Asp localizes to the minus ends of spindle microtubules at the pole and within the spindle and cross-links them to adjacent microtubules.
Abstract
Depletion of Drosophila melanogaster Asp, an orthologue of microcephaly protein ASPM, causes spindle pole unfocusing during mitosis. However, it remains unclear how Asp contributes to pole focusing, a process that also requires the kinesin-14 motor Ncd. We show that Asp localizes to the minus ends of spindle microtubule (MT) bundles and focuses them to make the pole independent of Ncd. We identified a critical domain in Asp exhibiting MT cross-linking activity in vitro. Asp was also localized to, and focuses the minus ends of, intraspindle MTs that were nucleated in an augmin-dependent manner and translocated toward the poles by spindle MT flux. Ncd, in contrast, functioned as a global spindle coalescence factor not limited to MT ends. We propose a revised molecular model for spindle pole focusing in which Asp at the minus ends cross-links MTs at the pole and within the spindle. Additionally, this study provides new insight into the dynamics of intraspindle MTs by using Asp as a minus end marker.
Introduction
Faithful chromosome segregation during mitosis is dependent on the integrity of the mitotic spindle. In animal cells, the mitotic spindle is diamond shaped, which helps ensure the equal segregation of sister chromatids into two daughter cells; defects in this process lead to aneuploidy and micronuclei formation, probable causes of human diseases such as cancer or microcephaly (Rujano et al., 2013; Maiato and Logarinho, 2014; Zhang et al., 2015). The centrosome is present at the spindle pole and functions as a microtubule (MT) organizing center. However, physical or genetic deletion of the centrosome does not significantly perturb spindle shape and function (Khodjakov et al., 2000; Megraw et al., 2001). The molecular players required for acentrosomal spindle formation are constitutively active in the presence of centrosomes (Moutinho-Pereira et al., 2013). MTs are generated within the spindle through augmin- and MT-dependent nucleation (branching nucleation) and in a chromatin-dependent manner, regardless of the presence or absence of centrosomes. Spindle MTs are focused by the action of minus end–directed motor proteins (Gaglio et al., 1996; Goshima et al., 2005a; Maiato and Logarinho, 2014; Baumbach et al., 2015).
Two motor proteins, cytoplasmic dynein and kinesin-14 (Ncd in Drosophila melanogaster and HSET in mammals), have been identified as the factors responsible for organizing the spindle poles (Endow et al., 1994; Heald et al., 1996, 1997; Walczak et al., 1998; Mountain et al., 1999; Morales-Mulia and Scholey, 2005; Goshima et al., 2005a). In Drosophila cells, depletion of Ncd or dynein primarily affects spindle MT focusing or centrosome association to the pole, respectively (Morales-Mulia and Scholey, 2005; Goshima et al., 2005a, 2007). Because the motors possess MT cross-linking activity and minus end–directed motility, a mechanistic model involving minus end–directed MT transport (by dynein) and MT minus end cross-linking (by Ncd) has been proposed (Endow et al., 1994; Maiato et al., 2004; Goshima et al., 2005a; Baumbach et al., 2015). In addition, two nonmotor proteins also play a role in spindle MT focusing. NuMA possesses MT cross-linking activity and is localized at the pole (Merdes et al., 1996). However, NuMA also binds to, and is transported by, dynein to the pole (Merdes et al., 2000). Therefore, it is possible that NuMA can also cross-link spindle MTs by interacting with the dynein motor (Radulescu and Cleveland, 2010).
Another factor is Drosophila Asp, an orthologue of the microcephaly protein ASPM, depletion of which results in severe spindle MT unfocusing (Ripoll et al., 1985; Saunders et al., 1997; Wakefield et al., 2001; Morales-Mulia and Scholey, 2005). In the absence of Asp, the centrosomes are detached from the main body of the spindle, and the spindle MTs are unfocused at the pole, similar to dynein and Ncd depletion, respectively. Immunofluorescence microscopy indicated that Asp is localized at the spindle pole, the area enriched with the spindle MT minus ends, as well as at the centrosome (Saunders et al., 1997; Wakefield et al., 2001; Morales-Mulia and Scholey, 2005). Interestingly, a recent study has shown that asp mutant flies have reduced brain size, which is at least partly attributed to chromosome missegregation associated with unfocused spindle poles (Rujano et al., 2013). However, the molecular activity of Asp/ASPM remains unclear. Asp immunoisolated from cell extracts was initially reported to possess MT organizing activity around the centrosome (do Carmo Avides and Glover, 1999). However, a subsequent phenotypic study was inconsistent with this biochemical activity; centrosomal MTs appear normal in the absence of Asp, and spindle MT focusing is defective regardless of the presence or absence of the centrosome (Wakefield et al., 2001). An additional possible activity of Asp is to regulate myosin and dynein motors (Morales-Mulia and Scholey, 2005; van der Voet et al., 2009; Rujano et al., 2013). The interaction with these motors may be critical for regulating spindle rotation and positioning (van der Voet et al., 2009; Rujano et al., 2013). However, because the pole-unfocusing phenotype is not observed (myosin) or distinct (dynein) from Asp, it is unlikely that these interactions play a role in spindle MT focusing at the pole, at least in the Drosophila S2 cell line (Goshima et al., 2007). Overall, the mechanism of action of Asp remains unclear in regard to pole organization.
In this study, we studied the mechanism of spindle MT focusing in S2 cells by determining the function and dynamics of Asp (note that RNAi depletion of Mud, the Drosophila NuMA orthologue, does not induce pole disorganization in this cell line; Goshima et al., 2007; Capalbo et al., 2011; Moutinho-Pereira et al., 2013). The majority of S2 cells have more than two centrosomes, and depletion of Asp or Ncd affects centrosome number and position in the spindle, which complicates quantitative assessment of the pole defect (Goshima and Vale, 2003; Morales-Mulia and Scholey, 2005; Goshima et al., 2007). However, a functional bipolar spindle with two focused poles efficiently forms without centrosomes in S2 cells (Mahoney et al., 2006; Moutinho-Pereira et al., 2013). Here, using this experimental system, we show that Asp, independently of Ncd, accumulates at the spindle poles from early prometaphase to metaphase and functions to focus spindle MTs at the pole. Depletion of Ncd had a more global effect on spindle coalescence. Asp was also found to localize to the minus ends of intraspindle MTs that were nucleated via the augmin-dependent mechanism and was translocated to the pole via spindle MT flux. Live imaging suggested that this population of Asp is critical for cross-linking intraspindle MT minus ends to other MTs in the body of the spindle. These results led to a revised model of how spindle poles are focused in animal cells.
Results
Asp is involved in spindle MT focusing independent of Ncd
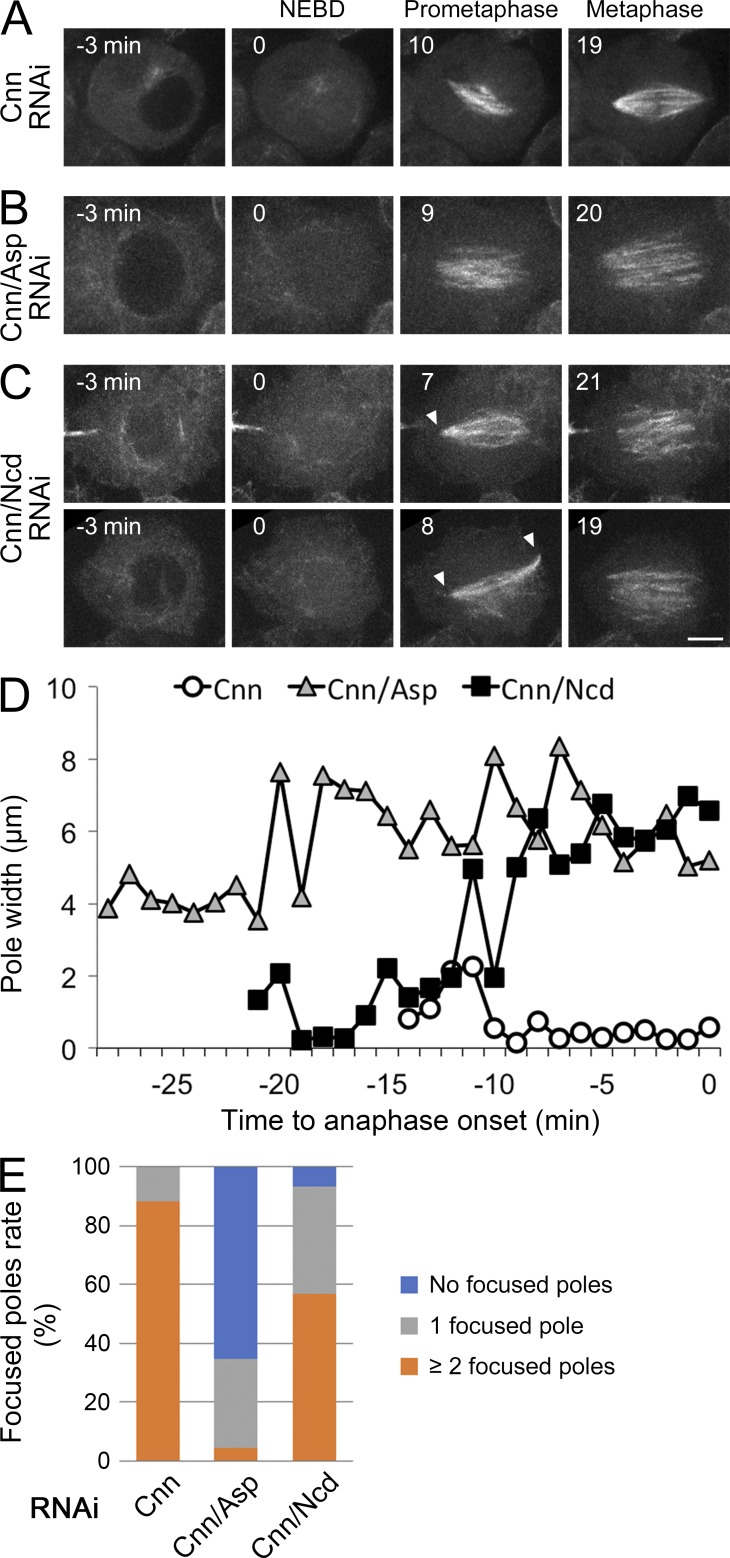
We first characterized the spindle MT–unfocusing phenotype in the absence of centrosomes and Asp using live cell microscopy. Cells were simultaneously subjected to RNAi for both Asp and centrosomin (Cnn), a centrosome maturation factor; GFP-tubulin was monitored in 3D using spinning-disk confocal microscopy (Fig. 1, Fig. S1 A, and Video 1). Control Cnn RNAi cells exhibited one or few MT clouds (two were predominant) before nuclear envelope breakdown (NEBD), and the focused spindle poles formed around the cloud upon NEBD in all 26 observed cells (Fig. 1, A, D, and E; Mahoney et al., 2006; Moutinho-Pereira et al., 2013). In contrast, pole focusing was not observed in the absence of Asp in 15/23 cells throughout prometaphase, indicating that Asp commences playing a critical role from early prometaphase (Fig. 1, B, D and E). In addition, we performed imaging after Ncd/Cnn RNAi compared with Asp/Cnn (Fig. 1 C); as expected, the cells had unfocused spindle poles in metaphase. Interestingly, 28/30 of the observed spindles had one or more transient focused poles in early prometaphase (Fig. 1 C, arrowheads, and D and E). Thus, the MT-focusing phenotype differs between Asp and Ncd in early prometaphase.
Figure 1.
Distinct pole-unfocusing phenotype after Asp or Ncd depletion. (A–C) GFP-tubulin imaging during acentrosomal spindle formation after Asp or Ncd RNAi. Cells were depleted of centrosomes by Cnn RNAi. In the absence of Asp, poles were unfocused throughout prometaphase and metaphase (9–20 min in B), whereas in the absence of Ncd, most cells contained transient focused poles during prometaphase (arrowheads in C). Bar, 5 µm. (D) Change in pole width during prometaphase and metaphase of a representative cell after each RNAi. The first time point represents the timing when overall bipolar spindles were first detected (a few minutes after NEBD). Time 0, anaphase onset. Pole width was measured after z-stack projection. (E) Quantification of the pole phenotype. A focused score was applied when one or more focused poles were transiently or continuously observed during prometaphase.
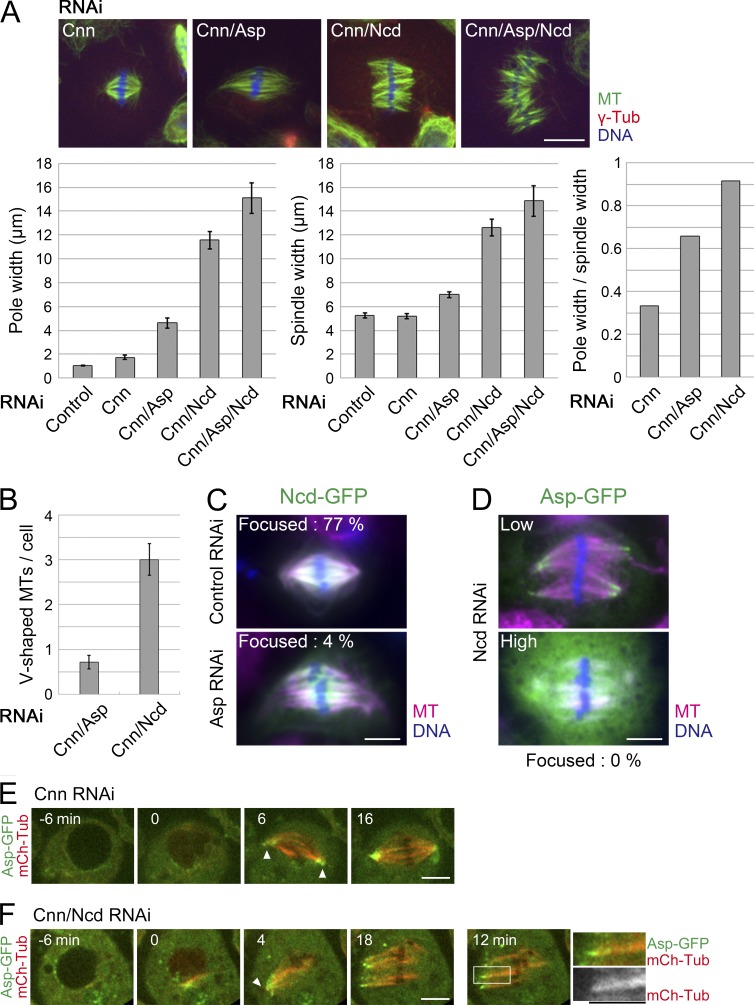
We next tested the relationship between Asp and Ncd by double RNAi and reciprocal rescue experiments. Spindle MT focusing, defined as the distance between the two farthest MT bundle ends, was more severely impaired in the double Asp/Ncd than in either single treatment (Fig. 2 A, left). Furthermore, overexpression of Ncd-GFP, which has been shown to rescue the Ncd RNAi phenotype (Goshima and Vale, 2005), could not rescue the Asp RNAi phenotype, although overexpressed Ncd induced increased MT bundling (Fig. 2 C; Goshima et al., 2005b). Conversely, ectopic expression of Asp-GFP, which is functional for spindle pole focusing (Fig. S1, B and C), failed to rescue the Ncd RNAi phenotype at the highest expression levels achieved in our cell line (Fig. 2 D). We conclude that Ncd and Asp cannot compensate for each other’s function in terms of spindle MT focusing at the pole.
Figure 2.
Asp functions independently of Ncd in acentrosomal pole focusing. (A) Quantification of spindle pole width (left), spindle width (middle), and their relative values (right) after Asp or Ncd RNAi in the absence of centrosomes. Error bars indicate SEM (n = 18–54). Bar, 10 µm. (B) Frequency of V-shaped MTs after Cnn/Asp (n = 32) or Cnn/Ncd (n = 38) RNAi. (C) Ectopic Ncd-GFP expression did not suppress the pole-unfocusing phenotype of Asp RNAi (n = 23; control n = 13). (D) Ectopic Asp-GFP expression (low and high expression levels) did not suppress the pole-unfocusing phenotype of Ncd RNAi (n = 25). (E and F) Asp-GFP and mCherry-tubulin were imaged in the presence or absence of Ncd. The cells were depleted of centrosomes using Cnn RNAi. In both cases, Asp-GFP was detected at the focused pole at prometaphase (arrowheads). Note that the right-side pole in F was out of microscopic focus. A magnification view is also displayed. Bars: (C–F) 5 µm.
We hypothesized that if Asp functions independently of Ncd, Asp-GFP would accumulate at the MT ends during early prometaphase, when MT focusing is observed regardless of the presence or absence of Ncd (Fig. 1). To test this, we performed simultaneous time-lapse imaging of Asp-GFP and mCherry-tubulin (Fig. 2, E and F; and Video 2). We observed that Asp-GFP was localized at the pole-proximal focusing points of the MTs in the Cnn RNAi background (68/71 poles had GFP signals). Interestingly, the 33/38 focusing points in early prometaphase had intense Asp-GFP signals in the absence of Ncd (Fig. 2 F, arrowhead). These data are consistent with a model according to which Asp localizes to the MT ends and ensures, at least transiently, MT cross-linking independent of Ncd.
These observations prompted us to reevaluate the spindle phenotype of Ncd. Previous studies measured the pole width after Ncd RNAi and concluded that Ncd is required for spindle MT focusing (Endow et al., 1994; Goshima et al., 2005a). However, the spindle was overall much wider in the absence of Ncd than in control or Asp RNAi–treated cells (Fig. 2 A, middle and right; the width of the spindle was quantified at the midzone). Moreover, although pole width was greater in the absence of Ncd, adjacent MTs were locally cross-linked more frequently (Fig. 2, A and B; some MTs exhibit a V shape by end connection), and the focal point had strong Asp-GFP signals (Fig. 2 D, fixed cell; F [right], living cell). These results suggest that Ncd possesses a more global spindle coalescence function, not limited to the pole region, and that pole-specific focusing is ensured by the action of Asp. In retrospect, this interpretation is more consistent with the localization data; Ncd is localized to the whole spindle and enriched at the growing plus ends of MTs (Hatsumi and Endow, 1992; Goshima et al., 2005a).
Identification of the Asp domain with MT cross-linking activity
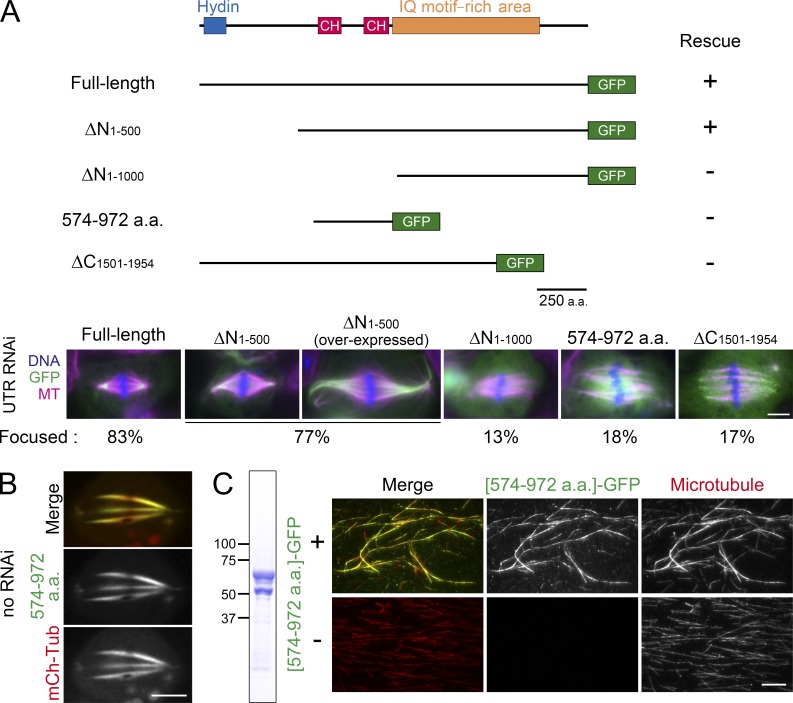
The observation described in the previous paragraph contradicts the hypothesis that Asp is merely a regulator of motors and suggests that it has its own molecular activity involving MTs. Thus far, our attempts to purify full-length recombinant Asp proteins (1,954 aa) have been unsuccessful, and therefore, the activity of full-length Asp could not be determined. To gain insight into Asp activity, we created several truncated versions of Asp and assessed their intracellular localization and function in cells depleted of endogenous Asp (Fig. 3 A).
Figure 3.
The middle region of Asp has MT cross-linking activity. (A) Rescue experiments using UTR RNAi and ectopic expression of GFP fusion proteins (n = 23–32). (B) Expression of Asp [574–972 aa]-GFP resulted in overbundling of spindle MTs and unfocused the poles. (C) Purified Asp [574–972 aa]-GFP demonstrated MT cross-linking activity in vitro. Taxol-stabilized, rhodamine-labeled MTs were mixed with (top) or without (bottom) Asp [574–972 aa]-GFP. We could not eliminate a smaller size band, possibly a degraded product of Asp [574–972 aa]-GFP. Bars: (A and B) 5 µm; (C) 10 µm.
The N-terminal region of Asp was previously assigned as the MT-binding domain based on the results of Far-Western analysis conducted with denatured protein (Saunders et al., 1997). However, we found that the Asp spindle MT–unfocusing phenotype could be rescued by a fragment deleted for the N-terminal 500 aa. This fragment localized to the pole region, and when overexpressed, the spindle had abnormally elongated, hyper-bundled MTs at the pole, suggesting that MT ends are stabilized and cross-linked by this fragment. An additional 500-aa deletion impaired Asp function and localization, indicating that the middle region of the protein is functionally critical. Interestingly, the 574–972-aa fragment, which contains two putative calponin homology domains (Rujano et al., 2013), decorated entire spindle MTs (but weakly at the midzone) and hyper-bundled MTs when overexpressed (Fig. 3 B). To test whether the Asp [574–972 aa] fragment possesses MT cross-linking activity, we expressed and purified the recombinant protein from bacteria (Fig. 3 C, left). We next mixed this protein with fluorescently labeled, stabilized MTs and observed that the protein decorated and bundled the MTs (Fig. 3 C). We conclude that the Asp [574–972 aa] fragment possesses MT cross-linking activity. However, this activity is insufficient to fulfill the full Asp function because expression of the fragment failed to rescue the spindle MT–unfocusing phenotype (Fig. 3 A). Instead, the fragment had a dominant-negative effect; the overbundled MTs were not well focused in the presence of endogenous Asp (Fig. 3 B). These results are consistent with the hypothesis that polar enrichment is a prerequisite for Asp function. Furthermore, they indicate that the IQ motifs following calponin homology domains and/or C-terminal regions are required for MT minus end targeting.
Spindle flux–dependent poleward motility of Asp within the spindle
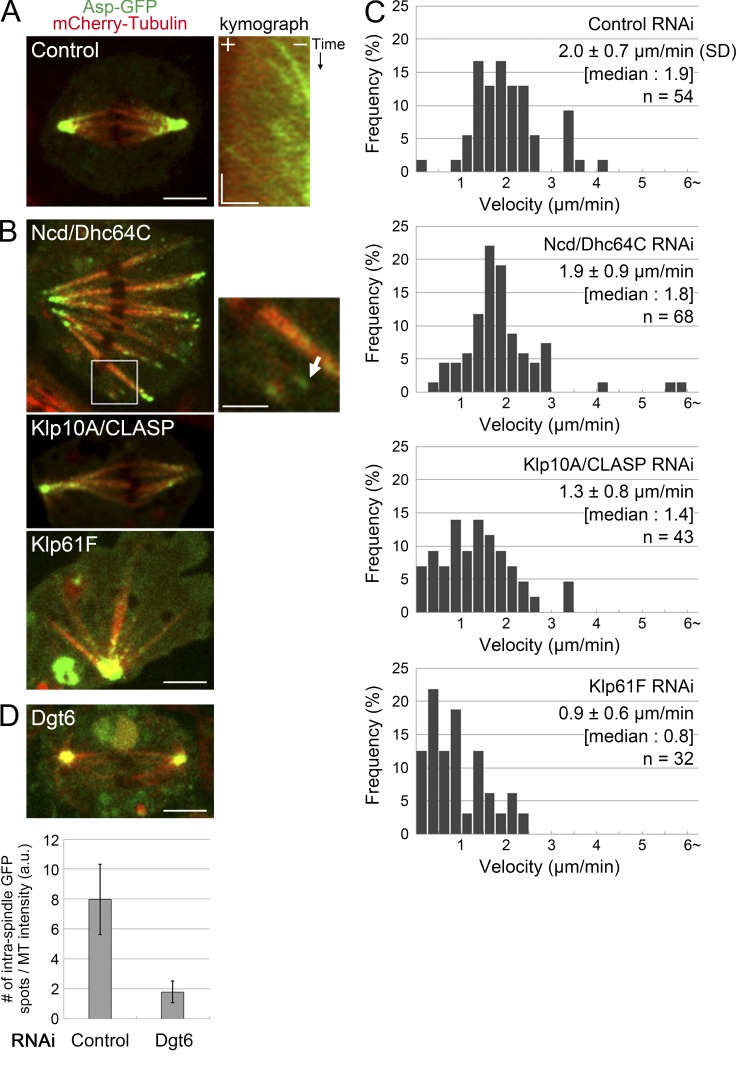
We next imaged Asp-GFP in metaphase with a higher acquisition rate. Interestingly, in addition to strong signals at the pole, we observed several Asp-GFP signals along the spindle MTs, many of which showed poleward movement at a velocity of 2.0 ± 0.7 µm/min (±SD; n = 54 GFP spots; Fig. 4 A and Video 3). Because our microscope was not sensitive enough to detect a single GFP molecule, the observed punctate signals must represent clusters of Asp-GFP. Intraspindle localization and poleward motility were similarly observed after Cnn RNAi, indicating that these are centrosome-independent events (Video 4).
Figure 4.
Asp-GFP at the intraspindle MT end is translocated to the pole by spindle flux. (A) Kymographs generated along spindle MTs indicate that Asp-GFP signals move poleward in control cells (a plus sign indicates the chromosomal side). Bars: (left) 5 µm; (right) 2 µm and 1 min. (B) Asp-GFP signals after Ncd/Dhc64C, Klp10A/CLASP, or Klp61F RNAi. An intraspindle MT bundle decorated with Asp-GFP in the Ncd/Dhc64C RNAi sample is magnified. Bars: 5 µm; (inset) 2 µm. (C) Velocity distribution of moving Asp-GFP signals. Mean velocities were significantly reduced after Klp10A/CLASP or Klp61F RNAi (P < 0.0001; Mann–Whitney U test), but not subsequent to Ncd/Dhc64C RNAi (P > 0.29). (D) Quantification of the Asp-GFP spot numbers in the absence of Dgt6, an augmin subunit (±SEM). We selected the first frame of the video in which a metaphase spindle was observed and counted the number of Asp-GFP spots in the spindle (a single focal plane). The numbers were then divided by the MT intensity in the central 60% area of the spindle (polar regions that had Asp-GFP clusters were excluded). Only the cells with similar total GFP intensity were selected for analysis (GFP intensity: 5.3 ± 1.4 [SD] for four control cells; 5.5 ± 1.5 [SD] for four Dgt6-depleted cells). Significant reduction (P < 0.021) was observed after Dgt6 RNAi. Bar, 5 µm. a.u., arbitrary units.
By analogy with NuMA, we hypothesized that the poleward motility of Asp-GFP requires the retrograde motors dynein and/or Ncd as transporters. To test this possibility, we depleted the dynein heavy chain subunit (Dhc64C) and/or Ncd by RNAi and observed Asp-GFP. Surprisingly, Asp-GFP motility was unaffected, although our RNAi constructs resulted in >90% target protein reduction (Fig. S1 A; Goshima and Vale, 2003) and induced clear spindle-unfocusing phenotypes (Fig. 4, B and C; and Video 3). Although we cannot rule out that the residual motor proteins might be sufficient for Asp transportation, these data suggest that other mechanisms are primarily responsible for Asp-GFP motility.
An additional poleward movement observed within the spindle is MT flux (Rogers et al., 2005). In S2 cells, kinetochore MTs are constantly polymerized at the plus end through the action of the CLIP-associated protein (CLASP), whereas the minus ends are depolymerized by the Klp10A/kinesin-13 MT depolymerase; thus, each tubulin subunit in the kinetochore MTs moves poleward (Rogers et al., 2004; Maiato et al., 2005; Laycock et al., 2006). Nonkinetochore MTs are also transported poleward via kinesin-5–dependent antiparallel sliding (Goshima et al., 2005b). We therefore performed double Klp10A/CLASP or single Klp61F (Drosophila kinesin-5) depletion and investigated Asp-GFP motility. Interestingly, the rate of Asp-GFP motility was significantly reduced in each of the depleted cell types (Fig. 4 C and Video 3). These results indicate that MT flux plays a role in Asp-GFP motility within the spindle.
Asp is localized to the minus end of MTs that are nucleated in the body of the spindle
Flux-based translocation of Asp-GFP in the metaphase spindle could be explained by two possible mechanisms: the Asp-GFP clusters are stably bound to the lattice of spindle MTs that are being fluxed, or alternatively, Asp-GFP is associated with the minus ends of MTs present in the spindle, and Asp-decorated MTs are being fluxed via, for example, kinesin-5–mediated sliding. Because the MTs are crowded, we were unable to visually determine whether each moving Asp-GFP is associated with the MT minus ends in the normal metaphase spindle. To distinguish between these two possibilities, we depleted augmin by RNAi. Augmin is required for MT-dependent MT nucleation within the spindle (Goshima et al., 2008). Moreover, in mammals, it has been demonstrated by electron microscopy that the minus ends are scarcely distributed within the spindle in the absence of augmin (Kamasaki et al., 2013). When Dgt6, a core subunit of augmin, was depleted, we observed in seven out of eight cells a clear reduction of Asp-GFP spots in the body of the metaphase spindle (Fig. 4 D and Video 3). This result supports the second model, according to which Asp-GFP signals in the spindle represent MT minus ends nucleated within the spindle in an augmin-dependent manner. In the absence of augmin, a few punctate GFP signals were observed at the polar region (Video 5), suggesting that Asp accumulation at the pole is not merely a result of poleward transport via flux, but Asp recognizes a specific feature on the minus ends. Consistent with these interpretations, we occasionally observed that particularly in unfocused spindles depleted of minus end–directed motors, MT bundles exhibited Asp-GFP signals at the pole-proximal end (Fig. 4 B, arrow in inset; and see the following paragraph).
Asp cross-links MT minus ends to other MTs at the pole and within the spindle
Finally, we aimed to reveal the role of Asp within the spindle, which decorates minus ends of intraspindle MTs. However, we did not have a tool with which intraspindle Asp is specifically inhibited. Then, we reasoned that valuable information could be obtained if the minus ends of MTs are marked and their behaviors are compared in the presence or absence of Asp.
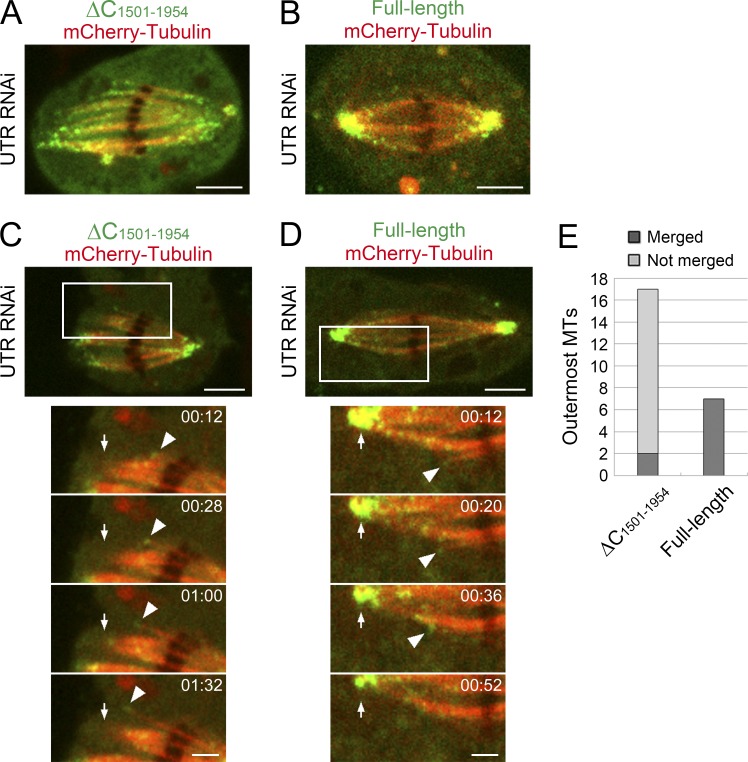
In the course of functional domain mapping, we constructed a truncated fragment, AspΔC (Asp [1–1,500 aa]), which lacks the C-terminal region and a part of the repeated IQ motifs that bind to calmodulin (Fig. 3 A; note that calmodulin RNAi phenocopies Asp in S2 cells; Goshima et al., 2007). This fragment could not rescue the spindle MT–unfocusing phenotype. However, this fragment was still enriched at the polar region (Fig. 3 A, right). Time-lapse imaging confirmed that AspΔC-GFP is localized to the minus ends of spindle MTs and translocated to the pole, similar to full-length Asp-GFP (Fig. 5, A and C; and Videos 6 and 7). Thus, by observing AspΔC-GFP in the absence of endogenous Asp, we could generate a condition in which minus ends of MTs are marked while the Asp depletion phenotype is exhibited. We compared the dynamics of AspΔC-GFP with full-length Asp(FL)-GFP in the absence of endogenous Asp.
Figure 5.
MT minus end dynamics in the absence of functional Asp. (A) Scattered MT minus ends at the polar region in the absence of functional Asp, as visualized by AspΔC-GFP expression. Endogenous Asp was depleted by RNAi. (B) Control full-length Asp(FL)-GFP at the pole. A giant cluster of GFP signals was observed at each pole because of MT focusing. (C–E) Behavior of the minus end of the outermost MT bundle in the spindle. In the absence of functional Asp (C and E), the minus end was translocated to the polar region without coalescence with other MTs. With functional Asp, such MTs were merged with adjacent MTs during translocation (D and E). Arrowheads indicate the minus ends of the outermost MT bundle, whereas arrows indicate the pole. Time is displayed in minutes/seconds. Bars: (A–D) 5 µm; (C and D, bottom) 2 µm.
In the absence of functional Asp, AspΔC-GFP signals were still mostly located at either edge of the spindle, suggesting that minus ends are enriched at the polar region similar to the wild type (Fig. 5 A). However, unlike control Asp(FL)-GFP that was visible as a single giant cluster at the pole, many dynamic GFP spots were detected in a more scattered manner (Fig. 5, A and B; and Video 6). Some of the AspΔC-GFP signals clearly decorated the minus ends of MTs. This observation directly demonstrated that the minus ends do not stably cohere to each other at the pole without functional Asp.
We next observed MT ends that were not located at the polar region. Unfortunately, it was unclear for most of the Asp(FL)-GFP or AspΔC-GFP spots in the body of the spindle whether they were in contact with other MTs or freed from them because MTs were crowded regardless of the presence or absence of functional Asp. However, the exception was the outermost MT in the spindle, which was occasionally observed as an individual MT bundle with GFP signals at the end (Fig. 5, C and D; and Video 7). We therefore quantified the frequency and destiny of those MT ends. In control cells that expressed Asp(FL)-GFP, we identified seven distinguishable outermost MTs in eight spindles during a total of 117 min of imaging. The MT ends moved poleward, and all these seven MTs were merged into the adjacent MTs before the minus ends reached the pole (Fig. 5, D and E). In contrast, we identified outermost MTs at about a twofold higher frequency in the absence of functional Asp (17 outermost MTs decorated with AspΔC-GFP were identified in 11 spindles during a total of 147 min of imaging). Furthermore, 15/17 outermost MTs did not merge with adjacent MTs during poleward movement and instead constituted unfocused MTs that ran uninterrupted from the pole to the spindle midzone (Fig. 5, C and E). These results suggest that Asp assists in organizing spindle MTs by cross-linking MT minus ends to other MTs not only at the pole but also within the spindle.
Discussion
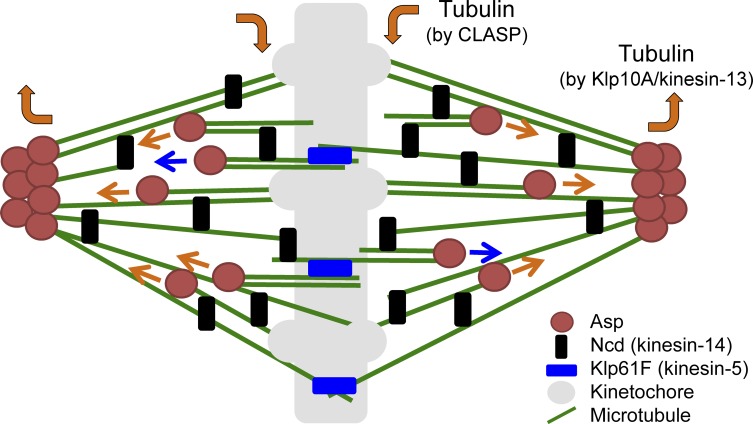
Taking into account the results of this and previous studies, we propose a revised model for how spindle MTs are focused in Drosophila cells (Fig. 6). Contrary to the prevailing model in which Ncd was considered to be the specific cross-linker of spindle MT minus ends, we propose that Asp plays this specific role from early prometaphase to metaphase. We postulate that Asp binds directly to MT minus ends and then stabilizes and cross-links them. In contrast, Ncd cross-links MTs through the entire lattice binding and plus end accumulation. However, when MTs were overly unfocused during prometaphase in the absence of Ncd, Asp could no longer recover the focusing of these bundles, although it transiently focused the pole in early prometaphase. Thus, the cross-linking function of Ncd is also critical for maintaining the focused pole. Centrosomes also play a supportive role in spindle MT focusing, perhaps by supplying additional MTs for the motors and Asp for interaction because simultaneous depletion of centrosomes with Ncd or Asp has a more profound effect on pole width than single depletion (Wakefield et al., 2001; Goshima et al., 2005a). In contrast, the basis of the centrosome detachment phenotype, commonly observed after dynein or Asp RNAi but not Ncd RNAi (Wakefield et al., 2001; Maiato et al., 2004; Morales-Mulia and Scholey, 2005; Goshima et al., 2005a), remains unclear. The MT cross-linking activity of Asp appears to be required for centrosome association with the spindle pole (Fig. 3 A). In addition, Asp might regulate dynein activity in this process.
Figure 6.
A model for the focusing of spindle MT minus ends by Asp. Asp binds to spindle MT ends from the early phases of mitosis and cross-links them. Ncd is a more general and dynamic spindle MT cross-linker and does not function specifically at the pole (the halftime of recovery after Ncd-GFP photobleaching was 2.5 s; Goshima et al., 2005a). When spindle MTs are nucleated at the pole, these MTs are immediately focused by the action of polar Asp. Spindle MTs are also nucleated in a polarized manner in the body of the spindle in an augmin-dependent manner. Those MT ends are also decorated by Asp, cross-linked with other MTs, and translocated to the pole by the action of spindle MT flux driven by the kinesin-5 sliding motor (blue arrows) or CLASP- and kinesin-13–dependent kinetochore MT treadmilling (orange arrows). Asp also ensures the focusing of these intraspindle MTs. Centrosomes would serve as an extra source of polar MTs on which Ncd and Asp function (not described in this diagram).
Another new element in our model is the incorporation of intraspindle MTs, which are nucleated via the augmin-mediated mechanism, decorated with Asp, and move poleward through MT flux. Without functional Asp, the “free” minus ends occasionally moved in an undesired direction and did not become cross-linked with other spindle MTs (Fig. 5 C). Because of resolution limits, our observation was restricted to the outermost MTs in the spindle. However, because intraspindle MTs are also generated via augmin-mediated, MT-dependent MT nucleation, we speculate that Asp cross-links newly nucleated short “daughter” MTs with long “mother” MTs such as kinetochore MTs or with other daughter MTs also in the MT-crowded region. Augmin itself may have MT bundling activity (Wu et al., 2008; Hsia et al., 2014) and therefore could also play a cross-linking role; however, a previous electron microscopy study suggests that the augmin-dependent link between mother and daughter MTs is not persistent (Kamasaki et al., 2013). We propose that Asp assists in maintaining the MTs inside the spindle during poleward movement.
A recent study in human cells used γ-tubulin within the spindle as a marker of the intraspindle MT minus ends and showed that they are transported poleward by the action of kinesin-14 and dynein (Lecland and Lüders, 2014). This dynamic differs from what we observed for Asp-GFP. However, the same study also identified kinesin-5 as an additional factor controlling movement, suggesting that the flux-mediated motility is a conserved mechanism for poleward translocation of intraspindle MTs. An alternative interpretation of this discrepancy is that the minus end–directed motors transport MT-free γ-tubulin, as not all γ-tubulin on the spindle may be associated with MT minus ends (Hallen et al., 2008). Conversely, Asp might decorate only a subset of intraspindle MTs; for example, the bundled MTs, which are moved exclusively by a flux-mediated mechanism. Thus far, human ASPM, mutations of which lead to microcephaly (Bond et al., 2002), has been poorly characterized in terms of intracellular dynamics, spindle pole organization, and molecular activity. Observing ASPM behavior in the spindle, as well as assessing its function in spindle pole focusing, would be an interesting future research direction.
Materials and methods
RNAi and cell line selection
Cell culture and RNAi were performed as previously described (Goshima et al., 2007; Bettencourt-Dias and Goshima, 2009). In brief, Schneider’s medium (Gibco) supplemented with 10% serum was used for cell culture. Cell lines were selected with hygromycin after plasmid transformation with the Cellfectin II reagent (Invitrogen). The primers used for Asp RNAi are listed in Table S1. Other double-stranded RNA (dsRNA) sequences used in this study are previously described (Goshima et al., 2007). For the RNAi experiments, cells were treated with dsRNAs for 4–7 d and then plated on ConA-coated glass-bottomed dishes for microscopy. Cdc16 or Cdc27 RNAi was used for the enrichment of metaphase-arrested cells (Goshima et al., 2007).
Microscopy
Live imaging was mostly performed using spinning-disk confocal microscopy with a 100× 1.40 NA objective lens (∼24°C). A confocal unit (CSU-X1; Yokogawa Electric Corporation) attached to an inverted microscope (TE; Nikon) and an electron-multiplying charge-coupled device (EMCCD) camera (ImagEM; Hamamatsu Photonics) were used for image acquisition. Some images, including those presented in Figs. 4 and 5, were obtained using another spinning-disk confocal microscope (Ti; Nikon) that has CSU-W1 (Yokogawa Electric Corporation) and an EMCCD camera (iXON DU888E; Andor Technology). Characterization of the phenotypes of fixed samples was performed with the Nikon Ti inverted microscope attached to the EMCCD camera (Evolve; Roper Scientific). Two objective lenses (100× 1.49 NA or 40× 1.30 NA) were used. The microscopes and attached devices were controlled using Micro-Manager (Vale lab, University of California, San Francisco) or NIS-Elements software (Nikon). Images were analyzed with ImageJ (National Institutes of Health). Asp-GFP movement was analyzed by generating a kymograph along the spindle MT. A total internal reflection fluorescence microscope (Ti; Nikon; 100× 1.49 NA lens) with an EMCCD camera was used for the in vitro MT cross-linking experiment. In Fig. 1 A and Video 1, z stacks (1 × 6 µm) were acquired, and the maximum projection images are presented. Immunostaining was performed with the standard procedure using 6.4% paraformaldehyde and anti–α-tubulin (1:500; YOL1/34; Serotec) or anti–γ-tubulin antibodies (1:500; GTU88; Sigma-Aldrich; Goshima et al., 2008). DNA was stained with 1 µg/ml DAPI. Images were acquired with Micro-Manager and processed with ImageJ.
Immunoblotting
Consistent with a recent study (Rujano et al., 2013), we also failed to detect the Asp protein using a standard procedure with SDS sample buffer. In this study, we prepared the sample in two ways. In Fig. S1 B, we treated cells with a urea-containing solution (62.5-mM Tris, pH 6.8, 10% glycerol, 2% SDS, 4-M urea, and 2-mercaptoethanol) at room temperature for >1 h. In Fig. S1 A, we first treated cells with the Cytobuster (EMD Millipore) solution that also contained benzonase, DTT, and protein inhibitors for 30 min on ice, followed by addition of the urea-containing solution. Rabbit polyclonal anti-Asp antibody was generated by using recombinant Asp [1–500 aa] as the antigen, and the crude serum was used for immunoblotting after a 1:500 dilution. Rabbit polyclonal anti-Ncd antibody was a gift from J. Scholey (University of California, Davis, Davis, CA; Morales-Mulia and Scholey, 2005).
Protein purification
GST-Asp [574–972 aa]-GFP-His fusion was constructed with a PreScission protease site inserted between the GST and Asp sequences. The fusion protein was expressed in Escherichia coli SoluBL21 cells (expression was induced by the addition of 0.2-mM IPTG at 18°C for 22 h). Cells were resuspended in the lysis buffer (50-mM Hepes-KOH, pH 7.6, 250-mM NaCl, 1-mM MgCl2, 2-mM CaCl2, 10-mM imidazole, 5-mM 2-mercaptoethanol, and protease inhibitors), sonicated, and then subjected to nickel–nitrilotriacetic acid purification at 4°C for 1 h followed by washing and elution with 350-mM imidazole. Next, the eluate was mixed with glutathione Sepharose beads for 1 h at 4°C, followed by PreScission protease treatment at 4°C overnight in GST cleavage buffer (50-mM Tris-Cl, pH 7.5, 0.5 M NaCl, 1-mM EDTA, 0.1% Tween 20, and 1-mM DTT). The buffer was then exchanged against the storage buffer (80-mM K-Pipes, pH 6.8, 4-mM MgCl2, 1-mM EGTA, 1-mM DTT, and 20% glycerol) using the PD MiniTrap G-25 column (GE Healthcare), and the protein solution was flash frozen.
In vitro MT cross-linking assay
The MT cross-linking assay was performed basically as previously described (Goshima, 2011). Tubulin derived from pig brains were polymerized in the presence of taxol (4% of the tubulin was rhodamine labeled). Next, 400-nM Asp [574–972 aa]-GFP-His protein was mixed with 4-µM taxol-stabilized MTs in 1× MRB80 (80-mM K-Pipes, pH 6.8, 4-mM MgCl2, and 1-mM EGTA), 50-mM KCl, 1-mM DTT, and 20-µM taxol for 30 min at room temperature. Specimens were then fixed with 1% glutaraldehyde for 3 min at room temperature and placed on a coverslip for total internal reflection fluorescence imaging.
Online supplemental material
Fig. S1 shows the efficiency of Ncd and Asp knockdown described in Fig. 1 (A) and the expression and functionality of Asp-GFP (B and C). Table S1 lists primers used for RNAi. Video 1 shows the spindle MT–unfocusing phenotype after Asp or Ncd RNAi. Video 2 shows localization of Asp-GFP in Ncd RNAi–treated cells. Video 3 shows flux-dependent poleward movement of Asp-GFP. Video 4 shows poleward movement of Asp-GFP in Cnn RNAi–treated cells. Video 5 shows localization of Asp-GFP in Dgt6 RNAi–treated cells. Video 6 shows scattered minus ends near the pole in the absence of functional Asp. Video 7 shows dynamics of minus ends of intraspindle MTs in the presence or absence of functional Asp. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201507001/DC1.
Supplementary Material
Acknowledgments
We wish to thank Tomoko Nishiyama for her assistance with microscopy, Momoko Nishina for technical assistance, and Tomomi Kiyomitsu for critical reading of the manuscript.
This work was supported by the Uehara Memorial Foundation, the Daiichi-Sankyo Foundation, and the Takeda Science Foundation (grant to G. Goshima). A. Ito is a recipient of a Japan Society for the Promotion of Science predoctoral fellowship.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- Cnn
- centrosomin
- dsRNA
- double-stranded RNA
- EMCCD
- electron-multiplying charge-coupled device
- MT
- microtubule
- NEBD
- nuclear envelope breakdown
References
- Baumbach J., Novak Z.A., Raff J.W., and Wainman A.. 2015. Dissecting the function and assembly of acentriolar microtubule organizing centers in Drosophila cells in vivo. PLoS Genet. 11:e1005261 10.1371/journal.pgen.1005261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M., and Goshima G.. 2009. RNAi in Drosophila S2 cells as a tool for studying cell cycle progression. Methods Mol. Biol. 545:39–62. 10.1007/978-1-60327-993-2_3 [DOI] [PubMed] [Google Scholar]
- Bond J., Roberts E., Mochida G.H., Hampshire D.J., Scott S., Askham J.M., Springell K., Mahadevan M., Crow Y.J., Markham A.F., et al. 2002. ASPM is a major determinant of cerebral cortical size. Nat. Genet. 32:316–320. 10.1038/ng995 [DOI] [PubMed] [Google Scholar]
- Capalbo L., D’Avino P.P., Archambault V., and Glover D.M.. 2011. Rab5 GTPase controls chromosome alignment through Lamin disassembly and relocation of the NuMA-like protein Mud to the poles during mitosis. Proc. Natl. Acad. Sci. USA. 108:17343–17348. 10.1073/pnas.1103720108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Carmo Avides M., and Glover D.M.. 1999. Abnormal spindle protein, Asp, and the integrity of mitotic centrosomal microtubule organizing centers. Science. 283:1733–1735. 10.1126/science.283.5408.1733 [DOI] [PubMed] [Google Scholar]
- Endow S.A., Chandra R., Komma D.J., Yamamoto A.H., and Salmon E.D.. 1994. Mutants of the Drosophila ncd microtubule motor protein cause centrosomal and spindle pole defects in mitosis. J. Cell Sci. 107:859–867. [DOI] [PubMed] [Google Scholar]
- Gaglio T., Saredi A., Bingham J.B., Hasbani M.J., Gill S.R., Schroer T.A., and Compton D.A.. 1996. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J. Cell Biol. 135:399–414. 10.1083/jcb.135.2.399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G. 2011. Identification of a TPX2-like microtubule-associated protein in Drosophila. PLoS ONE. 6:e28120 10.1371/journal.pone.0028120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., and Vale R.D.. 2003. The roles of microtubule-based motor proteins in mitosis: Comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 162:1003–1016. 10.1083/jcb.200303022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., and Vale R.D.. 2005. Cell cycle-dependent dynamics and regulation of mitotic kinesins in Drosophila S2 cells. Mol. Biol. Cell. 16:3896–3907. 10.1091/mbc.E05-02-0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Nédélec F., and Vale R.D.. 2005a Mechanisms for focusing mitotic spindle poles by minus end–directed motor proteins. J. Cell Biol. 171:229–240. 10.1083/jcb.200505107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Stuurman N., Scholey J.M., and Vale R.D.. 2005b Length control of the metaphase spindle. Curr. Biol. 15:1979–1988. 10.1016/j.cub.2005.09.054 [DOI] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S.S., Zhang N., Scholey J.M., Vale R.D., and Stuurman N.. 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 316:417–421. 10.1126/science.1141314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Mayer M., Zhang N., Stuurman N., and Vale R.D.. 2008. Augmin: A protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol. 181:421–429. 10.1083/jcb.200711053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallen M.A., Ho J., Yankel C.D., and Endow S.A.. 2008. Fluorescence recovery kinetic analysis of γ-tubulin binding to the mitotic spindle. Biophys. J. 95:3048–3058. 10.1529/biophysj.108.134593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsumi M., and Endow S.A.. 1992. The Drosophila ncd microtubule motor protein is spindle-associated in meiotic and mitotic cells. J. Cell Sci. 103:1013–1020. [DOI] [PubMed] [Google Scholar]
- Heald R., Tournebize R., Blank T., Sandaltzopoulos R., Becker P., Hyman A., and Karsenti E.. 1996. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 382:420–425. 10.1038/382420a0 [DOI] [PubMed] [Google Scholar]
- Heald R., Tournebize R., Habermann A., Karsenti E., and Hyman A.. 1997. Spindle assembly in Xenopus egg extracts: Respective roles of centrosomes and microtubule self-organization. J. Cell Biol. 138:615–628. 10.1083/jcb.138.3.615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia K.C., Wilson-Kubalek E.M., Dottore A., Hao Q., Tsai K.L., Forth S., Shimamoto Y., Milligan R.A., and Kapoor T.M.. 2014. Reconstitution of the augmin complex provides insights into its architecture and function. Nat. Cell Biol. 16:852–863. 10.1038/ncb3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamasaki T., O’Toole E., Kita S., Osumi M., Usukura J., McIntosh J.R., and Goshima G.. 2013. Augmin-dependent microtubule nucleation at microtubule walls in the spindle. J. Cell Biol. 202:25–33. 10.1083/jcb.201304031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A., Cole R.W., Oakley B.R., and Rieder C.L.. 2000. Centrosome-independent mitotic spindle formation in vertebrates. Curr. Biol. 10:59–67. 10.1016/S0960-9822(99)00276-6 [DOI] [PubMed] [Google Scholar]
- Laycock J.E., Savoian M.S., and Glover D.M.. 2006. Antagonistic activities of Klp10A and Orbit regulate spindle length, bipolarity and function in vivo. J. Cell Sci. 119:2354–2361. 10.1242/jcs.02957 [DOI] [PubMed] [Google Scholar]
- Lecland N., and Lüders J.. 2014. The dynamics of microtubule minus ends in the human mitotic spindle. Nat. Cell Biol. 16:770–778. 10.1038/ncb2996 [DOI] [PubMed] [Google Scholar]
- Mahoney N.M., Goshima G., Douglass A.D., and Vale R.D.. 2006. Making microtubules and mitotic spindles in cells without functional centrosomes. Curr. Biol. 16:564–569. 10.1016/j.cub.2006.01.053 [DOI] [PubMed] [Google Scholar]
- Maiato H., and Logarinho E.. 2014. Mitotic spindle multipolarity without centrosome amplification. Nat. Cell Biol. 16:386–394. 10.1038/ncb2958 [DOI] [PubMed] [Google Scholar]
- Maiato H., Rieder C.L., and Khodjakov A.. 2004. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J. Cell Biol. 167:831–840. 10.1083/jcb.200407090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H., Khodjakov A., and Rieder C.L.. 2005. Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat. Cell Biol. 7:42–47. 10.1038/ncb1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw T.L., Kao L.R., and Kaufman T.C.. 2001. Zygotic development without functional mitotic centrosomes. Curr. Biol. 11:116–120. 10.1016/S0960-9822(01)00017-3 [DOI] [PubMed] [Google Scholar]
- Merdes A., Ramyar K., Vechio J.D., and Cleveland D.W.. 1996. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 87:447–458. 10.1016/S0092-8674(00)81365-3 [DOI] [PubMed] [Google Scholar]
- Merdes A., Heald R., Samejima K., Earnshaw W.C., and Cleveland D.W.. 2000. Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J. Cell Biol. 149:851–862. 10.1083/jcb.149.4.851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Mulia S., and Scholey J.M.. 2005. Spindle pole organization in Drosophila S2 cells by dynein, abnormal spindle protein (Asp), and KLP10A. Mol. Biol. Cell. 16:3176–3186. 10.1091/mbc.E04-12-1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountain V., Simerly C., Howard L., Ando A., Schatten G., and Compton D.A.. 1999. The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J. Cell Biol. 147:351–366. 10.1083/jcb.147.2.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutinho-Pereira S., Stuurman N., Afonso O., Hornsveld M., Aguiar P., Goshima G., Vale R.D., and Maiato H.. 2013. Genes involved in centrosome-independent mitotic spindle assembly in Drosophila S2 cells. Proc. Natl. Acad. Sci. USA. 110:19808–19813. 10.1073/pnas.1320013110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radulescu A.E., and Cleveland D.W.. 2010. NuMA after 30 years: The matrix revisited. Trends Cell Biol. 20:214–222. 10.1016/j.tcb.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoll P., Pimpinelli S., Valdivia M.M., and Avila J.. 1985. A cell division mutant of Drosophila with a functionally abnormal spindle. Cell. 41:907–912. 10.1016/S0092-8674(85)80071-4 [DOI] [PubMed] [Google Scholar]
- Rogers G.C., Rogers S.L., Schwimmer T.A., Ems-McClung S.C., Walczak C.E., Vale R.D., Scholey J.M., and Sharp D.J.. 2004. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 427:364–370. 10.1038/nature02256 [DOI] [PubMed] [Google Scholar]
- Rogers G.C., Rogers S.L., and Sharp D.J.. 2005. Spindle microtubules in flux. J. Cell Sci. 118:1105–1116. 10.1242/jcs.02284 [DOI] [PubMed] [Google Scholar]
- Rujano M.A., Sanchez-Pulido L., Pennetier C., le Dez G., and Basto R.. 2013. The microcephaly protein Asp regulates neuroepithelium morphogenesis by controlling the spatial distribution of myosin II. Nat. Cell Biol. 15:1294–1306. 10.1038/ncb2858 [DOI] [PubMed] [Google Scholar]
- Saunders R.D., Avides M.C., Howard T., Gonzalez C., and Glover D.M.. 1997. The Drosophila gene abnormal spindle encodes a novel microtubule-associated protein that associates with the polar regions of the mitotic spindle. J. Cell Biol. 137:881–890. 10.1083/jcb.137.4.881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Voet M., Berends C.W., Perreault A., Nguyen-Ngoc T., Gönczy P., Vidal M., Boxem M., and van den Heuvel S.. 2009. NuMA-related LIN-5, ASPM-1, calmodulin and dynein promote meiotic spindle rotation independently of cortical LIN-5/GPR/Gα. Nat. Cell Biol. 11:269–277. 10.1038/ncb1834 [DOI] [PubMed] [Google Scholar]
- Wakefield J.G., Bonaccorsi S., and Gatti M.. 2001. The Drosophila protein asp is involved in microtubule organization during spindle formation and cytokinesis. J. Cell Biol. 153:637–648. 10.1083/jcb.153.4.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak C.E., Vernos I., Mitchison T.J., Karsenti E., and Heald R.. 1998. A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr. Biol. 8:903–913. 10.1016/S0960-9822(07)00370-3 [DOI] [PubMed] [Google Scholar]
- Wu G., Lin Y.T., Wei R., Chen Y., Shan Z., and Lee W.H.. 2008. Hice1, a novel microtubule-associated protein required for maintenance of spindle integrity and chromosomal stability in human cells. Mol. Cell. Biol. 28:3652–3662. 10.1128/MCB.01923-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.Z., Spektor A., Cornils H., Francis J.M., Jackson E.K., Liu S., Meyerson M., and Pellman D.. 2015. Chromothripsis from DNA damage in micronuclei. Nature. 522:179–184. 10.1038/nature14493 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.