Abstract
Previous analyses of both Thermus aquaticus MutS homodimer and Saccharomyces cerevisiae Msh2–Msh6 heterodimer have revealed that the subunits in these protein complexes bind and hydrolyze ATP asymmetrically, emulating their asymmetric DNA binding properties. In the MutS homodimer, one subunit (S1) binds ATP with high affinity and hydrolyzes it rapidly, while the other subunit (S2) binds ATP with lower affinity and hydrolyzes it at an apparently slower rate. Interaction of MutS with mismatched DNA results in suppression of ATP hydrolysis at S1—but which of these subunits, S1 or S2, makes specific contact with the mismatch (e.g., base stacking by a conserved phenylalanine residue) remains unknown. In order to answer this question and to clarify the links between the DNA binding and ATPase activities of each subunit in the dimer, we made mutations in the ATPase sites of Msh2 and Msh6 and assessed their impact on the activity of the Msh2–Msh6 heterodimer (in Msh2–Msh6, only Msh6 makes base specific contact with the mismatch). The key findings are: (a) Msh6 hydrolyzes ATP rapidly, and thus resembles the S1 subunit of the MutS homodimer, (b) Msh2 hydrolyzes ATP at a slower rate, and thus resembles the S2 subunit of MutS, (c) though itself an apparently weak ATPase, Msh2 has a strong influence on the ATPase activity of Msh6, (d) Msh6 binding to mismatched DNA results in suppression of rapid ATP hydrolysis, revealing a “cis” linkage between its mismatch recognition and ATPase activities, (e) the resultant Msh2–Msh6 complex, with both subunits in the ATP-bound state, exhibits altered interactions with the mismatch.
Keywords: Mismatch repair, MutS, Msh2–Msh6, ATPase kinetics
1. Introduction
DNA mismatch repair is an important, widely conserved mechanism for maintaining the integrity of genetic information over generations. This repair mechanism corrects base substitution and insertion/deletion mismatches that occur due to errors in DNA replication and recombination, as well as DNA lesions resulting from a variety of internal and external stresses. Repair initiates with MutS protein in prokaryotes, or MutS homologues in eukaryotes (e.g., Msh2–Msh6, Msh2–Msh3), binding the site of the mismatch in duplex DNA. This recognition event triggers excision of the error-containing DNA strand past the site of the mismatch, which is followed by DNA resynthesis and ligation to complete the repair process [1–3].
In addition to their mismatch recognition activity, MutS/Msh proteins also possess an ATPase activity that is essential for DNA repair [4–7]. ATP binding and hydrolysis appear to modulate the interactions between MutS/Msh and DNA as well as other proteins in the repair pathway; thus, understanding how MutS/Msh proteins utilize ATP is necessary for understanding how they function in DNA mismatch repair. Several model mechanisms have been proposed for MutS/Msh action upon mismatch recognition: (a) MutS/Msh proteins translocate on DNA, fuelled by ATP binding and hydrolysis, possibly to interact with other proteins on DNA and coordinate mismatch recognition with downstream events such as initiation of strand excision and DNA resynthesis [8–10]; (b) upon binding ATP MutS/Msh proteins form sliding clamps that diffuse freely on DNA, again, to contact downstream repair proteins and direct repair [11,12]; (c) MutS/Msh proteins utilize ATP binding and hydrolysis to modulate their interaction with DNA, while remaining at the mismatch to direct repair [13–17]. At present, experimental data are available in support of each of these very different model mechanisms, therefore the investigation into MutS/Msh DNA binding and ATPase activities continues.
Recent studies from several research groups, including our own, have revealed clear differences between the ATP binding and hydrolysis activities of the two subunits in the MutS/Msh dimer [18–21]. For instance, in Thermus aquaticus MutS, one subunit binds nucleotide (ATPγS) with about 10-fold higher affinity than the other subunit (KD = 3μM versus 27μM). Also, the high-affinity subunit hydrolyzes ATP at >30-fold faster rate than the low-affinity subunit (10 s−1 versus 0.2–0.3 s−1 at 40 °C) [18]. These differences are striking especially since MutS is a homodimer; however, they are in accord with known differences in the DNA binding activities of the two MutS subunits (e.g., conserved phenylalanine and glutamate residues from only one subunit undergo base stacking and hydrogen bonding interactions with the mismatch, respectively) [22,23]. In fact it appears that the asymmetry in the ATPase sites is linked to asymmetry in the interactions of the two subunits with DNA [24,25]. Consistent with this hypothesis, binding of mismatched DNA to MutS specifically suppresses the catalytic activity of the high-affinity subunit, such that the rate of ATP hydrolysis is reduced from 10 to 0.3 s−1 [18]. The exact nature and function of asymmetry in the MutS dimer is not clear as yet, but the characteristic appears to be important for DNA mismatch repair as it is conserved among a variety of organisms. For instance, subunits of the E. coli MutS homodimer also exhibit differences in their interactions with nucleotides and with mismatched DNA [20,21]. The eukaryotic Msh2–Msh6 heterodimer is no different, as the subunits bind nucleotides with differing affinities [19,26], only one subunit catalyzes rapid ATP hydrolysis (Saccharomyces cerevisiae Msh2–Msh6: 2–3 s−1 at 20 °C) [19], and only Msh6 contains the conserved phenylalanine residue that can make specific contact with the mismatch in DNA [27,28]. As in the case of T. aquaticus MutS, mismatched DNA binding strongly suppresses the activity of the rapid ATP-hydrolyzing subunit in S. cerevisiae Msh2–Msh6 (the rate constant decreases from 2–3 to 0.1–0.2 s−1 at 20 °C) [19]. It is not known yet which of the two subunits, Msh2 or Msh6, catalyzes rapid ATP hydrolysis and, therefore, which one's activity is altered so dramatically following mismatch recognition by Msh2–Msh6.
Previous studies have probed the ATPase activity of both Msh2 and Msh6 subunits by mutating conserved residues in their active sites for ATP binding and hydrolysis. The results confirmed that the ATPase activities of both Msh2 and Msh6 are required for DNA mismatch repair, and also highlighted differences between the two subunits [6,29,30]. Thus, the effects of mutating the Walker A motif (GxxxxGKS), which coordinates the phosphate groups of ATP, and Walker B motif (DExx), which coordinates the Mg2+ ion essential for catalysis, differed depending on whether Msh2 or Msh6 was changed. Substitution of the conserved Walker A glycine with aspartate, or Walker B glutamate with alanine, in Msh6 reduced the ATPase activity of S. cerevisiae Msh2–Msh6 to a greater extent than did identical mutations in Msh2 [6,29]. Similar results were obtained with a Walker A lysine to arginine mutation in human Msh6 versus Msh2 [30]. All these studies indicated that the Msh6 subunit contributes “more” than Msh2 to the overall ATPase activity of Msh2–Msh6. However, since the ATPase experiments were all performed in the steady state regime, i.e., they measured the rate-limiting step following ATP hydrolysis, the exact contribution and role of each subunit's ATP binding and hydrolysis activity in the Msh2–Msh6 ATPase mechanism, including the identity of the subunit that catalyzes rapid ATP hydrolysis, remain unknown.
Here we report pre-steady state analysis of the ATPase activities of wild type and mixed wild type-Walker A/B mutant heterodimers of Msh2–Msh6, carried out in order to answer questions such as: (a) which subunit catalyzes rapid ATP hydrolysis and which one has the apparently slower activity? (b) does ATP binding and/or ATP hydrolysis by Msh2 influence ATP binding and/or ATP hydrolysis by Msh6, and vice versa? (c) how is Msh2–Msh6 ATPase activity linked to mismatch recognition, given that only Msh6 makes base specific contacts with the mismatch? The answers reveal complex coordination between Msh2 and Msh6 activities that is likely important for Msh2–Msh6 function in DNA mismatch repair.
2. Materials and methods
2.1. DNA and nucleotides
Synthetic oligodeoxyribonucleotides (37-nucleotide template and G:T complement) were purchased from Integrated DNA Technologies, purified by denaturing polyacrylamide gel electrophoresis, and annealed to prepare a G:T mismatch-containing duplex, as described [19]. pET11a vector was purchased from Novagen and pLANT 2b/RIL was a gift from Michael O'Donnell (The Rockefeller University) [31]. Radioactive nucleotides [α-32P]-ATP, [γ-32P]-ATP, and [35S]-ATPγS were purchased from Perkin-Elmer Life Sciences, and non-radioactive nucleotides were purchased from Sigma Chemicals Co. DNA was labeled with 32P as described previously [19].
2.2. Proteins
Point mutations were introduced in MSH2 and MSH6 genes (contained in pET11a or pLANT2b/RIL vectors) using overlap-extension PCR or the QuikChange site-directed mutagenesis kit (Stratagene) and verified by sequencing the entire gene. Mixed wild type–mutant Msh2–Msh6 dimers were co-expressed and purified from E. coli as described previously for wild type Msh2–Msh6 [19]. Restriction enzymes and T4 polynucleotide kinase were purchased from New England BioLabs.
2.3. Nucleotide and DNA binding assays
ATPγS binding to Msh2–Msh6 was measured by nitrocellulose membrane binding assays as described previously [19]. Briefly, the membranes (Schleicher and Schuell) were washed with 0.5N NaOH and equilibrated in binding buffer (50 mM Tris–HCl, pH 8.0, 5 mM MgCl2, 5% glycerol). Msh2–Msh6 (2μM) was incubated with 0–200 μM ATPγS + 0.3 μCi [35S]-ATPγS for 15 min at 25 °C (15μl reactions in binding buffer; 110 mM final NaCl concentration). Ten microliters of each reaction was filtered through the membrane and 1 μl was spotted onto a separate membrane to measure total nucleotide in the reaction. The molar amount of nucleotide bound to protein was determined and plotted versus nucleotide concentration. The binding isotherms were fit to equations describing 1:1 or 2:1 binding of ligands to macromolecules [18].
Dissociation of ATP from Msh2–Msh6 was measured by incubating Msh2–Msh6 (2 μM) with 200 μM ATP + 0.3 μCi [α-32P]-ATP in the binding buffer for 30 s at 25 °C (110 mM final NaCl concentration), followed by addition of 5 mM Mg2+-ATP chase and filtration of 10 μl aliquots through the membrane at 30 s intervals (up to 5 min). The molar amount of nucleotide bound to the protein was determined and plotted versus time of chase. The data describing decay of the protein–nucleotide complex were fit to a single exponential equation.
Interaction between Msh2–Msh6 protein complexes and mismatched DNA was also measured by nitrocellulose membrane filtration assays as described previously [19]. Msh2–Msh6 (1 μM) was incubated with 32P-labeled DNA (0.1 μM) in the binding buffer (15 μl reaction) with varying NaCl concentration (0–300 mM), in the absence or presence of ATPγS (500 μM), for 10 min at 4 °C. Ten microliters of each reaction was filtered through the membrane and the molar amount of DNA bound to protein determined and plotted versus NaCl concentration.
2.4. ATPase assays
Steady state ATPase assays were performed with Msh2–Msh6 (1 μM) and 500 μM ATP + 2μCi [α-32P]-ATP in reaction buffer (50 mM Tris–HCl, pH 8.0, 5 mM MgCl2, 4 mM DTT) at 30 °C (30 μl reaction); all ATPase assays were performed at 110 mM final NaCl concentration. Five microliters of the reaction were quenched after varying times with 5 μl of 0.5 M EDTA, and the amount of [α-32P]-ADP formed was analyzed by PEI-cellulose TLC (EM Science) with 0.6 M potassium phosphate buffer, pH 3.4. The molar amount of ADP formed was plotted versus time and the data fit to a linear equation. The slope of the line divided by Msh2–Msh6 concentration yielded the kcat for the reaction.
Pre-steady state assays for ATP hydrolysis were performed on a quench-flow instrument (KinTek Corp., Austin, TX) as described previously [19]. Briefly, 16μl of 4μM Msh2–Msh6 (±6μM DNA) was mixed with 16 μl of 1 mM ATP + 2μCi [α-32P]-ATP and quenched with 35 μl of 0.7 M formic acid after varying times (0.08–15 s), followed by TLC and data analysis, as above (final concentrations: 2 μM Msh2–Msh6, 500 μM ATP, and 3 μM DNA). The data were fit to a linear equation or an exponential + linear equation, as appropriate.
Phosphate (Pi) release assays were performed on an SF-2001 stopped-flow instrument (KinTek Corp., Austin, TX) as described [18]. Briefly, 60 μl of 4μM Msh2–Msh6 (±6μM DNA) and 16 μM MDCC-PBP was mixed with 60μl of 1 mM ATP (final concentrations: 2 μM Msh2–Msh6, 8 μM MDCC-PBP, 500 μM ATP, and 3 μM DNA). The change in MDCC-PBP fluorescence upon binding to Pi (an average of at least five traces) was related to Pi concentration using a standard curve, and the data fit to a linear equation or an exponential + linear equation, as appropriate.
3. Results
3.1. Mutations in the conserved Walker A motif, but not Walker B motif, disrupt nucleotide binding to Msh2 and Msh6
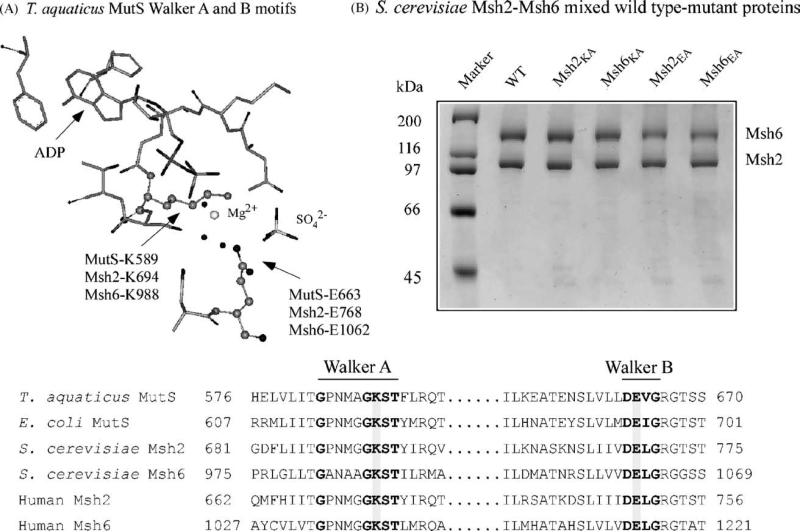
In order to quantify the contributions of Msh2 and Msh6 subunits to the ATPase activity of the S. cerevisiae Msh2–Msh6 dimer, we decided to prepare mutant versions of the proteins that were deficient in either ATP binding (and therefore hydrolysis) or only ATP hydrolysis activity. In doing so, we were guided by previous studies indicating that mutation of the Walker A lysine often disrupts ATP binding to proteins [30], while mutation of the Walker B glutamate appears to specifically disrupt ATP hydrolysis [32,33]. Four mixed wild type–mutant heterodimers – Msh2K694A–Msh6WT, Msh2WT–Msh6K988A, Msh2E768A–Msh6WT, and Msh2WT–Msh6E1062A – were over-expressed and purified from E. coli in milligram quantities, to enable accurate measurement of the stoichiometry of nucleotide binding and the kinetics of ATP binding, hydrolysis, and product release (Fig. 1).
Fig. 1.
Msh2, Msh6 Walker A and B motif mutants: (A) ADP, Mg2+, and SO42− bound in the conserved ATP binding and hydrolysis site of Thermus aquaticus MutS. (B) Purified mixed wild type–mutant Msh2–Msh6 heterodimers, containing mutations in Msh2 or Msh6 Walker A (Msh2: K694A, Msh6: K988A) and Walker B (Msh2: E768A, Msh6: E1062A) sites.
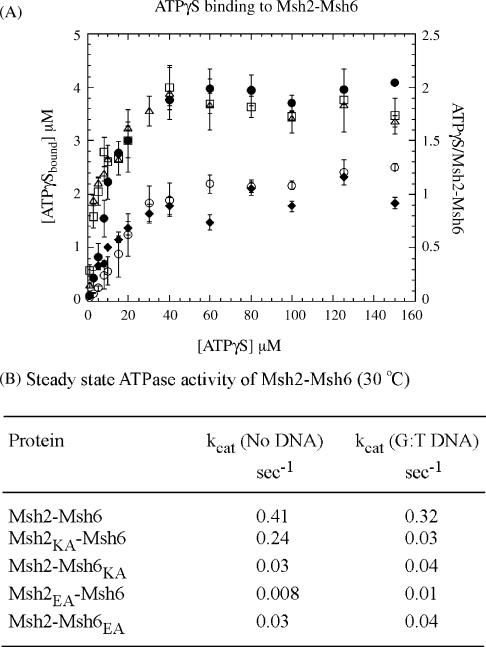
Previously, nitrocellulose membrane filtration assays had revealed that two molecules of ATPγS (a non-hydrolyzable ATP analog) bind per Msh2–Msh6 dimer, indicating that the recombinant protein purified from E. coli is fully active for nucleotide binding [19] (all protein preparations are free from nucleotide contaminants; Supplemental Fig. S1). Fig. 2A shows the results of similar experiments performed with the mixed heterodimers. In the case of wild type Msh2–Msh6, as well as Msh2E768A–Msh6WT and Msh2WT–Msh6E1062A, we detect 4 μM ATPγS bound to 2 μM dimer in the reaction (i.e., 2 ATPγS molecules per Msh2–Msh6). This stoichiometry clearly reveals that both Msh2 and Msh6 retain the ability to bind nucleotide even with mutation of the Walker B motif in the ATPase active site. All three isotherms, when fit to an equation describing binding of two ligands per macromolecule, yield KD1 = 2–3μM and KD2 = 8–12μM, consistent with previous reports of asymmetry in the interaction of the two subunits with nucleotides [19]. It should be noted that since the difference between the two apparent dissociation constants is quite small, the data are also fit quite well by a quadratic equation, which yields a single KD value of 4–7 μM. In any case, no significant difference can be detected between the stoichiometry or the affinity of wild type Msh2–Msh6 and mixed wild type-Walker B mutant Msh2–Msh6 dimers for binding ATPγS.
Fig. 2.
Effects of Walker A and B site mutations on Msh2–Msh6 ATP binding and steady state ATPase activities: (A) wild type Msh2–Msh6 (●), Msh2E768A–Msh6WT (Δ), and Msh2WT–Msh6E1062A (□), bind two ATPγS molecules per dimer, indicating that the Walker B site mutation does not disrupt nucleotide binding activity. In contrast, Msh2K694A–Msh6WT (○) and Msh2WT–Msh6K988A (◆) bind only one ATPγS molecule per dimer, indicating that the Walker A site mutation does disrupt nucleotide binding activity; (B) steady state assays indicate a substantial reduction of Msh2–Msh6 ATPase activity resulting from either mutation in Msh6; curiously, only the Walker B mutation in Msh2 appears to cause such a striking reduction in Msh2–Msh6 ATPase activity.
In contrast to the Walker B mutations, Walker A mutations knock out the nucleotide binding activity of both Msh2 and Msh6. As shown in Fig. 2A, Msh2K694A–Msh6WT and Msh2WT–Msh6K988A are capable of binding only one ATPγS molecule per dimer. The apparent binding affinities are similar for both mixed dimers: Msh2K694A–Msh6WT KD = 12μM and Msh2WT–Msh6K988A KD = 9μM. Since no additional nucleotide binding beyond one per dimer can be detected even at 250μM ATPγS concentration (data not shown), these mixed wild type-Walker A mutant dimers can be useful tools for examining Msh2–Msh6 function under conditions where one only one subunit, either Msh2 or Msh6, is active for ATP binding and hydrolysis.
All four protein complexes were also tested for ATPase activity under steady state conditions and, as expected from previous reports, each mutation resulted in a decrease in the overall activity of the dimer—the extent of which depended both on the subunit and the type of mutation (Fig. 2B). The Walker A lysine to alanine mutation in Msh6 (Msh2WT–Msh6K988A) lowers the kcat to 0.03 s−1 from the wild type Msh2–Msh6 level of 0.4 s−1 (30 °C), whereas the same mutation in Msh2 (Msh2K694A–Msh6WT) lowers kcat only to 0.24 s−1 (note: all experiments were performed at 500μM ATP, far in excess of reported KM values for these proteins; [29]). In contrast, the Walker B glutamate to ala-nine mutation appears to inactivate the Msh2–Msh6 ATPase, whether present in Msh6 (Msh2WT–Msh6E1062A kcat = 0.03 s−1) or in Msh2 (Msh2E768A–Msh6WT kcat = 0.008 s−1); although, Msh2WT–Msh6E1062A activity is still above the baseline whereas Msh2E768A–Msh6WT activity is at the baseline and appears to be shut down completely.
We see that both Walker A and B mutations in Msh6 almost completely inactivate Msh2–Msh6, suggesting that this subunit is the predominant ATPase in the dimer; it does remain possible that Msh2 is also a robust ATPase, but its optimal activity requires that Msh6 bind and/or hydrolyze ATP. It is also not clear why, if the ATP binding (and therefore hydrolysis) activity of Msh2 is knocked out (Msh2K694A–Msh6WT; Fig. 2A), Msh6 remains catalytically active, albeit to a lesser extent than wild type Msh2–Msh6 (Fig. 2B), but if Msh2 can bind ATP but not hydrolyze it (Msh2E768A–Msh6WT; Fig. 2A), Msh6 activity is affected much more severely (Fig. 2B). In order to understand the ATPase mechanisms of the two subunits, and how they might be linked, in greater detail than possible by steady state analysis, we assayed the mixed dimers under pre-steady state conditions, as described in the next section.
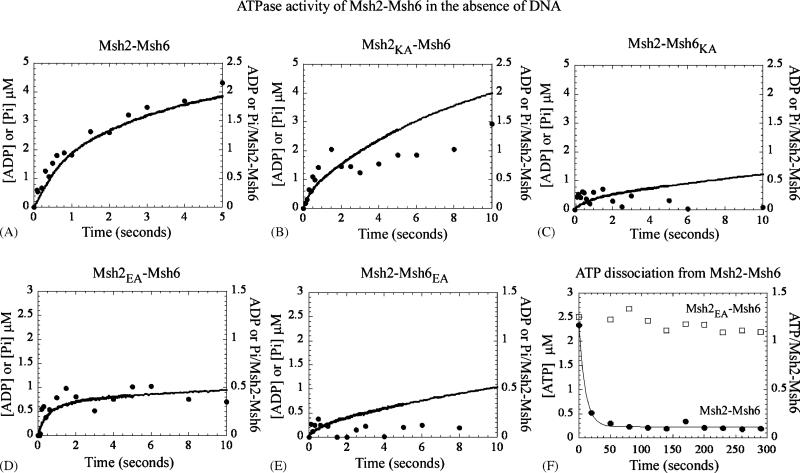
3.2. Msh6 is responsible for the rapid ATP hydrolysis activity of Msh2–Msh6 heterodimer
Wild type Msh2–Msh6 is known to bind at least one ATP molecule at a fast rate (0.1 μM−1 s−1), hydrolyze it and release the phosphate product at a fast rate (2 s−1), and then undergo slow catalytic turnover (0.1–0.2 s−1) [19], as shown here in Fig. 3A (2μM Msh2–Msh6 in the reaction; exponential burst amplitude = 2 μM, burst rate constant = 1.4 s−1, linear rate = 0.36μM s−1 or kcat = 0.18 s−1). The mixed wild type-Walker A mutant Msh2K694A–Msh6WT, in which the ATP binding activity of Msh2 is knocked out, still catalyzes a burst of ATP hydrolysis at a rate constant of 1.8 s−1, followed by a linear ATPase rate at 0.38 μM s−1 (Fig. 3B; Msh2K694A–Msh6WT ATPase rate can be expressed as kcat = 0.38 divided by 2 = 0.19 s−1, only if we make the possibly incorrect assumption that all of the protein in the reaction is fully active—as assumed for the kcat values reported in Fig. 2B). In contrast, Msh2WT–Msh6K988A, in which the ATP binding activity of Msh6 is knocked out, does not display any significant burst activity and the data are best fit by a linear ATPase rate at 0.1μM s−1 (Fig. 3C). Together, these data suggest that the Msh6 subunit is responsible for the rapid ATP hydrolysis activity of the Msh2–Msh6 dimer. Data from experiments with mixed wild type-Walker B mutant heterodimers support the above conclusion—Msh2E768A–Msh6WT catalyzes a burst of ATP hydrolysis at 2 s−1 (Fig. 3D); however, Msh2WT–Msh6E1062A hydrolyzes ATP at a much slower linear rate of 0.1μM s−1 (Fig. 3E). The residual ATPase rate of both Msh6 mutant-containing dimers may reflect inherently slow Msh2 activity or indicate that ATP binding and/or hydrolysis by Msh6 is necessary for optimal Msh2 activity.
Fig. 3.
Pre-steady state Msh2–Msh6 ATPase kinetics assayed by rapid quench (●) and phosphate release (—): (A) Msh2–Msh6 (2 μM dimer) hydrolyzes ATP asymmetrically, with a rapid burst of activity at one subunit of the dimer (burst rate = 1.4 s−1; amplitude = 2 μM) followed by slow turnover (rate = 0.36 μM s−1). (B) Msh2K694A–Msh6WT also exhibits a burst of ATP hydrolysis at 1.8 s−1 albeit with lower amplitude than wild type protein (0.8 μM), followed by slow turnover (rate = 0.38 μM s−1). (C) In contrast, Msh2WT–Msh6K988A displays significantly lower ATP hydrolysis activity (rate = 0.1 μM s−1). (D) Msh2 E768A–Msh6WT shows a partial burst of ATP hydrolysis (rate = 2 s−1; amplitude = 0.8 μM), but almost no turnover (rate = 0.02 μM s−1). (E) Like Msh2WT–Msh6K988A, Msh2WT–Msh6E1062A also has very low ATP hydrolysis activity (rate = 0.1 μM s−1). (F) In contrast to wild type Msh2–Msh6 (●; koff ≥0.1 s−1), Msh2 E768A–Msh6WT displays a very stable interaction with ATP (□).
Subtle differences among the four mixed heterodimers reveal some links between the ATPase mechanisms of Msh2 and Msh6. For instance, although Msh6 hydrolyzes ATP at a fast rate in both Msh2 mutant-containing dimers, Msh2K694A–Msh6WT can undergo catalytic turnover (Fig. 3B; linear rate = 0.38 μM s−1) while Msh2E768A–Msh6WT apparently cannot (Fig. 3D; linear rate = 0.02 μM s−1). This difference is likely related to the fact that the Msh2K694A mutant does not bind nucleotide and Msh2E768A clearly does (Fig. 2A). A filter binding chase assay measuring [α-32P]-ATP dissociation from these proteins shows that Msh2E768A–Msh6WT undergoes very stable binding to nucleotide, with no dissociation detectable over several minutes (Fig. 3F; wild type Msh2–Msh6 ATP koff ≥0.1 s−1). This nucleotide may be unhydrolyzed ATP retained at the Msh2E768A active site, possibly stabilized by the loss of negative charge repulsion between the wild type glutamate residue and γ phosphate of ATP. It is also possible that the stably bound nucleotide is ADP, produced by ATP hydrolysis at the Msh6 active site. We do not favor this possibility as extraction of the bound nucleotide from Msh2E768A–Msh6WT followed by chromatographic analysis indicates that it is mostly in ATP form (data not shown). In either case, it appears that the Msh2 active site must be empty (as in Msh2K694A–Msh6WT) for Msh6 to undergo catalytic turnover. It should be noted that for both Msh2K694A–Msh6WT and Msh2E768A–Msh6WT, the amplitude of the burst phase is about half that of wild type Msh2–Msh6 (0.8 μM instead of 2μM). The reason for this partial loss of ATP hydrolysis activity is not clear, as the proteins display the correct stoichiometry for ATP binding (Fig. 2A). The two subunits in a MutS dimer have composite catalytic sites with one subunit contributing residues to the active site of the other [13,22,25], and it is possible that the linkage is such that perturbation of the Msh2 site can potentially knock out Msh6 activity; so, while some fraction of the mixed wild type–mutant heterodimers can retain optimal Msh6 activity, the rest may well not.
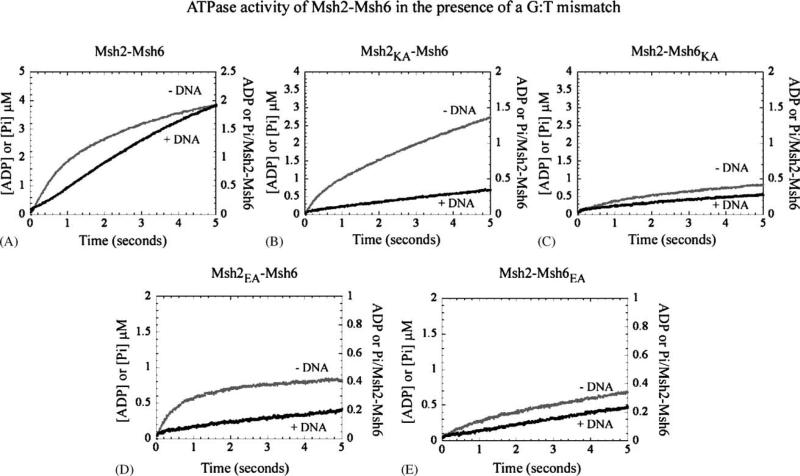
3.3. Contact between Msh6 and a mismatched base pair results in suppression of Msh6-catalyzed ATP hydrolysis
When Msh2–Msh6 is bound to mismatched DNA, its ATPase mechanism is altered such that instead of a step after ATP hydrolysis and phosphate release, a step before or at ATP hydrolysis becomes slow and rate limiting [19]. Thus, in the presence of a G:T mismatch there is no rapid burst of hydrolysis; instead, ATP is apparently hydrolyzed at a linear rate of 0.74μM s−1 (i.e., kcat = 0.37 s−1, Fig. 4A; an initial lag in the kinetic trace may be related to the length of the DNA substrate; Hingorani et. al., unpublished data). The same, slow ATPase rate is observed when Msh2–Msh6 interacts with G:T mismatch in an ADP-bound form (see Supplemental Fig. S2). The effect of mismatched DNA on Msh2 mutant–Msh6 wild type mixed dimers is similarly striking; the ATP hydrolysis rate constant of Msh2K694A–Msh6WT drops from 1.8 s−1 to 0.12μM s−1 (Fig. 4B), and that of Msh2E768A–Msh6WT from 2 s−1 to 0.04μM s−1 (Fig. 4D). These data confirm that mismatched DNA binding suppresses the activity of the subunit responsible for rapid ATP hydrolysis, which in this case is Msh6. In contrast, the residual activity of Msh2 wild type–Msh6 mutant mixed dimers does not change significantly in the presence of mismatched DNA. When bound to a G:T mismatch, Msh2WT–Msh6K988A and Msh2WT–Msh6E1062A hydrolyze ATP at 0.09μM s−1 (Fig. 4C) and 0.08μM s−1 (Fig. 4E), respectively, similar to their ATPase rates of 0.1 μM s−1 in the absence of DNA (Fig. 3C and E). If this rate reflects inherently slow ATPase activity of Msh2, this subunit does not appear to be affected by mismatched DNA.
Fig. 4.
Msh2–Msh6 binding to mismatched DNA results in suppression of ATP hydrolysis at the Msh6 subunit (— no DNA; — G:T): similar to (A) wild type Msh2–Msh6, the rapid ATP hydrolysis activity of mutant Msh2–wild type Msh6 dimers, (B) Msh2K694A–Msh6WT and (D) Msh2 E768A–Msh6WT, is inhibited by mismatched DNA. The residual activity of wild type Msh2–mutant Msh6 dimers, (C) Msh2WT–Msh6K988A and (E) Msh2WT–Msh6E1062A, is relatively insensitive to the presence of mismatched DNA.
3.4. ATP binding to both Msh2 and Msh6 is necessary to alter the interaction between Msh2–Msh6 and mismatched DNA
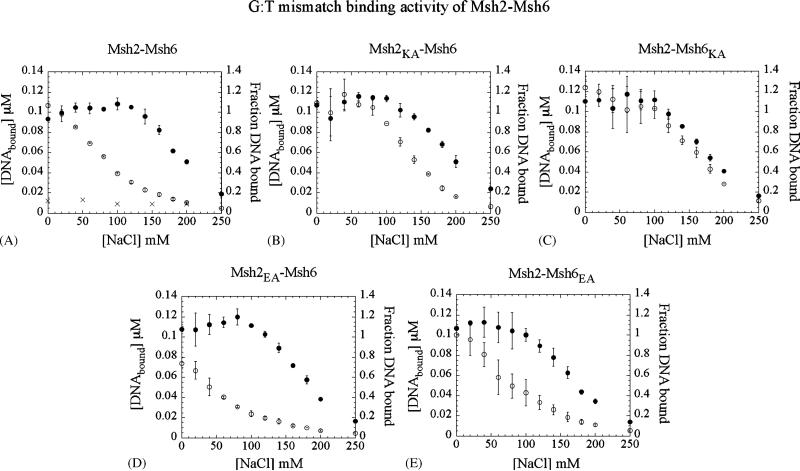
MutS/Msh proteins bind mismatched DNA with high affinity and stability, but in the presence of ATP (and ATPγS), the interaction is altered and the protein dissociates from DNA if its ends are left unblocked [9,11,15]. MutS/Msh binding to DNA appears also to be sensitive to the concentration of NaCl in the reaction, as expected for protein–DNA interactions that involve sequence non-specific contacts. Therefore, in order to assess the impact of ATP site mutations on Msh2–Msh6 interactions with DNA, we analyzed the activity of wild type and mixed wild type–mutant dimers in the absence and in the presence of ATPγS, as a function of NaCl concentration. In Fig. 5A, data from nitrocellulose membrane filtration experiments reveal that at low NaCl concentrations, wild type Msh2–Msh6 binds a G:T mismatch preferentially over fully matched DNA, with or without ATPγS present in the reaction. As NaCl is increased, however, the difference between Msh2–Msh6 and ATPγS-bound Msh2–Msh6 becomes obvious. Thus, at 120 mM NaCl, 100% of the DNA is bound by Msh2–Msh6 in the absence of ATPγS while only about 30% remains bound in the presence of ATPγS. The difference in the K1/2 values for the two isotherms provides a measure of the striking change in the interaction of Msh2–Msh6 with DNA upon binding ATP; K1/2 = 200 mM (−ATPγS) and 85 mM (+ATPγS). Disruption of nucleotide binding by a Walker A site mutation in either subunit impacts the link between the ATPase and mismatch binding activities of Msh2–Msh6. For Msh2K694A–Msh6WT, the K1/2 values are 200 mM (−ATPγS) and 145 mM (+ATPγS), indicating that the protein–DNA complex remains fairly resistant to NaCl if Msh2 is incapable of binding ATP (Fig. 5B). The effect is even more striking when Msh6 cannot bind ATP, as the binding isotherms for Msh2WT–Msh6K988A are virtually identical in the absence or presence of ATPγS, with K1/2 values of 190 mM (−ATPγS) and 175 mM (+ATPγS) (Fig. 5C). Experiments with the Walker B site mutants, which are capable of binding ATP (Fig. 2A), provide complementary results, as the interaction of these mixed wild type–mutant dimers with DNA is sensitive to NaCl in the presence of ATPγS, similar to wild type Msh2–Msh6; binding isotherms for Msh2E768A–Msh6WT have K1/2 values of 190 mM (−ATPγS) and 40 mM (+ATPγS) (Fig. 5D), and those for Msh2WT–Msh6E1062A have K1/2 values of 175 mM (−ATPγS) and 80 mM (+ATPγS) (Fig. 5E). Subtle differences between the data for Msh2 and Msh6 Walker A/B mutants suggest that the Msh6 subunit has a more critical role in the link between the mismatch recognition and ATPase activities of Msh2–Msh6 (e.g., the Walker A site mutation in Msh6 has a greater stabilizing effect on the protein–DNA complex than the same mutation in Msh2). Nonetheless, ATP binding to both Msh2 and Msh6 is necessary to trigger a substantial change in the interaction between the heterodimer and mismatched DNA.
Fig. 5.
Msh2–Msh6 binding to mismatched DNA is altered when both subunits bind ATP (ATPγS): interaction of a G:T mismatch with (A) Msh2–Msh6, (D) Msh2 E768A–Msh6WT, (E) Msh2WT–Msh6E1062A dimers (in which both subunits are capable of binding ATP) becomes highly sensitive to NaCl concentration in the presence of ATPγS (○) vs. in the absence of ATPγS (●). In contrast, there is no significant change in the NaCl sensitivity of G:T binding to (B) Msh2K694A–Msh6WT and (C) Msh2WT–Msh6K988A dimers (in which one subunit is mutated for ATP binding activity), in the presence of ATPγS. (A) A control experiment shows background level interaction between Msh2–Msh6 and matched DNA at all NaCl concentrations (×).
4. Discussion
Two key features of MutS/Msh function in DNA mismatch repair are the asymmetry and coordination between mismatch recognition and ATPase activities of the subunits in these dimeric proteins. The asymmetry is clearly evident from both structural and biochemical analyses of the proteins. Amino acids from each MutS subunit make distinctly different, sequence-independent contacts with mismatched DNA, and only one subunit uses a Phe-X-Glu motif for base-specific stacking and hydrogen bonding interactions with the mismatch [22,23,34]. Both subunits bind ATP, but with differing affinities, and their ATPase kineics are also very different [18–21]. Since prokaryotic MutS is a homodimer, it is not clear whether the asymmetry is an intrinsic property of the protein itself, or whether it arises following MutS interaction with nucleotide and/or DNA. For the same reason, it is difficult to identify exactly what each subunit contributes to the overall function of the dimer. Eukaryotic Msh2–Msh6, however, is composed of non-identical, homologous subunits, which facilitates examination of their individual activities and how they might be coordinated for DNA mismatch repair. In this context, two specific questions have been the focus of MutS/Msh studies in the past, and are addressed here in greater detail: (a) what is the mechanism of ATP binding, hydrolysis, and product release at each subunit, and are they/how are they linked? and (b) how is the ATPase mechanism of each subunit influenced by DNA binding, and vice versa?
Both subunits must be ATPase active for DNA mismatch repair, since mutation of a conserved Walker A site glycine in either S. cerevisiae Msh2 or Msh6 inactivates repair in vivo, and the same is true for mutation of a conserved Walker B site glutamate residue [6,29]. However, each subunit in the dimer appears to have a distinct ATPase mechanism (even in the prokaryotic MutS homodimer!), and likely a distinct function in the repair reaction. Thus, Msh6 (S1 in MutS) binds and hydrolyzes ATP rapidly (2 s−1), whereas Msh2 (S2 in MutS) apparently catalyzes ATP at a substantially slower rate (0.1 μM s−1); if we assume that Msh2 is fully active in Msh2WT–Msh6K988A or Msh2WT–Msh6E1062A dimers in the reaction (2 μM), then the rate constant for Msh2-catalyzed ATP hydrolysis is 0.05 s−1 or 40-fold slower than that for Msh6 (Fig. 3). This difference between Msh2 and Msh6 is consistent with the 30–50-fold difference in ATP hydrolysis rates observed between the S1 and S2 subunits of T. aquaticus MutS dimer. The data are also consistent with previous hypotheses, based on steady-state ATPase analysis of mixed wild type–mutant Walker A/B Msh2–Msh6 heterodimers, that Msh6 is a more efficient ATPase or that it makes a greater contribution to the ATPase activity of the dimer than Msh2 [6,29,30].
Nevertheless, the Msh2 subunit also makes several important contributions to the Msh2–Msh6 ATPase, through what appears to be a very particular link between the activities of the two subunits. Thus, when the Msh2 ATPase site is perturbed such that it does not bind nucleotide with the same affinity as the wild type protein (Msh2K694A–Msh6WT; Fig. 2A), we do not observe a full burst amplitude for Msh6-catalyzed rapid ATP hydrolysis (Fig. 3B). This result suggests that ATP binding to Msh2 is coupled to optimal ATP hydrolysis at Msh6; a similar link appears to exist between the T. aquaticus S2 and S1 subunits as well [18]. On the other hand, when the Msh2 ATPase site is perturbed such that the nucleotide remains tightly bound to it (Msh2E768A–Msh6WT; Fig. 3F and 2A), we do not observe any Msh6 ATPase activity beyond the first turnover (Fig. 3D); i.e., Msh6 appears to be trapped in an inactive state following ATP hydrolysis and phosphate release. Apparently, ATP (or ADP?) must dissociate from Msh2 for Msh2–Msh6 to continue to function as a catalytic ATPase (note: even though Msh6 appears only partially active in Msh2K694A–Msh6WT, it does undergo catalytic turnover; Fig. 2 and 3B). It is not surprising that the ATPase activities of the two subunits appear to be intimately linked, as crystal structures of MutS dimers indicate that significant portions of the C-terminal domains containing the ATPase sites are involved in the dimerization interface [22,23].
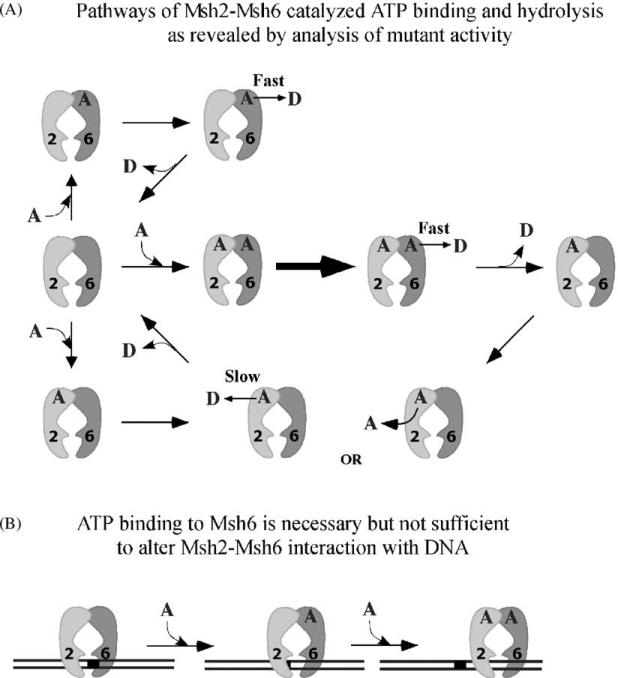
A model pathway for Msh2–Msh6 ATPase activity incorporating these new findings is shown in Fig. 6A. In the central pathway we see that upon ATP binding to both subunits, rapid ATP hydrolysis occurs at Msh6 and slow ATP hydrolysis occurs at Msh2 (alternately ATP could be released, unhydrolyzed, from Msh2). Once the nucleotide has dissociated from Msh2, the protein can undergo catalytic turnover. Alternative pathways show that Msh6 can catalyze rapid ATP hydrolysis if the Msh2 site is empty (top), and Msh2 can catalyze slow ATP hydrolysis if the Msh6 site is empty (bottom), but these reactions are not optimal (Fig. 6A). The reasons for this particular linkage mechanism are not clear yet, and they may range from a simple requirement that a significant population of Msh2–Msh6 be in such forms as Msh2ATP–Msh6ADP or Msh2ADP–Msh6ADP in steady state to more complex requirements such as a particular sequence of ATP binding and hydrolysis, and corresponding conformational changes in the two subunits, for optimal Msh2–Msh6 function.
Fig. 6.
Model pathways for Msh2–Msh6 ATPase activity and interaction with mismatched DNA. (A) The ATPase activities of mixed wild type–mutant Msh2–Msh6 heterodimers indicate that the Msh6 subunit catalyzes ATP hydrolysis at a fast rate while Msh2 hydrolyzes ATP at a relatively slow rate. However, Msh6 activity appears to be maximal only when Msh2 can also bind and hydrolyze ATP. Also, catalytic turnover of Msh6 requires that Msh2 be in a nucleotide-free state. (B) ATP binding to Msh6 is necessary but not sufficient to trigger a change in Msh2–Msh6 interaction with mismatched DNA.
This study also revealed some details of the link between the ATPase mechanisms of Msh2 and Msh6 and their DNA binding/mismatch recognition activities. We have found that the specific interaction between a mismatched base pair and the phenylalanine and glutamate residues of Msh6 results in 6–10-fold suppression of the ATP hydrolysis step in its ATPase mechanism (Fig. 4A). The ATPase activity of Msh2, which interacts only in a sequence non-specific manner with the DNA, appears not to be affected. Similar findings have been reported for T. aquaticus MutS, in which interaction with mismatched DNA suppresses the rapid ATP hydrolysis activity of S1 (30-fold) but does not appear to affect the activity of S2; of course in this case it was not known which subunit, S1 or S2, made specific contacts with the mismatch [18]; mismatched DNA inhibits rapid ATP hydrolysis catalyzed by E. coli MutS as well, although the stoichiometry of the reaction is not resolved [35]. We know now that Msh6 in S. cerevisiae Msh2–Msh6 is the equivalent of S1 in T. aquaticus MutS, and there exists a strong “cis” linkage between the mismatch recognition and ATPase activities of this subunit. Although the DNA binding and nucleotide binding sites within each subunit are separated by 60–70 Å, a long helical lever arm linking the two sites is thought to facilitate communication between them, and may be the crux of the linkage we have detected in this study [22,23].
In contrast to the apparently subunit-specific effect of mismatched DNA on the ATPase activity of the MutS/Msh dimer, ATP binding to both subunits is necessary to propagate a change in MutS/Msh interaction with mismatched DNA. If nucleotide binding to either Msh2 (Msh2K694A–Msh6WT) or Msh6 (Msh2WT–Msh6K988A) is disrupted, the protein–DNA complex becomes refractory to the effects of ATP/ATPγS relative to wild type Msh2–Msh6–DNA complex (Fig. 5). The data do suggest that ATP binding to Msh6 has a somewhat greater impact on Msh2–Msh6 interaction with mismatched DNA, and this is reflected in the schematic shown in Fig. 6B.
Several reports in the literature indicate that upon binding ATP, MutS/Msh proteins appear to release the mismatch by sliding away from the site [15,36]. But, ATP binding to MutS/Msh also facilitates formation of ternary complexes containing MutS and MutL proteins and the mismatch [12,15,16]. These two very different outcomes illustrate the dynamic character of MutS–DNA interactions, and highlight the need for continued kinetic analysis to better define the nature and timing of various MutS, MutL, and DNA binding/release events, and coupled ATP binding/hydrolysis events, in order to understand how MutS proteins signal DNA repair following mismatch recognition. Asymmetry within the MutS (and MutL) dimers adds yet one more layer of complexity to the mismatch repair system, but our study, along with others in the recent past, indicate that at least this aspect of the puzzle can be addressed effectively with the help of mixed wild type–mutant heterodimers [1].
Supplementary Material
Acknowledgement
This work was supported by a grant from the National Institutes of Health (GM64514).
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at 10.1016/j.dnarep.2005.08.016.
References
- 1.Kunkel TA, Erie DA. DNA mismatch repair. Annu. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 2.Schofield MJ, Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. Annu. Rev. Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- 3.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 4.Haber LT, Walker GC. Altering the conserved nucleotide binding motif in the Salmonella typhimurium MutS mismatch repair protein affects both its ATPase and mismatch binding activities. EMBO J. 1991;10:2707–2715. doi: 10.1002/j.1460-2075.1991.tb07815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu TH, Marinus MG. Dominant negative mutator mutations in the mutS gene of Escherichia coli. J. Bacteriol. 1994;176:5393–5400. doi: 10.1128/jb.176.17.5393-5400.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Studamire B, Quach T, Alani E. Saccharomyces cerevisiae Msh2p and Msh6p ATPase activities are both required during mismatch repair. Mol. Cell Biol. 1998;18:7590–7601. doi: 10.1128/mcb.18.12.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drotschmann K, Clark AB, Tran HT, Resnick MA, Gordenin DA, Kunkel TA. Mutator phenotypes of yeast strains heterozygous for mutations in the MSH2 gene. Proc. Natl. Acad. Sci. U.S.A. 1999;96:2970–2975. doi: 10.1073/pnas.96.6.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen DJ, Makhov A, Grilley M, Taylor J, Thresher R, Modrich P, Griffith JD. MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J. 1997;16:4467–4476. doi: 10.1093/emboj/16.14.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blackwell LJ, Martik D, Bjornson KP, Bjornson ES, Modrich P. Nucleotide-promoted release of hMutSalpha from heteroduplex DNA is consistent with an ATP-dependent translocation mechanism. J. Biol. Chem. 1998;273:32055–32062. doi: 10.1074/jbc.273.48.32055. [DOI] [PubMed] [Google Scholar]
- 10.Blackwell LJ, Bjornson KP, Allen DJ, Modrich P. Distinct MutS DNA-binding modes that are differentially modulated by ATP binding and hydrolysis. J. Biol. Chem. 2001;276:34339–34347. doi: 10.1074/jbc.M104256200. [DOI] [PubMed] [Google Scholar]
- 11.Gradia S, Subramanian D, Wilson T, Acharya S, Makhov A, Griffith J, Fishel R. hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol. Cell. 1999;3:255–261. doi: 10.1016/s1097-2765(00)80316-0. [DOI] [PubMed] [Google Scholar]
- 12.Acharya S, Foster PL, Brooks P, Fishel R. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol. Cell. 2003;12:233–246. doi: 10.1016/s1097-2765(03)00219-3. [DOI] [PubMed] [Google Scholar]
- 13.Junop MS, Obmolova G, Rausch K, Hsieh P, Yang W. Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol. Cell. 2001;7:1–12. doi: 10.1016/s1097-2765(01)00149-6. [DOI] [PubMed] [Google Scholar]
- 14.Wang H, Hays JB. Signaling from DNA mispairs to mismatch-repair excision sites despite intervening blockades. EMBO J. 2004 doi: 10.1038/sj.emboj.7600153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selmane T, Schofield MJ, Nayak S, Du C, Hsieh P. Formation of a DNA mismatch repair complex mediated by ATP. J. Mol. Biol. 2003;334:949–965. doi: 10.1016/j.jmb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 16.Habraken Y, Sung P, Prakash L, Prakash S. ATP-dependent assembly of a ternary complex consisting of a DNA mismatch and the yeast MSH2–MSH6 and MLH1-PMS1 protein complexes. J. Biol. Chem. 1998;273:9837–9841. doi: 10.1074/jbc.273.16.9837. [DOI] [PubMed] [Google Scholar]
- 17.Schofield MJ, Nayak S, Scott TH, Du C, Hsieh P. Interaction of Escherichia coli MutS and MutL at a DNA mismatch. J. Biol. Chem. 2001;276:28291–28299. doi: 10.1074/jbc.M103148200. [DOI] [PubMed] [Google Scholar]
- 18.Antony E, Hingorani MM. Asymmetric ATP binding and hydrolysis activity of the Thermus aquaticus MutS dimer is key to modulation of its interactions with mismatched DNA. Biochemistry. 2004;43:13115–13128. doi: 10.1021/bi049010t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antony E, Hingorani MM. Mismatch recognition-coupled stabilization of Msh2–Msh6 in an ATP-bound state at the initiation of DNA repair. Biochemistry. 2003;42:7682–7693. doi: 10.1021/bi034602h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bjornson KP, Modrich P. Differential and simultaneous adenosine di- and triphosphate binding by MutS. J. Biol. Chem. 2003;278:18557–18562. doi: 10.1074/jbc.M301101200. [DOI] [PubMed] [Google Scholar]
- 21.Lamers MH, Winterwerp HH, Sixma TK. The alternating ATPase domains of MutS control DNA mismatch repair. EMBO J. 2003;22:746–756. doi: 10.1093/emboj/cdg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 23.Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de Wind N, Sixma TK. The crystal structure of DNA mismatch repair protein MutS binding to a G × T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 24.Alani E, Lee JY, Schofield MJ, Kijas AW, Hsieh P, Yang W. Crystal structure and biochemical analysis of the MutS·ADP·beryllium fluoride complex suggests a conserved mechanism for ATP · interactions in mismatch repair. J. Biol. Chem. 2003;278:16088–16094. doi: 10.1074/jbc.M213193200. [DOI] [PubMed] [Google Scholar]
- 25.Lamers MH, Georgijevic D, Lebbink JH, Winterwerp HH, Agianian B, de Wind N, Sixma TK. ATP increases the affinity between MutS ATPase domains. Implications for ATP hydrolysis and conformational changes. J. Biol. Chem. 2004;279:43879–43885. doi: 10.1074/jbc.M406380200. [DOI] [PubMed] [Google Scholar]
- 26.Martik D, Baitinger C, Modrich P. Differential specificities and simultaneous occupancy of human MutSalpha nucleotide binding sites. J. Biol. Chem. 2004;279:28402–28410. doi: 10.1074/jbc.M312108200. [DOI] [PubMed] [Google Scholar]
- 27.Bowers J, Sokolsky T, Quach T, Alani E. A mutation in the MSH6 subunit of the Saccharomyces cerevisiae MSH2–MSH6 complex disrupts mismatch recognition. J. Biol. Chem. 1999;274:16115–16125. doi: 10.1074/jbc.274.23.16115. [DOI] [PubMed] [Google Scholar]
- 28.Dufner P, Marra G, Raschle M, Jiricny J. Mismatch recognition and DNA-dependent stimulation of the ATPase activity of hMutSalpha is abolished by a single mutation in the hMSH6 subunit. J. Biol. Chem. 2000;275:36550–36555. doi: 10.1074/jbc.M005987200. [DOI] [PubMed] [Google Scholar]
- 29.Drotschmann K, Yang W, Kunkel TA. Evidence for sequential action of two ATPase active sites in yeast Msh2–Msh6. DNA Rep. 2002;1:743–753. doi: 10.1016/s1568-7864(02)00081-2. [DOI] [PubMed] [Google Scholar]
- 30.Iaccarino I, Marra G, Palombo F, Jiricny J. hMSH2 and hMSH6 play distinct roles in mismatch binding and contribute differently to the ATPase activity of hMutSalpha. EMBO J. 1998;17:2677–2686. doi: 10.1093/emboj/17.9.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finkelstein J, Antony EA, Hingorani MM, O'Donnell M. Overproduction and analysis of eukaryotic multi-protein complexes in E. coli using a dual vector strategy. Anal. Biochem. 2003 doi: 10.1016/s0003-2697(03)00273-2. [DOI] [PubMed] [Google Scholar]
- 32.Zito CR, Antony E, Hunt JF, Oliver DB, Hingorani MM. Role of a conserved glutamate residue in the Escherichia coli SecA ATPase mechanism. J. Biol. Chem. 2005;280:14611–14619. doi: 10.1074/jbc.M414224200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iaccarino I, Marra G, Dufner P, Jiricny J. Mutation in the magnesium binding site of hMSH6 disables the hMutSalpha sliding clamp from translocating along DNA. J. Biol. Chem. 2000;275:2080–2086. doi: 10.1074/jbc.275.3.2080. [DOI] [PubMed] [Google Scholar]
- 34.Natrajan G, Lamers MH, Enzlin JH, Winterwerp HH, Perrakis A, Sixma TK. Structures of Escherichia coli DNA mismatch repair enzyme MutS in complex with different mismatches: a common recognition mode for diverse substrates. Nucl. Acids Res. 2003;31:4814–4821. doi: 10.1093/nar/gkg677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bjornson KP, Allen DJ, Modrich P. Modulation of MutS ATP hydrolysis by DNA cofactors. Biochemistry. 2000;39:3176–3183. doi: 10.1021/bi992286u. [DOI] [PubMed] [Google Scholar]
- 36.Mendillo ML, Mazur DJ, Kolodner RD. Analysis of the interaction between the Saccharomyces cerevisiae MSH2–MSH6 and MLH1-PMS1 complexes with DNA using a reversible DNA end-blocking system. J. Biol. Chem. 2005;280:22245–22257. doi: 10.1074/jbc.M407545200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.