Abstract
Mosquito blood meals taken from humans and animals potentially represent a useful source of blood for the detection of blood-borne pathogens. In this feasibility study, Anopheles stephensi mosquitoes were fed with blood meals spiked with dengue virus type 2 (DENV-2) and harvested at serial time points. These mosquitoes are not competent vectors, and the virus is not expected to replicate. Ingested blood was spotted on Whatman FTA cards and stored at room temperature. Mosquito abdomens were removed and stored at −80°C. Control blood meal aliquots were stored in vials or applied onto FTA cards. After 4 weeks of storage, the samples were extracted using beadbeating and QIAamp Viral RNA kit (Qiagen Sciences, Germantown, MD). Recovered viral RNA was analyzed by DENV-2 TaqMan RT-PCR assay and next-generation sequencing (NGS). Overall viral RNA recovery efficiency was 15% from the directly applied dried blood spots and approximately 20% or higher for dried blood spots made by blotting mosquito midgut on FTA cards. Viral RNA in mosquito-ingested blood decreases over time, but remains detectable 24 hours after blood feeding. The viral sequences in FTA-stored specimens can be maintained at room temperature. The strategy has the potential utility in expedited zoonotic virus discovery and blood-borne pathogen surveillance.
Introduction
Mosquitoes are major vectors of arboviruses, many of which are associated with epidemic illnesses.1–3 Virus isolation and molecular assay of targeted pathogens are common practices of arbovirus surveillance.4,5 Recently, the massively parallel high-throughput sequencing, that is, next-generation sequencing (NGS), has been applied for the discovery of novel arboviruses.6,7 However, captured blood-fed mosquitoes are not typically assayed for non-mosquito-vectored pathogens, even though their blood meals may contain a useful source of blood for the detection of pathogens in humans and animals.8–11 The practice of using blood-fed mosquitoes to conduct surveillance for human and livestock viral pathogens has been termed xenosurveillance.11 This study explored the pathogen nucleic acid sequence stability in host blood after mosquito ingestion to determine if blood-fed female mosquitoes could be used as a source for virus/pathogen surveillance and discovery and ultimately a surveillance tool. Specifically, the study explored viral stability in the mosquito gut, the time interval that a nonreplicating virus can be detected in the mosquito after blood feeding, and the efficiency of recovering viral RNA.
Materials and Methods
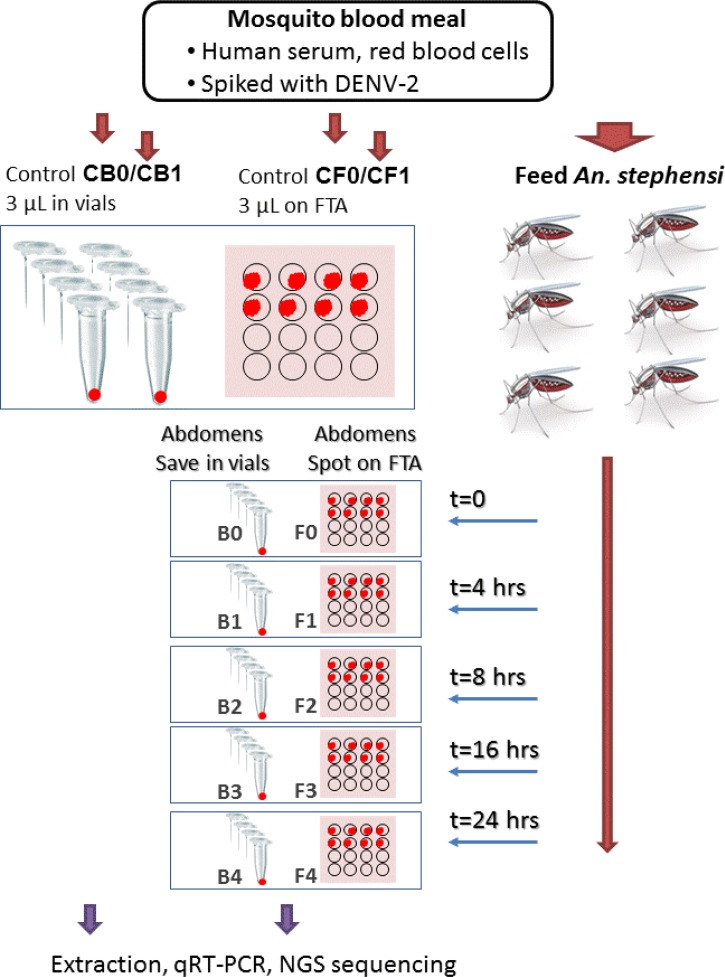
The experiment design is illustrated in Figure 1 and described in detail in the following sections.
Figure 1.
Experiment design for the feasibility study of using blood meal in mosquito abdomens for blood-borne virus surveillance. Blood spiked with dengue virus type 2 (DENV-2) was used to feed Anopheles stephensi mosquitoes. Controls and mosquito blood meal specimens, kept in vials at −80°C or on FTA cards at room temperature, were subjected to storage, nucleic acid extraction, and analyses by real-time reverse-transcription polymerase chain reaction (qRT-PCR) or next-generation sequencing.
Whole blood meal.
A virus-infected blood meal was prepared by mixing 900 μL of dengue virus serotype 2 (DENV-2) strain KDH0139A-02 AFRIMS SR-0570 diluted to a titer of 2 × 105 plaque-forming unit (PFU)/mL, 300 μL of packed human red blood cells (O+; Key Biologics, Memphis, TN), and 300 μL of healthy human serum (A+; Key Biologics). The blood meal was pre-warmed to 37°C and then pipetted into a water-jacketed glass feeder covered with a parafilm membrane and maintained at a constant temperature of 37°C.
Blood meal prior- and post-feeding controls.
Prior to the mosquitoes feeding, a 100 μL aliquot of the virus-infected blood meal was obtained and kept at 37°C for control purposes. To make pre-feeding controls, 3 μL was aliquoted into a vial (control CB0, N = 8) and 3 μL was spotted on Whatman FTA® CloneSaver™ Card (WB120028; GE Healthcare Bio-Sciences Corp., Piscataway, NJ) (control CF0, N = 8). The same procedure was repeated with the remaining blood meal mixture after mosquito feeding to make post-feeding controls (controls CB1 and CF1).
Mosquitoes blood feeding and sample collection.
Anopheles stephensi (Indian strain) mosquitoes were used in this study because anopheline mosquitoes are not competent vectors of DENV-2, and the virus is not expected to replicate in midgut tissue.2,12 Mosquitoes were reared under standard insectary conditions of 27°C, 80% relative humidity, and a 12:12 light/dark cycle. Larvae were fed a diet of ground and pelleted fish food and adults were maintained on 10% sucrose solution before experimentation. Five-day post-eclosion mosquitoes were allowed to feed on the DENV-2 spiked blood meal for 1 hour. After feeding, blood-fed females were separated from those that had not been blood fed. Sixteen blood-fed females were removed and frozen as t = 0 samples (occurred about 40 minutes after blood feed because of the time it took to separate blood-fed mosquitoes from non-blood fed). Remaining blood-fed mosquitoes were returned to standard insectary conditions, and subsequent samples were collected at 4, 8, 16, and 24 hours from the end of the feed, respectively. At each time point, mosquitoes were frozen at −20°C for 30 minutes. Eight individual mosquitoes had their midgut blood specimens blotted on FTA card spots, allowed to dry on the FTA cards, and stored at room temperature, and eight additional mosquitoes had their abdomens cut and saved in each of 1.5-mL tubes and stored at −80°C.
Sample storage, nucleic acids extraction, and quantification.
The vials of control blood meal aliquots and mosquito abdomens were stored at −80°C. All the FTA cards were kept in bags with desiccant packets stored in a Styrofoam pouch at room temperature for 4 weeks. All the stored samples were subjected to beadbeating and extraction to purify total nucleic acids. The FTA dried blood spot of approximate 5 mm diameter was cut into a 2-mL screw-cap microcentrifuge tube with one 3-mm glass bead and two 1-mm glass beads (Sigma-Aldrich, St. Louis, MO). After the addition of 560 μL AVL buffer and 5.6 μL of 1 μg/μL of RNA carrier (Qiagen Sciences, Germantown, MD), the microcentrifuge tube was vortexed by using BioSpec Mini-Beadbeater-16 (BioSpec Products Inc., Bartlesville, OK) for 45 seconds and then centrifuged at 14,000 × g for 5 minutes. The supernatant was collected, mixed with 560 μL of ethanol, and used in nucleic acids extraction with Qiagen QIAamp Viral RNA kit (Qiagen Sciences). The same protocol was used in the extraction of total nucleic acids from control blood meal aliquots and mosquito abdomens.
Quantification of DENV-2 RNA copy numbers.
TaqMan quantitative real-time reverse-transcription polymerase chain reaction assay (qRT-PCR) was used to quantify DENV-2 genome copy number in the purified RNA samples as described previously in a study.13 The instrument, Applied Biosystems (Foster City, CA) 7500 Fast Real-Time PCR System and software SDS version 1.4 were used in the analysis. DENV-2 RNA used as qRT-PCR standards was synthesized by in vitro transcription, purified and quantified by using BioAnalyzer 2100 (Agilent Technologies, Santa Clara, CA).
Random amplification, MiSeq sequencing, and data analysis.
The purified nucleic acids were subjected to reverse transcription using anchored random octamer and PCR amplification using the octamer and specific primer for the anchor sequence.14 The resulted amplicons were sequenced by using MiSeq benchtop sequencer (Illumina, Inc., San Diego, CA) with Nextera XT DNA Sample Preparation Kit and MiSeq Sequencing Kit v2 (500-cycle). The quality-filtered sequence reads were mapped against complete genome sequence for DENV-2 strain S16803 (GU289914) with software Roche (Branford, CT) GS Reference Mapper (Newbler) V2.8. The number of mapped DENV-2 reads was divided by the total number of MiSeq sequence reads to determine the sensitivity or limit of detection (LOD) for the sequence-based viral detection.
Results
Recovery of viral RNA from FTA card.
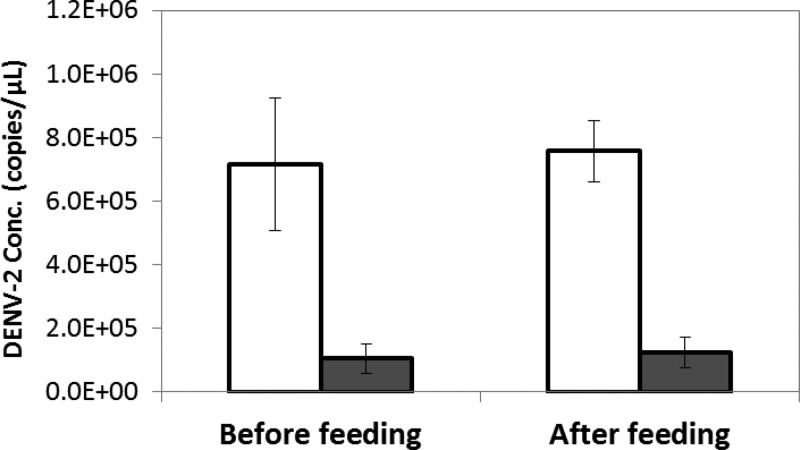
All RNA extracts from the 3-μL aliquots or the FTA cards were positive in DENV-2 qRT-PCR assay, with Ct value lower than the assay threshold Ct value of 40.13 The average concentrations (mean ± standard deviation) of DENV-2 extracted from the 3-μL aliquots of whole blood (N = 8 independent samples in parallel; Figure 1) stored under −80°C were 7.14 ± 2.08 × 105 copy/μL (Ct values = 21.36 ± 0.53) and 7.57 ± 0.97 × 105 copy/μL (Ct values = 21.20 ± 0.18) for sampling before (CB0) and after (CB1) blood meal feeding, respectively (Figure 1). The average concentrations of DENV-2 extracted from the FTA dried blood spots blotted with same aliquots (N = 8) and stored at room temperature for 4 weeks were 1.05 ± 0.46 × 105 copy/μL (Ct values = 23.90 ± 0.77) and 1.24 ± 0.48 × 105 copy/μL (Ct values = 23.69 ± 0.96) for sampling before (CF0) and after (CF1) blood meal feeding, respectively (Figure 2 ). The correspondent FTA extraction efficiency for DENV-2 was 14.8% (CF0/CB0) and 16.4% (CF1/CB1), respectively. The results suggested an overall recovery of viral RNA from stored FTA dried blood spots related to the whole blood was approximately 15%.
Figure 2.
Recovery of nucleic acids from Whatman FTA cards. Aliquots of artificial whole blood with dengue virus type 2 (DENV-2) were collected before and after feeding mosquitoes, kept in vial (empty bars) or spotted on FTA cards (gray bars), and subjected to storage at −80°C and room temperature for 4 weeks, respectively, nucleic acids extraction, and quantitation by real-time reverse-transcription polymerase chain reaction (qRT-PCR). The error bars are standard deviations of data for eight samples.
Viral RNA stability in mosquito midguts.
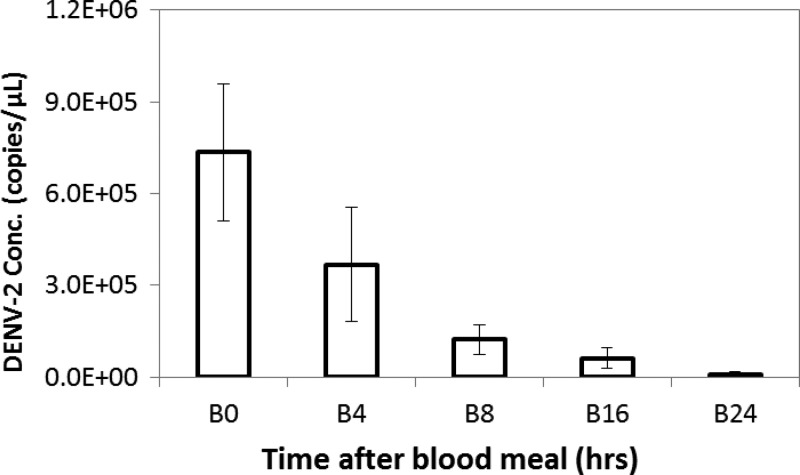
The stability of DENV-2 in mosquito midgut was assessed by TaqMan qRT-PCR assay of frozen midguts sampled at serial time points after blood meal (Figure 3 ). All RNA extracts from the 3-μL aliquots or the FTA cards were positive in DENV-2 qRT-PCR assay. When compared with DENV-2 quantity in midgut blood of time 0 (B0), which was 7.35 ± 2.24 × 105 copy/μL (set as 100%, Ct = 21.30 ± 0.49), DENV-2 level at 4, 8, 16, and 24 hours after blood meal was 3.68 ± 1.87 × 105 copy/μL (B4, 50.0% of B0; Ct = 22.42 ± 0.73), 1.23 ± 0.49 × 105 copy/μL (B8, 16.7% of B0; Ct = 24.05 ± 0.68), 6.20 ± 3.42 × 104 copy/μL (B16, 8.4% of B0; Ct = 25.06 ± 0.83), and 8.42 ± 8.35 × 103 copy/μL (B24, 1.1% of B0; Ct = 28.89 ± 2.34), respectively. All replicated samples were clearly positive for DENV-2, including those of B24 (8/8). The result indicated that in a non-susceptible mosquito species, such as An. stephensi used in this study, which does not allow DENV-2 to replicate,12 viral RNA titer will reduce gradually but remains detectable for at least 24 hours in the midgut.
Figure 3.
Dengue virus type 2 (DENV-2) in mosquito abdomens after blood feeding. Mosquito abdomen specimens, collected at 0–24 hours after blood feeding, were cut and kept in vial (empty bars) and subjected to storage at −80°C, nucleic acids extraction, and quantitation by real-time reverse-transcription polymerase chain reaction (qRT-PCR). The error bars are standard deviations of data for eight samples.
Detection of viral RNA from FTA dried blood spots.
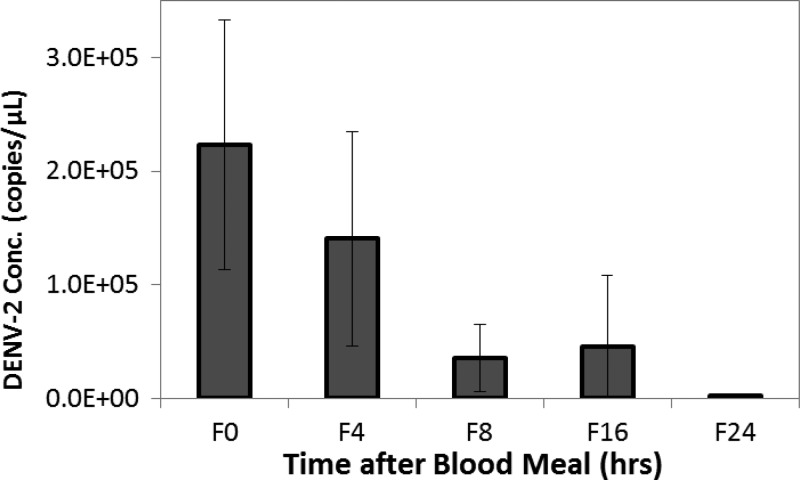
Blotting biological samples on FTA card inactivates potential infectious pathogens in the collected environmental and clinical specimens and allows storage and transportation of the samples in room temperature. Viral RNA from FTA dried blood spots stored in ambient conditions was extracted and quantified to assess the recovery of DENV-2 RNA (Figure 4 ). In this study, mosquito midgut specimens collected at sequential time points after blood feeding were blotted onto FTA cards and stored at room temperature for 4 weeks to test whether FTA card storage can stabilize viral RNA and preserve viral titer profile in the specimens. The DENV-2 concentration for FTA dried blood spots (N = 8) of midguts collected at time 0 was on average 2.23 ± 1.10 × 105 copy/μL (Ct = 22.86 ± 1.00). When compared with the concentration of time 0 FTA spots (F0, set as 100%), the average DENV-2 RNA concentrations were 1.40 ± 0.94 × 105 copy/μL (F4, 63.0% of F0; Ct = 24.24 ± 2.69), 3.55 ± 2.92 × 104 copy/μL (F8, 15.9% of F0; Ct = 25.92 ± 1.41), 4.58 ± 6.25 × 104 copy/μL (F16, 20.5% of F0; Ct = 27.70 ± 3.02), and 1.89 ± 1.39 × 103 copy/μL (F24, 0.8% of F0; Ct = 30.40 ± 1.36) for FTA dried blood spots of 4, 8, 16, and 24 hours, respectively, after the blood meal (Figure 4). All replicated samples were clearly positive for DENV-2, including those of F24 (8/8). Moreover, these concentrations were 30.34%, 38.22%, 28.93%, 73.86%, and 22.45% of correspondent RNA extracts of midgut samples collected and frozen at each time points from 0 to 24 hours.
Figure 4.
Dengue virus type 2 (DENV-2) in FTA dried blood meal spots for mosquito abdomens after blood feeding. Mosquito abdomen specimens, collected at 0–24 hours after blood feeding, were blotted on FTA cards (gray bars) and subjected to storage at room temperature, nucleic acids extraction, and quantitation by real-time reverse-transcription polymerase chain reaction (qRT-PCR). The error bars are standard deviations of data for eight samples.
Sequence data analysis.
The nucleic acids extracts (Table 1) were subjected to an unbiased sequencing using the previously described method14; however, the MiSeq was used in NGS of the random RT-PCR amplicons. The qRT-PCR assay specific for DENV-2 determined the absolute level, that is, the copy number concentration for viral RNA. In contrast, the sensitivity of viral detection by high-throughput NGS was expressed as viral sequence hits related to the total sequence reads, such as DENV reads per million MiSeq reads in this study (Table 1). This method readily detected DENV-2 in the whole blood (CB0 and CB1) with DENV reads constituting about half of the MiSeq reads (56.53% and 46.97% of all sequence reads, respectively). The DENV percentage was much lower for CF0 and CF1 (14.03% and 3.55% of all sequence reads, respectively), suggesting that for extraction from FTA dried blood spots, recovery of viral RNA was less efficient than recovery of blood RNA. It is interesting that this difference was not observed in the comparison of frozen mosquito midguts with FTA midgut spots (Table 1). Despite the high titer of DENV-2 in the blood meal, there were only a small and variable number of viral reads in each of the frozen or FTA specimens. Moreover, the relative DENV abundance, that is, DENV per million total reads, determined by random sequencing did not correlate with the sampling hours. Though the sequencing results were not as explicit as qPCR, with the large volume of sequence data from MiSeq, the number of DENV reads in all FTA midgut samples and frozen midguts, except for B24, was evidently above the background level (no template controls) (Table 1).
Table 1.
MiSeq sequencing of FTA dried blood spots and controls
| Sample* | Description | MiSeq reads | DENV reads | DENV (%) | DENV per million reads |
|---|---|---|---|---|---|
| CB0 | Artificial blood meal, DENV-2, before feeding | 165,220 | 93,393 | 56.526 | 565,264 |
| CF0 | CB0 on FTA | 65,458 | 9,183 | 14.029 | 140,288 |
| CB1 | Artificial blood meal, DENV-2, after feeding | 470,674 | 216,365 | 45.969 | 459,692 |
| CF1 | CB1 on FTA | 55,852 | 1,981 | 3.547 | 35,469 |
| B0 | Anopheles stephensi midgut, 0 hour after feeding | 279,856 | 15 | 0.005 | 54 |
| B4 | Midgut, 4 hours | 301,510 | 65 | 0.022 | 216 |
| B8 | Midgut, 8 hours | 277,882 | 48 | 0.017 | 173 |
| B16 | Midgut, 16 hours | 299,418 | 39 | 0.013 | 130 |
| B24 | Midgut, 24 hours | 173,214 | 2 | 0.001 | 12 |
| F0 | An. stephensi midgut blotted on FTA, 0 hour after feeding | 310,056 | 16 | 0.005 | 52 |
| F4 | Midgut on FTA, 4 hours | 134,162 | 23 | 0.017 | 171 |
| F8 | Midgut on FTA, 8 hours | 281,964 | 20 | 0.007 | 71 |
| F16 | Midgut on FTA, 16 hours | 147,968 | 97 | 0.066 | 656 |
| F24 | Midgut on FTA, 24 hours | 338,480 | 19 | 0.006 | 56 |
| NTC† | No template reaction and process control | 251,720 | 2 | 0.001 | 8 |
DENV = dengue virus; NTC = no template controls.
CB and B samples, aliquots stored in vials at −80°C; CF and F samples, dried blood spots on FTA cards stored at room temperature.
Water was used instead of nucleic acids to detect background contamination from the process in laboratory.
Discussion
In this study, we investigated the suitability of using the mosquito blood meal as a convenient and untapped source for the surveillance of pathogens in human and animal blood. This approach is novel and essentially expands the capabilities of surveillance of mosquito-borne viruses. In addition, this method can be used in conjunction with NGS to identify known arboviruses and discover new potential transmissible agents that exist in the mosquito blood meal. The practical importance is intriguing and suggestive of a promising means of utilizing mosquitos as a “mobile” microliter blood-sampling device. The qRT-PCR results (Figure 2) showed that the reduction of dengue viral RNA in the blood meal is gradual, and it takes more than 24 hours of digestion before the viral RNA signal is abolished. Therefore, there is a large time window lasting for hours and viral RNA of sufficient quantity and integrity for sensitive viral detection with molecular assays or NGS-based analysis.
FTA cards have been evaluated for their usefulness in easy preservation and room temperature storage of various biological specimens for the downstream analyses of their protein and nucleic acids contents.11,15–18 Ingredients on FTA cards inactivate potential biohazardous components in the sample applied on the card, allowing for safe and less expensive handling and shipping.16,19 In this study, we tested the technical aspects to verify the feasibility of using FTA cards to carry mosquito blood meal contents. By using the extraction procedure that included beadbeating and QIAamp viral RNA purification, we were able to obtain viral RNA recovery efficiency of approximately 15% from the dried blood spots and 20% or higher from the dried mosquito midgut blots on FTA. In a study by Stangegaard and others,20 four protocols for extraction of DNA from FTA dried blood spots were tested, and their results showed that 50% or greater, depending on which method was used, of DNA contents on FTA cards were not recovered in the first extraction. Interestingly, all these methods used buffer and high-temperature incubation with shaking for 15 minutes to 1 hour to release DNA from FTA cards before DNA extractions.20,21 We speculate beadbeating facilitated breaking up FTA fiber matrices and disrupting the hardened dried blood spots to release viral particles and components, leading to the good yields of nucleic acid from FTA dried blood spots.
Samples in this study were analyzed by qRT-PCR and NGS for comparison. As well established and widely used diagnostic techniques, molecular assays are target based, sensitive, specific and can be quantitative.22 In contrast, NGS is still relatively new for diagnostics use.23 Sequencing-based diagnosis has attracted more and more attention and is believed to be broadly and routinely used in clinical and public health areas in the near future.24,25 However, the comparison of results clearly suggested the complexity of NGS data and the technical challenges and potential risks that need to be recognized and addressed carefully. Defining sensitivity or LOD for a NGS-based pathogen identification can be very difficult. NGS has single-base resolution and is, in theory, extremely sensitive—a single copy of pathogen-specific gene can be amplified, sequenced, and identified by the robust NGS and data mining. When the random amplification and sequencing approach is used, the detection of viral sequences is in proportion to the volume of quality NGS data collected for the analysis. Sensitivity may be defined as the number of viral sequence reads per million reads. As shown in this study, quantitative analysis for NGS data was not necessarily in correlation with the quantification from qRT-PCR (Figure 3, Table 1). For the use of NGS in clinical, epidemiological, and ecological fields, more studies are required to establish standard laboratory protocols, data analyzing tools, and results interpretation guidelines.
NGS analysis using an unbiased approach has been shown to have the advantage of discovery of novel or uncommon viruses. In several published studies, NGS was successfully used in identification of a few sequence reads of a pathogen, allowing subsequent clinical diagnosis made by including additional clinical evidence and/or confirmatory tests.26–28 Our study showed that using recently blood-fed mosquitoes allows for detection of viruses that do not replicate and remain undestroyed inside the mosquito for sufficient and practical periods of time for collection. The procedure will be further tested and applied to field-captured mosquitoes. In addition to pathogen identification, sequence data analysis will be further developed to include genetic identification of mosquito species and host identity. If proven to be successful and informative, the method can be readily applied to other arthropods known to feed on humans or animals.
Conclusions
DENV-2 RNA in blood meal is stable for at least 24 hours in the mosquito midgut and, while it decreases over time, remains detectable for 24 hours after blood feeding. The FTA dried blood spots can be maintained at room temperature for weeks prior to PCR- or NGS-based diagnostics. The strategy has the potential to be used as a high-throughput tool for expedited zoonotic virus discovery and vector-borne disease surveillance.
ACKNOWLEDGMENTS
We thank John S. Lee and Leonard N. Binn for their technical advices and discussions.
Disclaimer: The views expressed here are those of the authors and do not reflect the official policy of the Department of the Army, the Department of Defense, or the U.S. Government.
Footnotes
Financial support: This work was supported by the Global Emerging Infections Surveillance and Response System (GEIS), a Division of the Armed Forces Health Surveillance Center and the Military Infectious Diseases Research Programs (MIDRP).
Authors' addresses: Yu Yang, Jun Hang, and Richard G. Jarman, Viral Diseases Branch, Walter Reed Army Institute of Research, Silver Spring, MD, E-mails: yu.yang2.ctr@mail.mil, jun.hang.civ@mail.mil, and richard.g.jarman.mil@mail.mil. Lindsey S. Garver and Silas A. Davidson, Entomology Branch, Walter Reed Army Institute of Research, Silver Spring, MD, E-mails: lindsey.s.garverbaldwin.ctr@mail.mil and silas.davidson.mil@afrims.org. Karen M. Bingham, Malaria Research Program, Walter Reed Army Institute of Research, Silver Spring, MD, E-mail: karen.m.bingham2.ctr@mail.mil. Ryan C. Jochim, Veterinary Diagnostic Technology, Inc., Wheat Ridge, CO, E-mail: ryan.jochim@gmail.com. Jason H. Richardson, Armed Forces Pest Management Board, Silver Spring, MD, E-mail: jason.h.richardson.mil@mail.mil.
References
- 1.Knope KE, Doggett SL, Kurucz N, Johansen CA, Nicholson J, Feldman R, Sly A, Hobby M, El Saadi D, Muller M, Jansen CC, Muzari OM. Arboviral diseases and malaria in Australia, 2011–12: annual report of the National Arbovirus and Malaria Advisory Committee. Commun Dis Intell Q Rep. 2014;38:E122–E142. doi: 10.33321/cdi.2014.38.21. [DOI] [PubMed] [Google Scholar]
- 2.Tolle MA. Mosquito-borne diseases. Curr Probl Pediatr Adolesc Health Care. 2009;39:97–140. doi: 10.1016/j.cppeds.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Guillaumot L. Arboviruses and their vectors in the Pacific—status report. Pac Health Dialog. 2005;12:45–52. [PubMed] [Google Scholar]
- 4.Johnson N, Voller K, Phipps LP, Mansfield K, Fooks AR. Rapid molecular detection methods for arboviruses of livestock of importance to northern Europe. J Biomed Biotechnol. 2012;2012:719402. doi: 10.1155/2012/719402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van den Hurk AF, Hall-Mendelin S, Johansen CA, Warrilow D, Ritchie SA. Evolution of mosquito-based arbovirus surveillance systems in Australia. J Biomed Biotechnol. 2012;2012:325659. doi: 10.1155/2012/325659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coffey LL, Page BL, Greninger AL, Herring BL, Russell RC, Doggett SL, Haniotis J, Wang C, Deng X, Delwart EL. Enhanced arbovirus surveillance with deep sequencing: identification of novel rhabdoviruses and bunyaviruses in Australian mosquitoes. Virology. 2014;448:146–158. doi: 10.1016/j.virol.2013.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall-Mendelin S, Allcock R, Kresoje N, van den Hurk AF, Warrilow D. Detection of arboviruses and other micro-organisms in experimentally infected mosquitoes using massively parallel sequencing. PLoS One. 2013;8:e58026. doi: 10.1371/journal.pone.0058026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crabtree MB, Kading RC, Mutebi JP, Lutwama JJ, Miller BR. Identification of host blood from engorged mosquitoes collected in western Uganda using cytochrome oxidase I gene sequences. J Wildl Dis. 2013;49:611–626. doi: 10.7589/2012-08-213. [DOI] [PubMed] [Google Scholar]
- 9.Rovie-Ryan JJ, Zainuddin ZZ, Marni W, Ahmad AH, Ambu LN, Payne J. Blood meal analysis of tabanid fly after it biting the rare Sumatran rhinoceros. Asian Pac J Trop Biomed. 2013;3:95–99. doi: 10.1016/S2221-1691(13)60031-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoppenheit A, Steuber S, Bauer B, Ouma EM, Diall O, Zessin KH, Clausen PH. Host preference of tsetse: an important tool to appraise the Nagana risk of cattle in the cotton zone of Mali. Wien Klin Wochenschr. 2010;122(Suppl 3):81–86. doi: 10.1007/s00508-010-1443-9. [DOI] [PubMed] [Google Scholar]
- 11.Grubaugh ND, Sharma S, Krajacich BJ, Fakoli Iii LS, Bolay FK, Diclaro Ii JW, Johnson WE, Ebel GD, Foy BD, Brackney DE. Xenosurveillance: a novel mosquito-based approach for examining the human-pathogen landscape. PLoS Negl Trop Dis. 2015;9:e0003628. doi: 10.1371/journal.pntd.0003628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carissimo G, Pondeville E, McFarlane M, Dietrich I, Mitri C, Bischoff E, Antoniewski C, Bourgouin C, Failloux AB, Kohl A, Vernick KD. Antiviral immunity of Anopheles gambiae is highly compartmentalized, with distinct roles for RNA interference and gut microbiota. Proc Natl Acad Sci USA. 2015;112:E176–E185. doi: 10.1073/pnas.1412984112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paudel D, Jarman R, Limkittikul K, Klungthong C, Chamnanchanunt S, Nisalak A, Gibbons R, Chokejindachai W. Comparison of real-time SYBR green dengue assay with real-time taqman RT-PCR dengue assay and the conventional nested PCR for diagnosis of primary and secondary dengue infection. N Am J Med Sci. 2011;3:478–485. doi: 10.4297/najms.2011.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hang J, Forshey BM, Kochel TJ, Li T, Solorzano VF, Halsey ES, Kuschner RA. Random amplification and pyrosequencing for identification of novel viral genome sequences. JBT. 2012;23:4–10. doi: 10.7171/jbt.12-2301-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanchez S, Sellers H. Biological specimen collection and processing for molecular analysis. Methods Mol Biol. 2015;1247:61–75. doi: 10.1007/978-1-4939-2004-4_5. [DOI] [PubMed] [Google Scholar]
- 16.Liang X, Chigerwe M, Hietala SK, Crossley BM. Evaluation of Fast Technology Analysis (FTA) Cards as an improved method for specimen collection and shipment targeting viruses associated with Bovine Respiratory Disease Complex. J Virol Methods. 2014;202:69–72. doi: 10.1016/j.jviromet.2014.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gan W, Zhuang B, Zhang P, Han J, Li CX, Liu P. A filter paper-based microdevice for low-cost, rapid, and automated DNA extraction and amplification from diverse sample types. Lab Chip. 2014;14:3719–3728. doi: 10.1039/c4lc00686k. [DOI] [PubMed] [Google Scholar]
- 18.Awad F, Baylis M, Jones RC, Ganapathy K. Evaluation of Flinders Technology Associates cards for storage and molecular detection of avian metapneumoviruses. Avian Pathol. 2014;43:125–129. doi: 10.1080/03079457.2014.885114. [DOI] [PubMed] [Google Scholar]
- 19.Abdelwhab EM, Luschow D, Harder TC, Hafez HM. The use of FTA(R) filter papers for diagnosis of avian influenza virus. J Virol Methods. 2011;174:120–122. doi: 10.1016/j.jviromet.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 20.Stangegaard M, Borsting C, Ferrero-Miliani L, Frank-Hansen R, Poulsen L, Hansen AJ, Morling N. Evaluation of four automated protocols for extraction of DNA from FTA cards. J Lab Autom. 2013;18:404–410. doi: 10.1177/2211068213484472. [DOI] [PubMed] [Google Scholar]
- 21.Dauner AL, Gilliland TC, Jr, Mitra I, Pal S, Morrison AC, Hontz RD, Wu SJ. Evaluation of nucleic acid stabilization products for ambient temperature shipping and storage of viral RNA and antibody in a dried whole blood format. Am J Trop Med Hyg. 2015;93:46–53. doi: 10.4269/ajtmh.15-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Josko D. Molecular virology in the clinical laboratory. Clin Lab Sci. 2010;23:231–236. [PubMed] [Google Scholar]
- 23.Firth C, Lipkin WI. The genomics of emerging pathogens. Annu Rev Genomics Hum Genet. 2013;14:281–300. doi: 10.1146/annurev-genom-091212-153446. [DOI] [PubMed] [Google Scholar]
- 24.Bibby K. Metagenomic identification of viral pathogens. Trends Biotechnol. 2013;31:275–279. doi: 10.1016/j.tibtech.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 25.Xie G, Yu J, Duan Z. New strategy for virus discovery: viruses identified in human feces in the last decade. Sci China Life Sci. 2013;56:688–696. doi: 10.1007/s11427-013-4516-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson MR, Naccache SN, Samayoa E, Biagtan M, Bashir H, Yu G, Salamat SM, Somasekar S, Federman S, Miller S, Sokolic R, Garabedian E, Candotti F, Buckley RH, Reed KD, Meyer TL, Seroogy CM, Galloway R, Henderson SL, Gern JE, DeRisi JL, Chiu CY. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med. 2014;370:2408–2417. doi: 10.1056/NEJMoa1401268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palacios G, Druce J, Du L, Tran T, Birch C, Briese T, Conlan S, Quan PL, Hui J, Marshall J, Simons JF, Egholm M, Paddock CD, Shieh WJ, Goldsmith CS, Zaki SR, Catton M, Lipkin WI. A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med. 2008;358:991–998. doi: 10.1056/NEJMoa073785. [DOI] [PubMed] [Google Scholar]
- 28.Yolken RH, Jones-Brando L, Dunigan DD, Kannan G, Dickerson F, Severance E, Sabunciyan S, Talbot CC, Jr, Prandovszky E, Gurnon JR, Agarkova IV, Leister F, Gressitt KL, Chen O, Deuber B, Ma F, Pletnikov MV, Van Etten JL. Chlorovirus ATCV-1 is part of the human oropharyngeal virome and is associated with changes in cognitive functions in humans and mice. Proc Natl Acad Sci USA. 2014;111:16106–16111. doi: 10.1073/pnas.1418895111. [DOI] [PMC free article] [PubMed] [Google Scholar]