Abstract
During a chikungunya fever outbreak in late 2014 in Chiapas, Mexico, entomovirological surveillance was performed to incriminate the vector(s). In neighborhoods, 75 households with suspected cases were sampled for mosquitoes, of which 80% (60) harbored Aedes aegypti and 2.7% (2) Aedes albopictus. A total of 1,170 Ae. aegypti and three Ae. albopictus was collected and 81 pools were generated. Although none of the Ae. albopictus pools were chikungunya virus (CHIKV)–positive, 18 Ae. aegypti pools (22.8%) contained CHIKV, yielding an infection rate of 32.3/1,000 mosquitoes. A lack of herd immunity in conjunction with high mosquito populations, poor vector control services in this region, and targeted collections in locations of human cases may explain the high infection rate in this vector. Consistent with predictions from experimental studies, Ae. aegypti appears to be the principal vector of CHIKV in southern Mexico, while the role of Ae. albopictus remains unknown.
Introduction
Chikungunya fever (CHIKF), an emerging arthropod-borne disease primarily transmitted by Aedes aegypti and Aedes albopictus mosquitoes, is currently threatening the American continent. The etiologic agent, chikungunya virus (CHIKV), is an enveloped positive-sense, nonsegmented, single-stranded RNA virus in the Togaviridae family, genus Alphavirus.1 CHIKF is a febrile disease accompanied by disabling polyarthralgia (foot, hands, wrists, ankles, and knees) and a generalized rash.2 Before 2013, the Americas had only registered imported cases, most of them in the United States. Then, in October 2013, CHIKV emerged in the Caribbean island of Saint Martin.3,4 Since this introduction, CHIKV has spread throughout the Caribbean, Central America, and parts of South America and has caused an estimated 1.6 million CHIKF cases.5 Although the urban vectors Ae. aegypti and Ae. albopictus have a wide distribution in the Americas and New World populations of both species are CHIKV transmission competent,6 no direct evidence of the role of either during the current epidemic has been presented. Identification of the mosquito species involved in transmission is critical to focus vector control efforts. We therefore performed entomovirological surveillance to incriminate the primary vector(s) of CHIKV transmission during an ongoing outbreak in Chiapas, Mexico.7
Methodology
In October 2014, before official announcement of first autochthonous Mexican CHIKF case on November 7, 2014,7 an unusual febrile outbreak with arthralgia was reported by a local physician in Ciudad Hidalgo, Chiapas State, and then confirmed as CHIKF months later.8 This small town is located near the Mexico–Guatemala border (14°40′45″ N 92°08′59″ W), with a population of 12,678 inhabitants. This region is characterized by a tropical climate with its rainy season occurring during the summer.9 From October 1 to 15, 2014, mosquito collections were carried out in 75 houses, both indoors and outdoors, using a CDC backpack aspirator. The houses were selected from San Caralampio, San Juan, 26 de Julio, and 15 de Enero neighborhoods, where a local physician reported possible cases of CHIKF (acute onset of fever > 38.5°C and severe arthralgia/arthritis not explained by other medical condition) (Figure 1 ).10 Specifically in San Caralampio, a small neighborhood of two blocks, 44 of 68 houses were sampled. After obtaining permission, a health technician aspirated dark and humid places in each house. Mosquitoes were placed into a cooler with icepacks and transported to the Centro Regional de Investigación en Salud Pública, Tapachula, Chiapas.
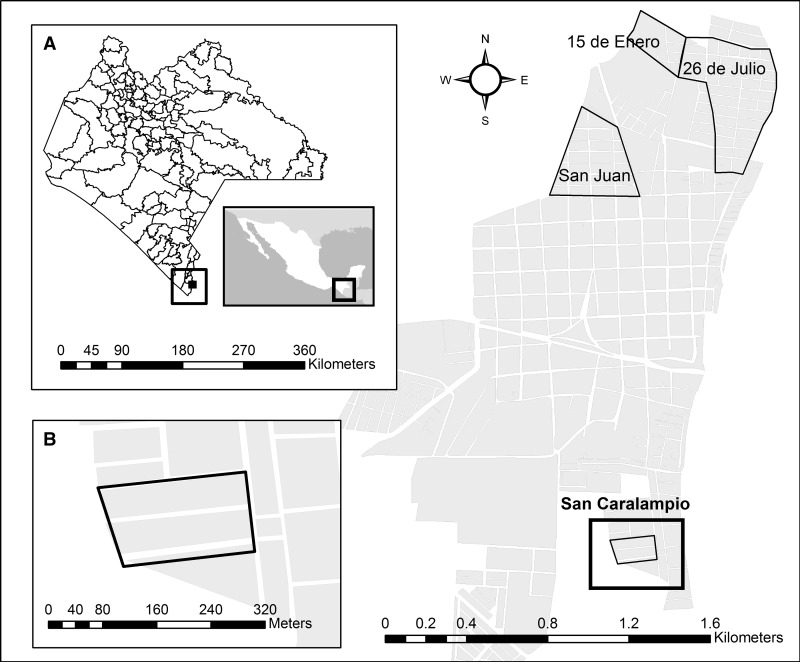
Figure 1.
Study area map from Ciudad Hidalgo, Chiapas, Mexico. Selected neighborhoods for mosquito sampling are framed by a black line. (A) Approximation to Chiapas State. (B) San Caralampio, a small neighborhood of two blocks, where most of the houses were sampled (44 of 68).
Mosquitoes were separated by sex and species on a Chill-Table® (Bioquip, Rancho Dominguez, CA) and later pooled with 1–20 mosquitoes per pool, and 1–4 pools per house. Mosquito pools were stored at −75°C, then homogenized in cell culture medium supplemented with 10% fetal bovine serum, gentamicin, and fungizone, and centrifuged to remove mosquito particulates. This supernatant was stored at −80°C.
Viral RNA extraction from each mosquito pool homogenate was performed using the ZR-96 Viral RNA Kit (Zymo Research, Orange, CA) according to the manufacturer's protocol. To determine the presence of CHIKV RNA, quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed in duplicate as previously described11 using the TaqMan® RNA-to-Ct™ 1-Step Kit (Applied Biosystems, San Francisco, CA) according to the manufacturer's protocol using 1 μL of extracted RNA. CHIKV strain Y011213 RNA (a 2014 isolate from the Caribbean) was used as a positive control. West Nile virus RNA isolated from human serum and a non-template control were used as negative controls.
Cytopathic effect (CPE) assays were used to detect infectious virus from qRT-PCR-positive mosquito pools. Confluent Vero cells were inoculated with 100 μL of the supernatant from each mosquito pool homogenate. Plates were incubated for 4 days at 37°C with 5% CO2 injection in Dulbecco's modified minimal essential medium (Gibco, Grand Island, NY) supplemented with gentamicin and 2% fetal bovine serum. At the completion of the incubation period, cells were fixed with 10% formaldehyde and stained with crystal violet to visualize CPE. CHIKV vaccine strain 181/25 was used as positive control and two CHIKV-negative mosquito pools were used as negative controls.
Viral RNA extracted from each mosquito pool was used to generate complementary DNA using the SuperScript® III First-Strand Synthesis Kit (Invitrogen, Waltham, MA) according to the manufacturer's protocol. The Phusion® High-Fidelity PCR Kit (New England Biolabs, Ipswich, MA) was then used to generate three overlapping PCR amplicons from E1/E2 regions, which were then purified using the QIAquick® PCR Purification Kit (Qiagen, Valencia, CA). Sequences were generated using a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) and sequenced using an ABI Prism model 3100 genetic analyzer (Applied Biosystems). Sequencher v5.0.1 (Gene Codes, Ann Arbor, MI) was used to edit and assemble the sequences. Primer sequences are available from the authors on request.
These E1/E2 partial sequences were compared with one partial sequence from a recent infection in Panama (courtesy of Jean Paul Carrera) and 27 representative CHIKV samples containing the complete open reading frames of the genome. In addition, Mexican CHIKV sequences CH0008, CH0045, CH0072, TA0031, and LI0006 (Genbank accession nos.: KT327163, KT327164, KT327165, KT327166, and KT3271678) were included. The first two Mexican sequences were obtained from two CHIKV-positive cases from San Juan and San Caralampio, respectively, in the first 2 weeks of October 2014.8 Sequence alignments were performed using MUSCLE (Drive, Mill Valley, CA).12 A maximum likelihood tree was reconstructed via PAUP* v4.0b package (Sinauer Associates Inc., Sunderland, MA)13 based on the best-fit nucleotide substitution model estimated via MODELTEST v3.7 (Brigham Young University, Provo, UT).14
For statistical analysis, pooled rate of mosquito infection by CHIKV was calculated using maximum likelihood estimation (MLE) methods for unequal pool sizes. We used PooledInfRate 4.0 software (CDC, Fort Collins, CO) to estimate MLE and 95% confidence intervals (CI) by bias-corrected MLE methods.15 Only for data from San Caralampio, a Mann–Whitney test was used with Statgraphics 16.1.11 software (StatPoint Technologies Inc., Warrenton, VA) to compare vector densities per house between houses with CHIKV-infected mosquitoes and those with uninfected mosquitoes.
Results
A total of 1,747 mosquitoes were collected, including 1,170 Ae. aegypti (67.0%), three Ae. albopictus (0.2%), 453 Culex quinquefasciatus (25.9%), and four Culex coronator (0.2%). Of these, 1,006 were females, consisting of 635 Ae. aegypti (63.1%), two Ae. albopictus (0.2%), 248 Cx. quinquefasciatus (24.7%), and four Cx. coronator (0.4%). Female Ae. aegypti mosquitoes were found in 80% of the sampled households, with an average of 8.5 females collected per house (Table 1). In contrast, Ae. albopictus numbers were very low, accounting for only three of the collected mosquitoes.
Table 1.
Female Aedes aegypti collections from Ciudad Hidalgo, Chiapas, Mexico, October–December 2014
| Neighborhood | Total households | Total female Ae. aegypti mosquitoes | Mean of female Ae. aegypti mosquitoes per house | % Houses with presence of female Ae. aegypti mosquitoes (n) | % Houses with CHIKV-positive mosquitoes (n) | Median of mosquitoes per house with CHIKV-positive mosquitoes (CI 95%) | % Houses with CHIKV-negative mosquitoes (n) | Median of mosquitoes per house with CHIKV-negative mosquitoes (CI 95%) |
|---|---|---|---|---|---|---|---|---|
| San Caralampio | 44 | 302 | 6.9 | 81.8 (36) | 25.0 (11) | 8.0 (4.9–21.3) | 56.8 (25) | 4.0 (1.2–8.2) |
| San Juan | 13 | 140 | 10.8 | 76.9 (10) | 38.5 (5) | 13.0 (3.0-40.0) | 38.5 (5) | 8.0 (1.0–14.3) |
| 15 de Enero | 10 | 155 | 15.5 | 80.0 (8) | 0.0 (0) | 0.0 | 80.0 (8) | 11.5 (8.5–24.9) |
| 26 de Julio | 8 | 38 | 4.8 | 75.0 (6) | 0.0 (0) | 0.0 | 75.0 (6) | 3.5 (1.4–16.3) |
| Total | 75 | 635 | 8.5 | 80.0 (60) | 21.3 (16) | 8.0 (5.0–13.0) | 58.7 (44) | 5.0 (3.4–10.0) |
CHIKV = chikungunya virus; CI = confidence interval.
From the 635 Aedes females collected in Ciudad Hidalgo, 79 pools of Ae. aegypti and two Ae. albopictus were tested by qRT-PCR for CHIKV RNA. Eighteen Ae. aegypti pools (22.8%) were positive, whereas both of the Ae. albopictus samples were negative. CPE assays were used to confirm that infectious virus was also present in 12 of the 18 qRT-PCR-positive pools. The samples negative for CPE may have had low amounts of virus that were further reduced by two freeze-thaw cycles: once to homogenize the pools and another time to isolate the RNA. One house from San Juan had three of the 18 positive pools, therefore, 16 (21.3%) of the 75 houses harbored CHIKV-infected mosquitoes (Table 1).
Using these data, we determined that the minimum field infection rate in Ciudad Hidalgo was 32.26/1,000 mosquitoes (95% CI = 20.06–49.62). There was no significant difference in the vector density between houses positive or negative for CHIKV (W = 82.5, P = 0.0594).
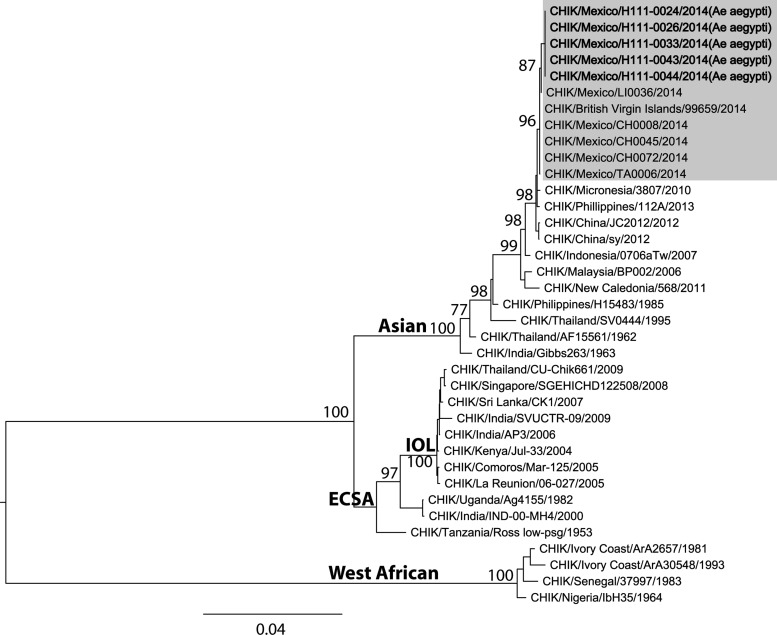
Five positive pools were selected for E1/E2 sequencing: two from San Caralampio, H111-0024 and H111-0026 (Genbank accession nos.: KT444685 and KT444686, respectively) and three from San Juan, H111-0033, H111-0043 and H111-0044 (Genbank accession nos.: KT444687, KT444688, and KT444689, respectively). The sequences were identical to previous CHIKV sequences isolated from humans during the ongoing CHIKV outbreak in Ciudad Hidalgo. These sequences belong to the Asian lineage and were closely related to others from the Caribbean and Central America, such as isolates from Panama (imported from the Caribbean) and the British Virgin Islands (Figure 2 ).
Figure 2.
Maximum likelihood phylogenetic tree based on E1/E2 sequences of CHIKV obtained from Aedes aegypti mosquitoes, October 2014.
Conclusions
The incrimination of CHIKV vectors in each regional outbreak plays an important role in vector control services. Although Ae. aegypti and Ae. albopictus may coexist in urban environments and use artificial containers as larval habitats, Ae. albopictus is more abundant in rural and semi-urban settlements, and its larvae are more localized in natural environments.16 Knowledge of the vectors involved in CHIKV transmission will allow control methods to focus on urban, semi-urban, and/or rural settlements and target natural oviposition sites. Our data indicate that Ae. aegypti was the principal epidemic vector of CHIKV in Suchiate, Chiapas, based on the following: 1) qRT-PCR and virus isolation data indicated a high rate of infected vectors temporally and spatially associated with CHIKF cases8 and 2) viral E1/E2 sequences from mosquitoes and humans from Ciudad Hidalgo were identical (Figure 2). The high infection rate of 32.26/1,000 mosquitoes reported here is similar to those previously reported in Comoros for Ae. aegypti during a CHIKF epidemic.17 Our vector density results suggest that the large numbers of females per house may have not necessary triggered the CHIKV outbreak in San Caralampio neighborhood. Instead, the outbreak may have been largely caused by the presence of an immunologically naive population,5 and therefore, even low densities of competent vectors could efficiently spread the virus.
Although few Ae. albopictus mosquitoes were collected, this species cannot be dismissed as a vector in other parts of Chiapas State and Mexico. Further collections in rural areas are needed to investigate this possibility. However, the inability of the Asian CHIKV genotype to adapt to Ae. albopictus,18 coupled with the lack of known Ae. albopictus-adaptive mutations19–22 in our sequences, suggests that Ae. aegypti was the principal vector in the locations that we sampled. Inefficient vector control activities may explain the presence of high densities of Ae. aegypti that were responsible for the CHIKV outbreak in Ciudad Hidalgo.
ACKNOWLEDGMENTS
A special thanks to Fernando García-Ordóñez, the physician who dared to see beyond and shared valuable information. We also thank Eufronio Diaz, Alejandro Gaitan, and Samanta del Rio for their endless support.
Footnotes
Financial support: This research was supported by Mexican CONACYT grant no. 214952.
Authors' addresses: Esteban E. Díaz-González, Facultad de Ciencias Biológicas, Universidad Autónoma de Nuevo León, Nuevo León, Mexico, and Centro de Investigación y Desarrollo en Ciencias de la Salud, Universidad Autónoma de Nuevo León, Nuevo León, Mexico, E-mail: esteban.diazgn@uanl.edu.mx. Tiffany F. Kautz and Rose M. Langsjoen, Department of Microbiology and Immunology, University of Texas Medical Branch, Galveston, TX, E-mails: tfkautz@utmb.edu and rolangsj@utmb.edu. Alicia Dorantes-Delgado, Iliana R. Malo-García, Maricela Laguna-Aguilar, Rogelio Danis-Lozano, and Ildefonso Fernández-Salas, Centro Regional de Investigación en Salud Pública, Instituto Nacional de Salud Pública, Chiapas, Mexico, E-mails: alice_dorantes@hotmail.es, malo@insp.mx, maricela.laguna@insp.mx, rdanis@insp.mx, and ildefonso.fernandez@insp.mx. Rubing Chen, Dawn I. Auguste, and Scott C. Weaver, Department of Pathology, University of Texas Medical Branch, Galveston, TX, E-mails: ruchen@utmb.edu, diaugust@utmb.edu, and sweaver@utmb.edu. Rosa M. Sánchez-Casas, Facultad de Medicina Veterinaria y Zootecnia, Universidad Autónoma de Nuevo León, Nuevo León, Mexico, and Centro de Investigación y Desarrollo en Ciencias de la Salud, Universidad Autónoma de Nuevo León, Nuevo León, Mexico, E-mail: sanchezcasasrossy@yahoo.com.
References
- 1.Weaver SC, Frey TK, Huang HV, Kinney RM, Rice CM, Roehrig JT, Shope RE, Strauss EG. Togaviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Eighth Report of the International Committee on Taxonomy of Viruses. London, United Kingdom: Elsevier Academic Press; 2005. pp. 999–1008. [Google Scholar]
- 2.Caglioti C, Lalle E, Castilletti C, Carletti F, Capobianchi MR, Bordi L. Chikungunya virus infection: an overview. New Microbiol. 2013;36:211–227. [PubMed] [Google Scholar]
- 3.Lindsey NP, Prince HE, Kosoy O, Laven J, Messenger S, Staples JE, Fischer M. Chikungunya virus infections among travelers—United States, 2010–2013. Am J Trop Med Hyg. 2015;92:82–87. doi: 10.4269/ajtmh.14-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leparc-Goffart I, Nougairede A, Cassadou S, Prat C, de Lamballerie X. Chikungunya in the Americas. Lancet. 2014;383:514. doi: 10.1016/S0140-6736(14)60185-9. [DOI] [PubMed] [Google Scholar]
- 5.Weaver SC, Lecuit M. Chikungunya virus and the global spread of a mosquito-borne disease. N Engl J Med. 2015;372:1231–1239. doi: 10.1056/NEJMra1406035. [DOI] [PubMed] [Google Scholar]
- 6.Vega-Rúa A, Zouache K, Girod R, Failloux A-B, Lourenço-de-Oliveira R. High level of vector competence of Aedes aegypti and Aedes albopictus from ten American countries as a crucial factor in the spread of chikungunya virus. J Virol. 2014;88:6294–6306. doi: 10.1128/JVI.00370-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centro Nacional de Programas Preventivos y Control de Enfermedades Declaratoria de Emergencia Epidemiológica EE-2-2014 para el estado de Chiapas ante el primer caso de transmisión autóctona de enfermedad por virus de Chikungunya. 2014. http://www.cenaprece.salud.gob.mx/programas/interior/emergencias/descargas/pdf/Declaratoria_Emergencia_Chiapas_Chikungunya.pdf Available at. Accessed April 17, 2015.
- 8.Kautz T, Diaz-Gonzalez EE, Erasmus J, Malo-Garcia IR, Langsjoen R, Patterson E, Auguste D, Forrester N, Sanchez-Casas RM, Hernandez-Avila M, Alpuche-Aranda C, Weaver S, Fernandez-Salas I. Chikungunya virus identified as the etiological agent of an outbreak of febrile illness in Chiapas, Mexico, 2014. Emerg Infect Dis. 2015;21 doi: 10.3201/eid2111.150546. doi: 10.3201/eid2111.150546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Insituto Nacional para el Federalismo y el Desarrollo Municipal . Enciclopedia de Los Municipios y Delegaciones de México. 2010. Suchiate.http://www.inafed.gob.mx/work/enciclopedia/EMM07chiapas/municipios/07087a.html Available at. Accessed April 17, 2015. [Google Scholar]
- 10.ECDC Mission Report Chikungunya in Italy. 2007. http://ecdc.europa.eu/en/healthtopics/Documents/0709_Chikungunya_fever_Mission_report.pdf Available at.
- 11.Powers AM, Roehrig JT. Alphavirus. In: Stephenson JR, Warnes A, editors. Diagnostic Virology Protocols. New Delhi, India: Humana Press; 2011. pp. 17–38. [Google Scholar]
- 12.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swofford DL. Phylogenetic Analysis using Parsimony (PAUP), Version 4. Sunderland, MA: Sinauer Associates; 1998. [Google Scholar]
- 14.Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 15.Biggerstaff B. PooledInfRate 4.0. Centers for Disease Control and Prevention; 2015. http://www.cdc.gov/westnile/resourcepages/mosqsurvsoft.html Available at. Accessed August 10, 2015. [Google Scholar]
- 16.Braks MAH, Honório NA, Lourenço-De-Oliveira R, Juliano SA, Lounibos LP. Convergent habitat segregation of Aedes aegypti and Aedes albopictus (Diptera: Culicidae) in southeastern Brazil and Florida. J Med Entomol. 2003;40:785–794. doi: 10.1603/0022-2585-40.6.785. [DOI] [PubMed] [Google Scholar]
- 17.Sang RC, Ahmed O, Faye O, Kelly CLH, Yahaya AA, Mmadi I, Toilibou A, Sergon K, Brown J, Agata N, Yakouide A, Ball MD, Breiman RF, Miller BR, Powers AM. Entomologic investigations of a chikungunya virus epidemic in the Union of the Comoros, 2005. Am J Trop Med Hyg. 2008;78:77–82. [PubMed] [Google Scholar]
- 18.Tsetsarkin KA, Chen R, Leal G, Forrester N, Higgs S, Huang J, Weaver SC. Chikungunya virus emergence is constrained in Asia by lineage-specific adaptive landscapes. Proc Natl Acad Sci USA. 2011;108:7872–7877. doi: 10.1073/pnas.1018344108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007;3:e201. doi: 10.1371/journal.ppat.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsetsarkin KA, Chen R, Yun R, Rossi SL, Plante KS, Guerbois M, Forrester N, Perng GC, Sreekumar E, Leal G, Huang J, Mukhopadhyay S, Weaver SC. Multi-peaked adaptive landscape for chikungunya virus evolution predicts continued fitness optimization in Aedes albopictus mosquitoes. Nat Commun. 2014;5:4084. doi: 10.1038/ncomms5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vazeille M, Moutailler S, Coudrier D, Rousseaux C, Khun H, Huerre M, Thiria J, Dehecq J-S, Fontenille D, Schuffenecker I, Despres P, Failloux A-B. Two chikungunya isolates from the outbreak of La Reunion (Indian Ocean) exhibit different patterns of infection in the mosquito, Aedes albopictus. PLoS One. 2007;2:e1168. doi: 10.1371/journal.pone.0001168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsetsarkin KA, Weaver SC. Sequential adaptive mutations enhance efficient vector switching by chikungunya virus and its epidemic emergence. PLoS Pathog. 2011;7:e1002412. doi: 10.1371/journal.ppat.1002412. [DOI] [PMC free article] [PubMed] [Google Scholar]