Abstract
We conducted the first survey of zoonotic risk of Hepatitis E virus (HEV) transmissions in Ouagadougou, Burkina Faso, through the direct contact with pork meat during professional activity. Anti-HEV antibodies were more prevalent in pork butchers, 76% than in the general population, which was 47.8% in 2013 (odds ratio = 3.46, 95% CI = 2.85–4.21, P < 0.001). Among slaughter-aged swine, HEV seroprevalence was of 80%, and HEV RNA was detected in 1% of pork livers. Phylogenetic analysis pointed out HEV genotype 3. Thus, in addition to possible HEV contamination through the water source, as in endemic region, zoonotic transmissions of HEV probably occur in west Africa.
Hepatitis E virus (HEV) is a causative agent of acute or fulminant hepatitis in humans occurring in many areas of the world.1 In most cases, it is a self-limited infection with a rapid viral clearance, but it can evolve into more severe forms as the mortality rate ranges from 1% to 4% in the general population and to nearly 20% in pregnancies during outbreaks. HEV epidemiology involves fecal and oral transmission either through the water source in endemic regions or through zoonotic exposure (direct contact with infected animal, meat or consumption of infected foods) in non-endemic areas.2 More recently, cases of chronic hepatitis E have been reported in patients under immunosuppressive treatment such as solid organ transplants, which has progressed to more serious conditions, including fibrosis, liver cirrhosis, and liver failure.3
In addition to humans, HEV has been identified in several other animal species, especially pigs where infections are asymptomatic.4 Hepatitis E is now a recognized zoonotic disease with swine being the main source of human infections.5 In addition, people working in contact with swine and at slaughterhouses are known to be at higher risk of HEV exposure.6,7
In Burkina Faso, there is very little epidemiological data available on HEV infection, and it is unknown if zoonotic transmissions occur. To assess if population in contact with pork meat are at higher risk of HEV infections, a survey was performed in butchers from Burkina Faso and in pigs at slaughterhouse. Thus the objectives of this study were to 1) determine HEV seroprevalence in pork butchers in comparison to the general population, 2) determine HEV serological and virological prevalences in slaughter-aged pigs, and 3) identify the HEV genotype present in pigs.
Human sera were collected during a serologic survey in Ouagadougou, the capital city of Burkina Faso, of blood donors (90 sera sampled in July 2013 in Ouagadougou blood bank) and among people working daily with pork meat and preparing street food, after obtaining informed consent (authorization no. 2014-12-128-3 December 2014). First, pork sale sites were recorded in the great Ouagadougou area in 2013. The sales sites were geographically displayed with ArcGIS 10.0 (Esri, Redlands, CA) (Figure 1 ). Behavior investigations were conducted in 155 sites from the 292 butchers' shop found. Thus 100 serum samples were collected on-site from volunteer butchers. Anti-HEV immunoglobulin G (IgG) were determined using HEV-IgG ELISA Dia.Pro (Diagnostic Bioprobes s.r.l, Milan, Italy) while anti-HEV IgM were determined using Wantai kit (Biological Pharmacy Enterprise Co., Beijing, China). The tests were carried out according to the manufacturer's instructions including positive and negative controls. Both tests detect IgG or IgM antibodies against the four major HEV genotypes. For the Burkinabe population, Dia.Pro and Wantai detection of IgG are correlated.8 The epidemiological characteristics of butchers are presented in Table 1.
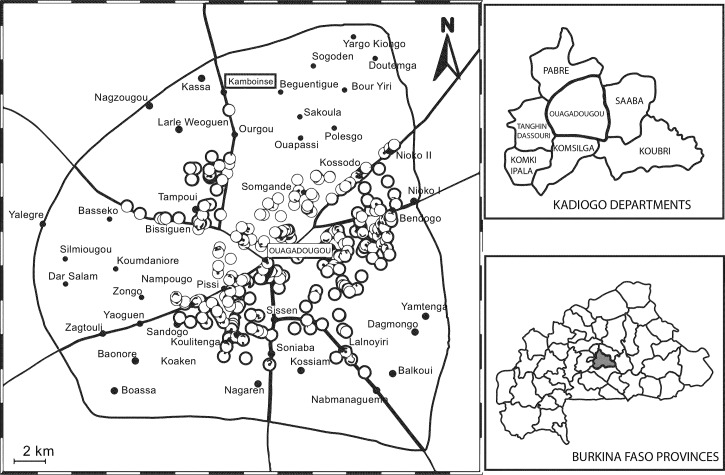
Figure 1.
Map of Ouagadougou (location 12°15′ N, 1°40′30″ W to 12°30′ N, 1°25′30″ W), showing location sales sites in 2013. Pork food points of sale are displayed as circles and borouth center in Ouagadougou black dots, Black lines are main roads. Kamboinse site of slauthering is within square.
Table 1.
Sociodemographic characteristics of the butchers
| Characteristics of individuals | No. (%) |
|---|---|
| Gender | |
| Male | 152 (98.1) |
| Female | 3 (1.9) |
| Age group (years) | |
| ≤ 25 | 48 (30.9) |
| 26–35 | 80 (51.6) |
| > 36 | 27 (17.4) |
| Duration of activity (years) | |
| < 5 | 90 (58.1) |
| 5–10 | 46 (29.7) |
| 10–20 | 19 (12.2) |
| Training on sanitation | 16 (10.3) |
| No knowledge of zoonotic diseases | 155 (100) |
Swine sera and liver samples were collected from November to December 2012 and July to August 2013 from 257 pigs of local herds. From the sale sites investigation, an average of 8,700–17,400 pigs were estimated to be slaughtered per month in Ouagadougou. The slaughter age of animals ranged from 6 to more than 18 months. About 100 samples of blood were collected during the bleeding at the slaughterhouse of Kambouinse, and 157 pieces of liver were bought from various sale sites. A commercial test validated for veterinary analysis was used to detect anti-HEV antibodies by enzyme-linked immunosorbent assay 4.0v (MP Diagnostics, Illkirch, France) according to the manufacturer's instructions except that 10 μL of sera was used.9
For each liver tissue sample, 30–50 mg were crushed and homogenized with ceramics beads in 1 mL of RNA lysis buffer (QIAGEN, Hilden, Germany) by using a Precellys®24 apparatus (Bertin Technology, Montigny-le-Bretonneux, France). Total RNA was extracted from 500 μL suspension by using the RNeasy Mini Kit (QIAGEN) according to the manufacturer's recommendations. HEV RNA was detected by using quantitative reverse transcription polymerase chain reaction (RT-PCR) adapted from the study of Barnaud and others.9,10 From the 157 liver tissues analyzed, one was found positive. For sequence analysis, a region from the open reading frame 2 (ORF-2) region (capsid protein) was amplified by nested RT-PCR from the positive sample. The PCR product (348 bp) was purified on a 1% (w/v) agarose gel electrophoresis with a PCR clean-up kit, extracted (MACHEREY-NAGEL, Düren, Germany), cloned in a pJET1.2 Easy vector (Thermo-Fisher Scientific, Illkirch, France), and then sequenced (Beckman Coulter Genomics Inc., Takeley, UK). HEV sequences with strong similarities with the HEV-BF strain (accession no. LN831924) were retrieved from databases using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The serological data from human and pigs were processed, analyzed, and plotted using Excel 2013 (Microsoft, Issy-les-Moulineaux, France), Statview (SAS Institute, Cary, NC), and Prism 6.0e (GraphPad Software, La Jolla, CA).
The human prevalence of IgG anti-HEV among blood donors in 2012 was 19.1% (95% confidence interval [CI] = 13.3–24.9) but reached 47.8% (95% CI = 37.5–58.1) in 2013 among the blood donor cohort sampled for this study during the wet season.8 This increase is not fully understood but might be associated with seasonal movement of population with poor hygienic status. In contrast, the global HEV prevalence among Ouagadougou butchers was estimated to be 76% (95% CI = 67–84). Thus, butchers had a significant risk factor 3.5 times higher compared with the general population sampled and tested in 2013 (odd ratio = 3.46, 95% CI = 2.85–4.21, P < 0.001). These findings suggest that HEV zoonotic transmissions occur through frequent contacts with biological samples (feces, blood) and organs of infected animals.11 These results are consistent with similar studies worldwide that had shown a high prevalence of HEV infections in people who work in direct contact with pigs or pig meat.12–14
Anti-HEV IgM were present in 3.19% (95% CI = 0.95–2.95) of 2013 blood donor population and in 1% (95% CI = 1.70–4.68) of butchers. However, this difference is not significant. The low percentage of recent infection (IgM positive) among butchers can be explained by the high percentage of anti-HEV IgG antibodies and, possibly, by the time spent in the activity (> 5 years). Despite the absence of registered HEV epidemics, these results indicate a high incidence of HEV infections. This is illustrated by the increase of IgG seroprevalence between 2011–2012 and 2013 in the general population and is probably associated with subclinical infections. As expected, we observed an increased prevalence with age among butchers (55% below 25 years of age, N = 40; 90% older than 25 years of age), which could be explained by a longer time of exposure.13
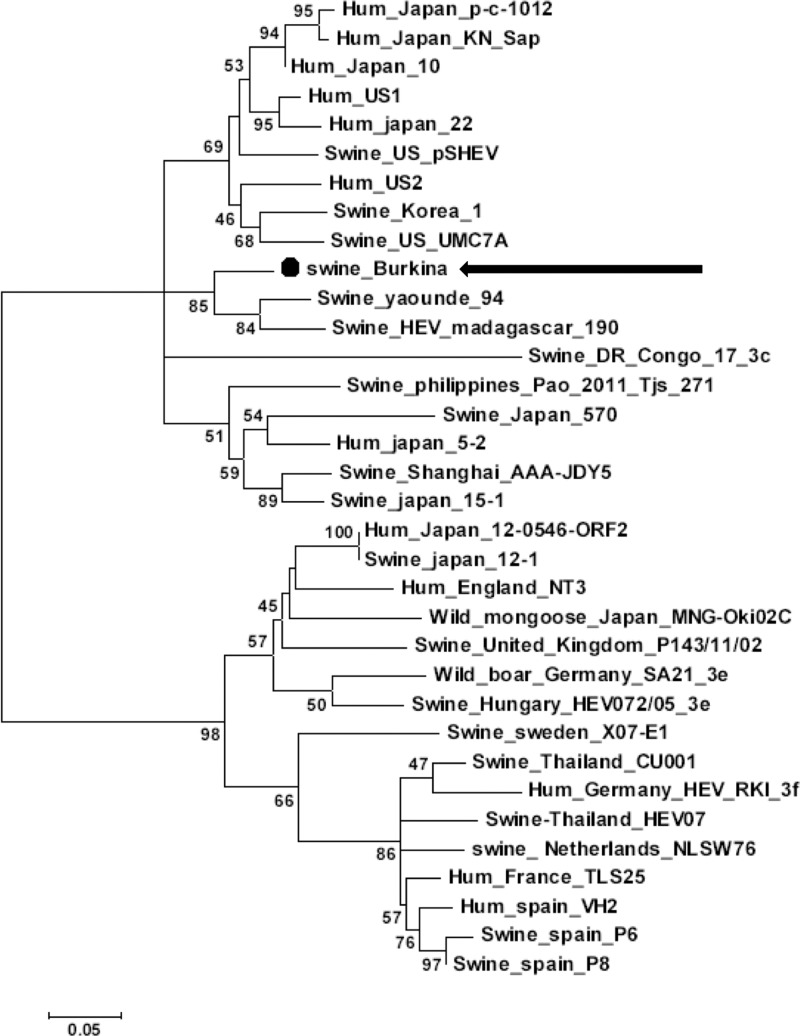
In the swine reservoir, a high seroprevalence rate of 80% (95% CI = 72–87) was found in pigs older than 6 months of age. These results show an active HEV circulation among domestic pigs in Burkina Faso. Furthermore, one liver sample was found positive for HEV RNA. Because the sampling of slaughter-aged pigs includes animals older than 12 months, this discrepancy between high seroprevalence and low HEV portage was not surprising.14 At that age animals had certainly cleared the infection. The HEV sequence, identified in this Burkinabe pig, was clustered with an African sequence of genotype 3 HEV from Yaounde (Cameroun) and Madagascar and had a > 98% similarity but was very different from the Nigerian strains as shown by BLAST analysis.15 The phylogenetic analysis of the swine HEV-Burkina sequence, together with retrieved sequences and reference sequences, showed that the Burkinabe sequence is related to swine HEV strains from Cameroun and Madagascar but not from Congo (bootstrap value = 85; Figure 2 ). Addition of other published swine HEV sequences showed that our strain is part of a group of sequence with no subtype or geographical origin cluster (Supplemental Figure 1). Thus, other factors, such as pork species and their international trade, should be investigated.
Figure 2.
Phylogenetic analysis from the partial hepatitis E virus open reading frame 2 (HEV-ORF2) sequence obtained from Burkinabe swine (swine Burkina AC#LN831924; indicated by a dot and an arrow) aligned with 33 reference sequences retrieved from Genbank using MEGA6 software.16 Maximum-likelihood method based on the Kimura two parameters model was used with a discrete gamma distribution (+G) and portion of invariable sites (+I) to model evolutionary rate differences among sites (+G = 0.4606; +I = 64.018% sites). The tree with the highest log likelihood (−2,304.6559) is shown. The robustness of the branching order was assessed by bootstrap method (200 replicates) as shown next to the branches (values below 45% were not shown). Branch lengths scale is the number of substitutions per site. Sequence name, given as species-HEV-country-strain-name, and accession number are given in Supplemental Table 1. All positions containing gaps and missing data were eliminated, leaving 276 positions in the final dataset.
In conclusion, this study showed the presence of HEV genotype 3 in sub-Sahel areas where only HEV genotype 1 had been identified or suspected until now. Thus the HEV from pigs may be a new source of human contamination by direct contact with infected animals (butchers, farmers) or consumption of infected meat that was not cooked properly.5 Additional studies are needed to explore the different HEV genotypes associated with clinical hepatitis E in human and to investigate the possible role of the pig reservoir and zoonotic transmission in HEV epidemiology in west Africa.
Supplementary Material
ACKNOWLEDGMENTS
We thank Essia Belarbi, Claire Torres, and Judicael Tarama for their help respectively in France and Burkina Faso.
Footnotes
Financial support: This study was supported by the French Government through the 3rd cycle university scholarship program cooperation of the French Embassy in Burkina Faso (www.burkina.campusfrance.org) and a cotutelle thesis grant from the University Paris-Sud.
Authors' addresses: Kuan Abdoulaye Traoré, Jean Bienvenue Ouoba, Alfred S. Traoré, and Nicolas Barro, LaBesta/CRSBAN, EDST, Université de Ouagadougou, Ouagadougou, Burkina Faso, E-mails: kuabtraore@live.fr, maitreouob@gmail.com, astraore@univ-ouaga.bf, and barronicolas@yahoo.fr. Sophie Rogée, Marine Dumarest, and Nicole Pavio, ANSES, UMR1161 Virologie, Maisons-Alfort, France, E-mails: rogeeso@aol.com, marinedumarest@gmail.com, and nicole.pavio@anses.fr. Nicolas Huot and Pierre Roques, Service d'Immuno-Virologie, CEA, Fontenay-aux-Roses, France, E-mails: nicolas.huot2@cea.fr and pierre.roques@cea.fr.
References
- 1.Liu P, Bu QN, Wang L, Han J, Du RJ, Lei YX, Ouyang YQ, Li J, Zhu YH, Lu FM, Zhuang H. Transmission of hepatitis E virus from rabbits to cynomolgus macaques. Emerg Infect Dis. 2013;19:559–565. doi: 10.3201/eid1904.120827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dai X, Dong C, Zhou Z, Liang J, Dong M, Yang Y, Fu J, Tian H, Wang S, Fan J, Meng J, Purdy MA. Hepatitis E virus genotype 4, Nanjing, China, 2001–2011. Emerg Infect Dis. 2013;19:1528–1530. doi: 10.3201/eid1909.130013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamar N, Abravanel F, Selves J, Garrouste C, Esposito L, Lavayssiere L, Cointault O, Ribes D, Cardeau I, Nogier MB, Mansuy JM, Muscari F, Peron JM, Izopet J, Rostaing L. Influence of immunosuppressive therapy on the natural history of genotype 3 hepatitis-E virus infection after organ transplantation. Transplantation. 2010;89:353–360. doi: 10.1097/TP.0b013e3181c4096c. [DOI] [PubMed] [Google Scholar]
- 4.Yugo DM, Meng XJ. Hepatitis E virus: foodborne, waterborne and zoonotic transmission. Int J Environ Res Public Health. 2013;10:4507–4533. doi: 10.3390/ijerph10104507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pavio N, Meng XJ, Doceul V. Zoonotic origin of hepatitis E. Curr Opin Virol. 2015;10:34–41. doi: 10.1016/j.coviro.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Di Martino B, Di Profio F, Martella V, Di Felice E, Di Francesco CE, Ceci C, Marsilio F. Detection of hepatitis E virus in slaughtered pigs in Italy. Arch Virol. 2010;155:103–106. doi: 10.1007/s00705-009-0544-0. [DOI] [PubMed] [Google Scholar]
- 7.Carpentier A, Chaussade H, Rigaud E, Rodriguez J, Berthault C, Boue F, Tognon M, Touzé A, Garcia-Bonnet N, Choutet P, Coursaget P. High hepatitis E virus seroprevalence in forestry workers and in wild boars in France. J Clin Microbiol. 2012;50:2888–2893. doi: 10.1128/JCM.00989-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Traore KA, Rouamba H, Nebie Y, Sanou M, Traore AS, Barro N, Roques P. Seroprevalence of fecal-oral transmitted hepatitis A and E virus antibodies in Burkina Faso. PLoS One. 2012;7:e48125. doi: 10.1371/journal.pone.0048125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnaud E, Rogee S, Garry P, Rose N, Pavio N. Thermal inactivation of infectious hepatitis E virus in experimentally contaminated food. Appl Environ Microbiol. 2012;78:5153–5159. doi: 10.1128/AEM.00436-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper K, Huang FF, Batista L, Rayo CD, Bezanilla JC, Toth TE, Meng XJ. Identification of genotype 3 hepatitis E virus (HEV) in serum and fecal samples from pigs in Thailand and Mexico, where genotype 1 and 2 HEV strains are prevalent in the respective human populations. J Clin Microbiol. 2005;43:1684–1688. doi: 10.1128/JCM.43.4.1684-1688.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Temmam S, Besnard L, Andriamandimby SF, Foray C, Rasamoelina-Andriamanivo H, Heraud JM, Cardinale E, Dellagi K, Pavio N, Pascalis H, Porphyre V. High prevalence of hepatitis E in humans and pigs and evidence of genotype-3 virus in swine, Madagascar. Am J Trop Med Hyg. 2013;88:329–338. doi: 10.4269/ajtmh.2012.12-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krumbholz A, Mohn U, Lange J, Motz M, Wenzel JJ, Jilg W, Walther M, Straube E, Wutzler P, Zell R. Prevalence of hepatitis E virus-specific antibodies in humans with occupational exposure to pigs. Med Microbiol Immunol (Berl) 2012;201:239–244. doi: 10.1007/s00430-011-0210-5. [DOI] [PubMed] [Google Scholar]
- 13.Adjei AA, Aviyase JT, Tettey Y, Adu-Gyamfi C, Mingle JA, Ayeh-Kumi PF, Adiku TK, Gyasi RK. Hepatitis E virus infection among pig handlers in Accra, Ghana. East Afr Med J. 2009;86:359–363. doi: 10.4314/eamj.v86i8.54155. [DOI] [PubMed] [Google Scholar]
- 14.Pavio N, Meng XJ, Renou C. Zoonotic hepatitis E: animal reservoirs and emerging risks. Vet Res. 2010;41:46. doi: 10.1051/vetres/2010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Owolodun OA, Gerber PF, Giménez-Lirola LG, Kwaga JKP, Opriessnig T. First report of hepatitis E virus circulation in domestic pigs in Nigeria. Am J Trop Med Hyg. 2014;91:699–704. doi: 10.4269/ajtmh.14-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.