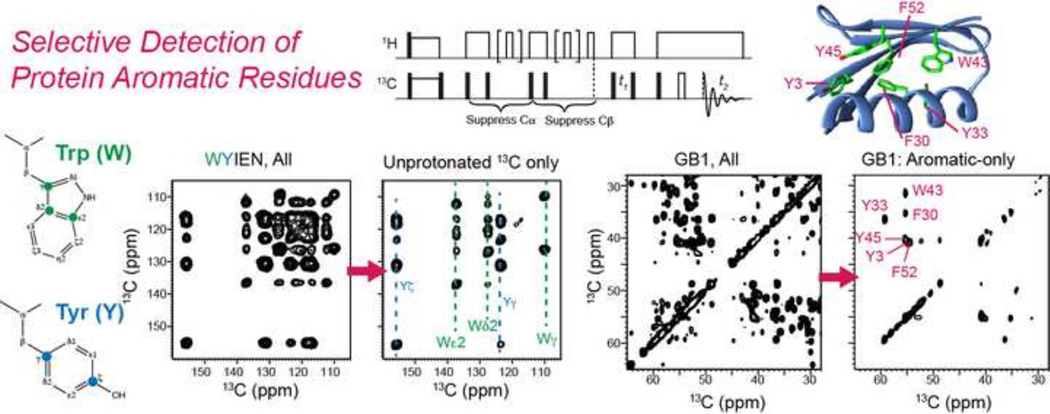
Abstract
The four aromatic amino acids in proteins, namely histidine, phenylalanine, tyrosine, and tryptophan, give highly overlapped 13C chemical shifts between 100 and 160 ppm, and have so far been largely neglected in solid-state NMR determination of protein structures. Yet aromatic residues play important roles in biology through π–π and cation-π interactions. To better resolve and assign aromatic residues’ 13C signals in magic-angle-spinning (MAS) solid-state NMR spectra, we introduce two spectral editing techniques. The first method uses gated 1H decoupling in a proton-driven spin-diffusion (PDSD) experiment to remove all protonated 13C signals and retain only quaternary 13C signals in the aromatic region of the 13C spectra. The second technique uses chemical shift filters and 1H-13C dipolar dephasing to selectively detect the Cα, Cβ and CO cross peaks of aromatic residues while suppressing the signals of all aliphatic residues. We demonstrate these two techniques on amino acids, a model peptide, and the microcrystalline protein GB1, and show that they significantly simplify the 2D NMR spectra and both reveal and permit the ready assignment of the aromatic residues’ signals.
Keywords: aromatic residues, gated decoupling, ASSET, PDSD
Graphical abstract
1. Introduction
Spectral overlap in solid-state NMR MAS spectra remains a challenge in structure determination of proteins and other biological macromolecules. While 2D and 3D correlation experiments are now routinely employed in structure determination [1–7], for dynamically or statically disordered or conformationally polymorphic proteins, the 13C and 15N linewidths often exceed 1 ppm [8], which make even 3D correlation spectroscopy often insufficient for complete resolution of the signals except for the smallest proteins [9]. A number of strategies have been proposed to address this challenge. Optimized sample preparation methods can produce conformational homogeneity, although the biological relevance of the selected conformation is usually unclear. Sparse isotopic labeling [10–13] and segmental labeling based on chemical ligation [14] decrease the complexity of the NMR spectra and thus increase the reliability of the assignment, but they contain lower information content per spectrum. Semi-automated resonance assignment protocols have been designed to explicitly address ambiguities in resonance assignment of broad-line spectra [15–18], but do not indicate to what extent the assignment ambiguity is due to incomplete information or due to structural polymorphism.
Spectral editing is a complementary approach to simplify and resolve protein solid-state NMR spectra without requiring the production of multiple samples. Spectral editing was first developed for natural abundance organic compounds. Opella and Frey described a modification of the 1D cross polarization (CP)-MAS experiment that selectively detects non-protonated 13C signals by inserting a period without 1H decoupling [19]. Zilm and coworkers distinguished between and assigned CH, CH2 and CH3 groups by combining CP with polarization inversion and depolarization, taking advantage of different polarization transfer rates of the differently protonated carbons [20–22]. Schmidt-Rohr and coworkers developed various spectral editing methods based on chemical shift filtering [23–25], multiple-quantum dipolar transfer [26; 27], and dipolar dephasing to assign the chemical shifts of materials such as soil organic matter and carbon materials.
Recently, spectral editing has also been introduced to facilitate the structure determination of uniformly 13C, 15N-labeled proteins. We showed how to selectively detect CH-containing Val, Leu and Ile signals using a dipolar DEPT (distortionless enhancement by polarization transfer) technique, distinguish carboxyl-containing Asp and Glu sidechains from amide-containing Asn and Gln by asymmetric 13C-15N REDOR, and selectively detect dynamic residues by gated 1H decoupling [28]. In addition to spectral editing based on chemical structure and molecular motion, differences in the Cα chemical shift anisotropies (CSAs) and backbone ϕ torsion angle between β-strand and α-helical residues have been exploited to simplify 2D correlation spectra and selectively detect helical or strand residues [29; 30]. Similarly, another structural property, which is the differential water contact of protein residues, has been used to edit the spectra of membrane proteins [8; 31–33].
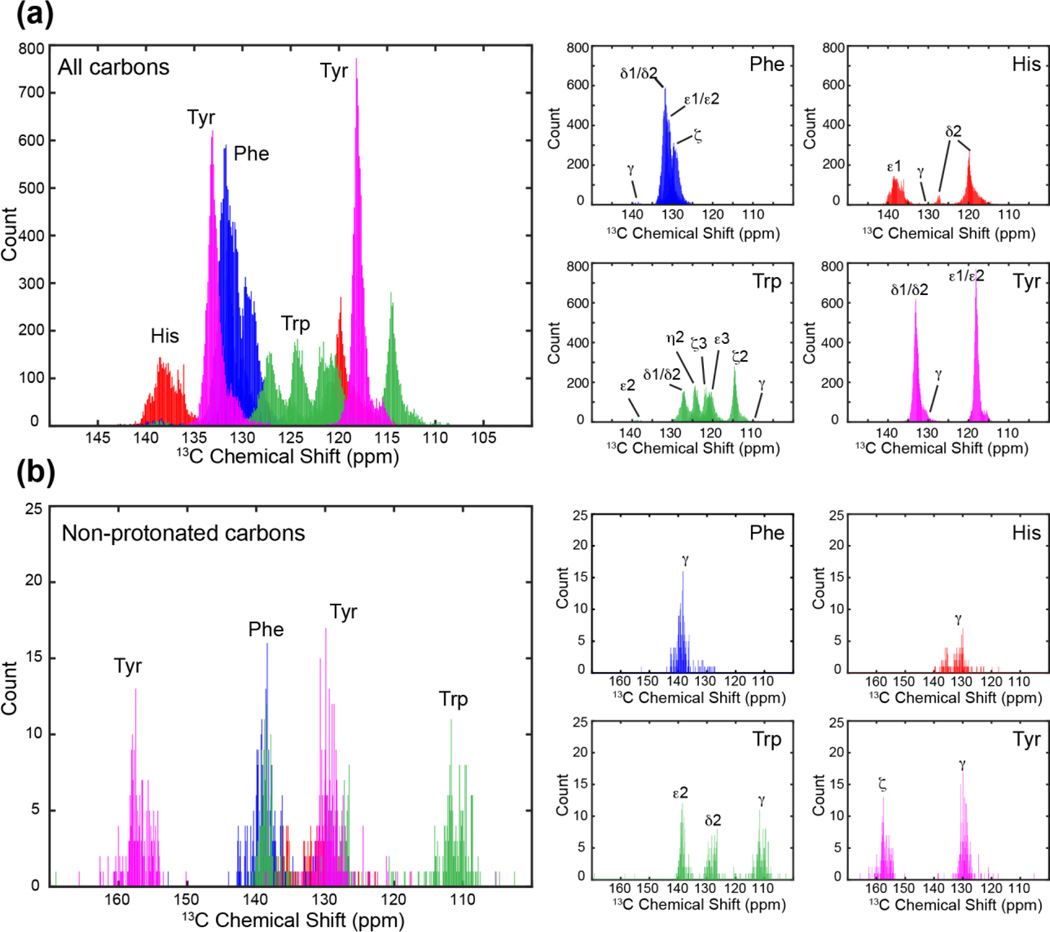
However, so far most protein spectral editing techniques target aliphatic residues, and no methods are yet available to enhance the resolution of aromatic residues. Trp, His, Tyr and Phe play important roles in biology through π–π and cation-π interactions among themselves and with polar residues [34–36] and by functionally relevant ring motions [37; 38]. Trp is also well known to stabilize membrane protein structure and topology by acting as an anchoring residue at the membrane-water interface [39]. Aromatic carbons in these four amino acids largely resonate between 140 and 100 ppm, giving rise to a highly congested spectral region. Fig. 1 shows the 13C chemical shift distribution, extracted from the BMRB database [40], of 23 aromatic carbons in proteins. For residue-type assignment, the only well resolved signals are the non-protonated Tyr Cζ peak at ~158 ppm and to some extent the Trp Cγ signal at ~110 ppm, which occupy the outer limits of the aromatic chemical shift range. The other non-protonated carbons, including Phe Cγ, Tyr Cγ, His Cγ, Trp Cδ2 and Trp Cε2, resonate between 140 and 130 ppm. For histidine, the protonation state of the sidechain significantly affects the 13C chemical shifts [41], causing the non-protonated Cγ chemical shift to vary between 128 and 138 ppm.
Figure 1.
13C chemical shift distributions of the four aromatic residues in proteins from the Biological Magnetic Resonance Data Bank. A large degree of resonance overlap exists in the 110–140 ppm region. (a) All aromatic 13C signals. (b) Only non-protonated aromatic 13C signals. Small panels on the right show the chemical shift dispersions of the individual amino acids.
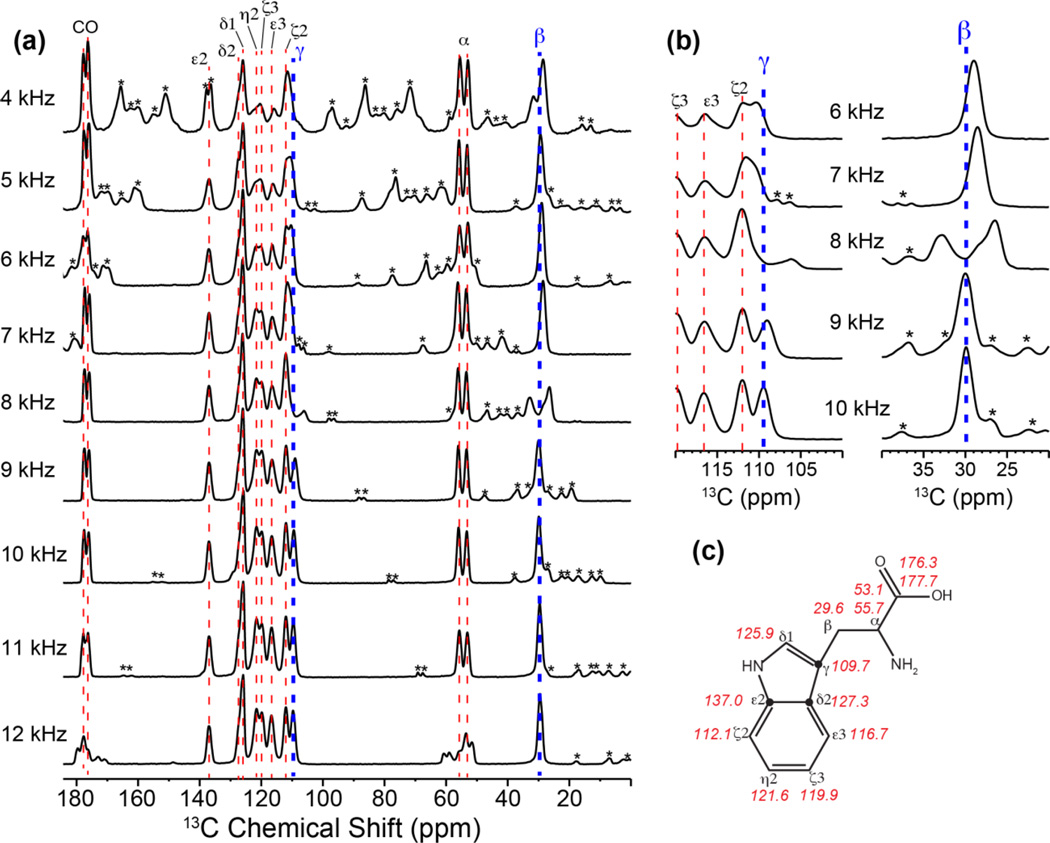
Two other factors that complicate the assignment of aromatic 13C signals are their large CSAs and the difficulty of avoiding rotational-resonance (R2) [42] line broadening that arises when the chemical-shift difference matches the MAS frequency νr or 2νr (n = 1 and n = 2 R2 condition). The large aromatic 13C CSAs give rise to spinning sidebands in the spectra that usually extend into the aliphatic region. More insidiously, the Cβ and Cγ isotropic chemical shift differences of aromatic residues are typically 80–100 ppm [15], which fall between the n = 1 and n = 2 R2 conditions of 120 ppm and 60 ppm for backbone Cα and CO chemical shifts. Thus, MAS frequencies are often accidentally close to some of the aromatic residues’ Cβ and Cγ isotropic shift differences, broadening both peaks. Fig. 2a shows the 13C CP-MAS spectra of uniformly 13C, 15N-labeled Trp under 4–12 kHz MAS at 1 kHz intervals. At the 13C Larmor frequency of 100 MHz used to measure these spectra, these MAS frequencies correspond to 40–120 ppm. The Cγ and Cβ isotropic shift difference for Trp is 80.1 ppm, which gives an n = 1 R2 condition of 8 kHz (80 ppm) MAS and an n = 2 R2 condition of 4 kHz (40 ppm) MAS. Thus, the Cγ and Cβ peaks are split and broadened at 4 kHz and 8 kHz MAS, as well as within 1 kHz windows of these two frequencies (Fig. 2b). Moreover, at MAS frequencies lower than 6 kHz (or 60 ppm), significant aromatic sidebands exist that overlap with the aliphatic signals. Under 12 kHz (or 120 ppm) MAS, the CO and Cα peaks are severely split by the R2 effect. These multiple R2 conditions restrict the choice of MAS frequencies to 11 kHz (or 110 ppm) for Trp, which gives the best compromise of avoiding R2 broadening as well as minimizing spinning sidebands (Fig. 2a, b). For uniformly 13C-labeled proteins with multiple aromatic residues, choosing an appropriate MAS frequency that avoids all R2 conditions is thus crucial for minimizing mis-assignment of aromatic residues’ Cβ and Cγ signals.
Figure 2.
1D CP-MAS spectra of U-13C,15N labeled Trp as a function of MAS frequencies. (a) Full 1D spectra measured at a 9.4 T magnetic field, corresponding to 100 MHz 13C Larmor frequency. At 8 kHz (80 ppm) and 12 kHz (120 ppm) MAS frequencies, rotational resonance broadening is observed for the Cβ-Cγ spin pair and the Cα-CO spin pair, respectively. (b) Expanded Cγ and Cβ regions of the 13C spectra, showing R2–induced line broadening. MAS frequencies that are within 1 kHz (or 10 ppm) of the R2 conditions are insufficient for removing line broadening. In all panels, spinning sidebands are marked with an asterisk. (c) Accurate 13C isotropic shifts of Trp, obtained from the 11 kHz MAS spectrum, which is free of R2 effects and spinning sideband overlap.
In this study, we describe two techniques for reducing spectral overlap and facilitating assignment of the 13C signals of aromatic residues in proteins. The first technique is a simple modification of the 2D 13C-13C PDSD experiment to include a gated 1H decoupling period before direct detection. The resulting 2D PDSD spectrum correlates all 13C signals in the indirect dimension with only the signals of non-protonated carbons in the direct dimension. The second technique is designed to suppress all Cα-Cβ cross peaks of aliphatic residues, leaving only the Cα, Cβ and CO cross peaks of aromatic residues. The Cα and Cβ magnetization of all residues is first removed by chemical shift filters and 13C-1H dipolar filters, then the remaining aromatic 13C magnetization is transferred to the Cα, Cβ and CO of only the aromatic residues by a short spin diffusion period. We demonstrate these two techniques on amino acids, a model peptide, and the microcrystalline model protein GB1.
2. Experimental conditions
Solid-state NMR experiments were carried out on Bruker 400 MHz (9.4 T) and 800 MHz (18.8 T) spectrometers using 4 mm or 3.2 mm MAS probes. The samples were spun at frequencies between 4.0 kHz and 16.5 kHz. Typical radiofrequency field strengths were 71–83 kHz for 1H and 50–71 kHz for 13C. 13C chemical shifts were referenced to the CH2 peak of adamantane at 38.48 ppm on the TMS scale [43]. Four uniformly 13C and 15N labeled model compounds were used to test the pulse sequences: amino acid Trp, a five-amino-acid mixture, WYIEN, the tripeptide formyl-Met-Leu-Phe (MLF) [2; 44; 45], and the microcrystalline protein GB1. These model systems provide increasing levels of complexity for spectral editing, culminating in GB1, which contains six aromatic residues out of a total of 56 residues [46].
3. Pulse sequences
3.1 1H gated decoupling PDSD (gPDSD) experiment
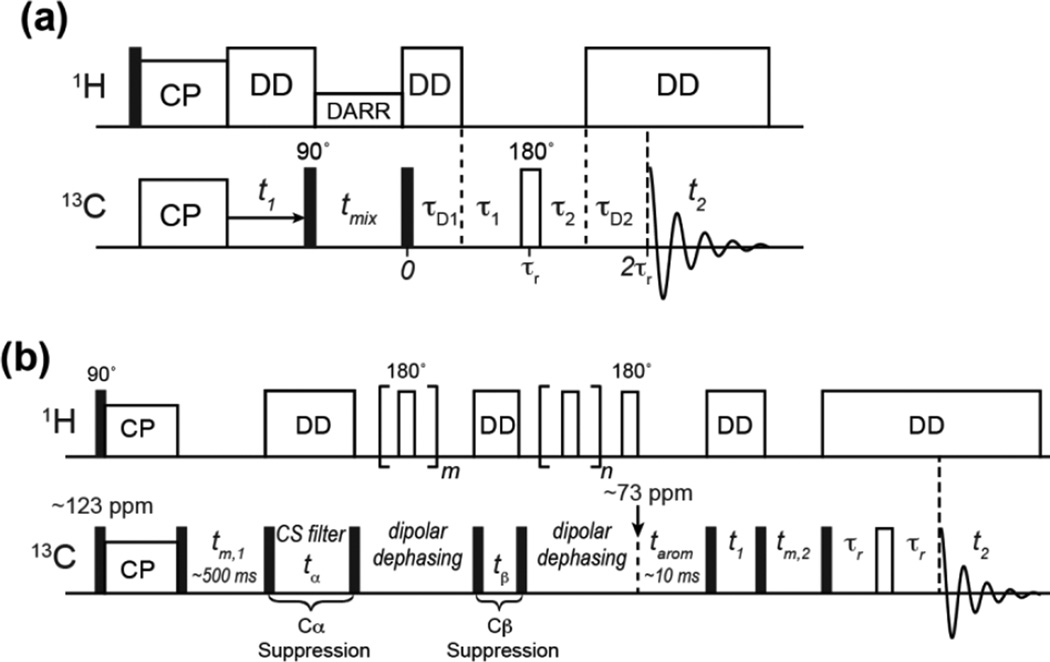
The conventional 2D PDSD experiment gives 13C-13C correlation peaks within an amino acid residue at short mixing times and both short and long-range correlations at long mixing times. With a T1-relaxation compensation scheme, difference spectra showing only long-range cross peaks can also be obtained [47]. To detect only non-protonated aromatic 13C signals in the aromatic region of the PDSD spectrum, we add a 1H gated decoupling period after the spin diffusion mixing time and before t2 detection (Fig. 3a). The gate during which 1H decoupling is turned off consists of a total delay of τ1 + τ2, which spans part of two rotor periods. They are demarcated by a 13C 180° pulse on the rotor echo to refocus the 13C chemical shift, and also recouple the 13C-1H dipolar coupling to better dephase the 13C magnetization. The remaining periods during which 1H decoupling is on are τD1 and τD2, where τD1 + τ1 and τ2 + τD2 equal one rotor period. τ1 and τ2 are in general not the same. In the current work, under 10.3 kHz MAS, we found that the optimal delays for maximal dephasing of the protonated aromatic signals are τ1 = 60 µs and τ2 = 40 µs.
Figure 3.
Pulse sequences developed in this work. (a) 2D PDSD with gated 1H decoupling. The gated decoupling period contains τ1 = 60 µs and τ2 = 40 µs for the MAS frequencies used in this work. (b) 2D Aromatic Selection by Spectral Editing (ASSET). The initial spin diffusion period is set relatively long (tm,1 = 500 ms) to ensure polarization of aromatic side chains. This is followed by two chemical shift filters, one for Cα suppression (tα) and one for Cβ suppression (tβ). Each chemical shift filter is followed by a 13C-1H dipolar recoupled z-filter period to destroy the transverse magnetization of other aliphatic carbons that are far off resonance from the aromatic region. A spin diffusion period, tarom, then transfers the remaining aromatic 13C magnetization to the Cβ and Cα of the aromatic residues. 2D correlation of Cβ and Cα resonances is then conducted using a typical PDSD module with Hahn echo detection.
3.2 The Aromatic Selection via Spectral Editing (ASSET) experiment
Fig. 3b shows the pulse sequence for obtaining only aromatic Cα-Cβ cross peaks in the aliphatic region of the 2D PDSD spectra. At the beginning of the pulse sequence, the 13C carrier frequency is set in the middle of the aromatic region, around 123 ppm. A 13C 90° pulse together with 1H-13C CP allow the detection of both dynamic and rigid functional groups. A subsequent spin diffusion period, tm,1, of ~500 ms further enhances the aromatic 13C intensities by equilibrating the aliphatic and aromatic 13C magnetization, since the low proton density of aromatic residues causes lower signals in a conventional 13C CP spectrum [25]. After the pre-equilibration period, a 90° pulse flips all 13C magnetization to the transverse plane, where chemical shift evolution with aromatic residues on or near resonance is allowed to occur for a period tα, which alternates between 0 and a finite time that creates antiparallel magnetization vectors between Cα and aromatic carbons. For experiments at a 13C Larmor frequency of 100 MHz (9.4 Tesla), the best tα is 0 and 83 µs. A subsequent 90° pulse with suitable phase cycling every two scans then stores the aromatic 13C magnetization along +z while the Cα magnetization alternates between +z and −z. Any transverse magnetization is then dephased by 13C-1H dipolar coupling, which is recoupled using a 1H 180° pulse every half rotor period. After this z-filter, the stored 13C z-magnetization is returned to the transverse plane by another 90° pulse, and a similar chemical shift filter together with a z-filter is used to destroy the Cβ magnetization. At 100 MHz 13C Larmor frequency, the optimal Cβ chemical shift filter time, tβ, is 0 and 47 µs. Due to the sufficiently high MAS frequency, tα and tβ need not be rotor-synchronized. The z-filter periods can also have different lengths, and were optimized to be 2 and 4 rotor periods at an MAS frequency of ~10 kHz. At this point, the remaining 13C magnetization is mostly aromatic, and is allowed to re-polarize the adjacent Cβ and Cα during a tarom period of 5–25 ms. The 13C carrier frequency is moved to ~73 ppm at the beginning of the tarom period to keep potential zero-frequency artifacts outside the relevant spectral region and avoid significant off-resonance effects. This is followed by a standard 2D PDSD module with a mixing time of tm,2, with optional echo detection.
4. Results and Discussion
We demonstrate these two spectral editing techniques on several model compounds. Fig. 4a, c show the full 2D PDSD spectra of Trp and WYIEN, where all intra-residue cross-peaks can be assigned. The gPDSD spectra show a greatly simplified aromatic region. For Trp, all protonated 13C signals are removed in the direct dimension, leaving only cross-peaks of the indole Cγ, Cδ2 and Cε2 (Fig. 4b). For the amino-acid mixture, Trp Cε, Cδ and Cγ and Tyr Cγ and Cζ signals are retained with efficiencies of 42–75% compared to the full spectrum while the protonated aromatic carbons are suppressed to 0–9% of the full intensity (Fig. 4d). In the aliphatic-aliphatic region of the 2D gPDSD spectrum (not shown), residual Ile Cδ and Cε methyl signals are also present due to their motionally averaged 13C-1H dipolar couplings, but the intensities are only ~6% of the full intensities. In the aliphatic-aromatic correlation region, the suppression of the protonated 13C signals significantly simplified the assignment of the Trp and Tyr Cβ and Cα chemical shifts.
Figure 4.
2D 13C-13C DARR (top) and gPDSD (bottom) spectra of U-13C Trp (a, b) and the amino acid mixture WYIEN (c, d). Spectra (a) and (b) were recorded under 11 kHz MAS using a mixing time of 2 ms and 1H gate times of τ1 = 60 µs and τ2 = 40 µs. Spectra (c) and (d) were measured under 10.3 kHz MAS using a mixing time of 10 ms and 1H gate times of τ1 = 60 µs and τ2 = 40 µs. The aromatic region in (b) and (d) retains only the non-protonated 13C signals in the direct dimension. For the amino acid mixture, the aliphatic – aromatic correlation region is also simplified after removal of the protonated aromatic 13C signals, facilitating assignment of the Cβ and Cα chemical shifts of Tyr and Trp.
The gated-decoupling sequence similarly simplified the aromatic region of the GB1 2D spectrum (Fig. 5a, b). Although the high resolution afforded by the 800 MHz spectrometer allowed us to resolve many aromatic 13C peaks of the three Tyr residues (Y3, Y33, Y45), two Phe residues (F30, F52) and one Trp (W43) [46], resonance overlap remains among Phe Cδ and Cε and Tyr Cγ and Cδ signals, which cluster around 130 ppm. With gated 1H decoupling, most of the F30 and F52 cross peaks in the 126–133 ppm region are suppressed, revealing the underlying Tyr Cγ and Cδ signals Fig. 5b). Compared to the regular PDSD spectrum, the non-protonated 13C signals are retained to an average of 86% of the full intensities while the protonated aromatic carbons’ signals are suppressed to an average of 18% of the full intensities. The slightly higher residual intensities of the protonated aromatic carbons in GB1 compared to the amino acids can be attributed to two reasons. The WYIEN sample was measured on a 400 MHz spectrometer under 10.3 kHz MAS, while GB1 was measured on a 800 MHz spectrometer under 16.5 kHz MAS. The higher MAS frequency was necessary to avoid overlap between the carbonyl spinning sidebands and the aromatic signals, and the resulting shorter rotor period caused a shorter gate, thus reducing the efficiency of dipolar dephasing. The GB1 gPDSD spectrum was measured using a total gating delay of τ1 + τ2 = 60 µs, while the amino acid mixture was measured with a total delay of 100 µs. The incomplete suppression under faster MAS can be remedied by extending the dipolar dephasing period. In addition, molecular motion, which may be present for the less hydrophobically embedded aromatic residues in GB1, can also reduce the efficiency of gated decoupling. For example, we observe 33% residual intensity of the Tyr Cδ and Tyr Cε peak in the gated decoupling spectrum (Fig. 5b). This can be assigned to one of the three Tyr residues, Y33, which is located on the surface of the protein with its sidechain pointed away from the hydrophobic pore. Indeed, Y33 was found to undergo two-site ring flips [46], which average the 13C-1H dipolar couplings, thus reducing the efficiency of gated decoupling.
Figure 5.
2D 13C PDSD (a) and gPDSD (b) spectra of GB1 measured at 18.8 T (800 1H Larmor frequency) under 16.5 kHz MAS with a mixing time of 100 ms. 1H gate times of τ1= 36 µs and τ2 = 24 µs were used for spectra (b). (c) GB1 structure (PDB: 2QMT) showing the positions of the six aromatic residues.
While gPDSD simplifies the aromatic region of 2D 13C PDSD spectra, the ASSET technique selectively detects the Cβ-Cα, Cα-Cβ, Cβ-CO and Cα-CO cross peaks of aromatic residues while suppressing those of aliphatic residues. Fig. 6a and 6d show the full 2D spectra of the WYIEN mixture and the tripeptide MLF. All intra-residue cross peaks are observed for WYIEN with a 20 ms mixing time (Fig. 6a) while intra- and inter-residue correlations are both observed in MLF with 30 ms mixing (Fig. 6d). When the ASSET technique is applied without the transfer, i.e. tarom = 0, the aliphatic signals are nearly completely suppressed, except for a few weak methyl signals (Fig. 6b, e) and Asn and Trp Cα-Cβ peaks, which may result from incomplete chemical shift filtering. When the aromatic transfer is turned on (tarom = 10 ms), the Cα and Cβ signals of Trp and Tyr in the mixture are selectively enhanced, as shown by Cα-Cβ and Cβ-Cα cross peaks on both sides of the diagonal (Fig. 6c, f). These Trp and Tyr cross peaks are 20–40% of the full intensities, compared to the incompletely suppressed Ile sidechain methyl signals, which are ~3% of the full intensities. A similar result is found for MLF. Here, the Phe shows a Cβ-Cα cross peak but not the Cα-Cβ peak, indicating that the 10 ms tarom is sufficiently short that polarization transfer is dominated by the Cγ to Cβ transfer while Cγ to Cα transfer is reduced. Such cross peak asymmetry can be helpful for spectral editing of larger proteins.
Figure 6.
2D 13C-13C DARR and ASSET spectra of the amino acid mixture WYIEN (a–c) and tripeptide MLF (d–f). (a, d) Full DARR spectra showing all cross peaks. (b, e) Spectra with tarom = 0, where most Cα and Cβ cross peaks are suppressed. (c, f) ASSET spectra with tarom = 10 ms, which exhibit mostly aromatic Cα and Cβ cross peaks. The spectra in (c) and (f) are reproduced in red in (a) and (d) to compare with the full DARR spectra.
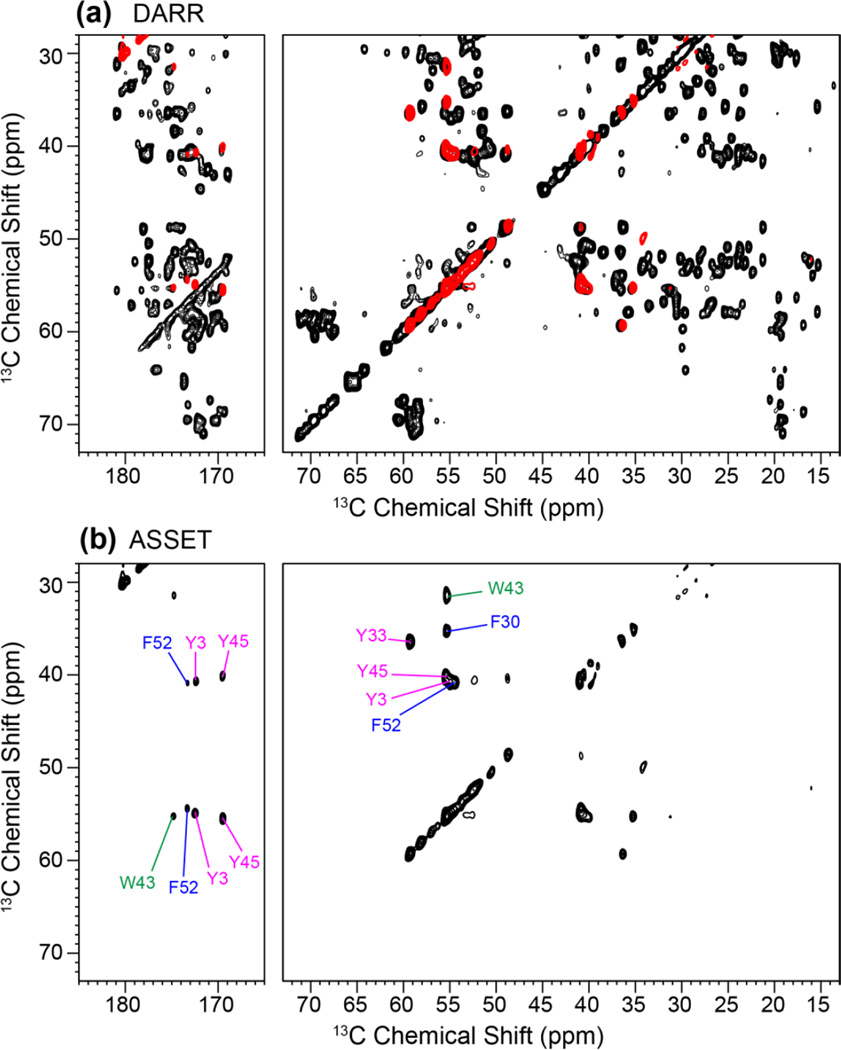
Fig. 7 shows the 2D DARR and ASSET spectra of GB1 using a mixing time of 100 ms between the t1 and t2 dimensions. The 2D spectra were measured on an 800 MHz spectrometer under 16.5 kHz MAS, and the DARR spectrum (Fig. 7a) exhibits comparable resolution as the published literature [46]. The ASSET sequence dramatically simplified the aliphatic and carbonyl regions of the spectrum (Fig. 7b), displaying only Cα/β-Cβ/α and Cα/β-CO cross peaks of the six aromatic residues with efficiencies of 10–20%. The Cβ-Cα cross peaks of W43, Y33, and F30 are well resolved, while the Cβ-Cα cross peaks of Y3, Y45 and F52 partially overlap. But the latter three residues are well dispersed by their carbonyl chemical shifts. The aliphatic region of the ASSET spectrum has moderate intensity asymmetry, with the Cβ-Cα cross peaks stronger than Cα-Cβ peaks, indicating that the aromatic carbon magnetization is transferred more readily to Cβ than to Cα with the tarom mixing time of 25 ms used here.
Figure 7.
2D 13C DARR (a) and ASSET (b) spectra of GB1. The ASSET spectrum was acquired with tarom = 25 ms and shows only the Cα - Cβ and Cα/β-CO cross peaks of the six aromatic residues while suppressing the signals of all aliphatic residues. The ASSET spectrum is reproduced in red in (a) and superimposed on the full PDSD spectrum.
5. Conclusions
We have demonstrated two pulse sequences for measuring simplified and higher-resolution 2D 13C-13C correlation spectra of aromatic residues in proteins. The gPDSD technique results in a non-protonated aromatic 13C sub-spectrum in the direct dimension, correlated with all carbons in the indirect dimension. The ASSET technique yields aromatic residues’ Cβ, Cα and CO cross peaks while suppressing all aliphatic residues’ cross peaks. The efficiency of the gPDSD technique for aromatic carbons is 62–86%, while the protonated 13C signals are suppressed to less than 20% of the original intensities. The efficiency of the ASSET experiment is 20–40% on small molecules and 10–20% on GB1. The lower efficiency of GB1 may be due to the inherent isotropic shift dispersion, which makes the chemical shift filter imperfect. Overall, the selectivity against non-aromatic residues is excellent for both techniques, thus increasing the repertoire of spectral editing techniques for complex and disordered proteins. In principle, the aromatic-residue-only Cα-Cβ sub-spectrum can also be obtained in a 3D 13C-13C-13C correlation experiment [48], provided that the aromatic 13C chemical shifts are reasonably well resolved, but the signal-averaging time for these 3D experiments is significantly longer than for the 2D spectrally edited ASSET experiment.
Highlights.
Two methods are described to selectively detect the signals of aromatic residues.
Gated 1H decoupling selectively detects unprotonated aromatic 13C signals.
Chemical shift filters select the Cα, Cβ and CO signals of aromatic residues.
We successfully demonstrated these techniques on model peptides and proteins.
Acknowledgements
This work is supported by NIH grant GM088204 to M. H. The authors would like to thank Dr. Robert Silvers for providing the GB1 sample used in this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hong M. Resonance Assignment of 13C/15N Labeled Proteins by Two- and Three- Dimensional Magic-Angle-Spinning NMR. J. Biomol. NMR. 1999;15:1–14. doi: 10.1023/a:1008334204412. [DOI] [PubMed] [Google Scholar]
- 2.Rienstra CM, Hohwy M, Hong M, Griffin RG. 2D and 3D 15N-13C-13C NMR chemical shift correlation spectroscopy of solids: assignment of MAS spectra of peptides. J. Am. Chem. Soc. 2000;122:10979–10990. [Google Scholar]
- 3.Baldus M, Petkova AT, Herzfeld J, Griffin RG. Cross polarization in the tilted frame: assignment and spectral simplification in heteronuclear spin systems. Mol. Phys. 1998;95:1197–1207. [Google Scholar]
- 4.Comellas G, Rienstra CM. Protein structure determination by magic-angle spinning solid-state NMR, and insights into the formation, structure, and stability of amyloid fibrils. Annu. Rev. Biophys. 2013;42:515–536. doi: 10.1146/annurev-biophys-083012-130356. [DOI] [PubMed] [Google Scholar]
- 5.McDermott AE. Structure and dynamics of membrane proteins by magic angle spinning solid-state NMR. Annu. Rev. Biophys. 2009;38:385–403. doi: 10.1146/annurev.biophys.050708.133719. [DOI] [PubMed] [Google Scholar]
- 6.Luca S, Heise H, Baldus M. High-resolution solid-state NMR applied to polypeptides and membrane proteins. Acc. Chem. Res. 2003;36:858–865. doi: 10.1021/ar020232y. [DOI] [PubMed] [Google Scholar]
- 7.Hong M. Solid-state NMR studies of the structure, dynamics, and assembly of β-sheet membrane peptides and α-helical membrane proteins with antibiotic activities. Acc. Chem. Res. 2006;39:176–183. doi: 10.1021/ar040037e. [DOI] [PubMed] [Google Scholar]
- 8.Liao SY, Fritzsching KJ, Hong M. Conformational analysis of the full-length M2 protein of the influenza A virus using solid-state NMR. Protein Sci. 2013;22:1623–1638. doi: 10.1002/pro.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tycko R. Prospects for resonance assignments in multidimensional solid-state NMR spectra of uniformly labeled proteins. J. Biomol. NMR. 1996;22:239–251. doi: 10.1007/BF00410323. [DOI] [PubMed] [Google Scholar]
- 10.Hong M. Determination of multiple ϕ torsion angles in solid proteins by selective and extensive 13C labeling and two-dimensional solid-state NMR. J. Magn. Reson. 1999;139:389–401. doi: 10.1006/jmre.1999.1805. [DOI] [PubMed] [Google Scholar]
- 11.Hong M, Jakes K. Selective and extensive 13C labeling of a membrane protein for solid-state NMR investigations. J. Biomol. NMR. 1999;14:71–74. doi: 10.1023/a:1008334930603. [DOI] [PubMed] [Google Scholar]
- 12.Loquet A, Lv G, Giller K, Becker S, Lange A. 13C spin dilution for simplified and complete solid-state NMR resonance assignment of insoluble biological assemblies. J. Am. Chem. Soc. 2011;133:4722–4725. doi: 10.1021/ja200066s. [DOI] [PubMed] [Google Scholar]
- 13.Castellani F, vanRossum B, Diehl A, Schubert M, Rehbein K, Oschkinat H. Structure of a protein determined by solid-state magic-angle spinning NMR spectroscopy. Nature. 2002;420:98–102. doi: 10.1038/nature01070. [DOI] [PubMed] [Google Scholar]
- 14.Kwon B, Tietze D, White PB, Liao SY, Hong M. Chemical Ligation of the Influenza M2 Protein for Solid-State NMR Characterization of the Cytoplasmic Domain Structure. Protein Sci. 2015;24:1087–1099. doi: 10.1002/pro.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fritzsching KJ, Yang Y, Schmidt-Rohr K, Hong M. Practical use of chemical shift databases for protein solid-state NMR 2D chemical shift maps and amino-acid assignment with secondary-structure information. J. Biomol. NMR. 2013;56:155–167. doi: 10.1007/s10858-013-9732-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu KN, Qiang W, Tycko R. A general Monte Carlo/simulated annealing algorithm for resonance assignment in NMR of uniformly labeled biopolymers. J. Biomol. NMR. 2011;50:267–276. doi: 10.1007/s10858-011-9517-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tycko R, Hu KN. A Monte Carlo/simulated annealing algorithm for sequential resonance assignment in solid state NMR of uniformly labeled proteins with magic-angle spinning. J. Magn. Reson. 2010;205:304–314. doi: 10.1016/j.jmr.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y, Fritzsching KJ, Hong M. Resonance Assignment of Disordered Proteins Using a Multi-Objective Non-Dominated Sorting Genetic Algorithm. J. Biomol. NMR. 2013;57:281–296. doi: 10.1007/s10858-013-9788-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Opella SJ, Frey MH. Selection of Nonprotonated Carbon Resonances in Solid-State Nuclear Magnetic Resonance. J. Am. Chem. Soc. 1979;101:5854–5856. [Google Scholar]
- 20.Wu X, Burns ST, Zilm KW. Spectral Editing in CPMAS NMR. Generating Subspectra Based on Proton Multiplicities. J. Magn. Reson. 1994;111:29–36. [Google Scholar]
- 21.Wu XL, Zilm KW. Complete spectral editing in CPMAS NMR. J. Magn. Reson. 1993;102:205–213. [Google Scholar]
- 22.Wu X, Zhang S, Wu X. Selective Polarization Inversion in Solid State High-Resolution CP MAS NMR. J. Magn. Reson. 1988;77:343–347. [Google Scholar]
- 23.Mao JD, Schmidt-Rohr K. Accurate Quantification of Aromaticity and Nonprotonated Aromatic Carbon Fraction in Natural Organic Matter by 13C Solid-State Nuclear Magnetic Resonance. Environ. Sci. Technol. 2004;38:2680–2684. doi: 10.1021/es034770x. [DOI] [PubMed] [Google Scholar]
- 24.Mao JD, Schmidt-Rohr K. Separation of aromatic-carbon 13C NMR signals from dioxygenated alkyl bands by a chemical-shift-anisotropy filter. Solid State Nuc. Magn. Reson. 2004;26:36–45. doi: 10.1016/j.ssnmr.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 25.Johnson RL, Anderson JM, Shanks BH, Fang X, Hong M, Schmidt-Rohr K. Spectrally edited 2D 13C-13C NMR spectra without diagonal ridge for characterizing 13C-enriched low-temperature carbon materials. J. Magn. Reson. 2013;234:112–124. doi: 10.1016/j.jmr.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt-Rohr K, Mao JD. Efficient CH-Group Selection and Identification in 13C Solid-State NMR by Dipolar DEPT and 1H Chemical-Shift Filtering. J. Am. Chem. Soc. 2002;124:13938–13948. doi: 10.1021/ja027362m. [DOI] [PubMed] [Google Scholar]
- 27.Mao J-D, Schmidt-Rohr K. Methylene Spectral Editing in Solid-State 13C NMR by Three-Spin Coherence Selection. J. Magn. Reson. 2005;176:1–6. doi: 10.1016/j.jmr.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt-Rohr K, Fritzsching KJ, Liao SY, Hong M. Spectral editing of two-dimensional magic-angle-spinning solid-state NMR spectra for protein resonance assignment and structure determination. J. Biomol. NMR. 2012;54:343–353. doi: 10.1007/s10858-012-9676-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong M. Solid-State NMR Determination of 13Cα Chemical Shift Anisotropies for the Identification of Protein Secondary Structure. J. Am. Chem. Soc. 2000;122:3762–3770. [Google Scholar]
- 30.Huster D, Yamaguchi S, Hong M. Efficient β-sheet Identification in Proteins by Solid-State NMR Spectroscopy. J. Am. Chem. Soc. 2000;122:11320–11327. [Google Scholar]
- 31.Ader C, Schneider R, Seidel K, Etzkorn M, Becker S, Baldus M. Structural rearrangements of membrane proteins probed by water-edited solid-state NMR spectroscopy. J. Am. Chem. Soc. 2009;131:170–176. doi: 10.1021/ja806306e. [DOI] [PubMed] [Google Scholar]
- 32.Kumashiro KK, Schmidt-Rohr K, Murphy OJ, Ouellette KL, Cramer WA, Thompson LK. A novel tool for probing membrane protein structure: solid-state NMR with proton spin diffusion and X-nucleus detection. J. Am. Chem. Soc. 1998;120:5043–5051. [Google Scholar]
- 33.Sergeyev IV, Bahri S, Day LA, McDermott AE. Pf1 bacteriophage hydration by magic angle spinning solid-state NMR. J. Chem. Phys. 2014;141:22D533. doi: 10.1063/1.4903230. [DOI] [PubMed] [Google Scholar]
- 34.Gallivan JP, Dougherty DA. Cation-π interactions in structural biology. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao SY, Yang Y, Tietze D, Hong M. The Influenza M2 Cytoplasmic Tail Changes the Proton-Exchange Equilibria and the Backbone Conformation of the Transmembrane Histidine Residue to Facilitate Proton Conduction. J. Am. Chem. Soc. 2015;137:6067–6077. doi: 10.1021/jacs.5b02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams JK, Zhang Y, Schmidt-Rohr K, Hong M. pH-Dependent Conformation, Dynamics, and Aromatic Interaction of the Gating Tryptophan Residue of the Influenza M2 Proton Channel from Solid-State NMR. Biophys. J. 2013;104:1698–1708. doi: 10.1016/j.bpj.2013.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu W, Lazo ND, Cross TA. Tryptophan dynamics and structural refinement in a lipid bilayer environment: solid state NMR of the Gramicidin channel. Biochemistry. 1995;34:14138–14146. doi: 10.1021/bi00043a019. [DOI] [PubMed] [Google Scholar]
- 38.Gall CM, Cross TA, DiVerdi JA, Opella SJ. Protein dynamics by solid-state NMR: aromatic rings of the coat protein in fd bacteriophage. Proc. Natl. Acad. Sci. U. S. A. 1982;79:101–105. doi: 10.1073/pnas.79.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Planque MR, Bonev BB, Demmers JA, Greathouse DV, Koeppe REn, Separovic F, Watts A, Killian JA. Interfacial anchor properties of tryptophan residues in transmembrane peptides can dominate over hydrophobic matching effects in peptide-lipid interactions. Biochemistry. 2003;42:5341–5348. doi: 10.1021/bi027000r. [DOI] [PubMed] [Google Scholar]
- 40.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Wenger RK, Yao HY, Markley JL. BioMagResBank. Nucleic Acids Res. 2007;35:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li S, Hong M. Protonation, Tautomerization, and Rotameric Structure of Histidine: A Comprehensive Study by Magic-Angle-Spinning Solid-State NMR. J. Am. Chem. Soc. 2011;133:1534–1544. doi: 10.1021/ja108943n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raleigh DP, Levitt MH, Griffin RG. Rotational resonance in solid state NMR. Chem. Phys. Lett. 1988;146:71–76. [Google Scholar]
- 43.Morcombe CR, Zilm KW. Chemical shift referencing in MAS solid state NMR. J. Magn. Reson. 2003;162:479–486. doi: 10.1016/s1090-7807(03)00082-x. [DOI] [PubMed] [Google Scholar]
- 44.Hong M, Griffin RG. Resonance Assignment for Solid Peptides by Dipolar-Mediated 13C/15N Correlation Solid-State NMR. J. Am. Chem. Soc. 1998;120:7113–7114. [Google Scholar]
- 45.Rienstra CM, Tucker-Kellogg L, Jaroniec CP, Hohwy M, Reif B, McMahon MT, Tidor B, Lozano-Perez T, Griffin RG. De novo determination of peptide structure with solid-state magic-angle spinning NMR spectroscopy. Proc. Natl. Acad. Sci. USA. 2002;99:10260–10265. doi: 10.1073/pnas.152346599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franks WT, Zhou DH, Wylie BJ, Money BG, Graesser DT, Frericks HL, Sahota G, Rienstra CM. Magic-Angle Spinning Solid-State NMR Spectroscopy of the b1 Immunoglobulin Binding Domain of Protein G (GB1): 15N and 13C Chemical Shift Assignments and Conformational Analysis. J. Am. Chem. Soc. 2005;127:12291–12305. doi: 10.1021/ja044497e. [DOI] [PubMed] [Google Scholar]
- 47.Wang T, Williams JK, Schmidt-Rohr K, Hong M. Relaxation-compensated difference spin diffusion NMR for detecting 13C-13C long-range correlations in proteins and polusaccharides. J. Biomol. NMR. 2014;61:97–107. doi: 10.1007/s10858-014-9889-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li S, Zhang Y, Hong M. 3D 13C-13C-13C correlation NMR for de novo distance determination of solid proteins and application to a human alpha defensin. J. Magn. Reson. 2010;202:203–210. doi: 10.1016/j.jmr.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]