Reversible ubiquitination strictly controls NLRC5 function: K63-linked ubiquitination of NLRC5 at lysine 1,178 mediated by TRAF2/6 generates a coherent feedforward loop to sensitize switch-like activation of NF-κB, whereas USP14 specifically removes the polyubiquitin chains from NLRC5 to enhance NLRC5-mediated inhibition.
Abstract
NLRC5 is an important regulator in innate immune responses. However, the ability of NLRC5 to inhibit NF-κB activation is controversial in different cell types. How dynamic modification of NLRC5 shapes NF-κB signaling remains unknown. We demonstrated that NLRC5 undergoes robust ubiquitination by TRAF2/6 after lipopolysaccharide treatment, which leads to dissociation of the NLRC5–IκB kinase complex. Experimental and mathematical analyses revealed that the K63-linked ubiquitination of NLRC5 at lysine 1,178 generates a coherent feedforward loop to further sensitize NF-κB activation. Meanwhile, we found USP14 specifically removes the polyubiquitin chains from NLRC5 to enhance NLRC5-mediated inhibition of NF-κB signaling. Furthermore, we found that different cell types may exhibit different sensitivities to NF-κB activation in response to NLRC5 ablation, possibly as a result of the various intrinsic levels of deubiquitinases and NLRC5. This might partially reconcile controversial studies and explain why NLRC5 exhibits diverse inhibitory efficiencies. Collectively, our results provide the regulatory mechanisms of reversible NLRC5 ubiquitination and its role in the dynamic control of innate immunity.
Introduction
Innate immune responses provide the first line of defense against invading microbes. The initiation of innate immune responses depends on the recognition of conserved patterns, including pathogen-associated molecular patterns (PAMPs) and danger-associated molecular patterns. Several classes of pattern recognition receptors, including Toll-like receptors (TLRs), NOD-like receptors (NLRs), RIG-I–like receptors, and certain DNA sensors, contribute to specificity during pathogen recognition (Price et al., 2008; Takeuchi and Akira, 2010; Orzalli and Knipe, 2014; Wu and Chen, 2014). Although these receptors recognize diverse ligands, they share common downstream pathways, such as NF-κB and type I interferon signaling, which may trigger adaptive immune responses (Akira et al., 2001). Stringent control of these common pathways is important for both innate and adaptive immunity.
Numerous PAMPs induce inflammation through activation of the NF-κB signaling pathway. TLR activation usually leads to the activation of the IκB kinase (IKK) complex, which is composed of IKK-α, IKK-β, and NEMO (Hayden and Ghosh, 2008). IKK-α and IKK-β both harbor a kinase domain, leading to subsequent phosphorylation of IκB-α, which results in ubiquitin (Ub)-mediated degradation and NF-κB activation with expression of proinflammatory factors (Hayden and Ghosh, 2008). Although many regulators have been identified to modulate the activity of the IKK complex (Vallabhapurapu and Karin, 2009; Chen, 2012), the molecular mechanisms regulating the switch-like behavior of NF-κB activation are not well understood.
Members of the NLR family feature a central nucleotide-binding and oligomerization domain (Wu and Chen, 2014). NLRs were originally thought to induce inflammatory responses by initiating large protein complexes, termed inflammasomes (Petrilli et al., 2005). However, recent evidence suggests that several NLRs (also known as regulatory NLRs), including NLRX1, NLRC5, NLRP4, and NLRC3, negatively regulate TLR and RIG-I–like receptor signaling (Cui et al., 2010, 2012; Xia et al., 2011; Zhang et al., 2014). NLRC5 (NLR family, CARD domain containing 5) has been demonstrated to function in both innate and adaptive immune signaling. Although it is quite clear that NLRC5 is a key regulator of major histocompatibility complex class I–dependent genes (Kobayashi and van den Elsen, 2012), its function in inflammation is still ambiguous. We have previously reported that NLRC5 inhibits NF-κB activation by interacting with IKK-α/-β and blocking IKK-α/-β phosphorylation (Cui et al., 2010). Similarly, Benko et al. (2010) also found a negative functional role for NLRC5 in dampening NF-κB signaling in RAW264.7 cells. Conversely, Kumar et al. (2011) revealed no functional role for NLRC5 in TLR4 signaling in bone marrow–derived dendritic cells (BMDCs; Tong et al., 2012). Meanwhile, Yao et al. (2012) also found that NLRC5 may not influence NF-κB–dependent transcription of proinflammatory cytokines in bone marrow–derived macrophages (BMMs). We previously generated Nlrc5−/− mice and found that NLRC5 ablation affected NF-κB signaling in a cell type–specific manner (Tong et al., 2012). NLRC5 deficiency markedly enhanced proinflammatory responses in peritoneal macrophages (pMs). However, little or no difference in proinflammatory cytokine secretion was observed in BMMs or BMDCs (Tong et al., 2012). Therefore, the exact role of NLRC5 in the regulation of NF-κB activation as well as in inflammatory responses remains controversial.
In this study, we demonstrate that NLRC5 undergoes K63-linked ubiquitination after lipopolysaccharide (LPS) treatment. Interestingly, we found that the Ub editing of NLRC5 determined NLRC5–IKK-β interaction dynamics and enhanced the activation of NF-κB signaling. The ubiquitination of NLRC5 at lysine 1,178 is catalyzed by TNF receptor–associated factor (TRAF) 2/6 and can be removed by several deubiquitinases (DUBs), including USP14, USP18, and USP22. Indeed, we showed that USP14 specifically enhanced the interaction between NLRC5 and IKK-β to inhibit NF-κB activation in an NLRC5-dependent manner. By incorporating NLRC5 ubiquitination into our mathematical model, we found that the simulation consistently replicates the experimental findings both qualitatively and quantitatively. Notably, the Ub editing of NLRC5 generates a coherent feedforward loop (CFL) to further sensitize NF-κB signaling. We experimentally verified that the cellular sensitivity and specificity to NLRC5 ablation on NF-κB activation varies with different levels of NLRC5 and DUBs. These data may help to explain previous controversial studies on the functional role of NLRC5 and may provide impetus to manipulate innate immune responses by changing the intracellular state of deubiquitination.
Results
Mathematical modeling reveals additional regulation of NLRC5 required for NLRC5–IKK-β interaction dynamics
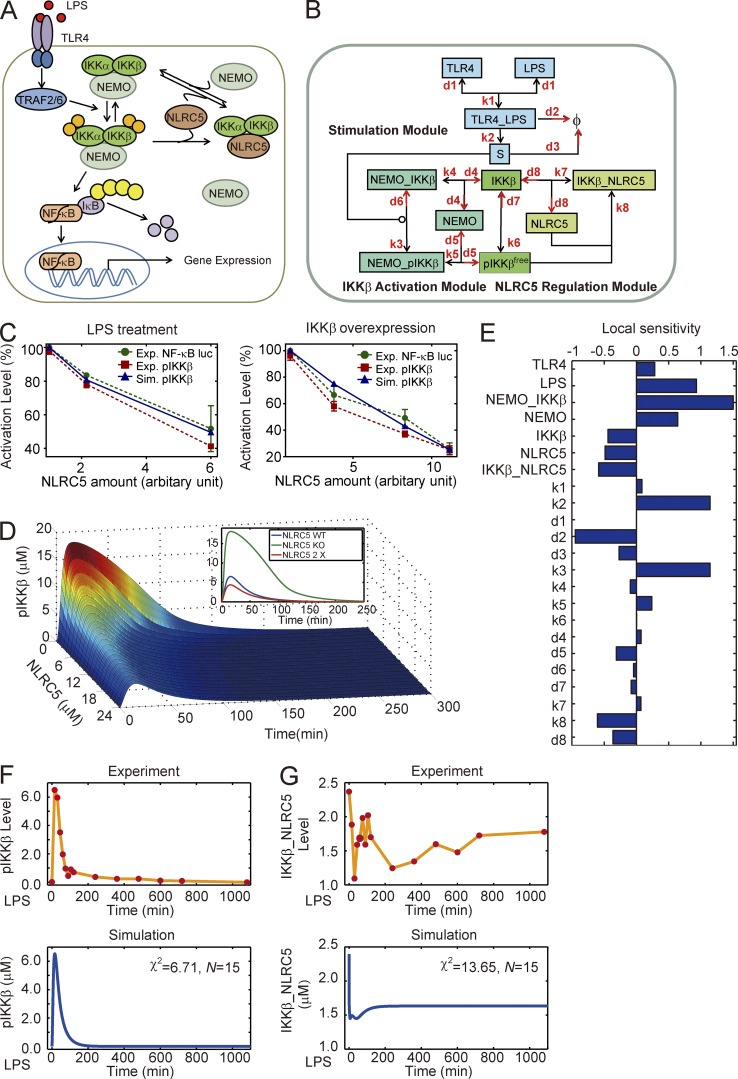
We have previously demonstrated that NLRC5 negatively regulates NF-κB signaling by interacting with both IKK-α and ΙΚΚ-β (Cui et al., 2010). Based on it (Fig. 1 A), we constructed a mathematical model that incorporated the competitive interactions between NLRC5 and NEMO binding to both IKK-β and pIKK-β (Fig. S1 A and Tables S1 and S2) to systematically determine the role of NLRC5 in NF-κB signaling (model I; Fig. 1 B). Indeed, we mainly considered LPS-induced phosphorylated IKK-β (pIKK-β) as a mark of NF-κB activation (Fig. S1 B) because several papers mentioned that IKK-β is indispensable for the activation of NF-κB induced by LPS treatment, whereas it has slight or no induced IKK-α phosphorylation following LPS treatment (Ghosh and Karin, 2002; Yang et al., 2003; Hayden and Ghosh, 2004; Birrell et al., 2006; Häcker and Karin, 2006). Model I was established based on our previous experimental data (Cui et al., 2010) using the PottersWheel toolbox (Maiwald and Timmer, 2008). Next, we verified our model by comparing the simulation results with our newly acquired experimental data (Fig. S1, B and C). We found that model I qualitatively described the inhibitory dose response for NLRC5 under both LPS treatment and IKK-β overexpression conditions (Fig. 1 C). Furthermore, the simulation data also showed that lower levels of NLRC5 significantly increased IKK-β phosphorylation (pIKK-β = pIKK-βfree + NEMO–pIKK-β) after LPS treatment (Fig. 1 D). A local sensitivity analysis was performed for all kinetic parameters and initial conditions. We found that NLRC5 may serve as a negative regulator with moderate sensitivity (Fig. 1 E). However, the local coefficient for IKK-β was also negative, which may contradict most current knowledge (Fig. 1 E). In addition, we observed reasonable χ2 values in the mathematical fitting of the measurements of both total activated IKK-β and IKK-β–NLRC5 complexes (Fig. 1, F and G). However, within the first couple of hours, the experimental temporal dynamics of IKK-β–NLRC5 complex formation did not precisely overlap with the mathematical model (Fig. 1 G). Therefore, we hypothesized that there might be additional modulation of NLRC5 required to control NLRC5–IKK-β interaction dynamics.
Figure 1.
Mathematical model of current NLRC5–IKK-β module cannot reproduce NLRC5–IKK interaction dynamics. (A) Schematic representation of TLR4-mediated NF-κB signaling regulated by NLRC5. (B) The mass action model considering only the competitive interactions between NLRC5 and NEMO to bind IKK-β (model I). (C) Comparison of experimental data from three independent experiments (n = 3; error bars indicate the SD) and simulation results of pIKK-β (the sum of pIKK-βfree and NEMO–pIKK-β; Tables S1 and S2) level from model I. (D) Temporal trajectories of total phosphorylated IKK (pIKK-β + NEMO–pIKK-β; also see Tables S1 and S2) with different levels of NLRC5. The inset denotes total pIKK-β dynamics with NLRC5 KO (no NLRC5 protein), WT, and a twofold elevation (2×) of WT. (E) Local sensitivity coefficients for all kinetic parameters and nonzero initial conditions. (F and G) Temporal curves of pIKK-β levels (F) and IKK-β–NLRC5 complex levels (G) upon treatment of RAW264.7 cells with 200 ng/ml LPS adapted from our previous measurements and simulation results by model I. Φ, null; χ2, the sum of deviations; N, number of experimental points.
NLRC5 undergoes robust ubiquitination upon LPS challenge
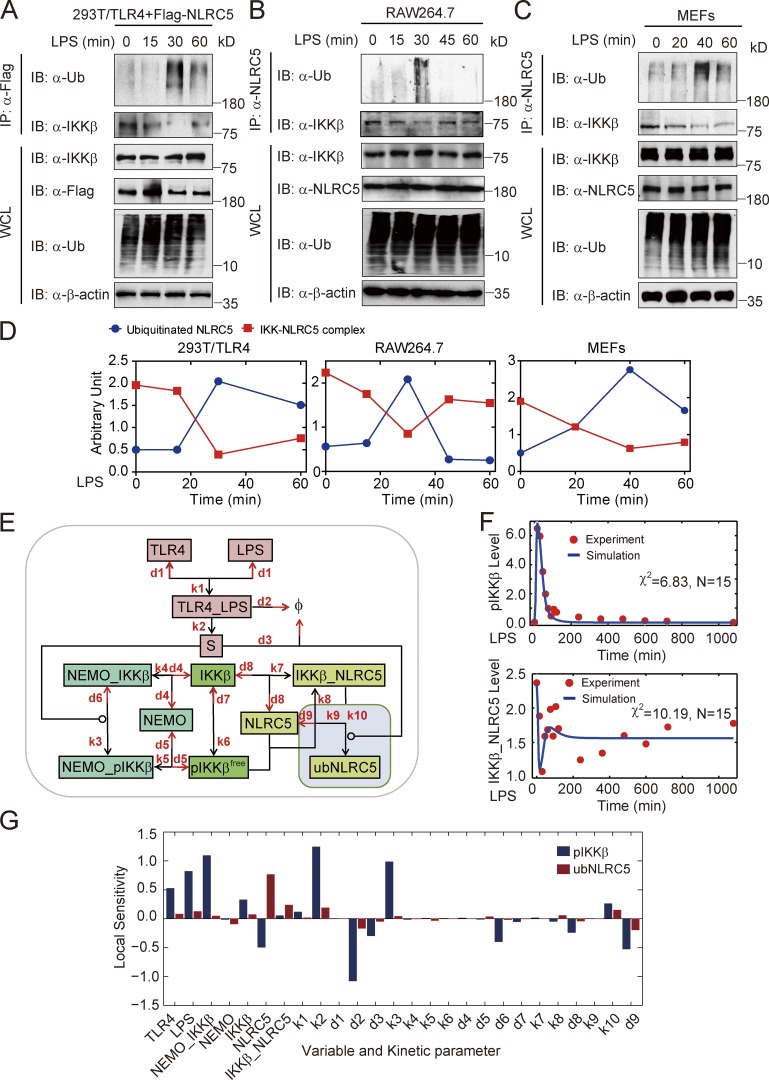
Posttranslational modification (PTM) is a common cellular process that modulates protein activities. Therefore, we tested whether the PTM, such as phosphorylation or ubiquitination, of NLRC5 occurred upon LPS treatment. To address this issue, we transfected HEK293T/TLR4 cells with Flag-tagged NLRC5 followed by LPS treatment. We did not observe any phosphorylation on NLRC5 after LPS treatment (Fig. S1 D). However, coimmunoprecipitation analyses revealed that Flag-tagged NLRC5 underwent dynamic ubiquitination, peaking ∼30 min after LPS treatment (Fig. 2 A). We further identified that NLRC5 can be ubiquitinated with K63 but not K48 linkage (Fig. S1 E). Through coimmunoprecipitation with 1% SDS, we confirmed that NLRC5 underwent direct ubiquitination after LPS treatment (Fig. S1 F). To test whether endogenous NLRC5 can be ubiquitinated in different cell types, we treated RAW264.7 cells and mouse embryonic fibroblasts (MEFs) with LPS at various time points. Coimmunoprecipitation analyses revealed that NLRC5 underwent polyubiquitination with different dynamic patterns after LPS treatment in RAW264.7 cells and MEFs (Fig. 2, B and C). In addition, we also observed colocalization of Ub and NLRC5 in BMMs after LPS treatment (Fig. S1 G). Collectively, these results indicate that NLRC5 undergoes robust ubiquitination upon LPS challenge in various cell types.
Figure 2.
NLRC5 ubiquitination upon LPS treatment is critical for NLRC5–IKK-β interaction dynamics. (A–C) Cell lysates of HEK293T/TLR4 cells (A), RAW264.7 cells (B), or MEFs (C) were collected at the indicated time after LPS (200 ng/ml) treatment and immunoprecipitated with anti-NLRC5 antibody, followed by immunoblotting with the indicated antibodies. (D) Quantification of A–C. IKK-β–NLRC5 complex (red) and ubiquitinated NLRC5 (blue) in HEK293T/TLR4, RAW264.7, and MEFs. (E) Proposed model II incorporating NLRC5 ubiquitination (shaded square). (F) Model fits to experimental data measured in our previous work (Cui et al., 2010). Blue curves indicate the Model II simulation. Red circles indicate experimental data. (G) Local peak sensitivities for all kinetic parameters and nonzero initial conditions in Model II for total pIKK-β (blue bars) and ubiquitinated NLRC5 (ubNLRC5, red bars). Φ, null; χ2, the sum of deviations; IB, immunoblotting; IP, immunoprecipitation; N, number of experimental points; WCL, whole cell lysate.
NLRC5 ubiquitination is critical for the dynamic interaction of NLRC5 and IKK-β
We next observed that the level of ubiquitinated NLRC5 is inversely correlated to the levels of the IKK-β–NLRC5 complex (Fig. 2 D), suggesting that NLRC5 ubiquitination may decrease its ability to bind IKK-β. Given the observation that NLRC5 can be ubiquitinated after LPS stimulation, we revised model I by incorporating NLRC5 ubiquitination to generate model II (Fig. 2 E, shaded box; and Table S1 and Table S2). The simulation results from model II can better replicate the dynamic patterns of IKK-β–NLRC5 complex formation (Fig. 2 F). We calculated the local sensitivities for all parameters and found that although a large fraction of parameters are not sensitive to IKK-β activation, NLRC5 levels and deubiquitination rate (d9) are among the most sensitive parameters for inhibition of IKK-β activity (Fig. 2 G).
In model II, the dose–response curve for NLRC5 also quantitatively matched our experimental data (Fig. S1 H). We altered NLRC5 expression in silico and found that although the total IKK-β–NEMO complex did not vary significantly, the levels of activated IKK-β in the NEMO–pIKK-β complex significantly declined (Fig. S1 I), which was qualitatively consistent with our previous results (Cui et al., 2010). The simulation by model II also confirmed the inverse correlation between IKK-β–NLRC5 and ubiquitinated NLRC5 (Fig. S1 J). In silico analyses showed that amplifying NLRC5 ubiquitination by increasing the value of k10 decreases the inhibitory effect of NLRC5 on IKK-β activation (Fig. S1 K). These results suggest that the dynamic Ub editing of NLRC5 determines its binding capacity to IKK-β and properly shapes its inhibitory function on NF-κB activation.
TRAF2 and TRAF6 ubiquitinate NLRC5 at lysine 1,178
It has been reported that unanchored K63 Ub chains act as critical mediators in the activation of NF-κB signaling (Xia et al., 2009). We examined whether NLRC5 is covalently attached to anchored Ub chains or noncovalently with unanchored Ub chains. We found that IsoT, an isopeptidase that can specifically degrade unanchored Ub chains (Rajsbaum et al., 2014), did not influence K63-linked ubiquitination of NLRC5, suggesting that NLRC5-associated Ub chains are primarily covalently attached (Fig. S2 A).
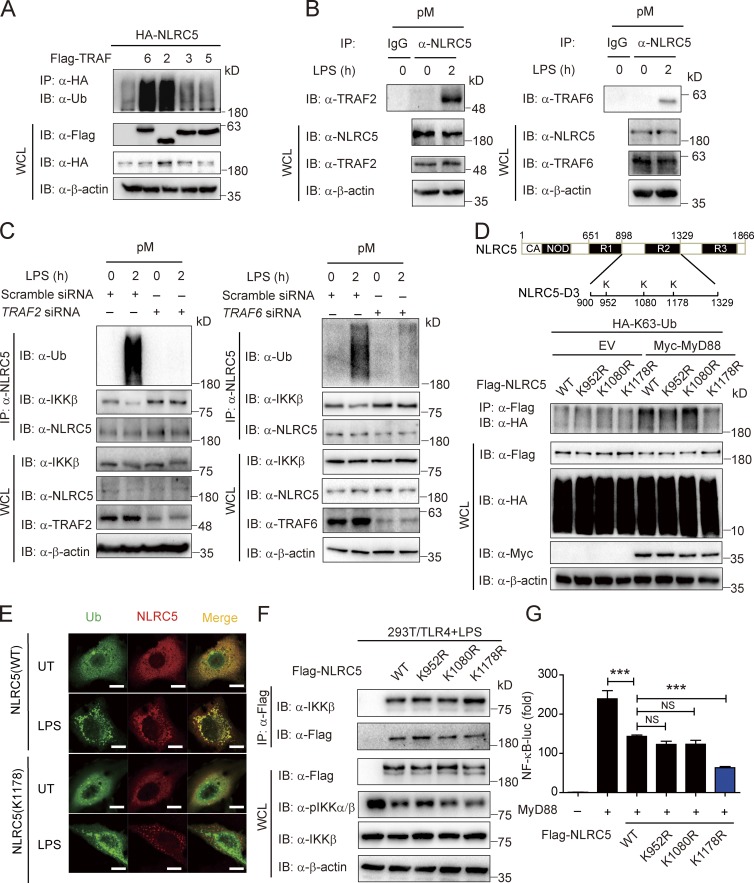
Next, we determined which specific E3 ligases are responsible for NLRC5 ubiquitination. The TRAF family members belong to the RING domain containing E3 ligases, which serve as adapters in NF-κB signaling (Häcker et al., 2011). Although several TRAFs interacted with NLRC5 (Fig. S2 B), only TRAF6 and TRAF2 mediated the ubiquitination of NLRC5 (Fig. 3 A) and decreased IKK-β–NLRC5 interaction (Fig. S2 C). Furthermore, we observed the enhanced interaction between NLRC5 and TRAF2 or TRAF6 in pMs upon LPS treatment (Fig. 3 B). To determine the role of endogenous TRAF2/6 in the ubiquitination of NLRC5 after LPS treatment, we knocked down endogenous TRAF2 or TRAF6 in pMs and assessed NLRC5 ubiquitination as well as the IKK-β–NLRC5 interaction. In cells transfected with control siRNA, we observed considerable polyubiquitination of NLRC5 after LPS treatment; however, such polyubiquitination of NLRC5 was completely abolished in cells transfected with TRAF2- or TRAF6-specific siRNA (Fig. 3 C). We also observed the decrease of IKK-β–NLRC5 interaction after LPS treatment was rescued by the knockdown of TRAF2/6 (Fig. 3 C).
Figure 3.
TRAF2 and TRAF6 ubiquitinate NLRC5 at lysine 1,178. (A) HEK293T cells were transfected with plasmids encoding Flag-tagged TRAF6, TRAF2, TRAF3, and TRAF5 and HA-tagged NLRC5. Cell lysates were immunoprecipitated with anti-HA beads followed by immunoblotting with anti-Ub antibody. (B) The interaction between NLRC5 and TRAF2 or TRAF6 in pMs by LPS treatment. (C) The ubiquitination level of NLRC5 and IKK-β–NLRC5 interaction by LPS treatment in WT or TRAF2/6 knockdown pMs. (D) A schematic representation of predicted ubiquitination sites (marked K) in NLRC5 (top). HEK293T cells were transfected with plasmids encoding Flag-NLRC5 or its mutants and HA-K63-Ub with or without Myc-MyD88. Cell lysates were immunoprecipitated with anti-Flag beads and immunoblotted with the indicated antibodies (bottom). (E) Fluorescence microscopy of NLRC5 or K1178R NLRC5 mutant (red) and Ub (green) in HeLa cells with or without LPS stimulation. Bars, 10 µm. (F) HEK293T/TLR4 cells were transfected with plasmids encoding Flag-tagged NLRC5 and its mutants 24 h before LPS (200 ng/ml) treatment. Cell lysates were immunoprecipitated with anti-Flag beads and immunoblotted with the indicated antibodies. (G) Luciferase activity of HEK293T cells transfected with plasmids encoding MyD88, with Flag-NLRC5 and its mutants, together with an NF-κB–luc reporter. ***, P < 0.001 versus the cells with MyD88 overexpression alone (two-tailed Student’s t test). Data are representative of at least three independent experiments. n = 3. Error bars indicate the SEM. IB, immunoblotting; IP, immunoprecipitation; UT, untreated; WCL, whole cell lysate.
To identify the functional ubiquitination sites on NLRC5, we first examined the interactions between TRAF2 and different domains of NLRC5 (Fig. S2, D and E). We found that TRAF2 strongly interacted with the functional domain NLRC5-D3, which is required for inhibition of NF-κB activity by NLRC5 (Cui et al., 2010). Furthermore, only the NLRC5-D3 construct, but not other NLRC5 deletions, can be efficiently ubiquitinated by TRAF2 (Fig. S2 F), suggesting that the TRAF-specific ubiquitination sites on NLRC5 are localized within the NLRC5-D3 region. Using computation-assisted algorithms (Xue et al., 2006; Moore et al., 2008), we identified three potential key ubiquitination sites in the D3 domain of NLRC5 and created K952R, K1080R, and K1178R mutants by substituting lysine 952, lysine 1,080, or lysine 1,178 with arginine (Fig. 3 D, top). The K1178R mutant, but not other mutants, almost completely abrogated the ubiquitination of NLRC5 induced by MyD88 (Fig. 3 D). Upon LPS treatment, the colocalization of Ub and the K1178R NLRC5 mutant was also markedly attenuated compared with that of wild-type (WT) NLRC5 in HeLa cells (Fig. 3 E). Consistent with our observed inverse correlation between NLRC5 ubiquitination state and IKK-β–NLRC5 binding activity (Fig. 2 D), the K1178R NLRC5 mutant showed enhanced binding ability with IKK-β as well as the enhanced inhibition of NF-κB activation compared with WT NLRC5 or other NLRC5 mutants (Figs. 3, F and G; and S2, G and H), although it can still bind to TRAF2/6 (Fig. S2 I). Collectively, these results suggest that lysine 1,178 on NLRC5 is essential for TRAF2/6-mediated ubiquitination and plays a crucial inhibitory role in NF-κB signaling.
TRAF2/6-mediated NLRC5 ubiquitination generates a feedforward loop to sensitize IKK-β signaling
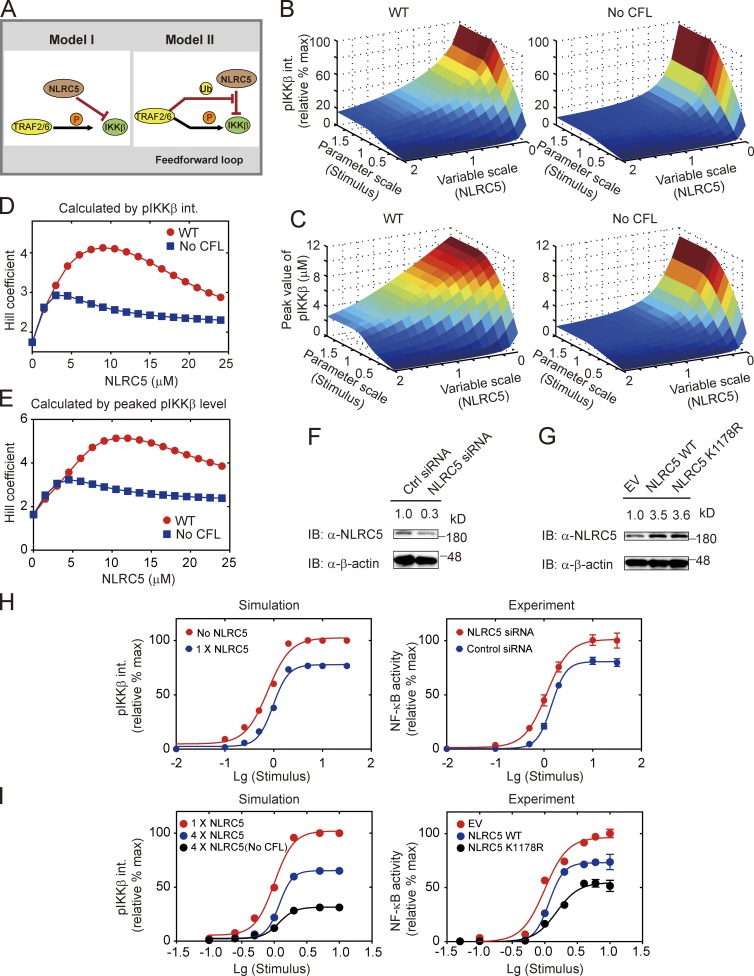
We found that TRAF2/6-mediated ubiquitination of NLRC5 generated a CFL upstream of IKK-β activation (Fig. 4 A). To analyze the effect of CFL on IKK-β activation, we varied LPS doses and NLRC5 levels in silico simultaneously to investigate the integrated or peaking levels of total pIKK-β under both WT and NLRC5 ubiquitination–deficient conditions. Analysis of the pIKK-β phase spaces showed that deficiency in NLRC5 ubiquitination severely inhibited IKK-β activation (Fig. 4, B and C). In addition, we found that cells with few NLRC5 proteins may compromise the ultrasensitivity of the dose–response curve for LPS (Fig. 4 D). The switch-like behavior for LPS-induced NF-κB activation became even more evident when ubiquitination of NLRC5 was considered in cells with intermediate levels of NLRC5 (Fig. 4 D). Qualitatively similar results were obtained when the systematic responses were evaluated with pIKK-β peak values (Fig. 4 E).
Figure 4.
Ubiquitination of NLRC5 by TRAF2/6 creates a feedforward loop to increase the ultrasensitivity of NF-κB signaling. (A) A proposed feedforward loop in model II generated by TRAF2/6-mediated NLRC5 ubiquitination. The circled Ub and P denote ubiquitination and phosphorylation, respectively. (B and C) The integrated pIKK-β (B) or peaked pIKK-β (C) responses with varying stimuli and NLRC5 under WT (left) and feedforward-deficient (right) conditions. Note that the stimuli and NLRC5 were rescaled to indicate relative changes to the reference values. The integrated pIKK-β responses were normalized by the maximum. (D and E) The dynamic changes for Hill coefficients of the integrated pIKK-β (D) or peaked pIKK-β (E) responses with varying NLRC5 levels under WT (red circle) and feedforward-deficient (blue squares) conditions. (F and G) HEK293T cells were transfected with NLRC5 siRNA or control siRNA (F) or with WT NLRC5 or K1178R NLRC5 mutations (G). Cell lysates were immunoblotted with anti-NLRC5 antibody. (H) Simulation of the dose response evaluated with integrated pIKK-β under either 1× NLRC5 (blue; 1.739 ± 0.243, the 95% confidence interval for Hill coefficient, the same for I) or NLRC5 KO (red; 2.569 ± 0.332) conditions in model II. The integrated pIKK-β value was normalized by the maximum (left). Experimental verification of the dose responses were performed by luciferase reporter assay (right). HEK293T cells were transfected with either control (blue; 2.929 ± 0.236) or NLRC5 siRNA (red; 1.816 ± 0.210). NF-κB–luc activity was determined and normalized by the maximum. (I) Simulation of the dose responses for LPS with 1× NLRC5 (red; 2.567 ± 0.391), 4× NLRC5 (blue; 3.934 ± 0.544), and 4× NLRC5 with no CFL (black; 2.805 ± 0.494). The integrated pIKK-β value was normalized by the maximum (left). NF-κB–luc activity was evaluated in HEK293T cells transfected with EV (red; 2.178 ± 0.226), NLRC5 plasmid (blue; 3.849 ± 0.444), or K1178R NLRC5 mutants (black; 2.827 ± 0.415). Data in H and I are represented in logarithm scale. Curves are fitted using the least squares method. Error bars indicate the SEM. IB, immunoblotting; Lg, Log10.
To systematically investigate the ultrasensitivity of NF-κB activation to NLRC5 or CFL, we knocked down endogenous NLRC5 or attenuated the CFL by coexpressing WT NLRC5 or K1178R NLRC5 mutant (Fig. 4, F and G). The simulation indicated that reducing NLRC5 levels promoted IKK-β activation but lowered the ultrasensitivity of IKK-β signaling (Fig. 4 H, left; Hill coefficient shifts from 2.569 to 1.739). The experimental data also showed similar results on NF-κB activity, using either scrambled siRNA or NLRC5 siRNA (Fig. 4 H, right; Hill coefficient shifts from 2.929 to 1.861). Meanwhile, elevating NLRC5 protein levels increased the ultrasensitivity of IKK-β or NF-κB signaling both theoretically (Fig. 4 I, left; Hill coefficient shifts from red 2.567 to blue 3.934) and experimentally (Fig. 4 I, right; Hill coefficient shifts from red 2.178 to blue 3.849). However, removing the CFL either in silico (Fig. 4 I, left; Hill coefficient shifts from blue 3.934 to black 2.805) or using the K1178R mutant (Fig. 4 I, right; Hill coefficient shifts from blue 3.849 to black 2.827) may decrease the ultrasensitivity of IKK-β or NF-κB signaling. Collectively, our results suggest that NLRC5 levels may determine NF-κB activation, and the CFL generated by TRAF2/6-mediated NLRC5 ubiquitination can sensitize systematic responses to LPS stimulation.
Identification of the DUBs responsible for NLRC5 deubiquitination
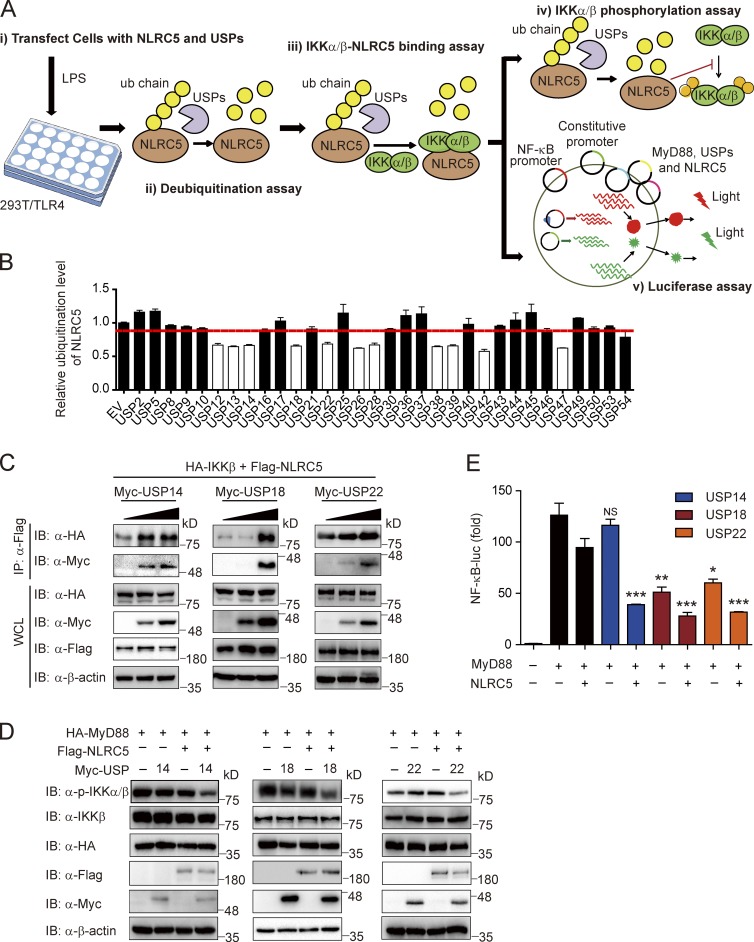
The simulation analysis suggested that deubiquitination of NLRC5 may contribute to the resolution of NF-κB signaling by restoring the pool of unmodified NLRC5 (Fig. 2 G). To determine which DUBs are responsible for restoring NLRC5 inhibitory function on NF-κB signaling, we screened 32 members of the Ub-specific protease (USP) family (Fig. 5 A). We discovered 11 USP members that down-regulated the ubiquitination of NLRC5 (Fig. 5 B and Fig. S3 A). Next, we tested whether these 11 USPs enhance the interaction between NLRC5 and IKK-β (Fig. S3 B). Three USPs, including USP14, USP18, and USP22, were found to bind with NLRC5 and to facilitate the interaction between NLRC5 and IKK-β (Fig. 5 C). USP14, USP18, and USP22 also enhanced the NLRC5-dependent inhibition of IKK-β phosphorylation as well as MyD88-induced NF-κB activation (Fig. 5, D and E).
Figure 5.
Identification of the specific DUBs responsible for NLRC5 deubiquitination. (A) The workflow showing experimental design to screen potential DUBs of NLRC5. (B) The quantified ubiquitination level of NLRC5 was regulated by USP family (related to Fig. S3 A). n = 3. The red line indicates the average NLRC5 ubiquitination level of all samples. (C) HEK293T/TLR4 cells were transfected with plasmids encoding HA-IKK-β, Flag-NLRC5, and increasing amounts of Myc-USP14, Myc-USP18, or Myc-USP22. Cell lysates were immunoprecipitated with anti-Flag antibodies followed by immunoblotting. (D) HEK293T cells were transfected with plasmids encoding HA-MyD88, Flag-NLRC5, Myc-USP14, Myc-USP18, or Myc-USP22 in the indicated combinations. Cell lysates were immunoblotted with the indicated antibodies. (E) HEK293T cells were transfected with NF-κB–luc and pRL-TK–luc reporters and the plasmids encoding MyD88, NLRC5, USP14, USP18, or USP22 in the indicated combinations and analyzed for NF-κB–dependent luciferase activity. *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus the cells with MyD88 and NLRC5 overexpression (two-tailed Student’s t test). Data are representative of at least three independent experiments. n = 3. Error bars indicate the SEM. IB, immunoblotting; IP, immunoprecipitation; WCL, whole cell lysate.
Importantly, we observed that USP18 and USP22, but not USP14, may directly inhibit IKK-β activation through an NLRC5-independent mechanism (Fig. S3 C). Therefore, we selected USP14 to further identify its regulatory mechanism in NLRC5 deubiquitination and NF-κB inhibition.
USP14 modulates NLRC5 function through its DUB activity
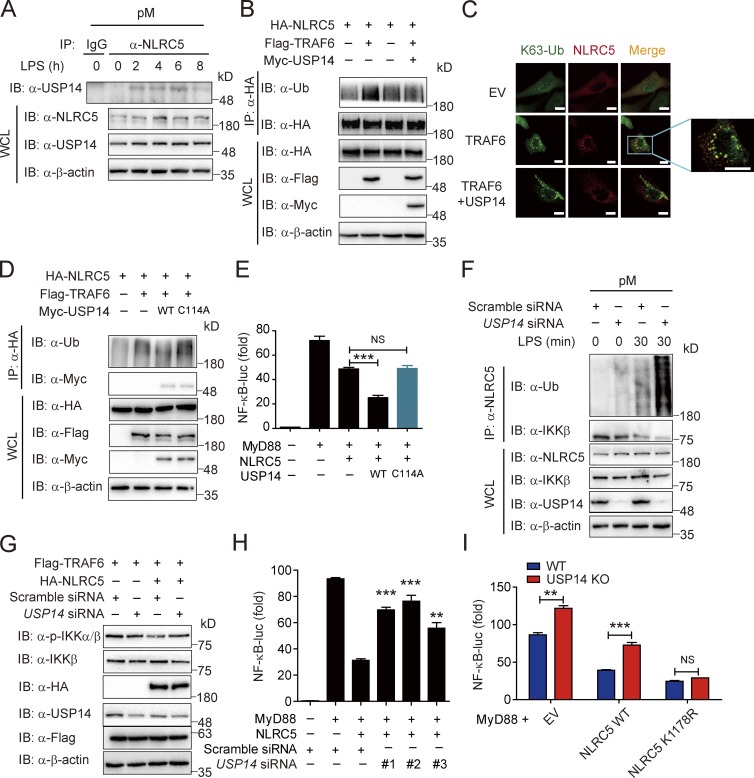
We next sought to determine whether USP14 directly interacts with NLRC5 to modulate its function and observed that the interaction between NLRC5 and USP14 is up-regulated by LPS treatment in pMs (Fig. 6 A). Without influencing the protein level of NLRC5 (Fig. S4 A), USP14 specifically decreased the K63- but not K48-linked ubiquitination of NLRC5 (Fig. S4 B). Furthermore, USP14 reversed TRAF2/6-mediated NLRC5 ubiquitination (Figs. 6 B and S4 C), and the puncta of ubiquitinated NLRC5 was diminished upon ectopic expression of USP14 in HeLa cells (Fig. 6 C). Next, we tested whether USP14 also enhanced NLRC5-mediated inhibition of TRAF2/6-independent noncanonical NF-κB signaling (Fig. S4 D). Our results showed that USP14 failed to enhance NLRC5 inhibition in response to NIK-induced NF-κB activation compared with MyD88-induced NF-κB activation (Fig. S4, E and F). These results suggest that USP14 specifically affects canonical NF-κB signaling by reversing TRAF2/6-mediated NLRC5 ubiquitination.
Figure 6.
USP14 modulates NLRC5 function through removal of polyubiquitin chains on NLRC5. (A) The interaction between NLRC5 and USP14 in pMs by LPS treatment at the indicated time points. (B) HEK293T cells were transfected with plasmids encoding HA-NLRC5 and Flag-TRAF6 with or without Myc-USP14. Cell lysates were immunoprecipitated with anti-HA beads and immunoblotted with the indicated antibodies. (C) Fluorescence microscopy of NLRC5 (red) and K63-Ub (green) in HEK293T cells transfected with TRAF6, together with USP14 or EV. Bars, 10 µm. (D) HEK293T cells were transfected with plasmids encoding HA-NLRC5, Flag-TRAF6, and Myc-USP14 (WT or C114A). Cell lysates were immunoprecipitated with anti-HA beads and immunoblotted with the indicated antibodies. (E) Luciferase activity of HEK293T cells transfected with plasmid for HA-NLRC5, Flag-MyD88, and Myc-USP14 (WT or C114A) together with an NF-κB–luc reporter. (F) pMs were transfected with scrambled siRNA or USP14 siRNA 36 h before LPS treatment at the indicated time points. Cell lysates were immunoprecipitated with anti-NLRC5 antibody and immunoblotted with anti-Ub antibody. (G) USP14 was knocked down in HEK293T cells by hUSP14 siRNA 12 h before transfection of HA-NLRC5 and Flag-TRAF6. Cell lysates were immunoblotted with the indicated antibodies. (H) Luciferase activity of HEK293T cells transfected with plasmids for Flag-NLRC5 and Myc-MyD88 as well as hUSP14-specific or control shRNA together with an NF-κB–luc reporter. (I) Luciferase activity of WT or USP14 KO cells transfected with plasmids for Myc-MyD88 and Flag-NLRC5 (WT or K1178R), together with an NF-κB–luc reporter. (E, H, and I) Data are representative of three independent experiments. n = 3. Error bars indicate the SEM. **, P < 0.01; ***, P < 0.001 versus the cells with MyD88 and NLRC5 overexpression or WT cells (two-tailed Student’s t test). Data are representative of at least three independent experiments. IB, immunoblotting; IP, immunoprecipitation; WCL, whole cell lysate.
Next, we determined whether USP14 inhibited NLRC5 ubiquitination through its DUB activity. We substituted a cysteine residue for arginine within the USP14 catalytic domain, thus generating an inactive USP14 mutant C114A (Lee et al., 2010). We found that USP14 (C114A) cannot remove the TRAF6-induced polyubiquitin chains on NLRC5 (Fig. 6 D). USP14 (C114A) also failed to enhance the inhibitory effect of NLRC5 on MyD88-induced NF-κB activation (Fig. 6 E). These results suggest that USP14 inhibits NLRC5 ubiquitination through its DUB activity.
Because USP14 specifically enhanced NLRC5-mediated inhibition of NF-κB activation, we reasoned that USP14 deficiency would result in the accumulation of ubiquitinated NLRC5 and increased NF-κB activation under physiological conditions. We first tested the knockdown efficiency of siRNAs on USP14 levels (Fig. S4 G). Next, we tested the effect of USP14 knockdown on the ubiquitination of NLRC5 in both pMs and BMMs (Figs. 6 F and S4 H). Ubiquitinated NLRC5 levels were higher in response to USP14 siRNA compared with scrambled siRNA transfection (control) at 30 min after LPS treatment. USP14 knockdown also reversed NLRC5-mediated inhibition of IKK phosphorylation (Fig. 6 G) as well as NF-κB–luc activity (Fig. 6 H).
To better understand the function of USP14 in NF-κB signaling, we knocked down USP14 in pMs and found that knockdown of USP14 enhanced IKK-β phosphorylation as well as TNF production after LPS treatment (Fig. S4, I and J). However, knockdown of NLRC5 significantly abrogated the inhibitory function of USP14 on NF-κB activation (Fig. S4 K). We next generated USP14 knockout (KO) cells using clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 technology (Fig. S4 L) and found that the up-regulation of NF-κB activation in USP14 KO cells is totally reversed by NLRC5 siRNA (Fig. S4 M). These results suggest that USP14 inhibits NF-κB signaling in an NLRC5-dependent manner. We also observed elevated NF-κB–luc activity in USP14 KO cells compared with WT cells after cotransfection with empty vector (EV) or WT NLRC5 (Fig. 6 I). However, the K1178R NLRC5 mutant showed similar inhibitory effects of NF-κB activation in both WT and USP14 KO cells (Fig. 6 I), suggesting that the function of USP14 in NF-κB signaling is dependent on the regulation of NLRC5 ubiquitination.
USP14 and NLRC5 abundance contribute to the differential sensitivities of NLRC5 in NF-κB activation in different cell types
Recent studies have shown that the effect of NLRC5 deficiency on NF-κB activation and inflammatory responses varies among immune cell types (Kumar et al., 2011; Tong et al., 2012; Yao et al., 2012). To identify potential factors contributing to this discrepancy, we varied each parameter and calculated the sensitivity coefficient of NLRC5. The results showed that variations in most parameters did not substantially influence the sensitivity to NLRC5 (Fig. S5 A). However, we noted that the NLRC5 deubiquitination rate (d9) and NLRC5 protein abundance were positively correlated with NLRC5 sensitivities (Fig. S5 A).
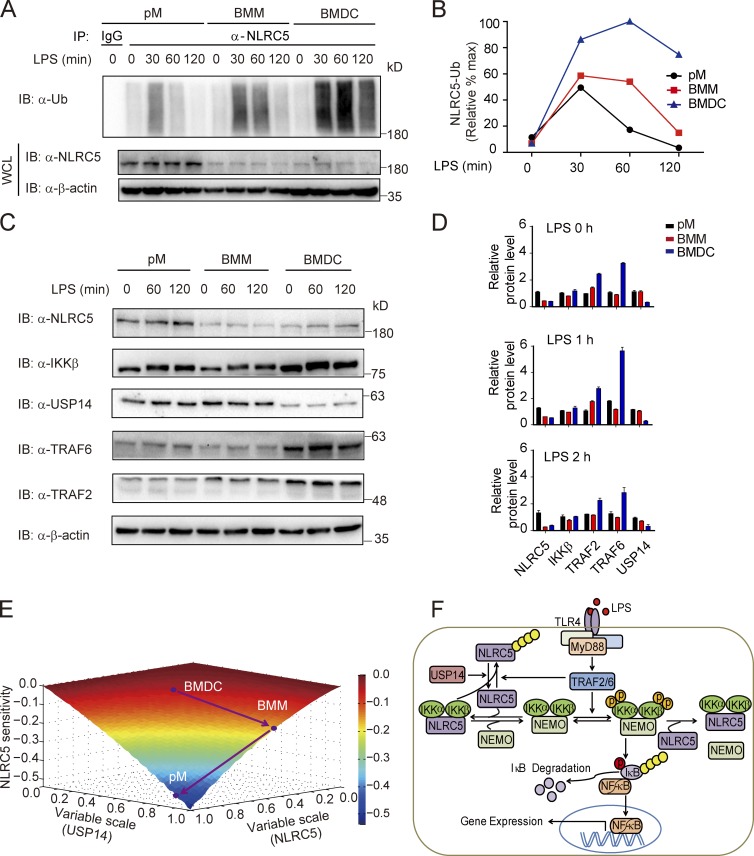
Based on the simulation, we explored whether NLRC5 ubiquitination state is varied upon LPS treatment in different cell types. We observed distinct ubiquitination patterns of NLRC5 in pMs, BMMs, and BMDCs with LPS treatment (200 ng/ml; Fig. 7, A, and B). Ubiquitination of NLRC5 in pMs appeared transient with a single peak at ∼30 min after stimulation. The duration of the peak was enhanced in BMMs (from 30 to 60 min after stimulation), with subsequent loss of ubiquitination ∼120 min after stimulation (Fig. 7, A and B). On the contrary, ubiquitination levels of NLRC5 were sustained in BMDCs up to at least 4 h after stimulation (Fig. S5 G), starting at 30 min after stimulation (Fig. 7 A). The distinct ubiquitination patterns of NLRC5 in pMs, BMMs, and BMDCs were also observed with a lower dose of LPS treatment (100 ng/ml; Fig. S5, B–E). Consistent with these results, we and others have reported that the production of proinflammatory cytokines is markedly enhanced in Nlrc5−/− pMs, moderately enhanced in Nlrc5−/− BMMs, and minimally different in Nlrc5−/− BMDCs compared with WT cells (Cui et al., 2010; Kumar et al., 2011; Tong et al., 2012). These results suggest the immune cell–specific differential sensitivities to Nlrc5 deficiency on NF-κB activation may partially be ascribed to diverse ubiquitination dynamics of NLRC5.
Figure 7.
USP14 and NLRC5 levels contribute to the differential NLRC5 ubiquitination dynamics as well as to the sensitivities of NLRC5 on NF-κB activation. (A) The ubiquitination level of NLRC5 by LPS treatment (200 ng/ml) at the indicated time from pMs, BMMs, and BMDCs. (B) The quantified ubiquitination level of NLRC5 in pMs, BMMs, or BMDCs. The ubiquitination level of NLRC5 was normalized by the maximum. (C) NLRC5, IKK-β, USP14, TRAF2, and TRAF6 protein abundance by LPS treatment at the indicated time points in pMs, BMMs, and BMDCs. (D) Densitometric measurement of band intensity in C. Data are representative of three independent experiments. n = 3. Error bars indicate the SEM. (E) Simulated effects of various degrees of the NLRC5 and USP14 on NLRC5 sensitivity. Dots denote three cell lines: BMDCs, BMMs, and pMs as indicated in the phase diagram. (F) Proposed model illustrating how the reversible Ub editing of NLRC5 by TRAF2/6 and USP14 regulates the NF-κB signaling pathway. The circled letter P denotes phosphorylation, and the yellow chain composed of several circles represents polyubiquitination. IB, immunoblotting; IP, immunoprecipitation; WCL, whole cell lysate.
Next, we experimentally tested whether the abundance of NLRC5 or DUBs was different among different types of cells. The results showed high levels of USP14 mRNA in pMs, intermediate levels of USP14 mRNA in BMMs, and minimal levels of USP14 mRNA in BMDCs (Fig. S5 F). Furthermore, pMs had the highest levels of several DUBs, including USP14, USP22, and CYLD, compared with those in BMMs and BMDCs (Fig. S5 F). We also examined the protein level of NLRC5 and other signaling proteins in pMs, BMMs, and BMDCs by LPS treatment and found that only the TRAF6 protein level was up-regulated in BMDCs, whereas the protein levels of NLRC5, IKK-β, TRAF2, and USP14 had little change after LPS treatment (Fig. 7, C and D). Consistently, we confirmed that the protein levels of USP14 were higher in macrophages (RAW264.7 cells, pMs, and BMMs) than in BMDCs (Figs. 7, C and D; and S5 H). In addition, we observed that NLRC5 protein abundance was higher in pMs, but relatively lower in BMMs and BMDCs (Fig. 7 D).
To reconcile current understanding of NLRC5 in innate immune regulation, we delineated NLRC5 sensitivities as a function of USP14 and NLRC5 to denote the relative locations of different cell lines based on our measurements (Fig. 7 D and Fig. S5 H; note that (1, 1) denotes RAW264.7 cells). The results showed that BMDCs, which expressed the lowest NLRC5 and USP14 levels overall, were most insensitive to NLRC5 variation (Fig. 7 E). Higher intrinsic USP14 levels in BMMs compared with BMDCs “forced” BMMs to move toward a state of higher USP14 values and subtly raised the sensitivity to NLRC5 in BMMs. At much higher levels of NLRC5 in pMs, relative to BMMs, pMs manifested in the valley, further increasing sensitivity to NLRC5 (Fig. 7 D). Collectively, our results suggest that differential sensitivities to NLRC5 variation in diverse cell types may partially result from the discrepancy between NLRC5 and USP14 expression.
Discussion
Specific recognition of PAMPs by innate immune receptors initiates immune responses, leading to the production of proinflammatory factors. Aberrant innate immune responses may result in inflammation-associated diseases and potential cancer development. Therefore, understanding how innate immunity is dynamically controlled by positive and negative regulators may shed light on effective therapeutics for inflammation-induced diseases. Accumulating evidence indicates that the roles of NLRs in innate immune regulation are versatile. NOD1, NOD2, NLRP3, and other inflammasome-associated NLRs act as microbial sensors or adapters to initiate inflammatory responses. However, several regulatory NLRs, including NLRC3, NLRC5, NLRX1, and NLRP4, function as negative regulators of innate immune responses, including inflammation, antiviral immunity, and autophagy (Cui et al., 2014).
Although it has been well demonstrated that PTM plays an important role in changing the functions of signaling proteins, only a few studies have mentioned the role of PTM for NLRs. It has been reported that deubiquitination of NLRP3, mediated by a DUB BRCC3, is a required step for activation of the NLRP3 inflammasome (Py et al., 2013). NLRP3 is also negatively regulated by NO-mediated S-nitrosylation (Mishra et al., 2013). In addition, phosphorylation of NLRC4 is related to NLRC4 inflammasome activity, although its exact role is still controversial (Qu et al., 2012). We previously determined that NLRX1, a member of regulatory NLRs, is rapidly ubiquitinated after LPS stimulation (Xia et al., 2011). However, the function of NLRX1 ubiquitination as well as its regulatory mechanism are still not clear. Besides NLRX1, the PTM of other regulatory NLRs is not known.
Our previous study demonstrated a role for NLRC5 in dampening NF-κB responses by blocking IKK-α/-β phosphorylation through interaction with IKK-α/-β (Cui et al., 2010). We also observed that the interaction between NLRC5 and IKK-α/-β appears to be dynamic during the early phase (1–2 h) of LPS stimulation (Cui et al., 2010). However, the mechanism responsible for the pattern of NLRC5–IKK-β interactions has not been well understood. In this study, we have combined experimental and computational methods to show that NLRC5 undergoes robust ubiquitination upon LPS treatment, which is responsible for the dynamic oscillatory pattern of NLRC5–IKK-β interactions. Furthermore, we defined a role for NLRC5 ubiquitination in the negative regulation of NF-κB signaling. We have shown that K63-linked ubiquitination of NLRC5 induced its dissociation from IKK-β, which abrogated the inhibitory function of NLRC5 in NF-κB signaling. Next, we identified specific E3 ligases and DUBs responsible for the reversible Ub editing of NLRC5. We found that TRAF2/6 ubiquitinated NLRC5 upon LPS stimulation, whereas several DUBs were able to deubiquitinate NLRC5 and enhance the interaction between NLRC5 and IKK-β. Unlike USP18 and USP22, we found that USP14 regulates NF-κB signaling in an NLRC5-dependent manner. USP14 specifically reversed TRAF2/6-mediated ubiquitination of NLRC5 and facilitated NLRC5 to negatively regulate canonical NF-κB signaling. However, USP14 failed to facilitate NLRC5 to inhibit noncanonical NF-κB signaling independent of TRAF2 and TRAF6. These results were further validated in USP14 KO cells. USP14 deficiency significantly attenuates the inhibitory ability of NLRC5 on NF-κB activation, whereas the K1178R NLRC5 mutant showed similar inhibition of NF-κB activity in both WT and USP14 KO cells. A recent study showed that USP18 attenuated NF-κB activation by targeting TAK1-TAB1 for deubiquitination (Liu et al., 2013). We also found that USP18 blocked IKK phosphorylation by inhibiting the ubiquitination of NEMO (Yang et al., 2015). These nonredundant deubiquitination events at multiple levels may help to efficiently terminate proinflammatory NF-κB signaling.
Based on these experiments, we propose a working model to illustrate how dynamic Ub editing of NLRC5 affects NF-κB signaling (Fig. 7 F). With LPS stimulation, TRAF2/6 are activated through MyD88 and in turn initiate a downstream kinase signaling cascade including IKK-β signaling. Meanwhile, active TRAF2/6 ubiquitinates NLRC5 at lysine 1,178. Ubiquitination of NLRC5 lowers its ability to bind to IKK-β, thereby freeing NLRC5-bound IKKs and increasing the pool of available IKK complex for efficient activation. The ubiquitination of NLRC5 could be reversely regulated by USP14. Once NLRC5 is deubiquitinated, it can bind to IKK-β again to terminate NF-κB signaling. Therefore, the abundance of USP14 may determine whether NLRC5 can be deubiquitinated in time to terminate NF-κB signaling. In addition, TRAF2/6-mediated NLRC5 ubiquitination may generate CFL, which may enhance the ultrasensitivity of NF-κB activation.
Dynamics in ultrasensitivity to protein signaling occurs naturally in biological systems, and direct experimental evidence has been shown in diverse signaling pathways (Ferrell and Ha, 2014). In contrast to Michaelian responses, which are usually generated by monomeric ligand/receptor binding, an ultrasensitive response may prevent signal degradation and establish thresholds (Ferrell and Ha, 2014). The NF-κB response has recently been shown to behave in a switch-like manner in individual cells, suggesting the existence of molecular thresholds (Tay et al., 2010). In B cell receptor signaling, engagement of CARMA1 may contribute to the switch-like activation of NF-κB (Shinohara et al., 2014). Additionally, a long-term effect involving NF-κB–induced Rela expression upon LPS stimulation may also evoke a feedback dominant switching and set a threshold for host pathogenic responses (Sung et al., 2014). Here, we identified a previously unrecognized CFL generated by NLRC5 ubiquitination. This CFL may provide a short-term effect upstream of NF-κB activation and generate another potential threshold mechanism for NF-κB signaling. In addition, we experimentally validated our simulation results, indicating that various NLRC5 abundances allow different thresholds of IKK activation to be set and thereby shape dynamic NF-κB signaling. Notably, even during CFL deficiency, there also exists a perceptible ultrasensitivity in dynamic IKK activation (Fig. 4 I). We reasoned that this might possibly be ascribed to competitive interactions among NLRC5, IKK-α/-β, and NEMO because inhibitor ultrasensitivity serves as another simple mechanism for generating ultrasensitive responses (Ferrell and Ha, 2014). These short-term and long-term mechanisms may possibly provide “fail-safe” redundancy (Kitano, 2004) and act in concert to ensure reliable decision making in inflammatory NF-κB responses.
Previous studies on the role of NLRC5 in NF-κB activation are controversial. We and others observed that NLRC5 deficiency markedly enhanced proinflammatory responses in NLRC5 knockdown RAW264.7 cells and Nlrc5−/− pMs, but only weakly enhanced proinflammatory responses in BMMs (Cui et al., 2010; Kumar et al., 2011; Tong et al., 2012). However, we and Kumar et al. (2011) revealed no functional role of NLRC5 in LPS-induced NF-κB signaling in BMDCs (Tong et al., 2012). Additionally, Yao et al. (2012) observed that NLRC5 ablation does not affect the NF-κB–dependent cytokine production in BMMs treated with a high dose of LPS. Therefore, in-depth understanding of NLRC5 regulation of NF-κB signaling is strongly needed to clarify the discrepancies of different sensitivities to NLRC5 variation on NF-κB activation in distinct cell types. We observed that cellular sensitivities to NLRC5 depend on only a subset of parameters or initial conditions. The simulation data implied that individual cells can use distinct strategies to fine-tune their sensitivities to NLRC5. Regulating only intrinsic NLRC5 expression seems to be a relatively simple mechanism. We observed that sensitivities to NLRC5 were generally negatively correlated with the ubiquitination level of NLRC5. Notably, the level of the IKK-β did not vary significantly among different cells. However, the NLRC5-related DUBs did show substantial variations, suggesting that different types of cells may modulate their sensitivities to NLRC5 ablation by adjusting the intracellular state of deubiquitination environment. The “two-dimensional control” (i.e., NLRC5 and DUB levels) may at least provide a plausible illustration about whether different types of cells show different sensitivities to NLRC5 ablation. However, we argue that there might be other unexploited dimensions that cells may use to alter their sensitivities to NLRC5 variation (e.g., by intricately modifying signaling generation; Fig. S5 A). These potential mechanisms will require future experimental investigation.
Collectively, the ubiquitination state of NLRC5 may specifically shape NF-κB signaling by multiple mechanisms: (a) allowing for rapid activation and efficient resolution of innate immune signaling; (b) creating a feedforward loop that sets a threshold to ensure robust innate immune responses; and (c) altering cellular sensitivity to NLRC5 ablation. Consequently, the Ub-linking status of NLRC5 may not simply affect the association between NLRC5 and IKK-β but also systematically redesign innate immune responses to invading pathogen.
Our findings have demonstrated that the reversible Ub editing of NLRC5 played a crucial role for precisely regulating NF-κB signaling. The CFL generated by NLRC5 ubiquitination must be tightly regulated to present the optimal threshold behavior according to diverse conditions. By this means, variations in the intracellular state of the deubiquitination environment may permit different thresholds of NF-κB activation and thus dynamically shape inflammatory responses.
Materials and methods
Cell culture and reagents
HEK293T, RAW264.7, and HeLa cells were cultured in DMEM (Hyclone) containing 10% FBS (Gibco) incubated in a 5% CO2 chamber (Thermo Fisher Scientific). BMMs, BMDCs, and mouse pMs were cultured in RPMI 1640 (Gibco) containing 10% FBS. BMMs or BMDCs were derived from bone marrow of 18–20 g C57BL/6 mice (Guangdong Medical Laboratory Animal Center) and cultured for 6–8 d with 100 ng/ml macrophage colony-stimulating factor (PeproTech) or 20 ng/ml granulocyte macrophage colony-stimulating factor (PeproTech). pMs were harvested with 5 ml PBS 3 d after intraperitoneal injection of 1 ml of 4% thioglycollate medium (Sigma-Aldrich). MEFs were derived from embryos staged at embryonic days (E) 13.5–14.5 of gestation of C57BL/6 WT mice. The embryos were cut and enzymatically disaggregated after removing blood and liver tissue to generate single MEF cells, which were then plated at 106 cells per 100-mm dish in DMEM. HEK293T, HEK293T/TLR4, RAW264.7, BMMs, BMDCs, and pMs were treated with 100–200 ng/ml LPS (Sigma-Aldrich). HeLa cells were treated with 1 µg/ml LPS.
Plasmids, antibodies, and reagents
USP14 KO plasmids were generated with lentiCRISPR v2. All of the following plasmids were generated with empty pcDNA3.1 vector, including Myc-MyD88, Myc-USP14, Myc-USP18, Myc-USP22, Myc-IsoT, HA-Ub, HA-Ub-K63, HA-Ub-K48, HA-NLRC5 (WT), HA-NLRC5 (D1), HA-NLRC5 (D2), HA-NLRC5 (D3), HA-NLRC5 (D4), HA-MyD88, Flag-TRAF2, Flag-TRAF3, Flag-TRAF5, Flag-TRAF6, Flag-NLRC5 (WT), Flag-NLRC5 (K952R), Flag-NLRC5 (K1080R), Flag-NLRC5 (K1178R), Flag-IKK-β (WT), Flag-IKK-β (SSEE), Flag-IKK-β (SSAA), and 32 USP family member plasmids. CF488A goat anti–rat IgG heavy and light chains (H+L), CF488A goat, anti–rabbit IgG (H+L), CF488A rabbit anti–HA IgG, CF568 donkey anti–goat IgG (H+L), and CF568 goat anti–mouse IgG (H+L) were purchased from Biotium. Anti-Flag M2-peroxidase and anti-USP14 mouse monoclonal antibodies were purchased from Sigma-Aldrich. Anti–IKK-β mouse monoclonal antibodies were from EMD Millipore. Anti–c-Myc-peroxidase mouse monoclonal antibodies and anti–HA-peroxidase rat monoclonal antibodies were purchased from Roche. Anti-NLRC5 rabbit monoclonal antibodies were from Abcam. Anti-NLRC5 goat monoclonal antibodies, anti-Ub mouse monoclonal antibodies, donkey anti–goat IgG-HRP, goat anti–mouse IgG-HRP, and goat anti–rabbit IgG-HRP antibodies were from Santa Cruz Biotechnology Inc. Mouse, goat, and rabbit IgG was from Beyotime. Antiphospho–IKK-α/-β rabbit monoclonal antibodies were obtained from Cell Signaling Technology. Antiphosphoserine/threonine antibodies were from BD. TRIzol, Lipofectamine 2000, and RNAiMAX were purchased from Invitrogen. ViaFect transfection reagent was from Promega. Protein G agarose and protein A agarose were purchased from Thermo Fisher Scientific. Anti-Flag M2 affinity gel and thioglycollate medium were purchased from Sigma-Aldrich. Point mutations, including Flag-NLRC5 (K952R), Flag-NLRC5 (K1080R), Flag-NLRC5 (K1178R), and Myc-USP14 (C114A), were generated by site-directed mutagenesis (Sbsgene) using the primers as follows: NLRC5 (K952R), forward (5′-AGCCAGAGGAGCAGAGGGGGCCCCAGGAGAG-3′) and reverse (5′-CTCTCCTGGGGCCCCCTCTGCTCCTCTGGCT-3′); NLRC5 (K1080R), forward (5′-TCCTGAGAGGGGACAGGACAAGCAGGGATAT-3′) and reverse (5′-ATATCCCTGCTTGTCCTGTCCCCTCTCAGGA-3′); NLRC5 (K1178R), forward (5′-CGGGACTGTCCCCGAACAGCCCCTTCCTGCT-3′) and reverse (5′-AGCAGGAAGGGGCTGTTCGGGGACAGTCCCG-3′); and USP14 (C114A), forward (5′-AAACCTTGGTAACACTCGTTACATGAATGCCAC-3′) and reverse (5′-GTGGCATTCATGTAACGAGTGTTACCAAGGTTT-3′).
Cell transfection and reporter assay
HEK293T, HEK293T/TLR4, and HeLa cells were transfected with Lipofectamine 2000 (Invitrogen) or ViaFect according to the manufacturers’ protocols, along with a plasmid encoding an NF-κB luciferase (luc) reporter (firefly luciferase) and a pRL-TK (renilla luciferase plasmid), together with plasmids encoding HA-NLRC5 and other indicated plasmids. Empty pcDNA3.1 vector was used to maintain equal amounts of DNA among wells. Cells were collected at 24 h after transfection, and luciferase activity was measured with a Dual-Luciferase assay (Promega) by a luminometer (Luminoskan Ascent; Thermo Fisher Scientific) according to the manufacturer’s protocol. Reporter gene activity was determined by normalization of the firefly luciferase activity to renilla luciferase activity.
Immunoprecipitation and immunoblotting
For immunoprecipitation, whole cell extracts were prepared after transfection or stimulation with appropriate ligands, followed by incubation overnight with the appropriate antibodies plus anti-Flag beads (Sigma-Aldrich), anti-HA beads (Sigma-Aldrich), or protein A/G beads (Thermo Fisher Scientific). Beads were washed three to five times with low-salt lysis buffer, and immunoprecipitates were eluted with 3× SDS loading buffer (Cell Signaling Technology) and resolved by SDS-PAGE. Proteins were transferred to polyvinylidene fluoride membranes (Bio-Rad Laboratories) and further incubated with the appropriate antibodies. LumiGlo Chemiluminescent Substrate System (KPL) was used for protein detection.
Immunofluorescence staining
BMMs, HeLa, or HEK293T cells were seeded on glass-bottomed culture dishes (Nest Scientific) and then fixed, permeabilized, blocked, and stained following the manufacturer’s instructions (Biotium). The secondary antibodies included Alexa Fluor 488– and Alexa Fluor 568–conjugated antibodies against mouse or rabbit IgG (Biotium). Confocal images were acquired using a microscope (LSM710; Carl Zeiss) equipped with 100× 1.40 NA oil objectives, with Immersol 518F (Carl Zeiss) as imaging medium and a camera (AxioCam HRc; Carl Zeiss) under the control of Zen 2008 software (Carl Zeiss). The images were processed for gamma adjustments using LSM Zen 2008 or ImageJ software (National Institutes of Health).
RNA extraction and quantitative PCR
Total RNA was isolated from primary cells with TRIzol reagent (Life Technologies), according to the manufacturer’s instructions. 2 µg RNA was reverse transcribed to cDNA with a PrimeScript RT reagent kit (Takara Bio Inc.). Quantitative real-time PCR was performed using Lightcyler 480 SYBR green I Master (Roche) with quantitative PCR primers as follows: mUsp14, forward (5′-TCCAGAAGAACCCTCTGCTAAA-3′) and reverse (5′-TCCATAGCGGTTGCTAACTGT-3′); mUsp18, forward (5′-CAGGAGTCCCTGATTTGCGTG-3′) and reverse (5′-CAGAGGCTTTGCGTCCTTATC-3′); mUsp22, forward (5′-TGGGAACCAACTAAACGGGAG-3′) and reverse (5′-TGCCTATCCGACAGAAAGAAGT-3′); mA20, forward (5′-ACCATGCACCGATACACGC-3′) and reverse (5′-AGCCACGAGCTTCCTGACT-3′); mCyld, forward (5′-GGACAGTACATCCAAGACCGT-3′) and reverse (5′-TCCCTCACAGTTGGTAATTGCC-3′); and mGapdh, forward (5′-AGGTCGGTGTGAACGGATTTG-3′) and reverse (5′-TGTAGACCATGTAGTTGAGGTCA-3′).
RNAi
hUSP14-siRNA, mUSP14-siRNA, mTRAF2-siRNA, mTRAF6-siRNA, and control (scramble) siRNA were obtained from Shanghai TranSheep Bio Co. Ltd. and transfected into HEK293T cells, pMs, and BMMs with RNAiMAX (Invitrogen) according to the manufacturer’s instructions. siRNA sequences were as follows: scramble-siRNA: forward (5′-UUCUCCGAACGUGUCACGUTT-3′) and reverse (5′-ACGUGACACGUUCGGAGAATT-3′); hUSP14-siRNA 1#, forward (5′-GCAAAGAAAUGCCUUGUAUTT-3′) and reverse (5′-AUACAAGGCAUUUCUUUGCTT-3′); hUSP14-siRNA 2#, forward (5′-GGCUCCAAUAAUUGUGGAUTT-3′) and reverse (5′-AUCCACAAUUAUUGGAGCCTT-3′); hUSP14-siRNA 3#, forward (5′-CCUCGCAGAGUUGAAAUAATT-3′) and reverse (5′-UUAUUUCAACUCUGCGAGGTT-3′); mUsp14-siRNA 1#, forward (5′-GGCUCCAAUAAUUGUGGCUTT-3′) and reverse (5′-AGCCACAAUUAUUGGAGCCTT-3′); mTRAF2-siRNA 1#, forward (5′-CCAUAACAACCGGGAGCAUTT-3′) and reverse (5′-AUGCUCCCGGUUGUUAUGGTT-3′); and mTRAF6-siRNA 1#, forward (5′-AUAUUUGCCAGAGGACAGCTT-3′) and reverse (5′-UUCCUCAUAAUUUGGGUCCTT-3′).
Generation of USP14 KO cells by CRISPR/Cas9 technology
USP14 KO cells were generated by a CRISPR/Cas9 system, and the sequence of target USP14–guide RNA (gRNA) is (5′-CACCGGAATGACTCTACTAATGAT-3′).
Computational modeling
The computational models were formulated using ordinary differential equations in MATLAB R2011a (MathWorks). The values of unfixed parameters were estimated with the MATLAB toolbox PottersWheel (Maiwald and Timmer, 2008). The local sensitivity coefficients of the nonzero variables and kinetic parameters were calculated using custom code in MATLAB. The Hill coefficient was calculated in Prism 5 (GraphPad Software).
Model construction
We developed a simplified model of the NLRC5-regulated IKK-β activation network using mass action kinetics. The reactions regarding LPS and TLR4 binding were adopted from Rivière et al. (2009); the first five reactions are shown in Table S1. It was reported that LPS induces a conformational change in MD-2 and enables each MD-2 molecule to make direct contacts with both TLR4-LRR domains (Shin et al., 2007). LPS signals via the TLR4–MD-2 complex; however, Visintin et al. (2006) have already indicated that LPS activates TLR4 signaling in an MD-2–independent manner, and these reactions can be interpreted as TLR4-mediated signaling for simplicity. Furthermore, we fixed the association and dissociation parameters (k1 and d1; Table S2) as 0.33 µM∙min−1 and 1.0 × 10−7 min−1, respectively, according to a published model (Rivière et al., 2009). We further fixed the degradation rate constant of TLR4-LPS as 0.2 min−1 (Rivière et al., 2009). There exists a series of reactions between LPS binding, IKK-β complex activation, and NLRC5 ubiquitination. According to experiments described in Lo et al. (2008), we fixed the association and dissociation rate of NEMO–IKK-β as 420 µM∙min−1 and 1.5 min−1. In addition, it was reported that activated IKK-β phosphorylates serine 68 in NEMO, which results in the dissociation of the NEMO–pIKK-β complex. Thus, NEMO–pIKK-β is easy to dissociate and difficult to associate (Hayden and Ghosh, 2008). k5 is comparatively less than k4, and d5 is relatively greater than d4. Incorporating these reactions into the model requires more parameters and gives rise to considerable uncertainties. To simplify our model, we took all the reactions between LPS binding and IKK-β activation as an activating signal (abbreviated as S in our model) to downstream pathways. The signal not only activates the IKK-β complex but also induces NLRC5 ubiquitination because TRAF2/6 contributes to both processes (Fig. 3, A–D; and Fig. S2, B and C). We supposed that S could ubiquitinate either free NLRC5 or that in a complex form with different efficiencies. Although specific recognition of (free) Ub chains (e.g., K63, mixed K11/K63, or Met1 linked) by NEMO is critically important in pathogen-induced IKK-β activation (Komander and Rape, 2012), this effect is not considered in our model. The signal-mediated ubiquitination of the IKK-β complex only introduces several direct IKK-β activation reactions and does not significantly affect the dynamic properties of the whole model, especially NLRC5-mediated IKK-β inhibition. In addition, discriminating between these NEMO ubiquitination events in the model would introduce much complexity and potentially mask the roles of NLRC5 in NF-κB regulation. We also considered the reversible interactions between NEMO and (activated) IKK-β. The competitive binding for IKK-β between NLRC5 and NEMO was also incorporated. We have previously identified that NLRC5 inhibited NF-κB–dependent responses by interacting with IKK-α and IKK-β with similar affinity (Cui et al., 2010). Therefore, we only consider one species (i.e., IKK-β) for simplicity. Furthermore, NLRC5 can bind constitutively active and inactive IKK-β with similar affinity and inhibit the kinase activity (Fig. S1 A). Therefore, we assumed that interaction between NLRC5 and pIKK-β contributed to the pool of IKK-β–NLRC5 complexes. For the kinetic reactions, refer to Table S1.
The ordinary equations governing the dynamics of the NLRC5 regulatory model were therefore given as follows:
and
The deterministic model was numerically simulated using the ode23s operator in MATLAB (version 7.12.0.635, R2011a; MathWorks). The model was fitted to experimental measurements using the PottersWheel MATLAB toolbox (Maiwald and Timmer, 2008). In brief, the deviation between the measurements (ŷ) and the simulation y was determined by
where h denotes the step size. In our model, there are two measurements: total phosphorylated IKK (pIKK-β) and total IKK-β–NLRC5. Total pIKK-β represents the sum of NEMO–pIKK-β and pIKK-βfree (corresponding species in Table S2), whereas total IKK-β–NLRC5 denotes IKK-β–NLRC5. Kinetic parameters and nonzero initial conditions were simultaneously estimated in a logarithmic space via a trust region optimization method with default settings. Note that several parameters were fixed according to experimental calculation before the fitting process. The amount of LPS was assigned as 2, corresponding to 200 ng/ml, as used in our previous experiments. Using arbitrarily reasonable initial values, we reached a parameter set that meets the χ2 rules (i.e., the reference parameter set; χ2 denotes the sum of deviations between experimental data and model simulations). (Given totally n experimental dots, the parameter set meets the χ2 rule if χ2/n < 1). Using this parameter as a seed (i.e., starting value), 2,000 plausible fits (χ2/n < 1) were generated to screen an optimized parameter set for further analysis.
In Fig. 7, we incorporated the DUB, USP14, into model II to investigate the effect of USP14 on NLRC5 sensitivity. We only considered the specific DUB USP14 in our model for simplicity. Other DUBs are not considered because incorporating these nonspecific effects adds much complexity to the model. The original parameter d9 in model II was modified to a mass action item d9′ × [USP14] for simplicity. For the modified parameter and initial condition, refer to Table S2.
Local sensitivity analysis
Local sensitivity or control coefficient evaluates the systematic response to an infinitesimal disturbance in nominal model parameters. The dynamic parameter sensitivity Si (Wu et al., 2008) is defined as follows:
where M denotes the peak amplitude (the maximum concentration), p (p1, p2 . . . pn) is the nonzero variable and kinetic parameters vector, and pi is the ith nonzero variable and kinetic parameter. The local sensitivity was calculated as the change in M aroused by a 1% change in each nonzero variable and kinetic parameter.
Calculation of the Hill coefficient
The normalized data (rescaled by the maximum) were analyzed by nonlinear regression (log [agonist] vs. normalized response-variable slope) in Prism 5. The Hill slope describes the steepness of the family of curves.
Statistical analysis
The results of all quantitative experiments are reported as mean ± SEM of three independent experiments. Comparisons between groups for statistical significance were assessed with a two-tailed Student’s t test.
Online supplemental material
Fig. S1 illustrates that NLRC5 ubiquitination is important for its inhibitory function on NF-κB activation. Fig. S2 presents data that TRAF2/6 targets the functional domain of NLRC5 for ubiquitination. Fig. S3 shows the identification of the potential DUBs for NLRC5. Fig. S4 shows the function of USP14 to deubiquitinate NLRC5 in canonical NF-κB signaling. Fig. S5 includes the data to demonstrate that diverse expression patterns of NLRC5 and USP14 determine the NLRC5 sensitivity on NF-κB activation. Table S1 lists the reactions and rates of model I and model II. Table S2 lists the parameters for model I and model II. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201505091/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201505091.dv.
Supplementary Material
Acknowledgments
This work was supported by the National Key Basic Research Program of China (grants 2014CB910800 and 2015CB859800), the National Natural Science Foundation of China (grants 31370869, 31522018, and 31400714), Guangdong Natural Science Funds for Distinguished Young Scholar (grant S2013050014772), and the Guangdong Innovative Research Team Program (grant 2011Y035). T. Sun. was partially supported by the Anhui Provincial Natural Science Foundation (grant 1408085QC50).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- BMDC
- bone marrow–derived dendritic cell
- BMM
- bone marrow–derived macrophage
- CFL
- coherent feedforward loop
- CRISPR
- clustered regularly interspaced short palindromic repeat
- DUB
- deubiquitinase
- EV
- empty vector
- IKK
- IκB kinase
- KO
- knockout
- LPS
- lipopolysaccharide
- MEF
- mouse embryonic fibroblast
- NLR
- NOD-like receptor
- PAMP
- pathogen-associated molecular pattern
- pM
- peritoneal macrophage
- PTM
- posttranslational modification
- TLR
- Toll-like receptor
- TRAF
- TNF receptor–associated factor
- Ub
- ubiquitin
- USP
- Ub-specific protease
- WT
- wild type
References
- Akira S., Takeda K., and Kaisho T.. 2001. Toll-like receptors: Critical proteins linking innate and acquired immunity. Nat. Immunol. 2:675–680. 10.1038/90609 [DOI] [PubMed] [Google Scholar]
- Benko S., Magalhaes J.G., Philpott D.J., and Girardin S.E.. 2010. NLRC5 limits the activation of inflammatory pathways. J. Immunol. 185:1681–1691. 10.4049/jimmunol.0903900 [DOI] [PubMed] [Google Scholar]
- Birrell M.A., McCluskie K., Hardaker E., Knowles R., and Belvisi M.G.. 2006. Utility of exhaled nitric oxide as a noninvasive biomarker of lung inflammation in a disease model. Eur. Respir. J. 28:1236–1244. 10.1183/09031936.00048506 [DOI] [PubMed] [Google Scholar]
- Chen Z.J. 2012. Ubiquitination in signaling to and activation of IKK. Immunol. Rev. 246:95–106. 10.1111/j.1600-065X.2012.01108.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Zhu L., Xia X., Wang H.Y., Legras X., Hong J., Ji J., Shen P., Zheng S., Chen Z.J., and Wang R.F.. 2010. NLRC5 negatively regulates the NF-κB and type I interferon signaling pathways. Cell. 141:483–496. 10.1016/j.cell.2010.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li Y., Zhu L., Liu D., Songyang Z., Wang H.Y., and Wang R.F.. 2012. NLRP4 negatively regulates type I interferon signaling by targeting the kinase TBK1 for degradation via the ubiquitin ligase DTX4. Nat. Immunol. 13:387–395. 10.1038/ni.2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Chen Y., Wang H.Y., and Wang R.F.. 2014. Mechanisms and pathways of innate immune activation and regulation in health and cancer. Hum. Vaccin. Immunother. 10:3270–3285. 10.4161/21645515.2014.979640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell J.E. Jr., and Ha S.H.. 2014. Ultrasensitivity part I: Michaelian responses and zero-order ultrasensitivity. Trends Biochem. Sci. 39:496–503. 10.1016/j.tibs.2014.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., and Karin M.. 2002. Missing pieces in the NF-κB puzzle. Cell. 109:S81–S96. 10.1016/S0092-8674(02)00703-1 [DOI] [PubMed] [Google Scholar]
- Häcker H., and Karin M.. 2006. Regulation and function of IKK and IKK-related kinases. Sci. STKE. 2006:re13 10.1126/stke.3572006re13 [DOI] [PubMed] [Google Scholar]
- Häcker H., Tseng P.H., and Karin M.. 2011. Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat. Rev. Immunol. 11:457–468. 10.1038/nri2998 [DOI] [PubMed] [Google Scholar]
- Hayden M.S., and Ghosh S.. 2004. Signaling to NF-κB. Genes Dev. 18:2195–2224. 10.1101/gad.1228704 [DOI] [PubMed] [Google Scholar]
- Hayden M.S., and Ghosh S.. 2008. Shared principles in NF-κB signaling. Cell. 132:344–362. 10.1016/j.cell.2008.01.020 [DOI] [PubMed] [Google Scholar]
- Kitano H. 2004. Biological robustness. Nat. Rev. Genet. 5:826–837. 10.1038/nrg1471 [DOI] [PubMed] [Google Scholar]
- Kobayashi K.S., and van den Elsen P.J.. 2012. NLRC5: A key regulator of MHC class I-dependent immune responses. Nat. Rev. Immunol. 12:813–820. 10.1038/nri3339 [DOI] [PubMed] [Google Scholar]
- Komander D., and Rape M.. 2012. The ubiquitin code. Annu. Rev. Biochem. 81:203–229. 10.1146/annurev-biochem-060310-170328 [DOI] [PubMed] [Google Scholar]
- Kumar H., Pandey S., Zou J., Kumagai Y., Takahashi K., Akira S., and Kawai T.. 2011. NLRC5 deficiency does not influence cytokine induction by virus and bacteria infections. J. Immunol. 186:994–1000. 10.4049/jimmunol.1002094 [DOI] [PubMed] [Google Scholar]
- Lee B.H., Lee M.J., Park S., Oh D.C., Elsasser S., Chen P.C., Gartner C., Dimova N., Hanna J., Gygi S.P., et al. . 2010. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 467:179–184. 10.1038/nature09299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Li H., Zhong B., Blonska M., Gorjestani S., Yan M., Tian Q., Zhang D.E., Lin X., and Dong C.. 2013. USP18 inhibits NF-κB and NFAT activation during Th17 differentiation by deubiquitinating the TAK1–TAB1 complex. J. Exp. Med. 210:1575–1590. 10.1084/jem.20122327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Y.C., Maddineni U., Chung J.Y., Rich R.L., Myszka D.G., and Wu H.. 2008. High-affinity interaction between IKKβ and NEMO. Biochemistry. 47:3109–3116. 10.1021/bi702312c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiwald T., and Timmer J.. 2008. Dynamical modeling and multi-experiment fitting with PottersWheel. Bioinformatics. 24:2037–2043. 10.1093/bioinformatics/btn350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra B.B., Rathinam V.A., Martens G.W., Martinot A.J., Kornfeld H., Fitzgerald K.A., and Sassetti C.M.. 2013. Nitric oxide controls the immunopathology of tuberculosis by inhibiting NLRP3 inflammasome–dependent processing of IL-1β. Nat. Immunol. 14:52–60. 10.1038/ni.2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C.B., Bergstralh D.T., Duncan J.A., Lei Y., Morrison T.E., Zimmermann A.G., Accavitti-Loper M.A., Madden V.J., Sun L., Ye Z., et al. . 2008. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 451:573–577. 10.1038/nature06501 [DOI] [PubMed] [Google Scholar]
- Orzalli M.H., and Knipe D.M.. 2014. Cellular sensing of viral DNA and viral evasion mechanisms. Annu. Rev. Microbiol. 68:477–492. 10.1146/annurev-micro-091313-103409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrilli V., Papin S., and Tschopp J.. 2005. The inflammasome. Curr. Biol. 15:R581 10.1016/j.cub.2005.07.049 [DOI] [PubMed] [Google Scholar]
- Price N.D., Foltz G., Madan A., Hood L., and Tian Q.. 2008. Systems biology and cancer stem cells. J. Cell. Mol. Med. 12:97–110. 10.1111/j.1582-4934.2007.00151.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Py B.F., Kim M.S., Vakifahmetoglu-Norberg H., and Yuan J.. 2013. Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Mol. Cell. 49:331–338. 10.1016/j.molcel.2012.11.009 [DOI] [PubMed] [Google Scholar]
- Qu Y., Misaghi S., Izrael-Tomasevic A., Newton K., Gilmour L.L., Lamkanfi M., Louie S., Kayagaki N., Liu J., Kömüves L., et al. . 2012. Phosphorylation of NLRC4 is critical for inflammasome activation. Nature. 490:539–542. 10.1038/nature11429 [DOI] [PubMed] [Google Scholar]
- Rajsbaum R., Versteeg G.A., Schmid S., Maestre A.M., Belicha-Villanueva A., Martínez-Romero C., Patel J.R., Morrison J., Pisanelli G., Miorin L., et al. . 2014. Unanchored K48-linked polyubiquitin synthesized by the E3-ubiquitin ligase TRIM6 stimulates the interferon-IKKε kinase-mediated antiviral response. Immunity. 40:880–895. 10.1016/j.immuni.2014.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivière B., Epshteyn Y., Swigon D., and Vodovotz Y.. 2009. A simple mathematical model of signaling resulting from the binding of lipopolysaccharide with Toll-like receptor 4 demonstrates inherent preconditioning behavior. Math. Biosci. 217:19–26. 10.1016/j.mbs.2008.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H.J., Lee H., Park J.D., Hyun H.C., Sohn H.O., Lee D.W., and Kim Y.S.. 2007. Kinetics of binding of LPS to recombinant CD14, TLR4, and MD-2 proteins. Mol. Cells. 24:119–124 [PubMed] [Google Scholar]
- Shinohara H., Behar M., Inoue K., Hiroshima M., Yasuda T., Nagashima T., Kimura S., Sanjo H., Maeda S., Yumoto N., et al. . 2014. Positive feedback within a kinase signaling complex functions as a switch mechanism for NF-κB activation. Science. 344:760–764. 10.1126/science.1250020 [DOI] [PubMed] [Google Scholar]
- Sung M.H., Li N., Lao Q., Gottschalk R.A., Hager G.L., and Fraser I.D.. 2014. Switching of the relative dominance between feedback mechanisms in lipopolysaccharide-induced NF-κB signaling. Sci. Signal. 7:ra6 10.1126/scisignal.2004764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O., and Akira S.. 2010. Pattern recognition receptors and inflammation. Cell. 140:805–820 10.1016/j.cell.2010.01.022 [DOI] [PubMed] [Google Scholar]
- Tay S., Hughey J.J., Lee T.K., Lipniacki T., Quake S.R., and Covert M.W.. 2010. Single-cell NF-κB dynamics reveal digital activation and analogue information processing. Nature. 466:267–271. 10.1038/nature09145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y., Cui J., Li Q., Zou J., Wang H.Y., and Wang R.F.. 2012. Enhanced TLR-induced NF-κB signaling and type I interferon responses in NLRC5 deficient mice. Cell Res. 22:822–835. 10.1038/cr.2012.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhapurapu S., and Karin M.. 2009. Regulation and function of NF-κB transcription factors in the immune system. Annu. Rev. Immunol. 27:693–733. 10.1146/annurev.immunol.021908.132641 [DOI] [PubMed] [Google Scholar]
- Visintin A., Halmen K.A., Khan N., Monks B.G., Golenbock D.T., and Lien E.. 2006. MD-2 expression is not required for cell surface targeting of Toll-like receptor 4 (TLR4). J. Leukoc. Biol. 80:1584–1592. 10.1189/jlb.0606388 [DOI] [PubMed] [Google Scholar]
- Wu J., and Chen Z.J.. 2014. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 32:461–488. 10.1146/annurev-immunol-032713-120156 [DOI] [PubMed] [Google Scholar]
- Wu W.H., Wang F.S., and Chang M.S.. 2008. Dynamic sensitivity analysis of biological systems. BMC Bioinformatics. 9:S17 10.1186/1471-2105-9-S12-S17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X., Cui J., Wang H.Y., Zhu L., Matsueda S., Wang Q., Yang X., Hong J., Songyang Z., Chen Z.J., and Wang R.F.. 2011. NLRX1 negatively regulates TLR-induced NF-κB signaling by targeting TRAF6 and IKK. Immunity. 34:843–853. 10.1016/j.immuni.2011.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z.P., Sun L., Chen X., Pineda G., Jiang X., Adhikari A., Zeng W., and Chen Z.J.. 2009. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 461:114–119. 10.1038/nature08247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Li A., Wang L., Feng H., and Yao X.. 2006. PPSP: Prediction of PK-specific phosphorylation site with Bayesian decision theory. BMC Bioinformatics. 7:163 10.1186/1471-2105-7-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Tang E., Guan K., and Wang C.Y.. 2003. IKKβ plays an essential role in the phosphorylation of RelA/p65 on serine 536 induced by lipopolysaccharide. J. Immunol. 170:5630–5635. 10.4049/jimmunol.170.11.5630 [DOI] [PubMed] [Google Scholar]
- Yang Z., Xian H., Hu J., Tian S., Qin Y., Wang R.F., and Cui J.. 2015. USP18 negatively regulates NF-κB signaling by targeting TAK1 and NEMO for deubiquitination through distinct mechanisms. Sci. Rep. 5:12738 10.1038/srep12738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Wang Y., Chen F., Huang Y., Zhu S., Leng Q., Wang H., Shi Y., and Qian Y.. 2012. NLRC5 regulates MHC class I antigen presentation in host defense against intracellular pathogens. Cell Res. 22:836–847. 10.1038/cr.2012.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Mo J., Swanson K.V., Wen H., Petrucelli A., Gregory S.M., Zhang Z., Schneider M., Jiang Y., Fitzgerald K.A., et al. . 2014. NLRC3, a member of the NLR family of proteins, is a negative regulator of innate immune signaling induced by the DNA sensor STING. Immunity. 40:329–341. 10.1016/j.immuni.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.