Abstract
Patients with triple-negative breast cancer (TNBC) have a poor prognosis because TNBC often metastasizes, leading to death. Among patients with TNBC, those with ERK2 (extracellular signal-regulated kinase 2)-overexpressing tumors were at higher risk of death than those with low-ERK2-expressing tumors (hazard ratio, 2.76; 95% confidence interval, 1.19–6.41). The MAPK pathway has been shown to be a marker of breast cancer metastasis, but has not been explored as a potential therapeutic target for preventing TNBC metastasis. Interestingly, when we treated TNBC cells with the allosteric MEK inhibitor selumetinib, cell viability was not reduced in 2-dimensional culture. However, in 3-dimensional culture, selumetinib changed the mesenchymal phenotype of TNBC cells to an epithelial phenotype. Cells that undergo epithelial-mesenchymal transition (EMT) are thought to contribute to the metastatic process. EMT leads to generation of mesenchymal-like breast cancer cells with stem cell-like characteristics and a CD44+CD24−/low expression pattern. We tested the hypothesis that targeted inhibition of the MAPK pathway by selumetinib inhibits acquisition of the breast cancer stem cell phenotype and prevents lung metastasis of TNBC. TNBC cells treated with selumetinib showed inhibition of anchorage-independent growth, an indicator of in vivo tumorigenicity (P<0.005), and decreases in the CD44+CD24−/low fraction, ALDH1 activity and mammosphere-forming efficiency. Mice treated with selumetinib formed significantly fewer lung metastases than control mice injected with vehicle (P<0.05). Our data demonstrate that MEK inhibitors can inhibit breast cancer stem cells and may have clinical potential for the prevention of metastasis to prevent metastasis in certain cases in which tumors are MAPK dependent.
Keywords: TNBC, cancer stem cell, selumetinib, metastasis AZD 6244
INTRODUCTION
Triple-negative breast cancer (TNBC), which lacks estrogen receptor and progesterone receptor expression and has low HER2 expression, is an aggressive, poorly understood disease that accounts for 20–25% of all breast cancers. Patients with TNBC can be treated with chemotherapeutic agents, but disease often recurs, resulting in rapid disease progression, metastasis, and death. TNBC is the only major type of breast cancer for which no specific targeted therapy is available to improve patient outcomes (1).
The MAPK pathway is known to promote cell proliferation, angiogenesis, differentiation, and survival. Many drugs that act on signaling pathways upstream of ERK, such as the HER2-targeting drugs trastuzumab and lapatinib, may exert their actions through the MAPK pathway. The MAPK pathway has been shown to be a marker of breast cancer metastasis and increased MAPK activity was associated with lymph node metastasis (2).The MAPK pathway also has oncogenic potential, as demonstrated by the findings that elevated ERK1/2 MAPK activity is associated with shorter disease-free survival and MAPK activity is prognostic for relapse-free survival in patients with breast cancer (2–5).
We recently showed that among patients with TNBC, those with ERK2-overexpressing tumors were at higher risk of death than those with low-ERK2-expressing tumors (6). This evidence suggests that ERK modulation may be a helpful therapeutic strategy for some subsets of TNBC. ERK activity is tightly regulated by MEK through phosphorylation. Many potential therapeutic MEK inhibitors have been investigated. One of the most widely studied of these inhibitors to date is selumetinib (AZD6244; ARRY-142886; AstraZeneca, Macclesfield, United Kingdom), a highly selective allosteric inhibitor of MEK1/2 (7). At present, multiple clinical trials of selumetinib are ongoing, and basal-type breast cancer cell lines, which accounts for the majority of cases of TNBC, showed sensitivity to MEK inhibitors (8). Selumetinib may become part of a paradigm shift in the treatment of TNBC, from nonspecific chemotherapy to more rational, targeted therapy.
Epithelial-mesenchymal transition (EMT) is an essential developmental process by which cells of epithelial origin lose epithelial characteristics and polarity and acquire a mesenchymal phenotype with fibroblast-like morphology, cytoskeleton reorganization, increased motility, invasiveness, and metastatic capability (9, 10). EMT is characterized by loss of epithelial cell junction proteins, e.g., E-cadherin, occludins, and cytokeratins, and gain of mesenchymal markers, e.g., vimentin, smooth muscle actin, and fibronectin (9, 10). It has been proposed that EMT-like processes allow tumor cells to disassemble and migrate to tissue or organ sites distant from the primary tumor (9, 10). The EMT state in breast cancer is associated with a cancer stem cell (CSC) phenotype (11). For instance, cells undergoing EMT acquire a CD44+CD24−/low expression pattern, have increased ability to form mammospheres, and are enriched in tumor-initiating cells. Furthermore, cancer cells that undergo EMT are more migratory and invasive and have the capacity to metastasize (12). A microarray analysis of cells from breast cancer tumors showed that the gene expression profile of the breast CSC fraction (CD44+CD24−/low) resembled a stem cell-like profile, and this subpopulation was able to form mammospheres in vitro (11). The induction of EMT in immortalized, nontumorigenic human mammary epithelial cells resulted in acquisition of the CD44+CD24−/low phenotype (11). The cell-adhesion molecule CD44 is involved in binding cells to hyaluronic acid, whereas CD24 is a negative regulator of the chemokine receptor CXCR4 and is involved in breast cancer metastasis. The prevalence of CD44+CD24−/low tumor cells in breast cancer may favor distant metastasis (13).
EMT is associated with breast cancer tumorigenicity. A study in FSP-cre transgenic mice, in which mammary epithelial cells were genetically marked with the BGH polyadenylate signal to visualize EMT during cancer progression in vivo, demonstrated that these cells underwent EMT during breast cancer development (14). Whether EMT actually occurs in human cancer and the relevance of EMT in human cancer—in particular, in metastasis—is a matter of intense debate (9, 15, 16). Recently, EMT has been suggested to be a reason for the aggressiveness of basal-like breast cancer. Analysis of a tissue microarray of 479 breast carcinomas showed that up-regulation of mesenchymal markers (vimentin, smooth muscle actin, N-cadherin, cadherin-11), overexpression of proteins involved in extracellular matrix remodeling and invasion (SPARC, laminin, fascin), and reduction of epithelial markers (E-cadherin, cytokeratins) preferentially occurred in basal-like breast tumors.
The current view is that the 2-dimensional (2D) culture system does not accurately reflect the 3-dimensional (3D) cellular microenvironment where cancer cells are located. Previously, it was shown that different types of cells cultured in a 3D culture system show different sensitivities to drugs, indicating that responses in 3D culture may be representative of the way cancer cells respond to chemotherapeutic drugs in vivo (17).
In most cancer cell lines, the MAPK pathway plays a crucial role in inducing EMT; in some cell lines, transient activation of Src, PI3K, and Rac has an effect on particular aspects of EMT (18). ERK and PI3K have been implicated in TGF-beta signaling through the conventional Smad pathways and other as yet unknown pathways (15). A role of these signaling pathways, particularly cooperation between ERK and TGF-beta signaling, is also observed in EMT in squamous carcinoma of the skin, which can progressively acquire a fibroblast-like morphology in later stages of the disease (16). These findings justify studying targeting of ERK as a potential strategy for inhibiting metastasis of TNBC.
We hypothesized that inhibition of the MAPK pathway inhibits acquisition of the CSC phenotype and suppresses metastasis in TNBC. To evaluate this hypothesis, we tested the impact of selumetinib on in vitro and in vivo inhibition of EMT in TNBC cells. We found that selumetinib did not reduce cell viability in 2D culture but did inhibit anchorage-independent growth and reverse EMT in 3D culture model system and inhibited ALDH1 activity and mammosphere-forming efficiency. Using the highly metastatic breast cancer cell line MDA-MB-231-LM, we found that treatment with selumetinib prevented lung metastasis in a xenograft model. Our findings indicate that the MAPK pathway may be a promising therapeutic target in TNBC.
MATERIALS AND METHODS
Cell lines and cell cultures
Three human TNBC cell lines (MDA-MB-231, MDA-MB-468, and SUM149), three non-TNBC (SUM190, KPL-4, and MDA-IBC-3) and one TNBC lung metastatic cell line (MDA-MB-231-LM2) were used. MDA-MB-231 and MDA-MB-468 cells were purchased from American Type Culture Collection in 2008 and SUM149 and SUM190 cells from Asterand in 2011 (Detroit, MI). KPL-4 cells were a kind gift in 2008 from Dr. Junichi Kurebayashi (Kawasaki Medical School, Kawasaki, Japan) and MDA-IBC-3 cells from Dr. WA Woodward in 2010 (The University of Texas MD Anderson Cancer Center, Houston, TX). MDA-MB-231-LM2 cells were obtained in 2010 from Dr. Joan Massague at Memorial Sloan-Kettering Cancer Center. The MDA-MB-231-LM2 cells, a subline of MDA-MB-231, is highly metastatic to lung transduced with HSV1-TK. MDA-MB-231, MDA-MB-231-LM, MDA-MB-468 and KPL-4 cells were maintained in Dulbecco's modified Eagle's medium/F12 medium (GIBCO) supplemented with fetal bovine serum (FBS; 10%) and penicillin-streptomycin (100 units/mL). SUM149, SUM190 and MDA-IBC-3 cells were maintained in F12 medium (GIBCO) supplemented with fetal bovine serum (FBS; 5%), penicillin-streptomycin (100 units/mL) insulin (5 μg/mL), and hydrocortisone (1 μg/mL). All cell lines used in this study were validated either on August 8th 2014, October 28, 2014 and January 2015 by the Characterized Cell Line Core Facility at MD Anderson Cancer Center by using a short tandem repeat method based on primer extension to detect single base deviations.
Drugs
Selumetinib was provided by AstraZeneca.
Western blot analysis
For western blot analysis, cell pellets were lysed as described previously (16). Primary antibodies were anti-phospho-p42/44 MAPK (Thr202/Tyr204) (1:1000 dilution; Cell Signaling), anti-α-tubulin (1:5000 dilution; Sigma-Aldrich), anti-β-actin (1:5000 dilution; Sigma-Aldrich), anti-fibronectin (1:500 dilution; BD Transduction), anti-vimentin (1:1000 dilution; Cell Signaling), anti-E-cadherin (1:1000 dilution; BD Transduction), anti-beta-catenin (1:1000 dilution; Cell Signaling), anti-Twist (1:1000 dilution; Santa Cruz) and anti-Slug (1:1000 dilution; Santa Cruz). Secondary antibodies were horseradish peroxidase-conjugated IgG (1:10,000 dilution; Invitrogen) for chemiluminescent signal detection and the corresponding Alexa Fluor-conjugated IgG (1:5000 dilution; Invitrogen) for fluorescence signal detection.
WST-1 assay
Cell viability was assayed using cell proliferation reagent WST-1 (Promega) as described previously (17). MDA-MB-231 cells (2 × 103/90 μL), SUM149 cells (3 × 103/90 μL), MDA-MB-468 (3 × 103/90 μL), SUM190 (2 × 103/90 μL), KPL-4 (2 × 103/90 μL), and MDA-IBC-3 (2 × 103/90 μL) were seeded into each well of a 96-well plate and treated the next day with selumetinib at a final concentration of 0.001, 0.01, 0.1, 1, or 10 μM for 72 hours.
3D Matrigel projection formation assays
The 3D Matrigel projection formation assays were performed in triplicate using μ-Slide plates (ibidi GmbH, Germany) as described previously (19). For the bottom layer, 10 μL of 100% Matrigel was added into each well of the μ-Slide plate, and the plate was incubated in a CO2 incubator for 10 minutes at 37°C. MDA-MB-231 and SUM149 cells (2 × 104 cells/50 μL) were resuspended in 2% Matrigel/complete medium and then overlaid onto the bottom layer. After a 6- to 24-hour incubation in the CO2 incubator, cell network images were captured and analyzed for tube formation using S.CORE analysis software (S.CO LifeSciences GmbH, Germany).
Migration assay
Migration assays were performed in triplicate using a 48-well micro-chemotaxis chamber. MDA-MB-231 and SUM149 cells (2.5 × 105/250 μL) were resuspended in FBS-free medium and added into each chamber. The bottom wells were filled with complete medium (500 μL) containing 10% FBS as an attractant. The cells were allowed to migrate for 6 hours and were then fixed and stained with hematoxylin and eosin. Migrated cells were scanned using the PathScan Enabler IV Histology Slide Scanner (Meyer Instruments, Inc., TX, USA) and then quantified using National Institutes of Health Image J software (http://rsb.info.nih.gov/ij/).
Invasion assay
Invasion assays were performed using a 48-well micro-chemotaxis chamber as described above for the migration assay with the following modifications. The micro-chemotaxis chambers were pre-filled with 100 μL of 90% Matrigel/FBS-free medium, and cells were allowed to invade for 24 hours. Invaded cells were fixed, stained, scanned and then quantified as described above for the migration assay.
Immunohistochemistry staining
Tumor tissues were fixed in formalin, embedded in paraffin, sectioned to 5 μm, and mounted on slides. The sections were deparaffinized in xylene, rehydrated in graded alcohols, and washed in distilled water. Antigens on sections were retrieved by boiling in 10 mM citric acid (pH 6.0) for 40 minutes. Endogenous peroxidases were quenched by incubation in 3% H2O2 for 10 minutes at room temperature. The slides were washed 3 times with PBS and blocked for 30 minutes with 10% normal goat serum in 1% bovine serum albumin/PBS. The slides were then incubated with the following antibodies: anti-Ki-67 (Lab Vision), anti-phospho-p42/44 MAPK (Thr202/Tyr204) (Cell Signaling), anti-fibronectin (BD Transduction), anti-vimentin (Cell Signaling), anti-E-cadherin (BD Transduction), and anti-β-catenin (Cell Signaling). Stained slides were visualized with an Eclipse 80i microscope (Nikon), and the images were captured at a 200× magnification.
Soft agar assay
Cells (2 × 103 cells/well) were resuspended in 2 mL of 0.4% agarose solution in complete medium and overlaid onto the bottom agarose layer (0.8%) in 6-well plates. After a 25-day incubation, colonies greater than 80 μm in diameter were counted using the GelCount system (Oxford Optronix, UK) according to the manufacturer's instructions.
Mammosphere formation assay
Single cell suspensions (SUM149 cells at 2 × 104 cells/well and MDA-MB-231 cells at 5000 cells/well) were seeded in 6-well ultra-low attachment plates (Corning Incorporated Costar) in serum-free DMEM supplemented with 1% L-glutamine, 1% penicillin/streptomycin, 30% F12 (Sigma), 2% B27 (Invitrogen), 20 ng/mL EGF (Sigma), and 20 ng/mL FGFβ (Invitrogen). After a 7-day incubation, mammospheres greater than 80 μm in diameter were counted using the GelCount system (Oxford Optronix, UK) according to the manufacturer's instructions.
Cell cycle distribution analysis
MDA-MB-231 and SUM149 cells were plated in 6-well plates, cultured overnight, and then treated or untreated with selumetinib for 48 and 72 hours at final concentrations of 0.1 or 1 μM. Cell cycle distribution was analyzed by flow cytometry as described previously (16).
CSC subpopulation analysis
MDA-MB-231 and SUM149 cells (3 × 105 cells) were seeded in 60-mm plates and the next day treated with selumetinib at 0 μM, 0.1 μM, and 1 μM. Forty-eight hours after selumetinib treatment, cells were harvested, incubated at 37°C with ALDEFLUOR reagent (STEMCELL Technologies Inc.) or at room temperature with anti-CD24 and anti-CD44 antibodies (BD Biosciences) for 30 minutes, and then subjected to flow cytometry analysis. For the CD24−/CD44+ subpopulation analysis, non-treated cells incubated with CD24-PE alone or CD44-FITC alone were used as controls to determine non-specific signal and to gain the gates for CD24+ and CD44+ subpopulations, respectively. For the ALDH1+ subpopulation analysis, diethylamino benzaldehyde (DEAB), an ALDH inhibitor, was used as a control for each treatment; for each sample, the cells treated with DEAB were used as a control to gain the gate for ALDH1+ subpopulations.
Selumetinib treatment in a TNBC xenograft model
Institutional Animal Care and Use Committee approval for this study was obtained. Female athymic nu/nu mice, 6–8weeks, average weight 25 g, were randomly divided into 2 groups of 15 mice each. Suspensions of MDA-MB-231-LM2 a derivative of MDA-MB-231 cells that was selected for its ability to metastasize to lung (luciferase-expressing cells) were resuspended (1 × 106 cells in 0.2 mL of PBS) and injected under aseptic conditions into the tail vein of each mouse. Six hours before tumor cell inoculation, group 1 was given vehicle (0.5% hydroxypropyl methyl cellulose and 0.1% Tween 80, 10 ml/kg mouse body weight) and group 2 was given selumetinib 50 mg/kg mouse body weight by oral gavage. The treatment was continued daily with weekends off for 3 weeks. Mice were subjected to whole-body luciferase imaging under an IVIS 100 Imaging System (Xenogen) at 1-week intervals for 3 weeks. Before imaging, mice were injected intraperitoneally with luciferin (Caliper Life Sciences) at 150 mg/kg body weight. Then mice were kept under anesthesia with isoflurane. Starting 5.5 minutes after the luciferin injection, images were collected for 30 seconds each with mice in the ventral and dorsal positions. Images and amounts of bioluminescent signals were analyzed using Living Image Software (Xenogen). At the end of the experiment, tissue samples were collected and processed for expression of target proteins as described above in the section Immunohistochemistry Staining.
Statistical analysis
Statistical analyses were performed with Prism, version 5 (GraphPad Software Inc). Data are presented as means ± standard error. Means for all data were compared by 1-way analysis of variance with post hoc testing or by unpaired t-test. P values of <0.05 were considered statistically significant.
RESULTS
Selumetinib does not inhibit cell proliferation but does inhibit anchorage-independent growth in TNBC cells
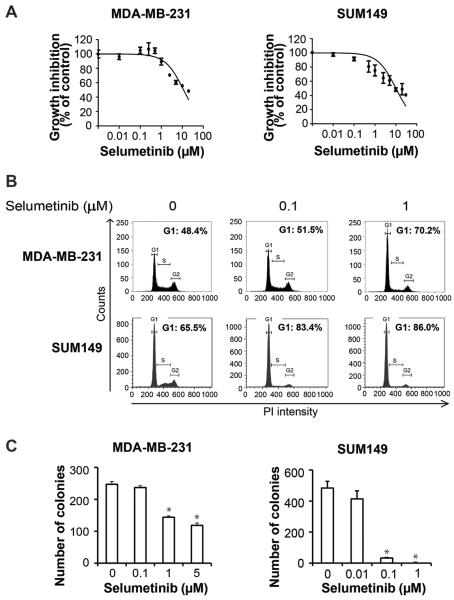
To determine whether selumetinib could suppress proliferation of TNBC cells, we treated a panel of breast cancer cells (MDA-MB-231, MDA-MB-468, SUM149, SUM190, KPL-4, and MDA-IBC-3) with different concentrations of selumetinib and assessed cell viability by WST-1 assay 72 hours later. We observed that in MDA-MB-468, SUM190, KPL-4, and MDA-IBC-3 cells, the 50% inhibitory concentration (IC50) was above 20 μM (Fig.S1A), and in MDA-MB-231 and SUM149 cells, the IC50 values were 8.6 μM and 10 μM, respectively (Fig. 1A).
Figure 1.
Selumetinib does not inhibit proliferation of TNBC cells in 2D culture but does inhibit anchorage-dependent growth. A, MDA-MB-231 and SUM149 cells were treated with selumetinib for 72 hours, and then cell viability was measured using the WST-1 assay. B, MDA-MB-231 and SUM149 cells were treated with selumetinib for 48 hours, and FACS analysis was performed to determine cell cycle distribution. PI, propidium iodide. C, MDA-MB-231 and SUM149 cells in soft agar were treated with selumetinib, and colonies were counted 3 weeks later. *, P<0.001; **, P<0.0001.
For further studies, we selected the TNBC cell lines MDA-MB-231 (which harbors BRAF and RAS mutations) and SUM149 (p53 mutant) on the basis of lower IC50 values, metastasis capability and/or mesenchymal features. We examined the effect of selumetinib on cell cycle distribution in the 2 cell lines. As shown in Fig. 1B, in both MDA-MB-231 and SUM 149 cells, selumetinib induced G1 arrest at 48 hours (denoted by an increased proportion of cells in G1 phase). This trend was maintained at 72 hours (Fig. S1B). Next, we assessed the impact of selumetinib treatment of these TNBC cells on anchorage-independent growth in soft agar, a hallmark of tumorigenicity. We observed that selumetinib inhibited colony formation in a dose-dependent manner (Fig. 1C). Selumetinib inhibited colony formation in MDA-MB-231 cells by 41.7% at 1 μM and 52.1% at 5 μM (P < 0.05) and in SUM149 cells by 93.1% at 0.1 μM and 100% at 1 μM (P < 0.05).
Selumetinib induces mesenchymal to epithelial transition in a 3D cell culture model of TNBC
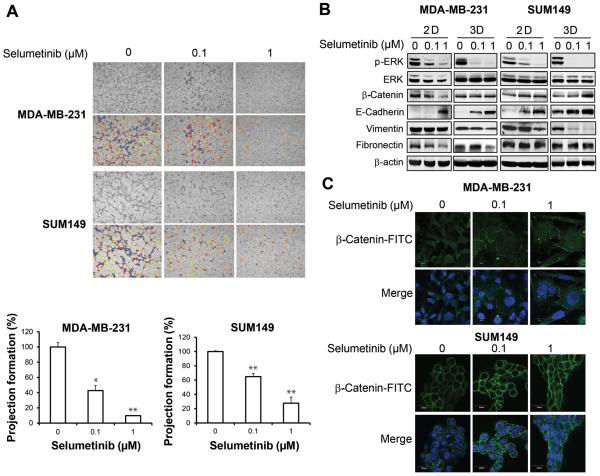
Three-dimensional cell culture models mimic the in vivo microenvironment, a feature that simple 2D cell monolayer culture systems lack. We observed that when MDA-MB-231 and SUM149 TNBC cells were grown in a 3D cell culture model, both types of cells exhibited a mesenchymal phenotype, including formation of projections/filopodia (Fig. 2A, upper panels). When cells were treated with selumetinib, their phenotype changed from mesenchymal to epithelial (inhibition of projections/filopodia formation by 57.3% at 0.1 μM [P < 0.05] and 90.2% at 1 μM [P < 0.01] in MDA-MB-231 cells and by 35.1% at 0.1 μM [P < 0.01] and 72.3% at 1 μM [P < 0.01] in SUM149 cells), suggesting reversal to a more epithelial phenotype (Fig. 2A, lower panels). We further confirmed mesenchymal to epithelial transition by determining the expression of epithelial and mesenchymal markers by western blot analysis. We observed that inhibition of ERK phosphorylation by selumetinib correlated with decreased expression of mesenchymal marker vimentin in 3D culture and increased expression of epithelial marker E-cadherin in both 2D and 3D cultures (Fig. 2B). We found that knocking down MEK1/2 with siRNA had a similar effect on levels of EMT markers as treatment with selumetinib in 3D culture, although treatment with selumetinib had a more robust effect on the EMT marker expression change (Fig S2). We also observed a change in the localization of β-catenin, which was diffusely expressed in both the cytoplasm and the nucleus before selumetinib treatment but was localized at the plasma membrane after treatment in 3D culture (Fig. 2C). These results suggested that treating TNBC with selumetinib induced mesenchymal to epithelial transition in both 2D and 3D cell culture models. Interestingly, the concentration of selumetinib that reversed the mesenchymal phenotype was low (0.1 μM), whereas a 10 fold higher concentration (1 μM) did not inhibit proliferation in a 2D cell culture model.
Figure 2.
Selumetinib reverses 3D mesenchymal phenotype in TNBC cells. A, MDA-MB-231 and SUM149 cells treated with selumetinib in a 3D cell culture model changed from a mesenchymal phenotype to an epithelial phenotype (upper panels). Images were taken under 100× magnification. Projection formation was measured using S.CORE analysis software (lower panels). *, P < 0.05; **, P < 0.01. B, MDA-MB-231 and SUM149 cells were treated with selumetinib for 48 hours, and western blot analysis was performed, with β-actin used as a loading control. C, MDA-MB-231 and SUM149 cells were treated with selumetinib in a 3D culture for 48 hours. Immunofluorescence staining was done to detect β-catenin-FITC (fluorescein isothiocyanate), and nuclei were stained with DAPI. Images were taken under 60× magnification.
Selumetinib inhibits the CSC-like cell population and mammosphere formation in TNBC cells
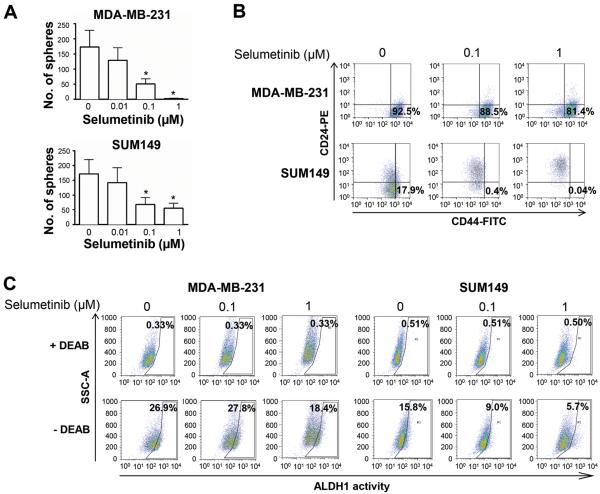
Previous studies have shown that EMT leads to generation of breast cancer cells with stem-cell-like characteristics (11). CSCs are capable of self-renewal. They can be enriched by growth as mammospheres (spheres that grow in suspension in serum-free and growth-factor-enriched medium) (20) and are characterized by high expression of cell surface marker CD44, low expression of cell surface marker CD24 (21), and high ALDH1 activity. To determine the effect of selumetinib on the self-renewal capacity of MDA-MB-231 and SUM149 TNBC cells, we examined the effects of selumetinib on mammosphere formation, expression of CD44 and CD24, and ALDH1 activity. We found that selumetinib significantly decreased the formation of mammospheres in both TNBC cell lines in a dose-dependent manner (Fig. 3A; 70.5% at 0.1 μM and 98.5% at 1 μM in MDA-MB-231 cells [P < 0.05] and 59.9% at 0.1 μM and 67.6% at 1 μM in SUM149 cells [P < 0.05]). Selumetinib decreased the proportion of cells with CD44+/CD24−/low surface marker expression pattern (Fig. 3B). Selumetinib also decreased the proportion of cells with ALDH1 activity in both MDA-MB-231 cells (control 26.9% vs selumetinib 18.4% at 1 μM) and SUM149 cells (control 15.8% vs selumetinib 5.69% at 1 μM) (Fig. 3C). These results suggested that selumetinib could exert its inhibitory effect on the self-renewal capacity of TNBC cells and thus that targeting the MAPK pathway may be a way to reduce the CSC population in TNBC, which is known to have a high potential to metastasize and to to be resistant to chemotherapy.
Figure 3.
Selumetinib reduces the CSC-like cell populations and inhibits mammosphere formation in TNBC cells. A, MDA-MB-231 and SUM149 cells were cultured in the presence of selumetinib for 6 days. Mammosphere formation is presented as the mean ± SEM number of spheres per 5000 (MDA-MB-231) or 20,000 (SUM149) cells plated. *, P < 0.05. B, MDA-MB-231 and SUM149 cells were treated with selumetinib for 24 hours, and CD44+/CD24−/low CSC subpopulations were was analyzed using FACS. Non-treated cells incubated with CD24-PE alone or CD44-FITC alone were used as controls. C, MDA-MB-231 and SUM149 cells were treated with selumetinib for 48 hours, and ALDH1activity was analyzed using FACS. DEAB (diethylamino benzaldehyde), an ALDH inhibitor, was used as a control for each treatment.
Selumetinib inhibits the motility and invasiveness of TNBC cells and prevents lung metastasis in a TNBC xenograft model
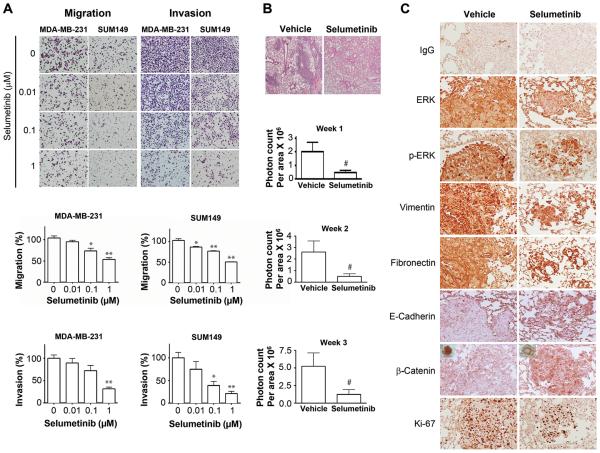
It was previously shown that CD44+CD24−/low enriched breast CSCs display more invasive properties than other cells do (22). Given our findings that selumetinib reversed the mesenchymal phenotype and inhibited the CSC subpopulation in TNBC cells and given the high propensity for early metastasis we investigated whether selumetinib can inhibit migration and invasion of TNBC cells. In MDA-MB-231 and SUM149 cells, compared to migration of untreated cells, migration of cells treated with selumetinib (1 μM) was reduced by 52.0% (P < 0.0001) and 50.1% (P < 0.0001), respectively (Fig. 4A). Similarly, selumetinib (1 μM) inhibited the invasion of both MDA-MB-231 and SUM149 cells, by 68.3% (P < 0.0001) and 78.6% (P < 0.0001), respectively. These results suggest that selumetinib inhibits both cell motility and invasion in TNBC cells.
Figure 4.
Selumetinib inhibits migration and invasion of TNBC cells and prevents lung metastasis in a TNBC xenograft model. A, MDA-MB-231 and SUM149 cells were treated with selumetinib, and migration and invasion assays were performed. *, P < 0.001; **, P < 0.0001. Images were taken under 100× magnification. B, In a TNBC xenograft model, selumetinib treatment significantly reduced lung metastases. Female athymic nude mice were injected intravenously with luciferase-transfected MDA-MB-231-LM2 cells. The experimental groups were treated with vehicle control (0.5% hydroxypropyl methyl cellulose and 0.1% Tween-80) at 10 mL/kg mouse body weight or selumetinib at 50 mg/kg mouse body weight daily for 3 weeks beginning 6 hours before cell injection. #, P < 0.05. The photon counts per area (monitored by IVIS) for each treatment group were measured at weeks 1, 2, and 3 after inoculation. C, Immunohistochemical stains of lung tissues for ERK1/2, pERK1/2, mesenchymal markers (vimentin, fibronectin), epithelial markers (E-cadherin, β-catenin localization), and proliferation (Ki-67) from control mice and mice treated with selumetinib. Images were taken under 200× magnification.
Given that selumetinib reversed the mesenchymal phenotype in TNBC cells and inhibited cell migration and invasion in vitro, we hypothesized that suppression of ERK by selumetinib prevents metastasis in a breast cancer xenograft model. We found that the incidence of lung metastasis (21% – 38%) was lower in the selumetinib-treated group than was in the vehicle-treated group (60% – 64%) (Table 1). Furthermore, bioluminescent signals (to assess metastasis) were significantly lower in selumetinib-treated mice (50 mg/kg/day) than in mice treated with vehicle control on days 7, 14, and 21 after TNBC cell inoculation, indicating that mice treated with selumetinib formed significantly fewer lung metastases than did control mice injected with a vehicle (P ≤ 0.05, 2-sided t-test.) (Fig. 4B).
Table 1.
Numbers of mice bearing MDA-MB-231–LM2-4175 xenografts that had lung metastases after treatment with vehicle (control) or selumetinib (50 mg/kg mouse body weight) daily for 3 weeks.
| Weeks of treatment | Group of treatment | Number of mice with metastatic tumors | Incidence of lung metastasis |
|---|---|---|---|
| Week 1 | Vehicle | 9/15 | 60 |
| Selumetinib | 3/14 | 21 | |
|
| |||
| Week 2 | Vehicle | 9/14 | 64 |
| Selumetinib | 3/13 | 23 | |
|
| |||
| Week 3 | Vehicle | 9/14 | 64 |
| Selumetinib | 5/13 | 38 | |
To further evaluate the effect of selumetinib on metastasis of TNBC, at the end of the animal study, we collected lung tissues from mice and confirmed lung metastasis by hematoxylin and eosin staining (Fig 4B; upper panel). We observed fewer lung metastatic tumors in the treatment group (Fig 4B; upper panel). Lung tissues were analyzed for ERK1/2, pERK1/2, mesenchymal markers (vimentin, fibronectin), epithelial markers (E-cadherin, β-catenin localization), and proliferation (Ki-67) in control mice and mice treated with selumetinib. As expected, expression levels of pERK1/2, ERK, and Ki-67 were reduced in the selumetinib-treated tumors compared to the control tumors. The expression of mesenchymal markers fibronectin and vimentin was also reduced in the treated group. More importantly, selumetinib increased the expression of E-cadherin and β-catenin as well as relocalization of β-catenin to the membrane from the nucleus and cytoplasm (Fig. 4C). These results indicate an inhibition of lung metastasis and reversal from mesenchymal to epithelial phenotype of tumor cells in the mice treated with selumetinib, confirming our in vitro findings in TNBC cells.
DISCUSSION
Our results demonstrate that selumetinib effectively reduced lung metastasis established using MDA-MB-231 cells in a xenograft model. We also showed that in a 3D cell culture model, selumetinib changed the mesenchymal phenotype of TNBC to an epithelial phenotype and decreased the proportion of CSCs, as shown by reduced proportions of cells expressing CSC surface markers and ALDH1, leading to inhibition of mammosphere formation. Through these findings, we have demonstrated that suppression of MAPK pathway signaling by selumetinib can reduce CSC subpopulations in TNBC, leading to inhibition of metastasis.
TNBC is characterized by a mesenchymal phenotype, which has features in common with those of CSCs. Cells undergoing EMT are thought to contribute to the metastatic process and have increased ability to form mammospheres in vitro. TNBC is now classified into 6 subtypes, including 2 basal-like subtypes, which have high expression of cell cycle and DNA damage response genes; an immunomodulatory subtype; a mesenchymal subtype and a mesenchymal stem-like subtype, both of which are enriched for EMT and growth factor pathways; and a luminal androgen receptor subtype, which is characterized by androgen receptor signaling. One of the top canonical pathways associated with the mesenchymal stem-like subtype is the MAPK pathway, as determined by enrichment of genes from samples of mesenchymal stem-like TNBC using mRNA microarray data (23). Our findings suggest that patients with the mesenchymal stem-like subset of TNBC could be effectively treated with therapy targeting the MAPK pathway.
One of the major problems in patients treated for TNBC is tumor recurrence and metastasis. This has been attributed to the concept that CSCs, which are known to be resistant to conventional chemotherapeutic agents and radiation therapy, remain dormant, and give rise to secondary tumors at a later stage. Since CSCs are resistant to chemotherapy and radiation therapy, it is important to target these cells. In our current study, we showed that inhibition of CSCs with MEK inhibitors inhibits lung metastasis in a TNBC xenograft model, which may reduce the frequency of TNBC recurrence and overcome resistance due to CSCs.
In metastatic melanoma with BRAFV600E mutation, MEK1 mutations conferred resistance to treatment with individual MEK or BRAF inhibitors, but the combination of MEK and BRAF inhibitors prevented the emergence of resistant clones (24). Other ways in which cells acquire resistance to MEK inhibition are amplification of BRAF (25, 26) and upregulation of STAT3 (27). Recently, MEK inhibition has been shown to cause loss of ERK activity and c-Myc degradation, which resulted in activation of many receptor tyrosine kinases; receptor tyrosine kinase activation overcame MEK2 inhibition and resulted in ERK activation and drug resistance (28). This shows that there maybe a need for combination treatment. Therefore, we are performing a synthetic lethality screen using the whole-genome siRNA library, and will identify genes whose silencing sensitizes TNBC cells to selumetinib. Small molecules that inhibit the activity of the protein products of the identified target genes will be used with selumetinib as combinatorial therapy against TNBC to overcome resistance.
In our future studies, we will conduct a phase II clinical trial in which MEK inhibitor will be administered to patients with primary TNBC after completion of all local and systemic therapies to determine whether we can prolong progression-free survival.
Supplementary Material
Acknowledgments
We thank Stephanie Deming of the Department of Scientific Publications at MD Anderson for her expert editorial assistance and Wendy Schober, Nalini Patel, and Dr. Jared Burks of the Flow Cytometry and Cellular Imaging Facility at MD Anderson for assistance with cell cycle and stem cell subpopulation analyses and confocal imaging.
Grant support: NIH grant R00CA139006 and MD Anderson Cancer Center Start-up funds (C. Bartholomeusz), NIH grants R01CA127562 (N.T. Ueno) and CA016672 (Cancer Center Support Grant to MD Anderson Cancer Center).
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
REFERENCES
- 1.Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: a critical review. J Clin Oncol. 2008;26:2568–81. doi: 10.1200/JCO.2007.13.1748. [DOI] [PubMed] [Google Scholar]
- 2.Adeyinka A, Nui Y, Cherlet T, Snell L, Watson PH, Murphy LC. Activated mitogen-activated protein kinase expression during human breast tumorigenesis and breast cancer progression. Clin Cancer Res. 2002;8:1747–53. [PubMed] [Google Scholar]
- 3.Gee JM, Robertson JF, Ellis IO, Nicholson RI. Phosphorylation of ERK1/2 mitogen-activated protein kinase is associated with poor response to anti-hormonal therapy and decreased patient survival in clinical breast cancer. International journal of cancer. 2001;95:247–54. doi: 10.1002/1097-0215(20010720)95:4<247::aid-ijc1042>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 4.McClelland RA, Barrow D, Madden TA, Dutkowski CM, Pamment J, Knowlden JM, et al. Enhanced epidermal growth factor receptor signaling in MCF7 breast cancer cells after long-term culture in the presence of the pure antiestrogen ICI 182,780 (Faslodex) Endocrinology. 2001;142:2776–88. doi: 10.1210/endo.142.7.8259. [DOI] [PubMed] [Google Scholar]
- 5.Mueller H, Flury N, Eppenberger-Castori S, Kueng W, David F, Eppenberger U. Potential prognostic value of mitogen-activated protein kinase activity for disease-free survival of primary breast cancer patients. International journal of cancer. 2000;89:384–8. doi: 10.1002/1097-0215(20000720)89:4<384::aid-ijc11>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 6.Bartholomeusz C, Gonzalez-Angulo AM. Targeting the PI3K signaling pathway in cancer therapy. Expert Opin Ther Targets. 2012;16:121–30. doi: 10.1517/14728222.2011.644788. [DOI] [PubMed] [Google Scholar]
- 7.Yeh TC, Marsh V, Bernat BA, Ballard J, Colwell H, Evans RJ, et al. Biological characterization of ARRY-142886 (AZD6244), a potent, highly selective mitogen-activated protein kinase kinase 1/2 inhibitor. Clin Cancer Res. 2007;13:1576–83. doi: 10.1158/1078-0432.CCR-06-1150. [DOI] [PubMed] [Google Scholar]
- 8.Mirzoeva OK, Das D, Heiser LM, Bhattacharya S, Siwak D, Gendelman R, et al. Basal subtype and MAPK/ERK kinase (MEK)-phosphoinositide 3-kinase feedback signaling determine susceptibility of breast cancer cells to MEK inhibition. Cancer Res. 2009;69:565–72. doi: 10.1158/0008-5472.CAN-08-3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–95. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Savagner P. Leaving the neighborhood: molecular mechanisms involved during epithelial-mesenchymal transition. Bioessays. 2001;23:912–23. doi: 10.1002/bies.1132. [DOI] [PubMed] [Google Scholar]
- 11.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Abraham BK, Fritz P, McClellan M, Hauptvogel P, Athelogou M, Brauch H. Prevalence of CD44+/CD24−/low cells in breast cancer may not be associated with clinical outcome but may favor distant metastasis. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11:1154–9. [PubMed] [Google Scholar]
- 14.Trimboli AJ, Fukino K, de Bruin A, Wei G, Shen L, Tanner SM, et al. Direct evidence for epithelial-mesenchymal transitions in breast cancer. Cancer Res. 2008;68:937–45. doi: 10.1158/0008-5472.CAN-07-2148. [DOI] [PubMed] [Google Scholar]
- 15.Brabletz T, Jung A, Kirchner T. Beta-catenin and the morphogenesis of colorectal cancer. Virchows Arch. 2002;441:1–11. doi: 10.1007/s00428-002-0642-9. [DOI] [PubMed] [Google Scholar]
- 16.Gavert N, Conacci-Sorrell M, Gast D, Schneider A, Altevogt P, Brabletz T, et al. L1, a novel target of beta-catenin signaling, transforms cells and is expressed at the invasive front of colon cancers. The Journal of cell biology. 2005;168:633–42. doi: 10.1083/jcb.200408051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serebriiskii I, Castello-Cros R, Lamb A, Golemis EA, Cukierman E. Fibroblast-derived 3D matrix differentially regulates the growth and drug-responsiveness of human cancer cells. Matrix Biol. 2008;27:573–85. doi: 10.1016/j.matbio.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larue L, Bellacosa A. Epithelial-mesenchymal transition in development and cancer: role of phosphatidylinositol 3' kinase/AKT pathways. Oncogene. 2005;24:7443–54. doi: 10.1038/sj.onc.1209091. [DOI] [PubMed] [Google Scholar]
- 19.Lee J, Bartholomeusz C, Krishnamurthy S, Liu P, Saso H, Lafortune TA, et al. PEA-15 unphosphorylated at both serine 104 and serine 116 inhibits ovarian cancer cell tumorigenicity and progression through blocking beta-catenin. Oncogenesis. 1:e22. doi: 10.1038/oncsis.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klopp AH, Lacerda L, Gupta A, Debeb BG, Solley T, Li L, et al. Mesenchymal stem cells promote mammosphere formation and decrease E-cadherin in normal and malignant breast cells. PLoS One. 2010;5:e12180. doi: 10.1371/journal.pone.0012180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Hajj M, Becker MW, Wicha M, Weissman I, Clarke MF. Therapeutic implications of cancer stem cells. Curr Opin Genet Dev. 2004;14:43–7. doi: 10.1016/j.gde.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 22.Sheridan C, Kishimoto H, Fuchs RK, Mehrotra S, Bhat-Nakshatri P, Turner CH, et al. CD44+/CD24− breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res. 2006;8:R59. doi: 10.1186/bcr1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–67. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emery CM, Vijayendran KG, Zipser MC, Sawyer AM, Niu L, Kim JJ, et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc Natl Acad Sci U S A. 2009;106:20411–6. doi: 10.1073/pnas.0905833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corcoran RB, Dias-Santagata D, Bergethon K, Iafrate AJ, Settleman J, Engelman JA. BRAF gene amplification can promote acquired resistance to MEK inhibitors in cancer cells harboring the BRAF V600E mutation. Sci Signal. 3:ra84. doi: 10.1126/scisignal.2001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Little AS, Balmanno K, Sale MJ, Newman S, Dry JR, Hampson M, et al. Amplification of the Driving Oncogene, KRAS or BRAF, Underpins Acquired Resistance to MEK1/2 Inhibitors in Colorectal Cancer Cells (vol 4, art no ra17, 2011) Science Signaling. 2011;4 doi: 10.1126/scisignal.2001752. [DOI] [PubMed] [Google Scholar]
- 27.Dai B, Meng J, Peyton M, Girard L, Bornmann WG, Ji L, et al. STAT3 mediates resistance to MEK inhibitor through microRNA miR-17. Cancer Res. 2011;71:3658–68. doi: 10.1158/0008-5472.CAN-10-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149:307–21. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.