S. stercoralis, including all 85 samples positive by APC and 32 more samples, which were negative both by parasitological methods (Table 1). Therefore, there were no parasitologically positive cases that the nested PCR failed to detect. No amplification was detected in negative controls (Figure 3 ). The sequencing results of six randomly selected nested PCR products showed that all of them were S. stercoralis, according to the alignment with sequences submitted in GenBank (Figure 4 ). Since, there is no sensitive gold standard for the laboratory detection of S. stercoralis, the combined results of FEC and APC were considered as diagnostic gold standard in this study. Therefore, diagnostic sensitivity and specificity of nested PCR test were calculated as 100% and 91.6%, respectively (Figure 5 ).
Figure 3.
Agarose-gel electrophoresis of nested polymerase chain reaction (PCR) products amplified with genomic DNA from stool samples. Lanes 1–5: nested PCR products of 5 stool samples positive for Strongyloides stercoralis; lanes 6–8: negative controls; and lane 9: positive control (S. stercoralis). M = 100-bp DNA marker.
Figure 4.
Nucleotide alignment of sequences generated from six nested polymerase chain reaction (PCR) products: the samples corresponding to Strongyloides stercoralis 1, 2, 3, and 4 were negative by parasitological methods and those corresponding to S. stercoralis 5 and 6 were positive by parasitological methods.
Figure 5.
Comparison of results obtained by parasitological methods and nested polymerase chain reaction (PCR) for the detection of Strongyloides stercoralis in human fecal samples.
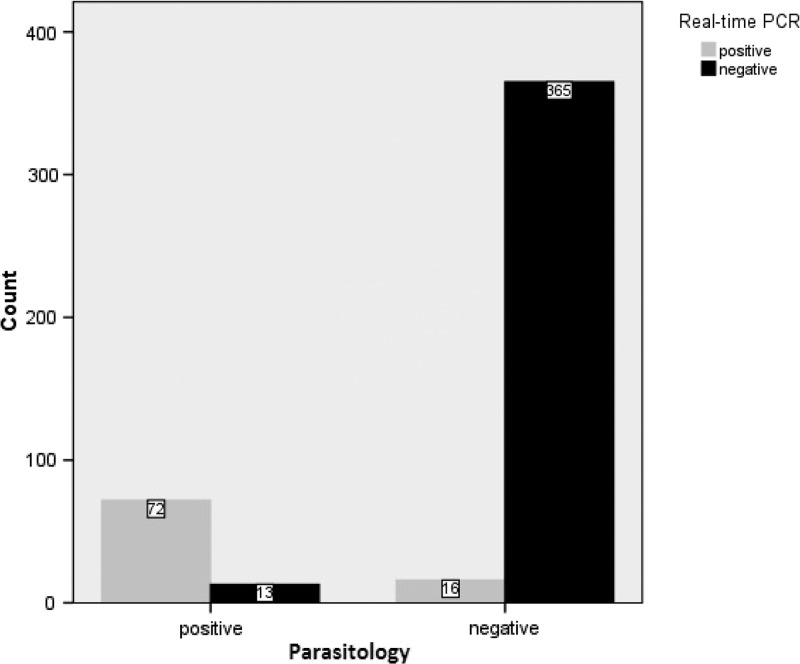
Real-time PCR assay detected S. stercoralis DNA in 88/466 samples (18.9%) (Table 1), all of which tested positive by nested PCR. However, real-time PCR failed to detect 13 parasitologically positive cases (Table 1), including 11 cases of low infection and two cases of moderate infection (data not shown). Considering the combination of FEC and APC as the diagnostic gold standard (Table 1), the sensitivity and specificity of real-time PCR were 84.7% and 95.8%, respectively (Figure 6 ). Strongyloides stercoralis threshold cycles (Ct values) in positive samples ranged between 19.55 and 37.60 cycles, with a median Ct value of 31.13 cycles. Among the 88 samples detected positive by real-time PCR, 72 samples were positive by parasitological methods. The Ct values of these samples ranged between 19.55 and 27.59 (median = 30.65). The Ct value of the 16 parasitologically negative but real-time PCR-positive stool samples ranged between 26.19 and 37.10 (median = 35.06).
Figure 6.
Comparison of results obtained by parasitological methods and real-time polymerase chain reaction (PCR) for the detection of Strongyloides stercoralis in human fecal samples.
With respect to the Ct values of real-time PCR-positive samples and the intensity of infections, all cases of high infection, 95.5% cases of moderate infection, and 59.9% cases of low infection samples could be detected by real-time PCR. The ranges in Ct values for low, moderate, and high infections were 28.16–37.59 (median = 35.29), 26.73–37.48 (median = 31.87), and 19.55–35.87 (median = 27.76), respectively. Statistically, there was a significant association between Ct value and parasitic load (P < 0.001).
Discussion
Strongyloidiasis is considered an important cause of morbidity and mortality in immunocompromised patients.20 Therefore, accurate diagnosis and treatment of S. stercoralis infection is necessary. Conventional parasitological methods are not sufficiently sensitive to detect strongyloidiasis, and repeated examinations of stools over a number of consecutive days are necessary.2 The agar plate culture method has been recognized as being more sensitive than other parasitological methods in the diagnosis of S. stercoralis.2 Similar to previous studies,8,21 the results of this study confirmed that the sensitivity of APC was higher than FEC, detecting 1.44 times more positive cases. Nevertheless, its efficacy was not 100%, therefore, implying the need for new methods with higher sensitivity. In addition, APC is time consuming, labor intensive, and requires multiple fresh stool samples and expert microscopists.8,22
PCR-based methods have shown variable sensitivity for detection of S. stercoralis in fecal samples.10–15,18 For detection of intestinal nematodes in stool samples by PCR-based techniques, optimization of DNA isolation method is essential, because of existence of large amounts of PCR-inhibitory substances in fecal samples such as bacterial proteases, nucleases, cell debris, bile acids, and so forth, and also the presence of a thick and complex cuticle that covers the worms.18,23,24 Repetto and others introduced an improved IH DNA extraction method based on efficient lysis of larvae and removal of inhibitors in the fecal samples.18 In this method, 1 g of stool is used for extraction, whereas routine commercial DNA extraction kits use only 200–300 mg stool. The DNA extraction process is a critical step affecting the efficacy of molecular methods. In this study, the modified IH method was efficiently applied for S. stercoralis DNA extraction from stool samples. In fact, utilizing 1 g stool combined with GTES buffer plus mechanical disruption enables successful DNA extraction, even in cases of low infection, in which only few larvae were present in the stool samples.
The cox1 gene was used as a conserved target for the S. stercoralis-specific nested PCR. Nested PCR not only identified all parasitologically positive samples (85/466 samples) as positive, but also 32 additional samples that could not be detected by parasitological methods. Considering parasitological methods as the gold standard, diagnostic sensitivity and specificity of nested PCR were 100% and 91.6%, respectively. However, the selected nested PCR products, including four parasitologically negative samples, were confirmed as S. stercoralis by sequence analysis. Therefore, deeming these 32 parasitologically negative but nested PCR-positive samples as true positive, the specificity of nested PCR assay is increased. In fact, nested PCR was the only method that did not miss any cases found positive by either of other three methods.
The target for the real-time PCR assay in this study, similar and in concordance with other studies,10–12,14,15 was the 18S ribosomal DNA region. This assay detected S. stercoralis DNA in 88 stool samples with a sensitivity of 84.7% and specificity of 95.8%. Different sensitivity and specificity have been reported in other studies according to sample size, DNA extraction methods, and parasitic load of stool samples. In a study performed on 115 stool samples, the sensitivity and specificity of real-time were reported as 100% and 94%, respectively.12 In a study in Cambodia, real-time PCR was evaluated for detection of S. stercoralis and hookworm in randomly collected stool samples from 218 asymptomatic school children, of whom 38 (17.4%) were positive for S. stercoralis. In that study, the sensitivity and specificity of S. stercoralis real-time PCR were 88.9% and 92.7%, respectively, compared with the combination of Baermann/Koga agar as gold standard.15 Knopp and others13 reported that the sensitivity of the Baermann method for detection of S. stercoralis was significantly higher than that of real-time PCR (47.1% versus 17.4%; P < 0.001), while the specificity of the Baermann method was lower than that of real-time PCR (78.4% versus 93.9%).
With respect to the two molecular methods compared in this study, the sensitivity of nested PCR was higher than that of real-time PCR, which is probably due to higher mitochondrial DNA copy numbers than rDNA (the target genes of nested PCR and real-time PCR, respectively) in nematodes.25,26 Studies on other microorganisms including Toxoplasma gondii and Histomonas meleagridis have also reported higher sensitivity of nested PCR compared with real-time PCR.27,28
In this study, 72/85 stool samples positive by parasitological methods were also identified as positive by real-time PCR. In a study by Verweij and others,10 S. stercoralis was detected in 54/212 stool samples by parasitological methods (Baermann and coproculture), but only 33 samples of them were positive using real-time PCR. In the study by Sultana and others,11 real-time PCR was able to detect 15/41 (36.6%) positive samples diagnosed with the Harada-Mori culture method. In another study,14 a pentaplex real-time PCR was evaluated for simultaneous detection of Ancylostoma duodenale, Necator americanus, A. lumbricoides, and S. stercoralis. In that study, 30/77 samples were positive for S. stercoralis by PCR, whereas only two samples were positive by microscopic examination. In two other studies, the sensitivity of real-time PCR compared with conventional methods was 61.0% (25/41)15 and 100% (18/18),12 respectively.
In this study, real-time PCR detected S. stercoralis DNA in 16/381 samples, which were not scored as positive by either APC or FEC methods. Similarly, in the studies by Rayan and others12 and Verweij and others,10 real-time PCR was positive in 5/97 and 12/158 samples, respectively, which were negative by parasitological methods. Failure of the parasitological methods to detect S. stercoralis in samples testing positive by real-time PCR may in part be due to the presence of nonviable larvae in those samples, and the necessity of the presence of living larvae in the APC method. On the other hand, the failure of real-time PCR to detect 13 cases detected by parasitological methods may be due to the low parasitic load in the samples (11/13 cases had low infection), an effect of fecal inhibitors and competing template in genomic DNA.
In this study, those samples that were positive by real-time PCR but negative by parasitological methods (16/466) had higher Ct values (median Ct value = 35.06), than those samples that were positive by both real-time PCR and parasitological methods (72/466) (median Ct value = 30.65). Similar results were reported by Rayan and others,12 indicating that the Ct values for samples positive by real-time PCR only were higher than those for samples positive by both parasitological methods and real-time PCR (Ct value = 36.3 versus 28.29). In low worm burden stool samples, detection of positive cases by parasitological methods is difficult, but in case of detection of DNA of such samples by real-time PCR, their Ct values would be higher than those of high larvae burden stool samples.
In this study, 100% of the cases with high infection, 95.5% of the cases with moderate infection, and 59.9% of the cases with low infection were scored positive by real-time PCR. Low larval infection samples showed higher Ct values, whereas high and moderate parasitic infection samples showed lower Ct values. There was statistically significant association between Ct values and parasitic load (P < 0.001). Similarly, Sultana and others11 reported that all culture-positive samples with high and moderate infections showed lower Ct values than low larval load infections. In accordance with the results of previous studies,11,12 Ct values are related to the amount of parasite-specific DNA in the samples, and therefore, correlates with the intensity of infections.
Conclusions
This is the first study simultaneously evaluating real-time PCR and nested PCR in comparison with parasitological methods for detection of S. stercoralis in fecal samples. The study was performed on a large sample size of an endemic indigenous population. In general, molecular methods were superior to parasitological methods. Of the two parasitological methods evaluated, APC was 1.44 times more efficient than FEC. Further comparative evaluations, applying more samples and samples of different populations, are necessary to develop and identify a reliable and sensitive diagnostic method for diagnosis of strongyloidiasis. In addition, efforts toward shortening the duration of S. stercoralis DNA extraction from stool samples using IH methods are warranted.
ACKNOWLEDGMENTS
We would like to thank all people who had contributed to the study, especially, Dr. S. Repetto from Departamento de Microbiología, Parasitología e Inmunología, Facultad de Medicina, Universidad de Buenos Aires for valuable advise, B. Kamranrashani, N. Jalalizand, and Z. Heidari from the Department of Medical Parasitology and Mycology, School of Public Health, Tehran University of Medical Sciences, Tehran and B. Rahmati from the Department of Medical Microbiology, School of Medicine, Giulan University of Medical Sciences for their kind assistance. The American Society of Tropical Medicine and Hygiene (ASTMH) assisted with publication expenses.
Disclaimer: The study was part of a PhD Thesis developed by the first author (M. Sharifdini) in the Department of Medical Parasitology and Mycology, School of Public Health, Tehran University of Medical Sciences.
Footnotes
Financial support: This study was supported through grant no. 91-01-160-17294 afforded by the Center for Research of Endemic Parasites of Iran (CREPI), Tehran University of Medical Sciences, Tehran, Iran.
Authors' addresses: Meysam Sharifdini, Mehdi Mohebali, and Eshrat Beigom Kia, Center for Research of Endemic Parasites of Iran (CREPI), Tehran University of Medical Sciences, Tehran, Iran, and Department of Medical Parasitology and Mycology, School of Public Health, Tehran University of Medical Sciences, Tehran, Iran, E-mails: sharifdini@razi.tums.ac.ir, mohebali@tums.ac.ir, and keiaeshr@tums.ac.ir. Hossein Mirhendi, Department of Medical Parasitology and Mycology, School of Medicine, Isfahan University of Medical Sciences, Isfahan, Iran, E-mail: mirhendi@tums.ac.ir. Keyhan Ashrafi, Department of Medical Microbiology, School of Medicine, Guilan University of Medical Sciences, Rasht, Iran, E-mail: k_ashrafi@gums.ac.ir. Mostafa Hosseini, Department of Biostatistics and Epidemiology, School of Public Health, Tehran University of Medical Sciences, Tehran, Iran, E-mail: hoseinim@tums.ac.ir. Hossein Khodadadi, Department of Medical Parasitology and Mycology, School of Medicine, Shiraz University of Medical Sciences, Shiraz, Iran, E-mail: h_khodadadi@sums.ac.ir.
References
- 1.Keiser PB, Nutman TB. Strongyloides stercoralis in the immunocompromised population. Clin Microbiol Rev. 2004;17:208–217. doi: 10.1128/CMR.17.1.208-217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siddiqui AA, Berk SL. Diagnosis of Strongyloides stercoralis infection. Clin Infect Dis. 2001;33:1040–1047. doi: 10.1086/322707. [DOI] [PubMed] [Google Scholar]
- 3.Segarra-Newnham M. Manifestations, diagnosis, and treatment of Strongyloides stercoralis infection. Ann Pharmacother. 2007;41:1992–2001. doi: 10.1345/aph.1K302. [DOI] [PubMed] [Google Scholar]
- 4.Grove DI. Human strongyloidiasis. Adv Parasitol. 1996;38:251–309. doi: 10.1016/s0065-308x(08)60036-6. [DOI] [PubMed] [Google Scholar]
- 5.Montes M, Sawhney C, Barros N. Strongyloides stercoralis: there but not seen. Curr Opin Infect Dis. 2010;23:500–504. doi: 10.1097/QCO.0b013e32833df718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marcos LA, Terashima A, Dupont HL, Gotuzzo E. Strongyloides hyperinfection syndrome: an emerging global infectious disease. Trans R Soc Trop Med Hyg. 2008;102:314–318. doi: 10.1016/j.trstmh.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 7.Arakaki T, Iwanaga M, Kinjo F, Saito A, Asato R, Ikeshiro T. Efficacy of agar-plate culture in detection of Strongyloides stercoralis infection. J Parasitol. 1990;76:425–428. [PubMed] [Google Scholar]
- 8.Kia EB, Mahmoudi M, Zahabiun F, Meamar AR. An evaluation on the efficacy of agar plate culture for detection of Strongyloides stercoralis. Iran J Parasitol. 2007;2:29–34. [Google Scholar]
- 9.Sithithaworn P, Srisawangwong T, Tesana S, Daenseekaew W, Sithithaworn J, Fujimaki Y, Ando K. Epidemiology of Strongyloides stercoralis in north-east Thailand: application of the agar plate culture technique compared with the enzyme-linked immunosorbent assay. Trans R Soc Trop Med Hyg. 2003;97:398–402. doi: 10.1016/s0035-9203(03)90069-1. [DOI] [PubMed] [Google Scholar]
- 10.Verweij JJ, Canales M, Polman K, Ziem J, Brienen EA, Polderman AM, van Lieshout L. Molecular diagnosis of Strongyloides stercoralis in faecal samples using real-time PCR. Trans R Soc Trop Med Hyg. 2009;103:342–346. doi: 10.1016/j.trstmh.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Sultana Y, Jeoffreys N, Watts MR, Gilbert GL, Lee R. Real-time polymerase chain reaction for detection of Strongyloides stercoralis in stool. Am J Trop Med Hyg. 2013;88:1048–1051. doi: 10.4269/ajtmh.12-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rayan HZ, Soliman RH, Galal NM. Detection of Strongyloides stercoralis in fecal samples using conventional parasitological techniques and real-time PCR: a comparative study. Parasitol United J. 2012;5:27–34. [Google Scholar]
- 13.Knopp S, Salim N, Schindler T, Karagiannis Voules DA, Rothen J, Lweno O, Mohammed AS, Singo R, Benninghoff M, Nsojo AA, Genton B, Daubenberger C. Diagnostic accuracy of Kato-Katz, FLOTAC, Baermann, and PCR methods for the detection of light-intensity hookworm and Strongyloides stercoralis infections in Tanzania. Am J Trop Med Hyg. 2014;90:535–545. doi: 10.4269/ajtmh.13-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Basuni M, Muhi J, Othman N, Verweij JJ, Ahmad M, Miswan N, Rahumatullah A, Aziz FA, Zainudin NS, Noordin R. A pentaplex real-time polymerase chain reaction assay for detection of four species of soil-transmitted helminths. Am J Trop Med Hyg. 2011;84:338–343. doi: 10.4269/ajtmh.2011.10-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schar F, Odermatt P, Khieu V, Panning M, Duong S, Muth S, Marti H, Kramme S. Evaluation of real-time PCR for Strongyloides stercoralis and hookworm as diagnostic tool in asymptomatic schoolchildren in Cambodia. Acta Trop. 2013;126:89–92. doi: 10.1016/j.actatropica.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 16.Sharifdini M, Kia EB, Ashrafi K, Hosseini M, Mirhendi H, Mohebali M, Kamranrashani B. An analysis of clinical characteristics of Strongyloides stercoralis in 70 indigenous patients in Iran. Iran J Parasitol. 2014;9:155–162. [PMC free article] [PubMed] [Google Scholar]
- 17.Inatomi S, Kamo H, Otsuru M, Suzuki T, Yoshida Y. Ova and larvae of the common helminthes of man. In: Yamaguchi T, editor. A Colour Atlas of Clinical Parasitology. Tokyo, Japan: Wolf Medical Publication; 1981. [Google Scholar]
- 18.Repetto SA, Alba Soto CD, Cazorla SI, Tayeldin ML, Cuello S, Lasala MB, Tekiel VS, Gonzalez Cappa SM. An improved DNA isolation technique for PCR detection of Strongyloides stercoralis in stool samples. Acta Trop. 2013;126:110–114. doi: 10.1016/j.actatropica.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 19.Dorris M, Viney ME, Blaxter ML. Molecular phylogenetic analysis of the genus Strongyloides and related nematodes. Int J Parasitol. 2002;32:1507–1517. doi: 10.1016/s0020-7519(02)00156-x. [DOI] [PubMed] [Google Scholar]
- 20.Boulware DR, Stauffer WM, Hendel-Paterson BR, Rocha JL, Seet RC, Summer AP, Nield LS, Supparatpinyo K, Chaiwarith R, Walker PF. Maltreatment of Strongyloides infection: case series and worldwide physicians-in-training survey. Am J Med. 2007;120(545):e1–8. doi: 10.1016/j.amjmed.2006.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Kaminsky RG. Evaluation of three methods for laboratory diagnosis of Strongyloides stercoralis infection. J Parasitol. 1993;79:277–280. [PubMed] [Google Scholar]
- 22.Anamnart W, Pattanawongsa A, Intapan PM, Maleewong W. Factors affecting recovery of Strongyloides stercoralis larvae: an approach to a newly modified formalin-ether concentration technique for diagnosis of strongyloidiasis. J Clin Microbiol. 2010;48:97–100. doi: 10.1128/JCM.01613-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abu Al-Soud W, Radstrom P. Effects of amplification facilitators on diagnostic PCR in the presence of blood, feces, and meat. J Clin Microbiol. 2000;38:4463–4470. doi: 10.1128/jcm.38.12.4463-4470.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson IG. Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bik HM, Fournier D, Sung W, Bergeron RD, Thomas WK. Intra-genomic variation in the ribosomal repeats of nematodes. PLoS One. 2013;8:e78230. doi: 10.1371/journal.pone.0078230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ventura N, Rea SL. Caenorhabditis elegans mitochondrial mutants as an investigative tool to study human neurodegenerative diseases associated with mitochondrial dysfunction. Biotechnol J. 2007;2:584–595. doi: 10.1002/biot.200600248. [DOI] [PubMed] [Google Scholar]
- 27.Hierl T, Reischl U, Lang P, Hebart H, Stark M, Kyme P, Autenrieth IB. Preliminary evaluation of one conventional nested and two real-time PCR assays for the detection of Toxoplasma gondii in immunocompromised patients. J Med Microbiol. 2004;53:629–632. doi: 10.1099/jmm.0.45566-0. [DOI] [PubMed] [Google Scholar]
- 28.Hafez HM, Hauck R, Luschow D, McDougald L. Comparison of the specificity and sensitivity of PCR, nested PCR, and real-time PCR for the diagnosis of histomoniasis. Avian Dis. 2005;49:366–370. doi: 10.1637/7341-020805R.1. [DOI] [PubMed] [Google Scholar]