Microtubule-dependent long-range motility of early endosomes supports directed motility of peroxisomes, lipid droplets, and endoplasmic reticulum, and this process is mediated by transient interaction between all three organelles and the endosomes.
Abstract
Intracellular transport is mediated by molecular motors that bind cargo to be transported along the cytoskeleton. Here, we report, for the first time, that peroxisomes (POs), lipid droplets (LDs), and the endoplasmic reticulum (ER) rely on early endosomes (EEs) for intracellular movement in a fungal model system. We show that POs undergo kinesin-3– and dynein-dependent transport along microtubules. Surprisingly, kinesin-3 does not colocalize with POs. Instead, the motor moves EEs that drag the POs through the cell. PO motility is abolished when EE motility is blocked in various mutants. Most LD and ER motility also depends on EE motility, whereas mitochondria move independently of EEs. Covisualization studies show that EE-mediated ER motility is not required for PO or LD movement, suggesting that the organelles interact with EEs independently. In the absence of EE motility, POs and LDs cluster at the growing tip, whereas ER is partially retracted to subapical regions. Collectively, our results show that moving EEs interact transiently with other organelles, thereby mediating their directed transport and distribution in the cell.
Introduction
Peroxisomes (POs), lipid droplets (LDs), and the ER perform important cellular functions in fatty acid metabolism and lipid homoeostasis (Wanders and Waterham, 2006; Hashemi and Goodman, 2015). They are randomly distributed in the cell but interact with each other for metabolic purposes, during their biogenesis, for lipid transfer and signal transduction (Kohlwein et al., 2013; Schrader et al., 2015). In mammals, ER tubules, POs, and LDs are highly dynamic, undergoing random motion and directed motility along microtubules (MTs) that is mediated by kinesin and dynein (Rapp et al., 1996; Wiemer et al., 1997; Waterman-Storer and Salmon, 1998; Targett-Adams et al., 2003; Kural et al., 2005; Larsen et al., 2008; Shubeita et al., 2008; Woźniak et al., 2009). To mediate intracellular transport, motors bind directly to their cargo (Kamal and Goldstein, 2002; Karcher et al., 2002; Welte, 2009; Akhmanova and Hammer, 2010; Fu and Holzbaur, 2014). Little is known about PO, LD, and ER motility in filamentous fungi. In Aspergillus nidulans, PO motility depends on MTs and associated motors (Egan et al., 2012). Interestingly, deletion of kinesin-3 results in clustering of POs at apical MT plus ends. A similar accumulation is found in the absence of HookA, a motor adapter that is located on early endosomes (EEs; Zhang et al., 2014). How deletion of a plus-end–directed kinesin, or its adapter on EEs, drives POs to MT plus ends is not clear.
In this study, we set out to elucidate the transport mechanism underlying PO, LD, and ER motility in the fungal model U. maydis. We report that all three organelles “hitchhike” on moving EEs that are transported by kinesin-3 and dynein. This mechanism spatially organizes POs, LDs, and ER in fungal cells.
Results and discussion
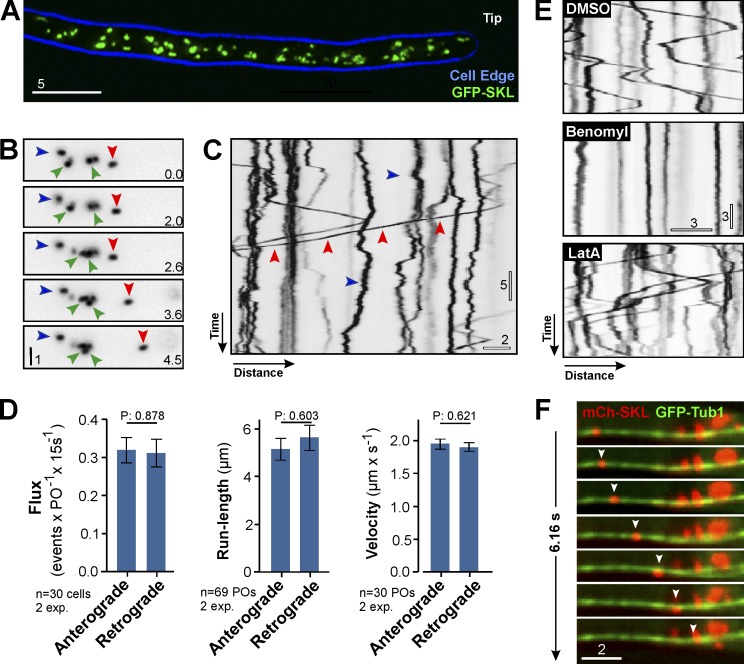
POs move long distances along MTs
We fused GFP, or mCherry, to a peroxisomal targeting signal SKL (GFP-SKL and mCherry-SKL). When coexpressed with a peroxisomal Pex3 homolog (Camões et al., 2015), the PO marker colocalized in POs (Fig. S1 A). The POs were evenly scattered in the hyphal cell (Fig. 1 A), with a majority undergoing nondirected short-range motility (Fig. 1, B and C, blue arrowheads) that led to occasional interaction between POs (Fig. 1 B, green arrowheads). Such behavior has been described in mammalian cells, where it is thought to allow lipid exchange between the organelles (Bonekamp et al., 2012). In U. maydis, in a time window of 15 s, 13.40 ± 1.93% (n = 30 cells) of the POs showed rapid and bidirectional motility over 5.50 ± 3.76 µm (n = 138; Fig. 1 C, red arrowheads; and Video 1), as is reminiscent in mammalian cells (Rapp et al., 1996; Wiemer et al., 1997). We compared motility toward the hyphal tip (anterograde) and away from the tip (retrograde) and found that neither flux nor the run-length or the velocity of PO motility differed between directions (Fig. 1 D; all, P > 0.6; Student’s t test; n = 30 cells). We tested for a role of the cytoskeleton in PO motility by applying the F-actin inhibitor latrunculin A and the MT inhibitor benomyl, which both are effective in U. maydis (Fuchs et al., 2005). Although the solvent DMSO and latrunculin A had no effect (Fig. 1 E, DMSO and LatA), directed PO motility stopped when MTs were absent (Fig. 1 E, Benomyl). This suggests that PO motility occurs along MTs. We tested this further by colocalizing mCherry-SKL–containing POs and MTs, labeled with GFP-αtubulin (GFP-Tub1; Steinberg et al., 2001). Indeed, long-range PO motility occurred exclusively along MTs (Fig. 1 F, arrowheads highlight moving PO; and Video 2). We conclude that PO motility is MT based. This is reminiscent of animal cells but different from budding yeast and plants, where PO motility is actin based (Hoepfner et al., 2001; Mathur et al., 2002).
Figure 1.
POs move along MTs in U. maydis. (A) POs in U. maydis. 2D-deconvolved maximum projection of a Z-axis stack, adjusted in brightness, contrast and gamma settings. Bar, 5 µm. (B) Dynamics of POs. POs are stationary (blue arrowheads), undergo short motions (green arrowheads), or switch to directed motility (red arrowheads). Images were contrast inverted and adjusted in brightness, contrast, and gamma settings. Bar, 1 µm. Time indicated in lower right corner. (C) Motility of POs. Most POs show random motion (blue arrowhead); few undergo directed motility (red arrowheads). Image contrast inverted. Bars, 5 s, 2 µm. See Video 1. (D) Flux, run length, and velocity of bidirectionally moving POs. Bars are mean ± SEM (n = 30–69 cells, two experiments). P-values of Student’s t tests shown above the bars. (E) Motility of POs in cells treated with DMSO, benomyl, or latrunculin A (LatA). PO motility stops when MTs are disrupted. Image contrast inverted. Bars, 3 s, 3 µm. (F) Motility of a PO (red; arrowhead) along a MT (green). Image series covers 6.16 s. Images adjusted in brightness, contrast, and gamma settings. Bar, 2 µm. See Video 2.
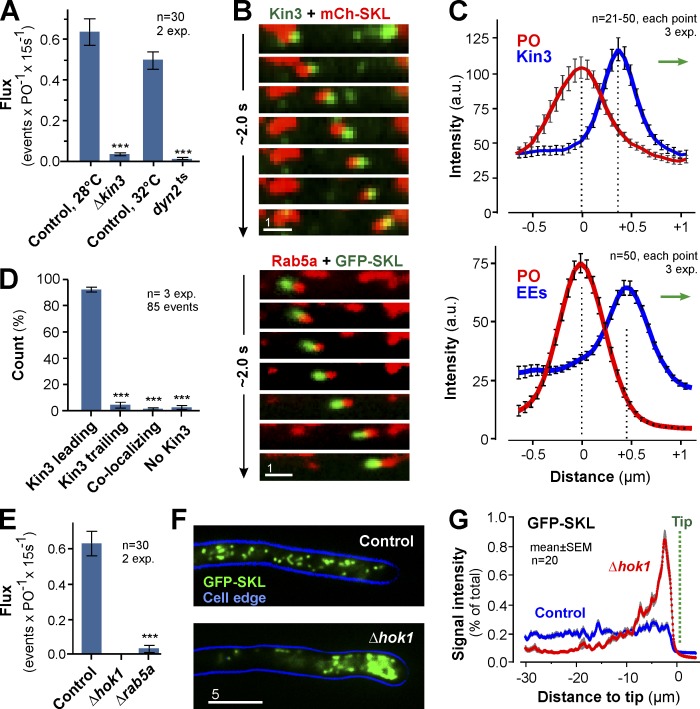
PO motility depends on EE transport
In A. nidulans, kinesin-3 and dynein support PO motility (Egan et al., 2012). We expressed GFP-SKL in a kinesin-3 null mutant (Δkin3) and a temperature-sensitive dynein mutant (dyn2ts) and found that directed PO motility was almost abolished in each mutant (Fig. 2 A). Thus, PO motility in U. maydis depends on kinesin-3 and dynein. We further tested this by colocalizing GFP-labeled Kin3 and mCherry-SKL–carrying POs. Indeed, both signals cotraveled in the cell (Fig. 2 B, top). Surprisingly, however, Kin3 usually did not localize on the organelles but rather led the moving POs (Fig. 2 C, top graph; and Fig. 2 D). In U. maydis, kinesin-3 mediates motility of EEs (Wedlich-Söldner et al., 2002; Schuster et al., 2011; Steinberg, 2014). This raised the possibility that EE-associated kinesin-3 led PO motions. We tested this by covisualization of EEs, labeled with mCherry fused to the EE-specific GTPase Rab5a (mCh-Rab5a; Fuchs et al., 2006), and GFP-SKL-containing POs. Indeed, EEs led in 97.37 ± 2.35% of all PO motions (n = 3 experiments, 85 events; Fig. 2, B and C, bottom; and Video 3). Furthermore, when EE motility was abolished in (a) mutants deleted in the EE-specific motor adapter hok1 (Bielska et al., 2014), (b) rab5a null mutants (Bielska et al., 2014), or (c) a yup1ts mutant that has reduced EE numbers because of defects in fusion with incoming transport vesicles (Wedlich-Söldner et al., 2000), directed PO motility also stopped (Figs. 2 E and S1 B). These results strengthen the conclusion that EE motility supports directed transport of POs. We further tested the involvement of EEs in PO motility by using a truncated Hok1 protein that lacks the C-terminal domain (Hok1ΔC). Hok1 is an adapter for motors on EEs, and it interacts with EEs via its C-terminal domain (Bielska et al., 2014). Hok1ΔC is unable to bind EEs and, when expressed in hok1 deletion mutants (Δhok1), did not restore EE motility (Fig. S1 C). However, when fused to a Phox domain, which targets the protein to phosphatidylinositol 3-phosphate–enriched EEs (Lemmon, 2003; Bielska et al., 2014), the resulting protein Hok1ΔC-PX protein restored EE motility (Fig. S1 C; Bielska et al., 2014). Consistent with a role of EEs in PO motility, PO motility was also restored (Fig. S1, C and D). Collectively, we provide several lines of evidence for an active involvement of EEs in PO motility: (a) in colocalization experiments, kinesin-3 and EEs lead PO motions; (b) the absence of EE motility in Δhok1, Δrab5a, or yup1ts mutants abolishes PO motility; and (c) when a nonfunctional truncated Hok1 motor adapter is targeted to EEs, it rescues EEs and PO motility. Interestingly, POs accumulated at the hyphal tip in Δhok1 (Fig. 2, F and G), and they redistribute in the presence of Hok1ΔC-PX (Fig. S1 E). This suggests that EE-mediated motility is required for even distribution of POs.
Figure 2.
EEs drive PO motility. (A) Flux of POs in controls, kinesin-3 null mutants (ΔKin3), and dynein mutants (Dyn2ts). Bars are mean ± SEM (n = 30 cells, 2 experiments). ***, P < 0.0001 versus control (Student’s t test). (B) Co-motility of a PO (top, red) and GFP-fusions to kinesin-3 (Kin3; top, green) and co-motility of a PO (green) and EE-associated mCherry-Rab5a (Rab5a; red). Image series covers 2.0 s. Images adjusted in brightness, contrast and gamma settings. Bar, 1 µm. See Video 3. (C) Intensity profiles of a co-migrating PO (red) and Kin3-GFP (Kin3; top, blue line) or EEs (bottom, blue line). Direction of transport indicated by green arrows. Data points are mean ± SEM of 21–50 cells from three experiments. (D) Position of Kin3 signals relative to co-moving POs. Bars are mean ± SEM (n = 3 experiments, 85 co-motility events). ***, P < 0.0001 versus control (Student’s t test). (E) Bidirectional flux of PO motility in control, Δhok1, and Δrab5a mutant. EE motility is almost abolished in both mutants (Bielska et al., 2014,). Bars are mean ± SEM (n = 30 cells, two experiments). ***, P < 0.0001 versus control (Student’s t test). (F) Distribution of POs, labeled with GFP-SKL, in control and Δhok1. POs form apical clusters in the absence of EE motility. Cell edge is indicated in blue. 2D-deconvolved maximum projection of a Z-axis stack, adjusted in brightness, contrast and gamma settings. Bar, 5 µm. (G) Fluorescent intensity profiles of GFP-SKL in control and Δhok1 cells. Position of cell tips is indicated (Tip). Each data point represents the mean ± SEM (20 cells, two experiments).
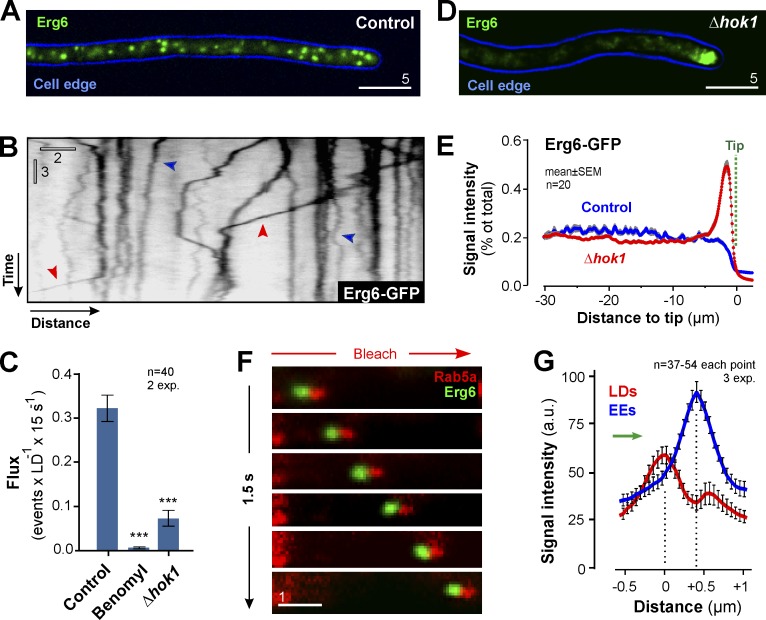
Motility of LDs, but not mitochondria, depends on EE transport
We next asked if motile EEs support motility of other organelles. LDs undergo directed MT-based transport in mammalian cells (Targett-Adams et al., 2003; Welte, 2009). To investigate LDs in U. maydis, we identified an U. maydis homologue of Erg6p, a sterol-24-C-methyltransferase in LDs in Saccharomyces cerevisiae (Zinser et al., 1993; Pu et al., 2011). U. maydis Erg6 and Erg6p share 39.4% amino acid sequence identity and a similar domain structure (Fig. S1, F and G). In addition, U. maydis Erg6 colocalizes with the neutral lipid dye LipidTOX in LDs (Fig. S1 H). We fused GFP to Erg6 and expressed the fusion protein in U. maydis. Erg6-GFP–positive LDs scattered along the length of the hyphal cell (Fig. 3 A), which corresponded well with LipidTOX staining in wild-type cells. In some cells, Erg6-GFP carrying LDs clustered in subapical regions, which was considered to be an artifact. Most LDs showed random motion (Fig. 3 B, blue arrowheads), but 7.26 ± 5.55% of all LDs underwent directed bidirectional motility (n = 30 cells; Fig. 3 B, red arrowheads; and Video 4), with an average run length of 4.78 ± 3.61 µm (n = 102 experiments, 30 cells). Such behavior was previously reported in mammalian cells (Targett-Adams et al., 2003). Similar to POs, directed movement of LDs was blocked when MTs were absent or in Δhok1 (Fig. 3 C), which led to polar accumulation of LDs in hyphal tips (Fig. 3, D and E). We covisualized mCherry-Rab5a–labeled EEs and Erg6-GFP–containing LDs and found that most organelles moved in pairs (63.64 ± 21.05%, n = 3 experiments, 78 LDs) with the EE leading (Fig. 3, F and G; and Video 5). We conclude that LDs also hitchhike on motile EEs. Interestingly, neither run length (P = 0.889.8, Student’s t test) nor distance to co-moving EEs differs significantly in POs and LDs (EEs-POs: 0.48 ± 0.15 nm, n = 131; EEs-LDs: 0.39 ± 0.34 nm, n = 94; P = 0.8453, Student’s t test; see Figs. 2 C and 3 G).
Figure 3.
EEs drive LD motility. (A) LDs in U. maydis. 2D-deconvolved maximum projection of a Z-axis stack, adjusted in brightness, contrast, and gamma settings. Bar, 5 µm. (B) Dynamic behavior of LDs. Organelles are stationary (blue arrowhead) or undergo random motility. Occasionally, directed long-range motility occurs (red arrowhead). Image was contrast inverted. Bars, 3 s, 2 µm. See Video 4. (C) Flux of LD motility in the presence of DMSO (control) or benomyl, and in Δhok1. LD motility depends on MTs and the EE-specific motor adapter. All bars are mean ± SEM (n = 40 cells, 2 experiments). ***, P < 0.0001 versus control (Student’s t test). (D) LDs in a Δhok1. 2D-deconvolved maximum projection of a Z-axis stack, adjusted in brightness, contrast, and gamma settings. Bar, 5 µm. (E) Fluorescent intensity profiles of Erg6-GFP in control and Δhok1 cells. Position of cell tips is indicated (Tip). Each data point represents the mean ± SEM (20 cells, two experiments). (F) Co-motility of EEs, labeled with mCherry-Rab5a (red) and LDs, labeled with Erg6-GFP (green). Background interference was reduced by photobleaching (red arrow). Image series covers ∼1.55 s and is adjusted in brightness, contrast, and gamma settings. Bar, 1 µm. See Video 5. (G) Intensity profiles of a co-migrating LD (red) and EEs (blue line). Direction of transport indicated by green arrow. Data points are mean ± SEM (n = 37–54 cells, three experiments).
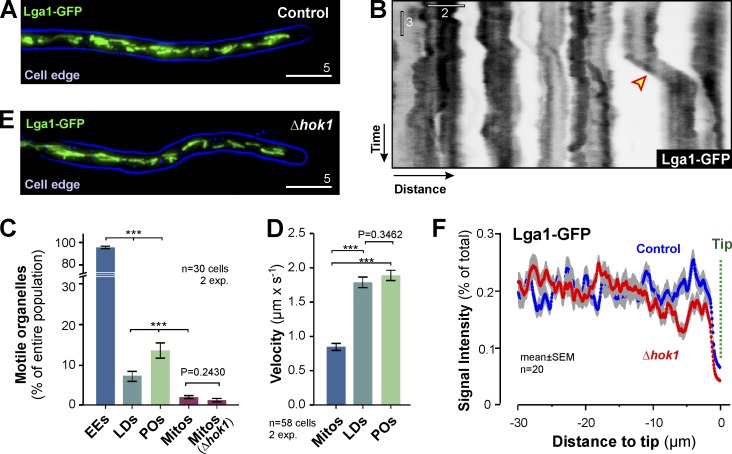
We next investigated the motility of mitochondria labeled with a fusion of GFP and mitochondrial protein Lga2 (Bortfeld et al., 2004). Mitochondria were evenly distributed within the hyphal cell (Fig. 4 A). In contrast to EEs, POs, or LDs, mitochondria showed rare short-range motility (Fig. 4, B and C), which was of significantly lower velocity (Fig. 4 D). In the absence of EE motility in Δhok1, neither the motility (Fig. 4 C) nor the distribution of mitochondria was remarkably affected (Fig. 4, E and F). Thus, we conclude that the rare mitochondrial motions are mediated by an EE-independent mechanism.
Figure 4.
Motility of mitochondria does not depend on EEs. (A) Distribution of mitochondria, labeled with Lga1-GFP, in U. maydis. 2D-deconvolved maximum projection of a Z-axis stack, adjusted in brightness, contrast, and gamma settings. Bar, 5 µm. (B) Contrast-inverted kymograph showing the dynamic behavior of mitochondria. The organelles show rare short-range motility (arrowhead). Bars, 3 s, 2 µm. (C) EE, PO, LD and mitochondria (Mitos) motility in U. maydis, given as the percentage of organelles that show motility within 15s observation time. Note that POs, LDs and mitochondria switch between stationary and directed transport, whereas EEs are constantly moving. All bars are mean ± SEM (n = 30 cells, two experiments). ***, P < 0.0001 versus control (Student’s t test). No difference in mitochondria motility was found in Δhok1 mutants (P = 0.2430; Student’s t test). (D) Velocity of bidirectionally moving mitochondria, LDs, and POs. Bars are mean ± SEM (n = 58 cells, two experiments). ***, P < 0.0001 (Student’s t test). P-value for nonsignificant difference shown above bars. (E) Mitochondria in a Δhok1 cell. 2D-deconvolved maximum projection of a Z-axis stack, adjusted in brightness, contrast, and gamma settings. Bar, 5 µm. (F) Fluorescent intensity profiles of mitochondria, labeled with Lga1-GFP, in control and Δhok1 mutant cells. The location of the hyphal tips is indicated (Tip). Each data point represents the mean ± SEM of 20 cells.
Organelle hitchhiking involves neither late endosomes nor the endosome-associated RNA-binding protein Rrm4
In mammalian cells, the Rab7-positive late endosomes (LEs) show rapid MT-dependent motility (Lebrand et al., 2002). U. maydis contains Rab7-positive LEs (Higuchi et al., 2014), and we tested for an involvement of these in PO and LD motility by expressing GFP-SKL and Erg6-GFP in Δrab7 mutants. We found that motility of POs and LDs was not significantly reduced (Fig. S2, A and B). This suggests that LEs are not involved in their motility.
On average, POs run for ∼5 µm (Fig. 1 D), whereas the average run-length of EEs is ∼22 µm (Schuster et al., 2011). This suggests that POs transiently interact with moving EEs. We tested this by co-observing mCherry-SKL–containing POs and GFP-Rab5a–positive EEs. We found that stationary POs begin their directed motility by interacting with moving EEs (Fig. S2 C, red arrowheads show EE trajectory). After detachment from the EEs, POs usually paused (Fig. S2 C, green arrowhead) before they interacted with other EEs, which could result in a turning of transport direction (Fig. S2 C, blue arrowheads). Thus, POs transiently bind to and are released from moving EEs. Such a mechanism was previously reported for polyribosomes in U. maydis (Higuchi et al., 2014), which link to EEs via the RNA-binding protein Rrm4 (Becht et al., 2006). We tested if Rrm4 is involved in the association of POs to EEs by investigating PO motility in Δrrm4 mutants. We found PO motility (Fig. S2, D and E) and co-migration with EEs (Fig. S2 F, arrowheads indicate trajectory of PO) in the absence of Rrm4. These results suggest that Rrm4 is not the linker between POs and EEs.
EEs support motility of the endoplasmic reticulum
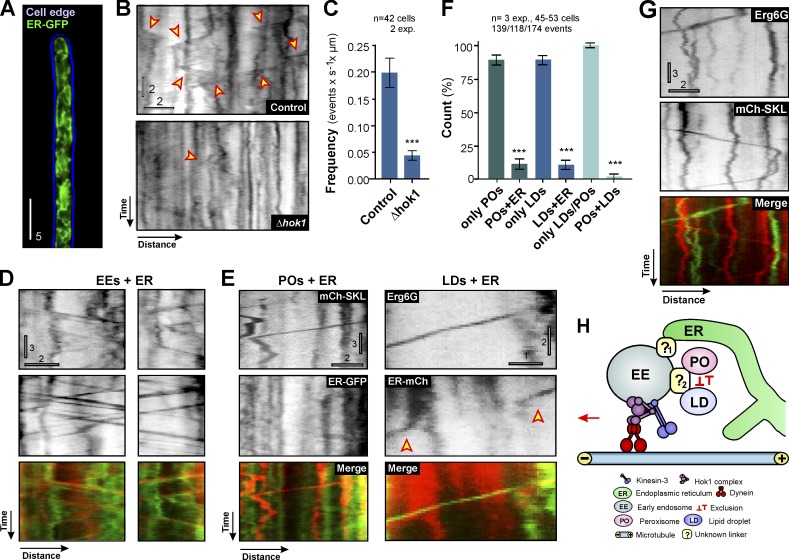
The ER plays a central role in biogenesis of POs and LDs. It also interacts with both organelles owing to its central role in lipid metabolism (Barbosa et al., 2015; Gao and Goodman, 2015; Shai et al., 2015). Furthermore, recent results show a significant interaction between ER and EEs during their maturation (Friedman et al., 2013). This raises the possibility that PO and LD motility could be mediated by EE-driven motility of the ER. We investigated this by labeling the ER using a specific fluorescent probe (ER-GFP, which is a fusion of EGFP and the ER-retention signal HDEL; Wedlich-Söldner et al., 2002). The ER was evenly distributed within the entire cell (Fig. 5 A), and ER tubules showed short-range directed motility (Fig. 5 B, arrowheads; and Video 6). The frequency of ER motility was significantly reduced in Δhok1 (Fig. 5, B [bottom] and C), suggesting that moving EEs mediate most ER movements. This conclusion was further supported by the finding that 85.01 ± 4.64% of all ER motions correlate with EE motility (n = 3 experiments, 109 events; Fig. 5 D and Videos 7 and 8), with EEs leading the EE-ER pairs in 85.00 ± 3.30% of all cases (n = 3 experiments, 52 events). We conclude that most ER tubule motions are mediated by EE motility.
Figure 5.
EE motility drives ER, PO, and LD motility independently. (A) ER in U. maydis. 2D-deconvolved maximum projection of a Z-axis stack, adjusted in brightness, contrast, and gamma settings. Bar, 5 µm. (B) Contrast-inverted kymographs showing ER motility (arrowheads) in control and Δhok1. Bars, 2 s, 2 µm. See Video 6. (C) Frequency of ER motility in control and Δhok1 mutants. Bars are mean ± SEM (n = 42 cells, 2 experiments). ***, P < 0.0001 versus control (Student’s t test). (D) Examples of co-motility of EEs (mCh-Rab5a) and ER (ER-GFP). Note that EEs lead the ER during co-motility. Top kymographs contrast inverted. Bars, 3 s, 2 µm. (E) ER-independent motility of POs (mCh-SKL) and LDs (Erg6-GFP). ER was labeled by ER-mCherry (ER-mCh) or ER-GFP. Note that right kymographs show transient interaction of ER (arrowheads) with a moving LD. Upper kymographs contrast inverted. Bars: (left) 3 s, 2 µm; (right) 2 s, 1 µm. (F) Graph showing the degree of co-motility among ER, LDs, and POs. Bars are mean ± SEM (n = 3 experiments, 45–53 cells). ***, P < 0.0001 versus control (Student’s t test). (G) Motility of POs (mChSKL) and LDs (Erg6G) in the same cell. Top kymographs contrast inverted. Bars, 3 s, 2 µm. (H) Proposed role of EEs in motility of POs, LDs and ER. ER interacts with EEs independently of POs and LDs, which appear to exclude each other during motility. The mode of interaction with EEs is unknown, but linker proteins are likely to exist (?1 and ?2). Most EEs are constantly moving because of the activity of dynein and kinesin-3, bound to the Hok1 adapter. Transient interaction with the organelles helps PO, LD, and ER distribution and may foster interorganelle communication.
We tested for a role of ER in bridging EEs and POs/LDs by covisualization of ER and POs/LDs during their motility in the cell. We found that 88.65 ± 6.37% (n = 3 experiments, 139 events) of the PO and 89.21 ± 6.14% (n = 3 experiments, 118 events) of the LD movements occur without corresponding ER motility (Fig. 5, E and F). If ER mediates contact between leading EEs and POs/LDs, each motion of POs/LDs should be accompanied by ER motility. Thus, we consider it highly unlikely that ER bridges between EEs and POs/LDs. However, we occasionally observed transient interaction of ER with moving organelles (Fig. 5 E, right, arrowheads indicate ER motility). This suggests that moving EE-PO/LD pairs collide with ER and passively drag membrane tubules. This may account for the ∼10% co-migration between ER and POs/LDs found in our experiments (Fig. 5 F).
Finally, we asked if POs and LDs simultaneously bind to EEs. LDs and POs are known to interact physically in mammalian cells (Schrader et al., 2015). We found such transient interaction when covisualizing both organelles in the same cell (Fig. S3 A). However, POs and LDs did not show co-motility (98.00 ± 3.46%, n = 3 experiments, 174 events; Fig. 5, F and G). Thus, POs and LDs appear to interact independently with moving EEs (Fig. 5 H).
We show here that motility of POs, LDs, and ER tubules depends on EE transport. Several lines of evidence support this conclusion: (a) the motility of these organelles significantly drops in Δhok1, where EE motility is abolished; (b) EEs and ER/POs/LDs move in pairs, with EEs usually leading; and (c) the even distribution of all organelles is affected in Δhok1. But is EE motility prominent enough to mediate motility of the other organelles? Together, POs and LDs slightly outnumber EEs in the cell (Fig. S3 B). However, given the disparity between the proportion of moving EEs (∼96%; Fig. 4 C), as compared with more rare motility of LDs and POs (together ∼21%, Fig. 4 C), coupled with the difference in run length (∼22 µm for EEs vs. ∼5 µm for POs and LDs), EEs have the capacity to move both compartments.
In conclusion, our results strongly suggest that moving EEs mediate motility of POs, LDs, and ER. We also show that this motility is required for cellular distribution of LDs, POs, and ER. Whereas POs and LDs cluster at the tip, ER shifts toward subapical parts when EE motility was abolished (Fig. S3, C and D). This reflects findings in neurons, where ER retracts when kinesin-1 activity is impaired (Feiguin et al., 1994). We currently do not know why POs, LDs, or ER are moving. Motility might be essential to permeate all parts of the cell and to deal with local lipid or fatty acid breakdown requirements or detoxification of reactive oxygen species (Schrader et al., 2013; Shai et al., 2015). In addition, POs, LDs, and ER motility could play an important role in their cooperative interplay in regulation of cellular lipid homeostasis (Chu et al., 2015; Schrader et al., 2015). Finding physiological conditions under which motility is increased could help understanding the role of this organelle motility.
In mammals, moving endosomes support various functions, including long-range signaling, cytokinesis, cell polarization and cell migration (Miaczynska et al., 2004; Gould and Lippincott-Schwartz, 2009). We do not know if mammalian EE are also involved in a “piggy-back” mechanism of organelle transport. In fungi, however, the mechanism of organelle hitchhiking may be conserved, as POs accumulate in A. nidulans mutants, deleted in their EE-associated hook motor adapter (Zhang et al., 2014). Fungal EEs support long-range signaling (Bielska et al., 2014) and septin delivery (Baumann et al., 2014) and distribute the protein translation machinery (Baumann et al., 2012; Higuchi et al., 2014). How EEs perform such functions simultaneously is not understood. POs and LDs share a similar run length and distance to co-moving EEs. Furthermore, they exclude each other during motion. Thus, they may share a common linker on EEs (Fig. 5 H). Identifying such cargo adapters will be key to understand the role of EEs as “master organizers” of the fungal cell.
Materials and methods
Molecular cloning and strain generation
U. maydis transformation techniques are described elsewhere (Schulz et al., 1990). All plasmids were generated by standard cloning techniques or in vivo recombination in the S. cerevisiae strains DS94 (MATα ura3-52 trp1-1 leu2-3 his3-111 lys2-801; Tang et al., 1996; Raymond et al., 1999) and FY834 (MATα his3 Δ200 ura3-52 leu2Δ1 lys2Δ202 trp1Δ63; Winston et al., 1995; Knop et al., 1999). Strains AB33GRab5, AB33ERG, and AB33LgaG were published previously (Wedlich-Söldner et al., 2000; Steinberg and Schuster, 2011). Newly generated strains are described below. Their genotypes and introduced plasmids are summarized in Table S1, their experimental usage is detailed in Table S2, and all sequences of cloning primers are provided in Table S3.
AB33GSKL
The strain allowed visualization of GFP-labeled POs in hyphal cells of U. maydis. poCGFP-SKL (Steinberg and Schuster, 2011) was digested using EcoRV and ectopically integrated into the strain AB33 (Brachmann et al., 2001), resulting in AB33GSKL.
AB33GTub1_mChSKL
The strain allowed visualization of PO motility along MTs. poHmChSKL was linearized with EcoRV and ectopically integrated into AB33GTub1 (Schuster et al., 2011), resulting in strain AB33GTub1_mChSKL. poHmCh-SKL encodes the PO marker mCh-SKL, expressed under the constitutive otef promoter (Spellig et al., 1996). It carries a hygromycin resistance cassette.
AB33ΔKin3_GSKL
In this strain, kinesin-3 is deleted and POs are labeled by GFP-SKL. It was obtained by linearizing poHGSKL with KpnI and ectopically integrated in AB33ΔKin3 (Higuchi et al., 2014). The plasmid was obtained by digestion of poCGSKL (Steinberg and Schuster, 2011) with SphI and NdeI, and insertion of a hygromycin cassette (2,642 bp) by ligation resulting in poHGSKL.
AB5Dyn2ts_GSKL
This strain was used for motility studies of POs in the absence of functional dynein. The strain was obtained by ectopic integration of poHGSKL, linearized with EcoRV, into strain AB5Dyn2ts.
AB33Kin3G_mChSKL
This strain allowed covisualization of Kin3 and POs was obtained by ectopic integration of poCmChSKL, digested with SspI, into AB33Kin3G (Schuster et al., 2011).
AB33mChRab5_GSKL
This strain allowed the observation of PO co-migration with EEs that carried mChRab5a. It was obtained by ectopic integration of plasmid poHGSKL, linearized with AgeI, into strain AB33mChRab5a (Bielska et al., 2014).
AB33GRab5a_mChSKL
To allow visualization of EEs and POs, mCherry-SKL was introduced into AB33GRab5a (Schuster et al., 2011). poCmChSKL was digested with EcoRV and ectopically integrated into AB33GRab5a.
AB33GRab5ΔHok1_mChSKL
This strain allowed motility analysis of EEs, labeled with GFP-Rab5a, and POs in a Δhok1 background. It was obtained by linearizing poGmChSKL with ScaI, followed by ectopic integration into AB33GRab5ΔHok1 (Bielska et al., 2014). The plasmid poGmchSKL was obtained by digesting poCmCh-SKL, using KpnI and PshAI, and subsequent replacement of the carboxin cassette for a neomycin resistance (G418).
AB33ΔRab5a_Yup1G_mChSKL
This strain was used to determine the motility of POs in the absence of the small endosomal GTPase Rab5a. It was obtained by ectopic integration of poGmChSKL, digested with ScaI, into AB33ΔRab5a_Yup1G (Bielska et al., 2014).
AB33ΔHok1_GSKL
This strain allowed motility analysis of POs in Δhok1 mutant. It was generated by ectopic integration of poCGSKL, digested with EcoRV into AB33ΔHok1 (Bielska et al., 2014).
AB33_Erg6G
This strain contains the LD marker Erg6-GFP. It was obtained by integration of AgeI-digested poErg6G into the succinate dehydrogenase locus of strain AB33. The gene for the Erg6 homolog was amplified from genomic DNA of U. maydis strain 521 using primers BM3 and BM4. The poNLSNESG, containing a carboxin resistance cassette, otef promoter, egfp, and nos terminator, was digested with MscI and HindIII, and the full-length erg6 gene (XP_011389634) was cloned by yeast recombination.
AB33ΔHok1_Erg6G
To study LD dynamics and distribution in the absence of Hok1, poErg6G was digested with AgeI and integrated into the succinate dehydrogenase locus of AB33ΔHok1 (Bielska et al., 2014).
AB33mChRab5_Erg6G
This strain allows covisualization of EEs and LDs. It was obtained by integration of AgeI-digested plasmid poErg6G into the succinate dehydrogenase locus of AB33mChRab5 (Bielska et al., 2014).
AB33LgaG_mChSKL_ΔHok1
To observe distribution of mitochondria in ΔHok1, pNΔHok1 plasmid (Bielska et al., 2014) was linearized with AlwNI and HpaI and integrated into AB33LgaG_mChSKL, resulting in AB33LgaG_mChSKL_ΔHok1. The strain AB33LgaG_mChSKL was generated by ectopical integration of poCmChSKL, digested with EcoRV, into AB33LgaG (Steinberg and Schuster, 2011). The correct deletion of Hok1 was confirmed by Southern blot.
AB33ΔHok1_ERG
To study ER dynamics and distribution in the absence of Hok1, poERG (Wedlich-Söldner et al., 2002) was digested with AgeI and integrated into the succinate dehydrogenase locus of AB33ΔHok1 (Bielska et al., 2014).
AB33ERG_mChRab5a
To perform co-migration studies of EE and ER, plasmid pomChRab5a (Schuster et al., 2011) was digested with ScaI and transformed into strain AB33ERG (Wedlich-Söldner et al., 2002), resulting in the strain AB33ERG_mChRab5a.
AB33ERG_mChSKL
For colocalization studies of ER and POs, strain AB33ERG_mChSKL was generated by ectopical integration of plasmid pGmChSKL, digested with DraI, into strain AB33ERG (Wedlich-Söldner et al., 2002).
AB33Erg6G_mChSKL
For colocalization studies of lipid droplets and POs, strain AB33Erg6G_mChSKL was generated by introducing ectopically the DraI-digested plasmid pGmChSKL into the strain AB33Erg6G.
AB33Erg6_Eca1mCh
To visualize co-migration events between LDs and ER, plasmid poNmGFP-Eca1 (Eca1 is an endoplasmic/sarcoplasmic calcium ATPase; Adamíková et al., 2004) was digested with DraI and transformed into the strain AB33Erg6G, resulting in the strain AB33Erg6G_Eca1mCh. poNEca1mCh was obtained by yeast homologous recombination using PCR-amplified fragments of the nat resistance cassette (using SG22 and SG24 primers) from the cloning vector pNEB193 vector (New England BioLabs) and mCherry, using YH166 and YH167 primers. The resulting fragments were recombined with BsiWI-digested pEca1GFP (Adamíková et al., 2004), using homologous recombination in yeast. Correct insertion was confirmed by PCR and restriction digestion.
AB33Pex3G_mChSKL
This strain allowed covisualization of the PO biogenesis factor Pex3 (Höhfeld et al., 1991) and the putative PO marker mCherry-SKL. The plasmid Pex3G contains egfp fused to partial U. maydis pex3 gene (NCBI accession number XP_011392566.1) for targeted integration in to the pex3 locus of U. maydis. A 10,294-bp fragment of the plasmid Kin3-GFP (Schuster et al., 2011; digested with SacI and SgrAI), 3′ end of 937-bp pex3 gene (without stop codon; amplified with GD92 and GD93), and 1,025-bp right flank covering the downstream of the pex3 gene (amplified with GD94 and GD95) were recombined in yeast S. cerevisiae to obtain the vector pPex3G. The resultant plasmid was digested with DraI and integrated into the pex3 locus of AB33 strain, resulting in AB33_Pex3G, and correct insertion was confirmed by Southern blot. Next, poCmChSKL was digested with EcoRV and ectopically inserted into AB33_Pex3G resulting in AB33Pex3G_mChSKL.
AB33GRab5a_ΔHok1_mChSKL_Hok1ΔC
In this strain, the EE-specific motor adapter Hok1 is deleted and EEs are labeled with GFP, fused to the small GTPase Rab5a. In addition, a truncated Hok1ΔC protein, missing the C-terminal 378 aa that confer binding to EEs (Bielska et al., 2014), is expressed under the control of the native promoter. It was obtained by integration of plasmid pHok11-624-HA, linearized with AgeI, into the succinate dehydrogenase locus of strain AB33GRab5a_ΔHok1_mChSKL, resulting in strain AB33GRab5a_ΔHok1_mChSKL_Hok1ΔC. Plasmid pHok11–624-HA was generated by amplification of a 2,970-bp region, encoding the hok1 promoter (−1,002 bp) and the first 624 aa of hok1 from genomic DNA of U. maydis strain 521, using primers EB408-EB410. A 391-bp region encoding the HA sequence (MVYPYDVPDYA), nos terminator, and a fragment of the carboxin resistance cassette was amplified from the plasmid pHok11-624GFP (Bielska et al., 2014), using primers EB412-EB14, and was cloned into the pNEBcbx-yeast-SspI plasmid (Bielska et al., 2014), digested with EcoRI and SacI, by in vivo recombination in the yeast S. cerevisiae FY834.
AB33GRab5a_ΔHok1_mChSKL_Hok1ΔCPX
Hok1ΔCPX, linearizing with AgeI, was integrated into the succinate dehydrogenase locus of AB33GFPAB33G-Rab5a_ΔHok1_mChSKL. This plasmid (Hok1ΔCPX) contains a region encoding the first 624 aa from Hok1 and the PX domain from Yup1 (Wedlich-Söldner et al., 2000), fused to the HA tag. To obtainplasmid pHok1ΔCPX, a 2,935-bp region encoding the hok1 promoter (−1,002 bp) and the first 624 aa of Hok1, was amplified from the genomic DNA of U. maydis strain 521, using primers EB408-EB402. A 546-bp region, encoding the PX domain from the endosomal t-SNARE Yup1 (aa 4–148) and flanking overhangs including the HA sequence, was amplified from the pHok11-624PXG (Bielska et al., 2014) using primers EB403-EB404. A region encoding the nos terminator and a fragment of the carboxin resistance cassette was amplified from the pHok11–624PXGFP (Bielska et al., 2014), using primers EB409-EB14, and cloned into the pNEBcbx-yeast-SspI (Bielska et al., 2014), digested with EcoRI and SacI, by in vivo recombination in the S. cerevisiae strain FY834. The plasmid was linearized with AgeI and integrated into the succinate dehydrogenase locus of AB33GFP-Rab5a_ΔHok1_mChSKL, resulting in the strain AB33GFP-Rab5a_ΔHok1_mChSKL_Hok1ΔCPX.
AB33Yup1ts_GSKL
This strain allowed visualization of POs in yup1ts mutants. It was generated by ectopic integration of AgeI-digested plasmid poCGSKL, into strain AB33Yup1ts (Wedlich-Söldner et al., 2000).
SG200_Erg6G
This strain contains the LD marker Erg6-GFP. It was obtained by integration of AgeI-digested plasmid poErg6G into the succinate dehydrogenase locus of strain SG200 (Bölker et al., 1995).
SG200ΔRab7_GSKL
For PO motility analysis in Δrab7 cells, the plasmid pNΔRab7 was generated by cloning a 1,073-bp fragment covering the upstream of the rab7 gene (left flank) and 972-bp fragment covering the downstream of the rab7 gene into the cloning vector pNEB193 vector (New England BioLabs), containing the nourseothricin resistance cassette. To the left flank, EcoRI and NotI restriction sites were added, and to the right flank, NotI and BamHI were integrated. Conventional ligation was performed with the obtained flanks into the digested pNEB193. The plasmid pNΔRab7 was digested with DraI and integrated into the rab7 locus of U. maydis strain SG200, resulting in strain SG200ΔRab7. Deletion of the rab7 gene was further confirmed by Southern blot. The plasmid poCGSKL was digested with AgeI and integrated into the succinate dehydrogenase locus of SG200ΔRab7, resulting in SG200ΔRab7_GSKL.
SG200ΔRab7_Erg6G
To visualize LD motility in the absence of Rab7, the plasmid poErg6G were digested with AgeI and integrated into the succinate dehydrogenase locus of SG200ΔRab7, resulting in SG200ΔRab7_Erg6G.
AB33ΔRrm4_GRab5_mChSKL
Co-migration of POs and EEs in Δrrm4 background was investigated in this strain. It was obtained by ectopic integration of poGmChSKL, digested with SspI, into AB33ΔRrm4-GRab5a (Higuchi et al., 2014).
FB1ERG_mChRab5a
This strain was used to visualize ER and EE motility. To obtain the strain, plasmid pomChRab5a (Schuster et al., 2011) was digested with ScaI and ectopically integrated into strain FB1ERG (Wedlich-Söldner et al., 2002).
Growth conditions
Cultures of U. maydis were grown at 28°C, shaking at 200 rpm, for 8–12 h in complete medium (CMgluc; Holliday, 1974), supplemented with 1% glucose. To induce hyphal growth cells were washed two to three times with nitrate minimal medium (NM; Brachmann et al., 2001) and finally the culture was transferred to NM, containing 1% glucose, followed by 8–14 h of growth at 28°C and 200 rpm. Cultures of temperature-sensitive Dyn2ts and Yup1ts mutants were grown in CMgluc overnight at 22°C and hyphal growth was induced at this permissive temperature. Before microscopy, mutant cells and control strains were preincubated at 32°C for 2 h (Dyn2ts) or 5 h at 34°C (Yup1ts). Motility of POs in yup1ts and dyn2ts mutants was analyzed under temperature-controlled conditions, using a water-heated objective, linked to a water bath (Huber) at 32°C (Dyn2ts) or 34°C (Yup1ts).
Laser-based epifluorescence microscopy
U. maydis microscopy was performed as previously described (Schuster et al., 2011, 2012). In brief, cells from a liquid culture (CMgluc) were placed on a 2% agarose cushion, covered with a coverslip and immediately observed at room temperature using a motorized inverted microscope (IX81; Olympus) with Plan-Apochromat 100×/1.45 NA oil TIRF or UPlan-SApochromat 60×/1.35 NA oil objective lenses (Olympus) and a VS-LMS4 Laser Merge System (Visitron Systems) with 70-mW observation solid-state lasers at 488 and 561 nm. Photobleaching experiments were performed using a 405-nm/60-mW diode laser, which was decreased by a neutral density 0.6 filter, resulting in 15-mW output power, coupled into the light path by an OSI-IX 71 adaptor (Visitron Systems). The 405-nm laser was controlled by a controller (UGA-40; Rapp OptoElectronic) and VisiFRAP 2D FRAP control software for Meta Series 7.5.x (Visitron Systems). Simultaneous observation of mCherry and GFP fluorescence was performed using a dual imager (Dual-View Micro; Photometrics) equipped with a dual-line beam splitter (z491/561; Chroma Technology) with an emission beam splitter (565 DCXR; Chroma Technology), an ET-Band pass 525/50 (Chroma Technology), and a single band pass filter (BrightLine HC 617/73; Semrock). Images were acquired using a black and white CCD camera (CoolSNAP HQ2; Photometrics/Roper Scientific). All parts of the system were under the control of the software package MetaMorph (Molecular Devices), which was also used for fluorescence measurements and image processing (gamma- contrast- and brightness-adjustment, Nearest Neighbor 2D deconvolution, generation of kyomographs, generation of linescans of average intensities; movie preparation). For temperature-dependent experiments, the objective lenses were cooled or heated using a metal hull connected to a water bath (Huber). For colocalization of LDs and EEs, a region of variable length was photobleached by a 100-ms light pulse using a solid-state 405-nm laser at 100% laser power with a beam diameter of 10 pixels. Subsequently, 100 frames were taken using the 488- and/or 561-nm lasers at exposure times of 150 ms. Statistical analysis was performed using Student’s t test, and all values given in the text are means ± SEM of at least two experiments. All statistical analysis was performed using Prism 5.03 (GraphPad Software).
Colocalization experiments
For an accurate alignment of different cell components, data acquisition was performed after calibration of the system. TetraSteck fluorescent microspheres (Thermo Fisher Scientific) of 0.2-µm size were diluted 1:10 in water. 1 µl of this suspension was placed on an agar pad and two or three images were taken, using the dual imager (10% output power of 488- and 561-nm lasers, at 150 ms). Both channels were aligned using the “Split View” function in MetaMorph using the defined parameters to align the acquired data. AB33Kin3G_mChSKL, AB33_mChRab5_GSKL, and AB33_GRab5_mChSKL strains were used for covisualization of POs with Kin3 and EEs. Colocalization was investigated by acquiring, containing 100 to 150 frames, which were taken using a Dual-View Micro imager, with the 488-nm laser (80% output for Kin3 and 20% output for GRab5a) and the 561-nm laser (25% output power) at a 150-ms exposure time. In cases of colocalization of EEs and LDs, an area of 10–20 µm at 5 µm behind the hyphal tip was photobleached by a 100-ms light pulse of the 405-nm laser (60 mW) at 80% laser power. Subsequently, 100 frames were taken using a Dual-View Micro imager, with the 488-nm laser (80% output for Kin3 and 20% output for GRab5a) and the 561-nm laser (25% output power) at a 150-ms exposure time. An merged movie was generated and a line, 2–3 pixels wide, was drawn over the moving organelle pair, following the path of motion. Intensity profiles of both signals were measured and transferred to Excel (Microsoft). All profile pairs were aligned on the peak of the cargo (POs, LDs, and ER) and the localization and distance of the EE or Kin3 was determined. To show POs motility along MTs in strain AB33G-Tub1_mChSKL, movie streams of 100 frame at 150-ms exposure were taken and merges image series were generated using the MetaMorph software. For colocalization of Erg6G and mCh-SKL 100 planes at 150 ms were acquired with the 488-nm laser at 40% output and the 561-nm laser at 50% output.
Organelle motility analysis
For organelle flux measurements, image series of 100 to 300 frames were taken at 100- to 150-ms exposure time. Flux analysis and additional PO and LD motility parameters (velocity, run length, and/or flux) was performed using kymographs that were generated with the software MetaMorph. Anterograde and retrograde signals were determined ∼10 µm behind the tip. To analyze the motility of the ER, image series of 100 to 300 frames were taken at 150 ms exposure time. The motility events were counted in a 5 µm region within the unipolar region of the hyphal cell. All experiments were performed at least twice. Co-motility of ER and LD with EE was analyzed in movies of 100 frames that were acquired using the Dual-View Micro Imager, with 488-nm laser (10% output power for ER-GFP and 20% output power for Erg6G) and the 561 nm (30% output). Movies were acquired after alignment of both channels using TetraSteck fluorescent microspheres, using the “Split View” function in MetaMorph. Events of PO, LD, or ER motility were identified in one of the resulting image stacks, a kymograph was created using 1 pixel line width, and the line was copied into the corresponding second image stack (depending on the experiment showing ER or EEs). A second kymograph was created over this region, and both were overlaid to identify motility in both channels. Only those motilities that lasted for >5 µm and showed colocalization for >80% of the time where considered as co-migration.
Inhibitor experiments and organelle staining
500-µl cultures were incubated in 2-ml reaction tubes at 28°C and 200 rpm for 30 min in the presence of Benomyl (30 µM; stock: 30 mM in DMSO; Fluka; Sigma-Aldrich) or latrunculin A (20 µM; stock: 20 mM in DMSO; Life Technologies). Control cells were treated with equivalent amounts of the solvent DMSO. Cells were placed onto a 2% agar cushion, supplemented with the respective inhibitor or DMSO at equal concentrations, followed by immediate microscopic analysis. Image series of 100 frames at 150 ms were acquired for further analysis of motility. All experiments were performed at least twice. Neutral lipids were stained using LipidTOX (Life Technologies), by adding 1 µl of stock solution to 200 µl of cell suspension. Cells were directly placed on an agar cushion and microscopically analyzed.
Analysis of organelle distribution
To measure POs (AB33_GSKL, AB33ΔHok1_GSKL, AB33GRab5a_ΔHok1_mChSKL_Hok1ΔC, and AB33GRab5a_ΔHok1_mChSKL_Hok1ΔCPX), ER (AB33ERG and AB33ΔHok1-ERG), LDs (AB33Erg6G and AB33ΔHok1-Erg6G), and mitochondria (AB33LgaG and AB33ΔHok1_mChSKL_LgaG) distribution in hyphal cells, Z-axis image stacks were acquired at an exposure time of 150 ms and 200 nm steps in z-direction. From these stacks, maximum projections were generated using MetaMorph and the mean fluorescent intensity over the length of individual hyphal cells was measured using the line-scan function in MetaMorph. Measurements for each cell were transferred into Excel (Microsoft) and the mean intensity was calculated. The intensity at each data point was calculated relative to the total fluorescent intensity in the measured area.
Bioinformatics
The putative sterol-24-C-methyltransferase UmErg6 and additional sequences used in the phylogenetic tree (accession numbers are given in Fig. S2 B) were identified using the S. cerevisiae protein sequence of Erg6p (accession number CAA89944.1) and BLAST. Sequences were obtained from the NCBI server and comparisons were made with EMBOSS Needle. Domain analysis was performed using PFAM (http://www.pfam.xfam.org/search). The phylogenetic tree was generated in MEGA6.0, using a Maximum Likelihood method, followed by 1,000 bootstrap cycles (http://www.megasoftware.net/).
Online supplemental material
Fig. S1 shows mCherry-SKL and Erg6-GFP as markers for POs and LDs, respectively, and PO motility in Yup1ts mutants and Δhok1 cells that express Hok1 mutant proteins. Fig. S2 shows organelle motility in Δrab7 and Δrrm4 mutants. Fig. S3 shows EE motility and ER, interaction between POs and LDs, and organelle numbers. Table S1 lists strains and plasmids. Table S2 shows experimental usage of strains. Table S3 lists cloning primers used in this study. Video 1 shows directed motility of POs in U. maydis. Video 2 shows motility of a PO along MTs. Video 3 shows co-motility of an EE and a PO. Video 4 shows motility of LDs in U. maydis. Video 5 shows co-motility of an EE and a LD. Video 6 shows motility of ER tubules in U. maydis. Video 7 shows co-motility of an EE and ER in a hyphal cell of U. maydis. Video 8 shows co-motility of an EE and ER in a yeast-like cell of U. maydis). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201505086/DC1.
Supplementary Material
Acknowledgments
We thank Dr. N. Clark for generating strain SG200ΔRab7 and Dr. A. Eskova for generating plasmid pNΔRab7. We are grateful to Prof. S.J. Gurr for helpful comments on the manuscript. We also wish to thank the anonymous reviewers, whose constructive criticism helped us improve the manuscript.
This work was supported by the Portuguese Foundation for Science and Technology and FEDER/COMPETE (SFRH/BD/73532/2010 to S.C. Guimaraes) and CRUP/Treaty of Windsor (ACÇÕES INTEGRADAS 2009, B-33/09 to G. Steinberg and M. Schuster). G. Steinberg acknowledges support from the Biotechnology and Biological Sciences Research Council.
The authors declare no competing financial interest.
Author contributions: S.C. Guimaraes generated strains and, with M. Schuster, performed experiments and analyzed data; G. Dagdas, E. Bielska, and S. Kilaru generated strains; B.R.A. Meadows generated strains and performed experiments; M. Schrader discussed data and concepts; and G. Steinberg conceived the project, analyzed data, and wrote the manuscript.
Footnotes
Abbreviations used in this paper:
- EE
- early endosome
- LD
- lipid droplet
- LE
- late endosome
- MT
- microtubule
- PO
- peroxisome
References
- Adamíková L., Straube A., Schulz I., and Steinberg G.. 2004. Calcium signaling is involved in dynein-dependent microtubule organization. Mol. Biol. Cell. 15:1969–1980. 10.1091/mbc.E03-09-0675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhmanova A., and Hammer J.A. III. 2010. Linking molecular motors to membrane cargo. Curr. Opin. Cell Biol. 22:479–487. 10.1016/j.ceb.2010.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa A.D., Savage D.B., and Siniossoglou S.. 2015. Lipid droplet-organelle interactions: emerging roles in lipid metabolism. Curr. Opin. Cell Biol. 35:91–97. 10.1016/j.ceb.2015.04.017 [DOI] [PubMed] [Google Scholar]
- Baumann S., Pohlmann T., Jungbluth M., Brachmann A., and Feldbrügge M.. 2012. Kinesin-3 and dynein mediate microtubule-dependent co-transport of mRNPs and endosomes. J. Cell Sci. 125:2740–2752. 10.1242/jcs.101212 [DOI] [PubMed] [Google Scholar]
- Baumann S., König J., Koepke J., and Feldbrügge M.. 2014. Endosomal transport of septin mRNA and protein indicates local translation on endosomes and is required for correct septin filamentation. EMBO Rep. 15:94–102. 10.1002/embr.201338037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht P., König J., and Feldbrügge M.. 2006. The RNA-binding protein Rrm4 is essential for polarity in Ustilago maydis and shuttles along microtubules. J. Cell Sci. 119:4964–4973. 10.1242/jcs.03287 [DOI] [PubMed] [Google Scholar]
- Bielska E., Higuchi Y., Schuster M., Steinberg N., Kilaru S., Talbot N.J., and Steinberg G.. 2014. Long-distance endosome trafficking drives fungal effector production during plant infection. Nat. Commun. 5:5097 10.1038/ncomms6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielska E., Schuster M., Roger Y., Berepiki A., Soanes D.M., Talbot N.J., and Steinberg G.. 2014. Hook is an adapter that coordinates kinesin-3 and dynein cargo attachment on early endosomes. J. Cell Biol. 204:989–1007. 10.1083/jcb.201309022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bölker M., Genin S., Lehmler C., and Kahmann R.. 1995. Genetic regulation of mating and dimorphism in Ustilago maydis. Can. J. Bot. 73:320–325. 10.1139/b95-262 [DOI] [Google Scholar]
- Bonekamp N.A., Sampaio P., de Abreu F.V., Lüers G.H., and Schrader M.. 2012. Transient complex interactions of mammalian peroxisomes without exchange of matrix or membrane marker proteins. Traffic. 13:960–978. 10.1111/j.1600-0854.2012.01356.x [DOI] [PubMed] [Google Scholar]
- Bortfeld M., Auffarth K., Kahmann R., and Basse C.W.. 2004. The Ustilago maydis a2 mating-type locus genes lga2 and rga2 compromise pathogenicity in the absence of the mitochondrial p32 family protein Mrb1. Plant Cell. 16:2233–2248. 10.1105/tpc.104.022657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann A., Weinzierl G., Kämper J., and Kahmann R.. 2001. Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol. Microbiol. 42:1047–1063. 10.1046/j.1365-2958.2001.02699.x [DOI] [PubMed] [Google Scholar]
- Camões F., Islinger M., Guimarães S.C., Kilaru S., Schuster M., Godinho L.F., Steinberg G., and Schrader M.. 2015. New insights into the peroxisomal protein inventory: Acyl-CoA oxidases and -dehydrogenases are an ancient feature of peroxisomes. Biochim. Biophys. Acta. 1853:111–125. [DOI] [PubMed] [Google Scholar]
- Chu B.B., Liao Y.C., Qi W., Xie C., Du X., Wang J., Yang H., Miao H.H., Li B.L., and Song B.L.. 2015. Cholesterol transport through lysosome-peroxisome membrane contacts. Cell. 161:291–306. 10.1016/j.cell.2015.02.019 [DOI] [PubMed] [Google Scholar]
- Egan M.J., Tan K., and Reck-Peterson S.L.. 2012. Lis1 is an initiation factor for dynein-driven organelle transport. J. Cell Biol. 197:971–982. 10.1083/jcb.201112101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiguin F., Ferreira A., Kosik K.S., and Caceres A.. 1994. Kinesin-mediated organelle translocation revealed by specific cellular manipulations. J. Cell Biol. 127:1021–1039. 10.1083/jcb.127.4.1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J.R., Dibenedetto J.R., West M., Rowland A.A., and Voeltz G.K.. 2013. Endoplasmic reticulum-endosome contact increases as endosomes traffic and mature. Mol. Biol. Cell. 24:1030–1040. 10.1091/mbc.E12-10-0733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M.M., and Holzbaur E.L.. 2014. Integrated regulation of motor-driven organelle transport by scaffolding proteins. Trends Cell Biol. 24:564–574. 10.1016/j.tcb.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs U., Manns I., and Steinberg G.. 2005. Microtubules are dispensable for the initial pathogenic development but required for long-distance hyphal growth in the corn smut fungus Ustilago maydis. Mol. Biol. Cell. 16:2746–2758. 10.1091/mbc.E05-03-0176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs U., Hause G., Schuchardt I., and Steinberg G.. 2006. Endocytosis is essential for pathogenic development in the corn smut fungus Ustilago maydis. Plant Cell. 18:2066–2081. 10.1105/tpc.105.039388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., and Goodman J.M.. 2015. The lipid droplet-a well-connected organelle. Front. Cell Dev. Biol. 3:49 10.3389/fcell.2015.00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould G.W., and Lippincott-Schwartz J.. 2009. New roles for endosomes: from vesicular carriers to multi-purpose platforms. Nat. Rev. Mol. Cell Biol. 10:287–292. 10.1038/nrm2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemi H.F., and Goodman J.M.. 2015. The life cycle of lipid droplets. Curr. Opin. Cell Biol. 33:119–124. 10.1016/j.ceb.2015.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi Y., Ashwin P., Roger Y., and Steinberg G.. 2014. Early endosome motility spatially organizes polysome distribution. J. Cell Biol. 204:343–357. 10.1083/jcb.201307164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner D., van den Berg M., Philippsen P., Tabak H.F., and Hettema E.H.. 2001. A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J. Cell Biol. 155:979–990. 10.1083/jcb.200107028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhfeld J., Veenhuis M., and Kunau W.H.. 1991. PAS3, a Saccharomyces cerevisiae gene encoding a peroxisomal integral membrane protein essential for peroxisome biogenesis. J. Cell Biol. 114:1167–1178. 10.1083/jcb.114.6.1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. 1974. Ustilago maydis. In Handbook of Genetics. Vol. 1 King R.C., editor. Plenum Press, New York: 575–595. [Google Scholar]
- Kamal A., and Goldstein L.S.. 2002. Principles of cargo attachment to cytoplasmic motor proteins. Curr. Opin. Cell Biol. 14:63–68. 10.1016/S0955-0674(01)00295-2 [DOI] [PubMed] [Google Scholar]
- Karcher R.L., Deacon S.W., and Gelfand V.I.. 2002. Motor-cargo interactions: the key to transport specificity. Trends Cell Biol. 12:21–27. 10.1016/S0962-8924(01)02184-5 [DOI] [PubMed] [Google Scholar]
- Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., and Schiebel E.. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 15:963–972. [DOI] [PubMed] [Google Scholar]
- Kohlwein S.D., Veenhuis M., and van der Klei I.J.. 2013. Lipid droplets and peroxisomes: key players in cellular lipid homeostasis or a matter of fat--store ‘em up or burn ’em down. Genetics. 193:1–50. 10.1534/genetics.112.143362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kural C., Kim H., Syed S., Goshima G., Gelfand V.I., and Selvin P.R.. 2005. Kinesin and dynein move a peroxisome in vivo: a tug-of-war or coordinated movement?. Science. 308:1469–1472. 10.1126/science.1108408 [DOI] [PubMed] [Google Scholar]
- Larsen K.S., Xu J., Cermelli S., Shu Z., and Gross S.P.. 2008. BicaudalD actively regulates microtubule motor activity in lipid droplet transport. PLoS One. 3:e3763 10.1371/journal.pone.0003763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrand C., Corti M., Goodson H., Cosson P., Cavalli V., Mayran N., Fauré J., and Gruenberg J.. 2002. Late endosome motility depends on lipids via the small GTPase Rab7. EMBO J. 21:1289–1300. 10.1093/emboj/21.6.1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon M.A. 2003. Phosphoinositide recognition domains. Traffic. 4:201–213. 10.1034/j.1600-0854.2004.00071.x [DOI] [PubMed] [Google Scholar]
- Mathur J., Mathur N., and Hülskamp M.. 2002. Simultaneous visualization of peroxisomes and cytoskeletal elements reveals actin and not microtubule-based peroxisome motility in plants. Plant Physiol. 128:1031–1045. 10.1104/pp.011018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miaczynska M., Pelkmans L., and Zerial M.. 2004. Not just a sink: endosomes in control of signal transduction. Curr. Opin. Cell Biol. 16:400–406. 10.1016/j.ceb.2004.06.005 [DOI] [PubMed] [Google Scholar]
- Pu J., Ha C.W., Zhang S., Jung J.P., Huh W.K., and Liu P.. 2011. Interactomic study on interaction between lipid droplets and mitochondria. Protein Cell. 2:487–496. 10.1007/s13238-011-1061-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp S., Saffrich R., Anton M., Jäkle U., Ansorge W., Gorgas K., and Just W.W.. 1996. Microtubule-based peroxisome movement. J. Cell. Sci. 109:837–849. [DOI] [PubMed] [Google Scholar]
- Raymond C.K., Pownder T.A., and Sexson S.L.. 1999. General method for plasmid construction using homologous recombination. Biotechniques. 26:134–138: 140–141. [DOI] [PubMed] [Google Scholar]
- Schrader M., Grille S., Fahimi H.D., and Islinger M.. 2013. Peroxisome interactions and cross-talk with other subcellular compartments in animal cells. Subcell. Biochem. 69:1–22. 10.1007/978-94-007-6889-5_1 [DOI] [PubMed] [Google Scholar]
- Schrader M., Godinho L.F., Costello J.L., and Islinger M.. 2015. The different facets of organelle interplay-an overview of organelle interactions. Front. Cell Dev. Biol. 3:56 (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz B., Banuett F., Dahl M., Schlesinger R., Schäfer W., Martin T., Herskowitz I., and Kahmann R.. 1990. The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell. 60:295–306. 10.1016/0092-8674(90)90744-Y [DOI] [PubMed] [Google Scholar]
- Schuster M., Kilaru S., Ashwin P., Lin C., Severs N.J., and Steinberg G.. 2011. Controlled and stochastic retention concentrates dynein at microtubule ends to keep endosomes on track. EMBO J. 30:652–664. 10.1038/emboj.2010.360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M., Kilaru S., Fink G., Collemare J., Roger Y., and Steinberg G.. 2011. Kinesin-3 and dynein cooperate in long-range retrograde endosome motility along a nonuniform microtubule array. Mol. Biol. Cell. 22:3645–3657. 10.1091/mbc.E11-03-0217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M., Lipowsky R., Assmann M.A., Lenz P., and Steinberg G.. 2011. Transient binding of dynein controls bidirectional long-range motility of early endosomes. Proc. Natl. Acad. Sci. USA. 108:3618–3623. 10.1073/pnas.1015839108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M., Treitschke S., Kilaru S., Molloy J., Harmer N.J., and Steinberg G.. 2012. Myosin-5, kinesin-1 and myosin-17 cooperate in secretion of fungal chitin synthase. EMBO J. 31:214–227. 10.1038/emboj.2011.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shai N., Schuldiner M., and Zalckvar E.. 2015. No peroxisome is an island - Peroxisome contact sites. Biochim. Biophys. Acta. S0167-4889(15)00309-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shubeita G.T., Tran S.L., Xu J., Vershinin M., Cermelli S., Cotton S.L., Welte M.A., and Gross S.P.. 2008. Consequences of motor copy number on the intracellular transport of kinesin-1-driven lipid droplets. Cell. 135:1098–1107. 10.1016/j.cell.2008.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellig T., Bottin A., and Kahmann R.. 1996. Green fluorescent protein (GFP) as a new vital marker in the phytopathogenic fungus Ustilago maydis. Mol. Gen. Genet. 252:503–509. [DOI] [PubMed] [Google Scholar]
- Steinberg G. 2014. Endocytosis and early endosome motility in filamentous fungi. Curr. Opin. Microbiol. 20:10–18. 10.1016/j.mib.2014.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg G., and Schuster M.. 2011. The dynamic fungal cell. Fungal Biol. Rev. 25:14–37. 10.1016/j.fbr.2011.01.008 [DOI] [Google Scholar]
- Steinberg G., Wedlich-Söldner R., Brill M., and Schulz I.. 2001. Microtubules in the fungal pathogen Ustilago maydis are highly dynamic and determine cell polarity. J. Cell. Sci. 114:609–622. [DOI] [PubMed] [Google Scholar]
- Tang X., Halleck M.S., Schlegel R.A., and Williamson P.. 1996. A subfamily of P-type ATPases with aminophospholipid transporting activity. Science. 272:1495–1497. 10.1126/science.272.5267.1495 [DOI] [PubMed] [Google Scholar]
- Targett-Adams P., Chambers D., Gledhill S., Hope R.G., Coy J.F., Girod A., and McLauchlan J.. 2003. Live cell analysis and targeting of the lipid droplet-binding adipocyte differentiation-related protein. J. Biol. Chem. 278:15998–16007. 10.1074/jbc.M211289200 [DOI] [PubMed] [Google Scholar]
- Wanders R.J., and Waterham H.R.. 2006. Biochemistry of mammalian peroxisomes revisited. Annu. Rev. Biochem. 75:295–332. 10.1146/annurev.biochem.74.082803.133329 [DOI] [PubMed] [Google Scholar]
- Waterman-Storer C.M., and Salmon E.D.. 1998. Endoplasmic reticulum membrane tubules are distributed by microtubules in living cells using three distinct mechanisms. Curr. Biol. 8:798–806. 10.1016/S0960-9822(98)70321-5 [DOI] [PubMed] [Google Scholar]
- Wedlich-Söldner R., Bölker M., Kahmann R., and Steinberg G.. 2000. A putative endosomal t-SNARE links exo- and endocytosis in the phytopathogenic fungus Ustilago maydis. EMBO J. 19:1974–1986. 10.1093/emboj/19.9.1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Söldner R., Schulz I., Straube A., and Steinberg G.. 2002. Dynein supports motility of endoplasmic reticulum in the fungus Ustilago maydis. Mol. Biol. Cell. 13:965–977. 10.1091/mbc.01-10-0475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Söldner R., Straube A., Friedrich M.W., and Steinberg G.. 2002. A balance of KIF1A-like kinesin and dynein organizes early endosomes in the fungus Ustilago maydis. EMBO J. 21:2946–2957. 10.1093/emboj/cdf296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte M.A. 2009. Fat on the move: intracellular motion of lipid droplets. Biochem. Soc. Trans. 37:991–996. 10.1042/BST0370991 [DOI] [PubMed] [Google Scholar]
- Wiemer E.A., Wenzel T., Deerinck T.J., Ellisman M.H., and Subramani S.. 1997. Visualization of the peroxisomal compartment in living mammalian cells: dynamic behavior and association with microtubules. J. Cell Biol. 136:71–80. 10.1083/jcb.136.1.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Dollard C., and Ricupero-Hovasse S.L.. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 11:53–55. 10.1002/yea.320110107 [DOI] [PubMed] [Google Scholar]
- Woźniak M.J., Bola B., Brownhill K., Yang Y.C., Levakova V., and Allan V.J.. 2009. Role of kinesin-1 and cytoplasmic dynein in endoplasmic reticulum movement in VERO cells. J. Cell Sci. 122:1979–1989. 10.1242/jcs.041962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Qiu R., Arst H.N. Jr, Peñalva M.A., and Xiang X.. 2014. HookA is a novel dynein-early endosome linker critical for cargo movement in vivo. J. Cell Biol. 204:1009–1026. 10.1083/jcb.201308009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser E., Paltauf F., and Daum G.. 1993. Sterol composition of yeast organelle membranes and subcellular distribution of enzymes involved in sterol metabolism. J. Bacteriol. 175:2853–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.