Abstract
There is mounting evidence that noncoding microRNAs (miRNA) are modulated by select chemoprotective dietary agents. For example, recently we demonstrated that the unique combination of dietary fish oil (containing n-3 fatty acids) plus pectin (fermented to butyrate in the colon) (FPA) up-regulates a subset of putative tumor suppressor miRNAs in intestinal mucosa, and down-regulates their predicted target genes following carcinogen exposure as compared to control (corn oil plus cellulose (CCA)) diet. To further elucidate the biological effects of diet and carcinogen modulated miR’s in the colon, we verified that miR-26b and miR-203 directly target PDE4B and TCF4, respectively. Since perturbations in adult stem cell dynamics are generally believed to represent an early step in colon tumorigenesis and to better understand how the colonic stem cell population responds to environmental factors such as diet and carcinogen, we additionally determined the effects of the chemoprotective FPA diet on miRNAs and mRNAs in colonic stem cells obtained from Lgr5-EGFP-IRES-creERT2 knock-in mice. Following global miRNA profiling, 26 miRNAs (P <0.05) were differentially expressed in Lgr5high stem cells as compared to Lgr5negative differentiated cells. FPA treatment up-regulated miR-19b, miR-26b and miR-203 expression as compared to CCA specifically in Lgr5high cells. In contrast, in Lgr5negative cells, only miR-19b and its indirect target PTK2B were modulated by the FPA diet. These data indicate for the first time that select dietary cues can impact stem cell regulatory networks, in part, by modulating the steady-state levels of miRNAs. To our knowledge, this is the first study to utilize Lgr5+ reporter mice to determine the impact of diet and carcinogen on miRNA expression in colonic stem cells and their progeny.
Introduction
miRNAs are a class of noncoding RNAs involved in the regulation of several key cellular processes such as signal transduction, apoptosis, cell proliferation, morphogenesis and the pathogenesis of cancer (1, 2). Structurally, these are 19–24 nucleotide RNAs which are first transcribed as long primary miRNAs, and then processed into 60–70 nt miRNA precursors (pre-miRNA) by nuclear RNAse III Drosha and its partner proteins. The pre-miRNA complex is subsequently transported to the cytoplasm where it is further processed by Dicer into a miRNA duplex, taken up by the RISC complex, which can bind to complementary sequences at the 3′ untranslated region (UTR) of target mRNA. This results in the blockade of mRNA translation and/or induces mRNA degradation (2, 3).
A plethora of studies have demonstrated that miRNAs and their targets are aberrantly expressed in colon cancer (4–8). For example, expression of miR-21, miR-143 and miR-145 have been shown to be differentially expressed in human colonic tumors as compared to normal colon. These miRNAs have been shown to target several key signal transduction pathways (e.g., Notch signaling) that directly modulate malignant transformation and have been suggested to be utilized as biomarkers for detection of colorectal cancer (CRC) (9–13). For example, Iliopoulos et al reported an inverse correlation between miR-193a and K-RAS, one of the oncogenes commonly mutated in CRC (14).
There is emerging evidence suggesting a role for miRNAs in regulating the post-transcriptional genetic programming of stem and progenitor cells (15). A recent study systematically examined miRNA expression profiles in adult tissue-specific stem cells and their differentiated counterparts (16). A common signature of miRNA expression in blood, muscle and neural stem cell populations was detected. This suggests that miRNA signatures mark the transition from self-renewing and quiescent stem cells to proliferative and differentiating progenitor cells, implying an extensive role of miRNAs in regulating self-renewal, proliferation, and quiescence programs in the cells (17). In recent years, it has been suggested that intestinal tumors arise from a population of ‘cancer stem cells (CSCs)’, which exhibit stem-like features (e.g., self-renewal and pluripotency) and contribute to metastasis and resistance to treatment (18, 19). In addition, changes in miRNA expression levels may contribute to the generation and maintenance of cancer stem cells. Emerging data support this hypothesis and suggest that changes in specific miRNAs may be diagnostic indicators of cancer related stemness (12, 13). Moreover, miRNA profiling in cancer stem cells isolated from human colon cancer cell lines (e.g., HT-29) using the CD133 surface marker revealed 11 overexpressed miRNAs and eight underexpressed miRNAs such as miR-429, miR-155, and miR-320d, some of which may be involved in the regulation of stem cell differentiation (20). With the identification of Lgr5 as a marker of fast-cycling intestinal stem cells, it is now possible to determine the role of miRNAs in this important cell population (21).
Over several decades, there is substantial experimental, epidemiological and clinical evidence indicating that fish oil containing diets, rich in n-3 polyunsaturated fatty acids (PUFAs), e.g., docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA), are protective against colon tumorigenesis (22–25). It is likely that EPA and DHA function through multiple pathways to decrease colon cancer risk, including modulation of cyclooxygenase (COX) activity, changes in cellular oxidative stress, modification of gene expression and alterations in membrane dynamics and cellular surface receptor function (23). Work from our laboratory has demonstrated a role for n-3 PUFA in increasing the level of oxidative stress in colonocytes to activate apoptosis (26, 27). We have also demonstrated fundamental differences in gene expression profiles in rats fed a fish oil enriched diet compared to corn oil or olive oil enriched diets (28). In addition, the chemoprotective effect of fish oil is enhanced when a highly fermentable fiber (pectin), capable of elevating colonic luminal butyrate levels, is added to the diet (29–32). Recently, we examined the effects of colon carcinogenesis on non-coding RNAs in a rat model by cataloguing mucosal miRNA expression following diet and carcinogen exposure (33). For this purpose, global miRNA and target mRNA profiling analyses were carried out. We demonstrated that the unique combination of chemoprotective dietary fish oil (containing EPA and DHA) and pectin (fermented to butyrate in the large intestine) modulated a subset of mucosal miRNAs-miR-19b, miR-26b and miR-203 and their predicted target genes -IGF1R, PTK2B, PDE4B and TCF4. Specifically, miR-19b, miR-26b and miR-203 were significantly down-regulated by the control (corn oil and cellulose) diet in presence of carcinogen as compared to fish oil pectin diet in the presence of carcinogen, whereas their predicted targets PTK2B, IGF1R, PDE4B and TCF4 were inversely up-regulated. Immunoblotting analyses further revealed that the targets were also modulated at the protein level (34). It is noteworthy that these diet-modulated miRNAs are also aberrantly expressed in colon tumorigenesis and other types of cancers (35). Collectively, these data demonstrate that the novel combination of fish oil/pectin protects the colon from carcinogen-induced miRNA dysregulation (34). However, to date, the miRNA expression profile in colon stem cells has not been documented, nor has the effects of chemoprotective dietary components on colonic stem cell non-coding miRNA signatures have not been determined. Hence, with the identification of Lgr5 as the first definitive intestinal stem cell marker in mice, we used the Lgr5-EGFP-IRES-cre ERT2 knock in mouse model to visualize and isolate intestinal stem cells, establish their stem cell-specific miRNA expression profile, and examine their response to chemotherapeutic agents by measuring miRNA expression. This was contrasted with components of the stem cell “niche” (i.e., daughter cells), which contribute to the physiological microenvironment consisting of specialized cells that physically anchor the stem cell and provide the necessary factors to maintain stemness. We hypothesized that a chemoprotective diet containing n-3 PUFA (fish oil) and butyrate (pectin) would modulate the stem cell miRNA profile. Our overall goal is to better understand how the colonic stem cell population responds to environmental factors such as diet and carcinogen. Hence, we investigated the effects of disease progression on miRNAs in colonic stem cells by feeding mice injected with azoxymethane (AOM, a colon-specific carcinogen) a diet containing n-3 PUFA and a fermentable fiber (fish oil + pectin) diet (FP) or n-6 PUFA and a poorly fermentable fiber (corn oil + cellulose) (CC).
For the purpose of validating the predicted miRNA:mRNA associations of miR-19b, 26b and 203 (34), we also conducted a series of loss and gain of function analyses by knocking down or overexpressing select miRNAs in human colon cancer cell line (HCT116). The global abundance of protein levels in the HCT116 model was subsequently assessed. In addition, luciferase reporter assays were utilized and miRNA physiological function was assessed by examining effects on apoptosis and cell proliferation. Our results demonstrate that PDE4B and TCF4 are direct targets of miR-26b and miR-203, respectively; while PTK2B is an indirect target of miR-19b and that these important mediators exhibit chemoprotective properties. The role of these miRNAs and their validated targets in intestinal stem cells was also investigated.
Materials and methods
Cell culture
HCT116 cells (p53+/+) were originally obtained from Dr. Bert Vogelstein, Johns Hopkins University, Baltimore, MD (36). These cells were cultured in McCoy’s 5A medium supplemented with 10% defined fetal bovine serum (Thermo Scientific, Wilmington, DE) and 2 mM glutamax (Gibco, Carlsbad, CA) at 37°C in 5% CO2.
Transfection
Cells were seeded at 1–2×105 cells per well in a 12-well plate, and upon reaching 70% confluence were transfected with 5′ FITC-labeled miRNA inhibitors (Exiqon, Denmark) anti-miR-19b, anti-miR-26b or anti-miR-203 (30 nM) using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA). Scrambled miR was utilized as a negative control. Following transfection (12 h), the media was changed and the efficiency was assessed by imaging with a Zeiss 510 META NLO Multiphoton Microscope System with an Axiovert 200 MOT microscope (Carl Zeiss Microimaging, Thornwood, NY). For overexpression of miRNAs, cells were transfected with miRNA mimics (30 nM) (Dharmacon, Lafayette, CO) for miR-19b, miR-26b, miR-203 or scrambled miR. Each experiment was performed in triplicate. The sequences of the miRNA mimics, inhibitors and scrambled controls are described in Supplementary Table I.
Total RNA isolation and Real Time PCR
Forty-eight hr following transfection with miRNA inhibitors in HCT116 cells, media was removed and cells were washed with phosphate buffered saline (PBS) (Gibco, Carlsbad, CA). RNA lysis buffer provided with miRVana miRNA isolation kit (Ambion, Austin) was added to each well and monolayers were removed using a cell scraper. Total RNA was isolated using the miRVana miRNA isolation kit as per the manufacturer’s protocol. RNA quantity and quality were measured using an Nanodrop (Thermo Scientific, Wilmington, DE) and an Agilent 2100 Bioanalyzer (Agilent Technologies, CA), respectively. Real time PCR was carried out using miRNA Taqman PCR assays (Applied Biosystems) to determine the expression of mature miR-19b, miR-26b or miR-203 in treated and untreated samples. Normalization was carried out using the 2ΔΔCT method relative to 18S rRNA.
Western blotting
In order to determine the change in protein expression associated with miRNA knockdown, western blotting was carried out. Cells (2–4×106) were seeded in 100 mm dishes on the day of transfection. Transfection with miRNA inhibitors (30 nM) was carried out as described above. Forty-eight hr following transfection, media was removed and cells were washed with PBS. Subsequently, lysis buffer containing 50 mM Tris-HCl (pH 7.2), 250 mM sucrose, 2 mM ethylenediaminetetraacetic acid (pH 7.6), 1 mM ethyleneglycol-bis(aminoethylether)-tetraacetic acid (pH 7.5), 50 μM NaF, 1% Triton-X, 100 μM sodium orthovanadate, protease inhibitor cocktail and 10 mM β-mercaptoethanol was added to the cells. The lysate was then passed through a 27 gauge needle and incubated on ice for 30 min. Subsequently, lysates were centrifuged at 16,000g for 20 min. Supernatants were collected and protein concentration determined by the Bradford method (37). Samples (20–80 μg) were loaded onto 4–20% Tris-Glycine gels (Invitrogen, Carlsbad, CA). After blotting, membranes were incubated overnight with rabbit polyclonal PDE4B antibody at 1:200 (Abcam, Cambridge, MA), rabbit TCF4 monoclonal antibody at 1:1000 (Cell signaling Technology, Boston, MA) or rabbit PTK2B antibody at 1:1000 (Cell Signaling Technology, Boston, MA) and horseradish peroxidase linked (Jackson Immunoresearch Laboratories, West Groove, PA) secondary antibody at 1:7000 dilution and chemoluminescent detection was performed. Unless noted, all other reagents were from Sigma-Aldrich, MA.
Dual luciferase reporter assay
Cells were plated at 0.2 × 105 cells per well in a 96 well plate and transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) with a reporter plasmid containing a Renilla luciferase coding region followed by the 3′ UTR of PTK2B or PDE4B (Switchgear Genomics, Menlo Park, CA) or dual luciferase reporter plasmid containing wild type or mutated 3′ UTR of TCF4 (Genecopoeia, Rockville, MD). For PTK2B and PDE4B constructs, a firefly reporter plasmid was cotransfected for normalization purposes. A control 3′ UTR plasmid with a random non-conserved and non-repetitive 3′ UTR sequence (scrambled 3′UTR) was utilized as a negative control for reporter plasmids. miR-19b, miR-26b, miR-203 or scrambled controlled miRNA mimics (30 nM) were subsequently co-transfected with cells according to the manufacturer’s instructions. Twenty-four hr after transfection, dual luciferase reporter assays (Promega) were performed according to the manufacturer’s instructions. The Renilla luciferase activity was measured using a SpectraMax L Luminescence Microplate Reader (Molecular Devices Sunnyvale, CA) and normalized to the firefly luciferase activity. To assess the direct binding of PDE4B with miR-26b, morpholinos (0.5 μM, classified as target protectors) (Gene Tools, Philomath, OR) against PDE4B were also co-transfected along with a reporter plasmid containing 3′ UTR of PDE4B. Results are presented as the average of three sets of replicates. The sequences of the miRNA inhibitors, miRNA mimics and 3′ UTR constructs are described in Supplemental Table I.
Animals
All procedures involving animals followed guidelines approved by the Institutional Animal Care and Use Committee at Texas A&M University in accordance with EU Directive 2010/63/EU for animal experiments. Lgr5-EGFP-IRES-creERT2 originally described by Barker et al (10–15 wk of age) were used (21). The study utilized two types of dietary fat and fiber – (control diet) corn oil (n-6 PUFA) + cellulose compared to (chemoprotective diet) fish oil (n-3 PUFA) + pectin and two injection treatments (colon carcinogen, AOM or saline). Animals (n=5 per group) were terminated eight wks after the last AOM injection. Immediately after termination, 1 cm of distal colon was rapidly removed, flushed with ice-cold saline and immediately fixed in 4% paraformaldehyde for immunofluorescence (IF) analyses. Supplemental Figures 1 and 2 show the timeline of the treatments and the experimental design.
Experimental diets
Lgr5-EGFP-IRES-creERT2 mice were assigned to one of the two diet groups (fish oil/pectin or corn oil/cellulose), which differed only in the type of fat and fiber. Diets contained (g/100 g diet): dextrose, 51.00; casein, 22.40; D,L-methionine, 0.34; American Institute of Nutrition (AIN)-76 salt mix, 3.91; AIN-76 vitamin mix, 1.12; choline chloride, 0.13; pectin or cellulose, 6.00. The total fat content of each diet was 15% by weight with the n-6 PUFA diet containing 15.0 g corn oil/100 g diet (Dyets, Bethlehem, PA) and the n-3 PUFA diet containing 11.5 g fish oil/100g diet (Omega Protein, Houston, TX) plus 3.5 g corn oil/100 g diet to prevent essential fatty acid deficiency. All diet ingredients except oils were obtained from Bio-serv (Frenchtown, NJ). To prevent the formation of oxidized lipids, diets were stored at −20°C and provided fresh to animals every day. Fluorescence activated cell sorting of colonic stem cells. Colonic crypts from individual mice were isolated as previously described Sato et al. (38) with minor modification. The intact colons were everted on a disposable mouse gauge needle (Instech Laboratories) and incubated with 20 mM EDTA in PBS at 37°C for 30 min. Following transfer to chilled Ca/Mg free HBSS, colons were vigorously vortexed to release crypts. The crypts were then incubated with 50 ul of DNase (stock concentration- 20 units/ml) in 10 ml of trypsin solution and single cells were then passed through a 40 micron cell strainer. Cells were counted and resuspended to a final cell density of 2 ×106 cells/mL. FACS (Fluorescence activated cell sorting) was then carried out to isolate the Lgr5high expressing stem cells, Lgr5low expressing daughter cells and Lgr5negative cells isolated from the colon using a BD FACS Aria II cytometer/sorter (BD Biosciences). Cells from wild type mice were used to set the gates for sorting (Supplemental Figure 2).
RNA analyses
Total RNA from Lgr5high, Lgr5low and Lgr5negative sorted cells was isolated. For this purpose, cells from individual mice from all 4 groups (total of 60 samples) were separately processed using the mirVana miRNA Isolation Kit according to manufacturer’s instructions (Ambion, Austin, TX). Expression of 368 mature miRNAs was determined using TaqMan Rodent MicroRNA A Array 2.0 (Life Technologies, Grand Island, NY) as we have previously described (33, 34). For mRNA profiling, samples were randomized prior to RNASeq library preparation. Sequencing libraries from RNA (10 ng) were generated using the TruSeq RNA Sample Preparation kit (Illumina, San diego, CA). ERCC (Life Technologies, Grand Island, NY) was added at the appropriate level as per manufacturer instructions. The libraries were pooled and sequenced using an Illumina HiSeq 2500 at SeqWright Genomic Services (Houston, TX). Sequencing data was provided demultiplexed and aligned using STAR with default parameters (39) and referenced against Mus musculus (UCSC version mm10).
In vivo apoptosis measurement
To investigate whether alkylating agent-induced DNA damage induces apoptotic cell death in colonic epithelial cells, fragmented DNA was visualized using the TACS 2 TdT-Fluor in situ apoptosis detection kit (Trevigen) as per the manufacturer’s recommended protocol for staining. Negative control slides were incubated without TdT enzyme.
Slide scoring
Images of colonic crypts were captured using an inverted TE 300 Nikon Eclipse fluorescence microscope equipped with a Photometrics Cool snap EZ cooled digital CCD camera. Images were processed using NIS Image software, version 3.2. A total of 68 (CCS, corn oil+cellulose+saline), 75 (CCA, corn oil+cellulose+AOM), 51 (FPS, fish oil+ pectine+saline) and 141 (FPA, fish oil+pectine+AOM) GFP+ crypts from 5 mice were counted. All images were captured at a magnification of 40X.
High multiplicity ACF assay
To determine whether the chemoprotective (fish oil+pectin) diet was able to suppress formation of early preneoplastic lesions of colon cancer (aberrant crypt foci, ACF) compared with the control (corn oil+cellulose) diet, we collected distal colon 8 wk after the last AOM injection. Colons were opened and placed flat onto folded Whatman #1 paper, followed by fixation in 70% ethanol for 24 hr. To identify aberrant crypts, tissue was stained in a 0.5% solution of methylene blue for 45 s. The total number of high multiplicity ACF (HM ACF) (foci containing 4 or more aberrant crypts) were counted using a 40× objective.
Statistical Analyses
For target validation experiments, the effect of more than two independent variables (treatment effects) was assessed using the one-way analysis of variance test (ANOVA), and differences among the means were evaluated using Tukey’s and Bonferroni post-hoc test of contrast. P values <0.05 were considered to be statistically significant. Graphs were plotted using means and standard errors of 6 to 8 cell culture wells (data points) obtained from each treatment, collected from three different experiments (performed in triplicate). Standard error bars were plotted in order to document the variation in the population mean. For miRNA and mRNA profiling, two sided t-tests with Welch correction for unequal variance were performed on select miRNA across the specific treatment comparisons of interest. Mann-Whitney U nonparametric tests were also performed as a control against non-normal data and similar P-values were obtained. Standard error bars were plotted in order to document the variation in the population mean. P values <0.05 were considered to be statistically significant, and genes were selected for analysis using prior knowledge without considering P-values. Therefore, no multiple testing correction procedure was used. Standardized differences for the miRNAs and mRNAs were computed and a two-sample t-test was utilized to compare them. Small p-values indicated strong evidence of the hypothesized trend.
For phenotypic analyses, cells were plated at 0.2 × 105 cells per well in 96 well plates and miRNA inhibitors or control treatments were transfected as described above. Approximately 48 hr later, apoptosis was measured using a cellular fragmentation enzyme linked immune-sorbent assay (ELISA) (Roche, IN). Floating cells were harvested, washed, lysed and centrifuged to sediment nuclei. Supernatants containing mono- and oligonucloesomes were incubated with substrate and subsequently analyzed by ELISA as previously described (40). To determine the effect of treatment on cell proliferation, equal numbers of cells were seeded in a 96 well plate and 48 hr later, the number of cells was counted using a hemocytometer. All experiments were performed in triplicate.
Results
Since the unique combination of chemoprotective dietary fish oil and pectin modulates a subset of mucosal miRNAs and their predicted target genes in the presence of carcinogen (33, 34), we further elucidated the biological properties of miR-19b, miR-26b and miR-203 miRNAs with respect to their target genes. In addition, the modulatory properties of diet with respect to miRNAs and their target genes in intestinal stem cells were investigated.
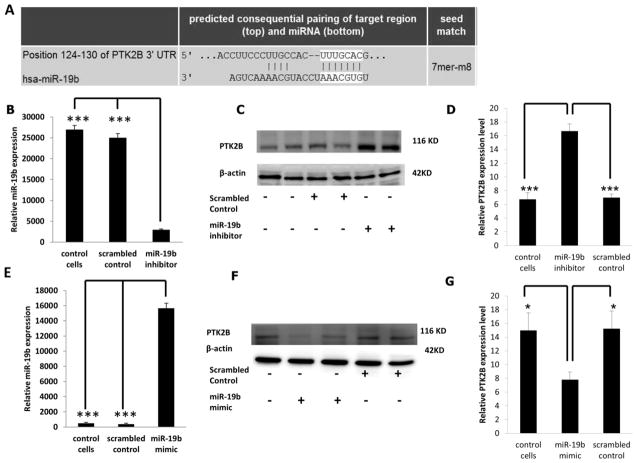
PTK2B is an indirect target of miR-19b
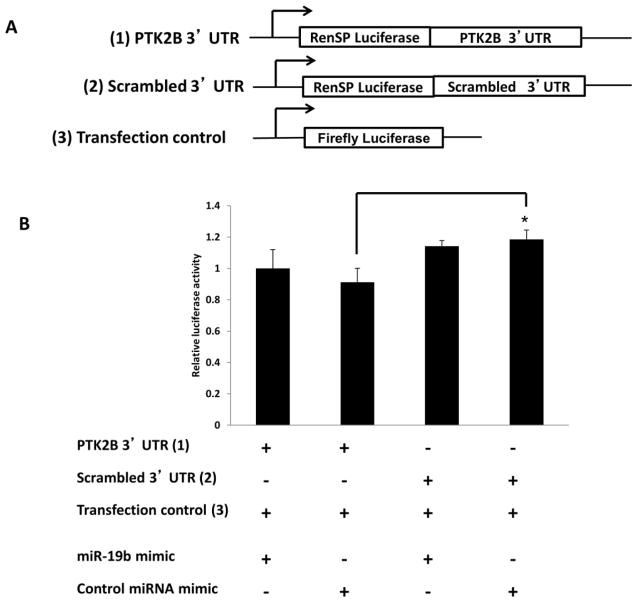
As shown in Supplemental Table II, miR-19b, miR-26b and miR-203 share 100% homology between human (hsa), mouse (mmu) and rat (rno). The sequences were obtained from the miRBase database (www.mirbase.org/). To determine if PTK2B levels are modulated following miR-19b knockdown, HCT116 cells were transfected with anti-miR-19b inhibitor or with a scrambled miRNA inhibitor control (negative control). The predicted pairing of the target 3′ UTR gene region with miR-19b is shown in Figure 1A. With a transfection efficiency of >70%, 48 hr following knockdown, miR-19b levels were significantly decreased by ~90% as compared to scrambled control and non-transfected control (Figure 1B). Dose dependent knockdown of miR-19b was carried out to determine the optimal concentration of miR-19b inhibitor (Supplemental Figure 3). Correspondingly, PTK2B protein levels were significantly increased by approximately 200% in miR-19b knockdown samples compared with negative control and non-transfected control (Figures 1C and D). Alternatively, HCT116 cells were transfected with miR-19b mimic or scrambled miR mimic control and 48 hr later, expression of miR-19b was significantly (P <0.05) increased (Figure 1E). Dose dependent overexpression of miR-19b was initially carried out to determine the optimal concentration of miR-19b mimic (Supplemental Figure 4). Following transfection, PTK2B protein levels were significantly (P <0.05) decreased by approximately 60% in cells overexpressing miR-19b compared to the negative control and non-transfected control (Figures 1F and G). These results indicate that PTK2B was significantly inhibited by miR-19b. In order to further determine if PTK2B is a direct target of miR-19b, a 3′UTR reporter plasmid containing the predicted miR-19b binding site of PTK2B (Figures 1A and 2A) was utilized. HCT116 cells were co-transfected with miR-19b mimic and 3′UTR PTK2B Renilla reporter plasmid (PTK2B 3′ UTR) and the Renilla luciferase signal was normalized to control-firefly activity (transfection control). Separately, a control plasmid, containing a 3′UTR with a scrambled sequence (scrambled 3′ UTR) that was non-conserved and non-repetitive, was co-transfected with miR-19b mimic in HCT116 cells. This served as a negative control. Twenty-four hr later, luciferase activity was unaffected relative to the negative control (Figure 2B). These data indicate that PTK2B is not a direct target of miR-19b.
Figure 1. PTK2B expression following knockdown or overexpression of miR-19b.
(A) Predicted binding of miR-19b to the 3′ UTR of PTK2B as assessed by Target Scan (http://www.targetscan.org/). (B) Knockdown of miR-19b was carried out by transfecting miR-19b inhibitor or scrambled miRNA inhibitor (control) in HCT116 cells. Forty-eight h following transfection, miR-19b expression was measured by qRT-PCR. (C) PTK2B protein levels were measured by Western blotting after miR-19b knockdown and β-actin was used as a loading control as described in the Materials and Methods. (D) Quantification of PTK2B levels in control (no transfection), scrambled control or miR-19b knockdown samples from immunoblot images. (E) Overexpression of miR-19b was carried out by transfecting miR-19b mimic or scrambled miRNA mimic control and 24 h later, miR-19b expression was measured by qRT-PCR. (F) PTK2B protein levels were measured by western blotting following miR-19b overexpression, and β-actin was used as a loading control. (G) Quantification of PTK2B levels in control (no transfection), scrambled control or miR-19b overexpressed samples from immunoblot images. Data represent means ± S.E. from six-eight replicate values obtained from three separate experiments. Significant differences between groups are indicated as ***P < 0.001, and *P < 0.05.
Figure 2. Validation of PTK2B as a gene target of miR-19b.
(A) Schematic representation of the 3′UTR PTK2B Renilla reporter plasmid, scrambled 3′UTR Renilla reporter plasmid and normalization using a control-firefly reporter plasmid (transfection control). (B) Luciferase activity was determined 24 h following transfection. All luciferase values were normalized to firefly luciferase (transfection control). Data represent means ± S.E. from nine replicate values obtained from three separate experiments. Significant differences between groups are indicated as *P < 0.05.
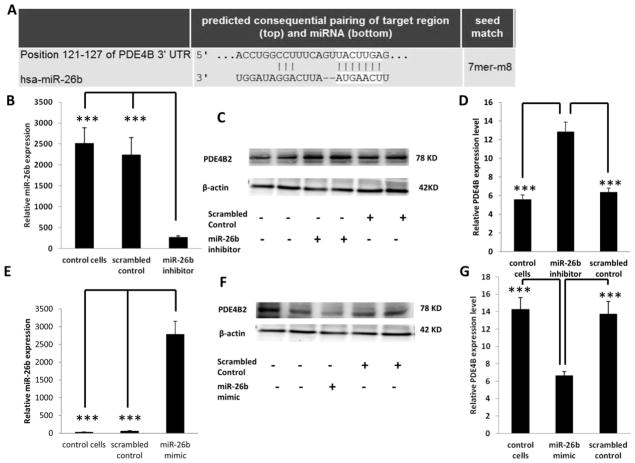
PDE4B is a direct target of miR-26b
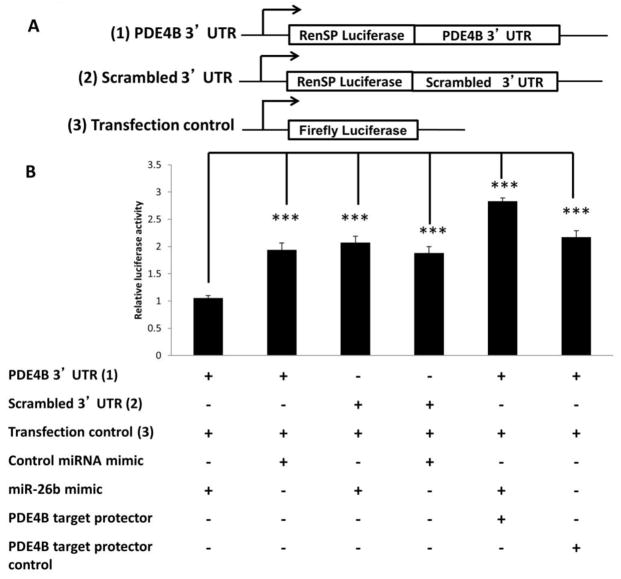
To determine if PDE4B is directly targeted by miR-26b, HCT116 cells were transfected with an anti-miR-26b inhibitor or with scrambled control miRNA inhibitor (negative control). Dose dependent knockdown of miR-26b was carried out to determine the optimal concentration of miR-26b inhibitor (Supplemental Figure 5). Forty-eight hr following knockdown, miR-26b levels were significantly decreased (P <0.05) by 75% as compared to control (Figure 3B), and correspondingly, PDE4B protein levels were significantly (P <0.05) increased by 100% in miR-26b knockdown samples compared with negative control and non-transfected control (Figure 3C and D). In addition, in HCT116 cells transfected with miR-26b mimic or scrambled miR mimic control for 48 hr, miR-26b levels were significantly (P <0.05) increased (Figure 3E) and PDE4B protein levels were significantly (P <0.05) decreased by 55% in miR-26b overexpressed samples compared to the negative control and non-transfected control (Figures 3F and G). Dose dependent overexpression of miR-26b was initially carried out to determine the optimal concentration of miR-26b mimic (Supplemental Figure 6). These results indicate that PDE4B is downregulated by miR-26b. In order to determine if PDE4B is a direct target of miR-26b, a 3′UTR reporter plasmid containing the miRNA binding site of PDE4B (Figure 3A and 4A) was utilized. HCT116 cells were cotransfected with miR-26b mimic, 3′UTR PDE4B Renilla reporter plasmid (PDE4B 3′ UTR) and a normalization control-firefly reporter plasmid (transfection control). Separately, a control 3′ UTR plasmid with a random sequence (scrambled 3′UTR) or a non-conserved and non-repetitive 3′ UTR sequence were co-transfected with miR-26b. Twenty-four hr later, luciferase activity was significantly (P <0.05) decreased by 50% relative to the negative control (Figure 4B). These data demonstrate that PDE4B is a direct target of miR-26b. To further confirm the miR-26b regulation on PDE4B, a target protection analysis was utilized. Predicted target sites on mRNAs were protected by the complimentary binding of morpholinos, thereby reducing miRNA suppression on luciferase activity (41). HCT116 cells were co-transfected with a morpholino targeting PDE4B mRNA, PDE4B 3′ UTR Renilla reporter plasmid, a normalization control-firefly reporter plasmid (transfection control) and miR-26b mimic. Significantly increased luciferase activity (P <0.05) was observed in HCT116 cells co-transfected with morpholino compared to cells with no protective oligos (Figure 4B). Separately, as a negative control, PDE4B target protector control was co-transfected with PDE4B 3′ UTR plasmid. The morpholino masked the 3′ UTR miRNA binding site, blocking the mimic, thereby increasing luciferase expression. This confirmed that PDE4B is a direct target of miR-26b.
Figure 3. PDE4B expression following knockdown or overexpression of miR-26b.
(A) Predicted binding of miR-26b to the 3′ UTR of PDE4B as assessed by Target Scan (http://www.targetscan.org/). (B) Knockdown of miR-26b was carried out by transfecting HCT116 cells with miR-26b inhibitor or scrambled miRNA inhibitor (control) for 48 h. miR-26b expression was subsequently measured by qRT-PCR. (C) PDE4B protein levels were measured by Western blotting following miR-26b knockdown. β-actin was used as a loading control as described in the Materials and Methods. (D) Quantification of PDE4B protein levels in control (no transfection), scrambled control or miR-26b knockdown cultures from immunoblot images. (E) Overexpression of miR-26b was carried out by transfecting miR-26b mimic or scrambled miRNA mimic control. 24 h post transfection, miR-26b expression was measured by qRT-PCR. (F) PDE4B protein levels were measured by Western blotting following miR-26b overexpression and β-actin was used as a loading control. (G) Quantification of PDE4B protein levels in control (no transfection), scrambled control or miR-26b overexpressed samples from immunoblot images. Data represent means ± S.E. from six-eight replicate values obtained from three separate experiments. Significant differences between groups are indicated as ***P < 0.001.
Figure 4. Validation of PDE4B as a gene target for miR-26b.
(A) Schematic representation of the 3′UTR PTK2B Renilla reporter plasmid, scrambled 3′UTR Renilla reporter plasmid and normalization control-firefly reporter plasmid used in the study. (B) Luciferase activity was determined 24 h after transfection. All luciferase values were normalized to firefly luciferase (transfection control). Data represent means ± S.E. from nine values obtained from three separate experiments. Significant differences between groups are indicated as ***P < 0.001.
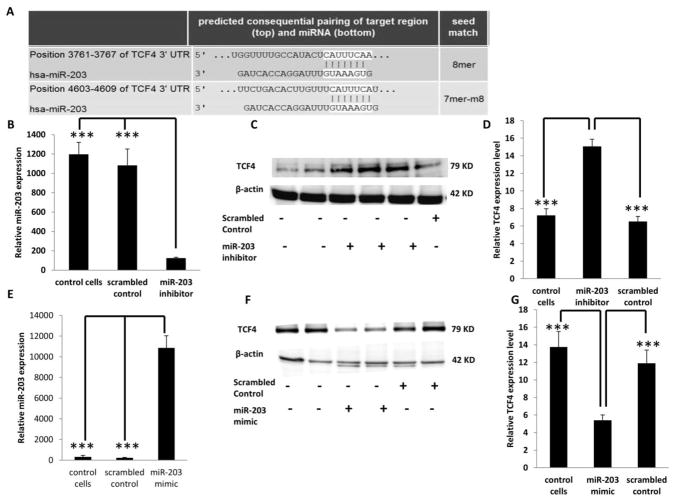
TCF4 is a direct target of miR-203
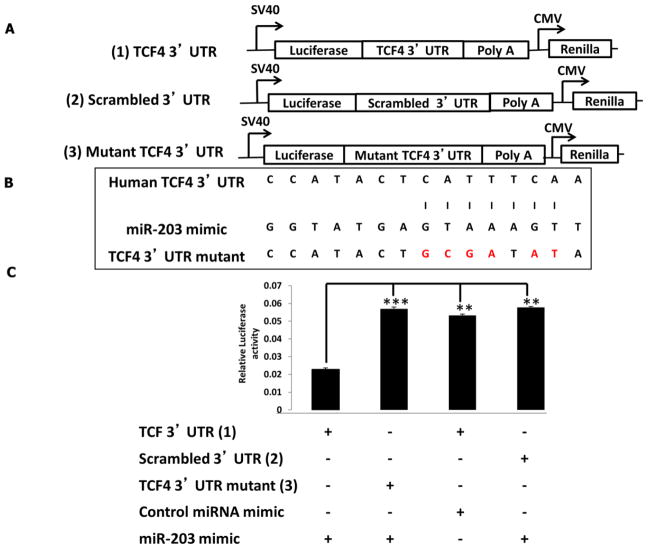
To determine if TCF4 levels are modulated following miR-203 knockdown, HCT116 cells were transfected with anti-miR-203 inhibitor or with a scrambled control miRNA inhibitor (negative control). Dose dependent knockdown of miR-203 was carried out to determine the optimal concentration of miR-203 inhibitor (Supplemental Figure 7). Forty-eight hr following knockdown, miR-203 levels were significantly (P <0.05) decreased as compared to the controls (Figure 5B). Correspondingly, TCF4 protein levels were significantly increased (P <0.05) by an average of 100% in miR-203 knockdown samples compared with the negative control and non-transfected control (Figures 5C and D). In comparison, in HCT116 cells transfected with miR-203 mimic or scrambled miR mimic control, miR-203 levels were significantly (P <0.05) increased (Figure 5E). Dose dependent overexpression of miR-203 was initially carried out to determine the optimal concentration of miR-203 mimic (Supplemental Figure 8). Correspondingly, TCF4 protein levels were significantly decreased (P <0.05) by an average of 180 % in miR-203 overexpressed samples compared with negative control and non-transfected control (Figures 5F and G). These results indicate that TCF4 expression is inversely correlated with miR-203 abundance. To determine if TCF4 is a direct target of miR-203, a 3′UTR reporter plasmid containing the miRNA binding site of TCF4 (Figures 5A and 6A) was utilized. HCT116 cells were cotransfected with miR-203 mimic and the 3′UTR TCF4 Firefly reporter plasmid. In separate experiments, as a negative control, a control 3′ UTR plasmid with a random sequence (scrambled 3′UTR) was co-transfected with miR-203 mimic. Twenty-four hr later, luciferase activity was significantly decreased (P <0.05) by ~120% relative to the negative control. In order to confirm this result, the binding sites of miR-203 in 3′ UTR of TCF4 were mutated. The miR-203 binding site in the 3′ UTR of TCF4, i.e., “catttcaa”, was mutated to “gcgatatc” (Figure 6B). When a mutated 3′ UTR version of TCF4 was co-transfected with the miR-203 mimic, there was no change in luciferase activity compared to the miRNA target clone control vector (Figure 6C), confirming the specificity of miR-203 binding. These findings demonstrate that TCF4 is a direct target of miR-203.
Figure 5. TCF4 expression following knockdown or overexpression of miR-203.
(A) Predicted binding of miR-203 to the 3′ UTR of TCF4. (B) Knockdown of miR-203 was carried out by transfecting miR-203 inhibitor or scrambled miRNA inhibitor (control) in HCT116 cells. 48 h following transfection, miR-203 expression was measured by qRT-PCR. (C) TCF4 protein levels were measured by western blotting after miR-203 knockdown and β-actin was used as a loading control as described in the Materials and Methods. (D) Quantification of TCF4 levels in control (no transfection), scrambled control or miR-203 knockdown samples from immunoblot images. (E) Overexpression of miR-203 was carried out by transfecting miR-203 mimic or scrambled miRNA mimic control. miR-203 expression was measured by qRT-PCR 24 h later. (F) TCF4 protein levels were measured by western blotting after miR-203 overexpression and β-actin was used as a loading control. (G) Quantification of TCF4 levels in control (no transfection), scrambled control or miR-203 overexpressed samples from immunoblot images. Data represent means ± S.E. from six - eight replicate values obtained from three separate experiments. Significant differences between groups are indicated as ***P < 0.001.
Figure 6. Validation of TCF4 as a target gene for miR-203.
(A) Schematic representation of the 3′UTR TCF4 Renilla reporter plasmid, scrambled 3′ UTR Renilla reporter plasmid and 3′UTR mutant TCF4 Renilla reporter plasmid. (B) Schematic representation of 3′ UTR of TCF4 showing the putative miR-203 target site. Letters highlighted in red indicate the mutated base pairs. (C) Luciferase activity was determined 24 h after transfection. All luciferase values were normalized to Renilla luciferase. Data represent means ± S.E. from nine values obtained from three separate experiments. Significant differences between groups are indicated as ***P < 0.001, **P < 0.01.
miR-19b and miR-26b induce apoptosis whereas miR-203 reduces cell proliferation
Studies have shown that dietary fish oil and pectin combination is uniquely chemoprotective, in part, because it promotes apoptosis in the colon (29, 42). We have previously demonstrated that the levels of miR-26b and miR-203 were reduced by feeding the control diet consisting of corn oil and cellulose (CCA) as compared to the chemoprotective fish oil and pectin (FPA) combination diet, in the presence of carcinogen (34). Hence, in order to elucidate how alterations in miRNAs in the colon alter cell growth and viability, HCT116 cells were transfected with miR-19b, miR-26b mimic or scrambled control miRNA. A 100% increase (P <0.05) in apoptotic index was observed when miR-19b and miR-26b were overexpressed compared to untreated control or scrambled control (Figure 7), whereas there was a modest increase of ~40% (P <0.05) in apoptotic index and a significant decrease in cell number when miR-203 was overexpressed (Figure 7). Cell proliferation was not determined for miR-19b and miR-26b since previous studies have demonstrated that these miRNAs do not alter cell division (43, 44).
Figure 7. Effects of miR-19b, miR-26b and miR-203 on colonocyte phenotype.
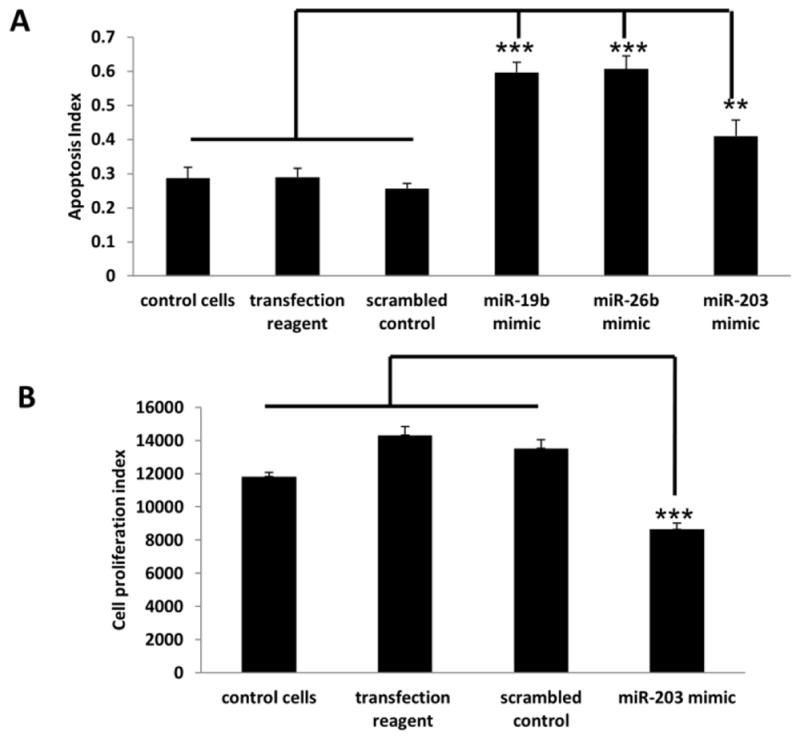
(A) HCT116 cells were transfected with miR-19b, miR-26b, miR-203 (30 nM) or scrambled control mimic. After 72 h, the apoptosis index was measured. (B) HCT116 cells were transfected with either miR-203 mimic or scrambled control mimic for 72 h and total cell number was counted. Data represent means ± S.E. from six culture wells obtained from two separate experiments. Significant differences between groups are indicated as ***P < 0.001, **P < 0.01.
Effect of diet on aberrant crypt foci (ACF) formation in the colon
To determine the role of miR-19b, miR-26b and miR-203 in the colonic stem cell niche and to identify a putative miRNA signature in vivo, Lgr5-EGFP-IRES-creERT2 knock in mice were fed with either a (control) corn oil + cellulose diet (CCA) or a (chemoprotective) fish oil+ pectin (FPA) diet in the presence of carcinogen. AOM injected mice fed the CCA diet exhibited a greater number of ACF compared to the FPA chemoprotective diet; [44.7±6.6 versus 28.4 ± 3.6, P <0.05] and high multiplicity-ACF; [6.4± 0.9 versus 4.3 ± 0.8, P =0.06] (Supplemental Figure 9).
Assessment of global miRNA expression in colon adult stem cells
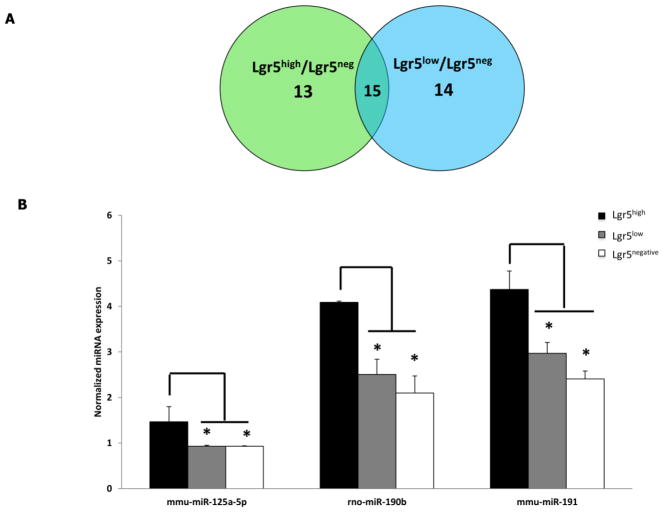
To globally assess miRNA profiles in stem/progenitors cells, colonic crypt cells isolated from carcinogen or saline (control) injected mice were FACS sorted to obtain stem cells (Lgr5high), progenitor/daughter cells (Lgr5low) and differentiated cells (Lgr5negative) (Supplemental Figure 2). Following global miRNA profiling, 103 mature miRNAs (critical threshold (ct) <35) were detected in the two cell populations (n=20 samples per cell type) (see Table 1 in (99)). Using pooled miRNA data from all 4 groups (i.e., CCA, FPA, CCS and FPS), 26 miRNAs (P <0.05) were differentially expressed in Lgr5high as compared to Lgr5negative cells (see Table 2 in (99)). In addition, comparisons between Lgr5high vs Lgr5negative, and Lgr5low vs Lgr5negative populations revealed that seven miRNAs were commonly down-regulated and ten up-regulated in both stem cells and daughter cells compared to differentiated cells (Figure 8A). These data suggest that stem cells and their daughter cells exhibit a somewhat similar miRNA profile. In contrast, miRNA profiles in stem cells and differentiated cells were quite distinct.
Figure 8.
Expression of miRNAs in Lgr5high, Lgr5low and Lgr5negative populations. (A) Common differentially expressed miRNAs in Lgr5high and Lgr5low cells. The Venn diagram indicates the number of differentially expressed miRNAs in Lgr5high vs. Lgr5negative cells and Lgr5low vs. Lgr5negative cells (n=20 mice per cell type). (B) miR-125a-5p, miR-190b and miR-191 are specific miRNAs enriched in intestinal stem cells. Data represent normalized miRNA expression (means ± SEM, for details, see Materials and methods). miRNA profiling in stem cells (Lgr5high- black bar), daughter cells (Lgr5low- grey bar) and differentiated cells (Lgr5negative- white bar) by real time PCR was used to quantify miR-125a-5p, miR-190b and miR-191 expression (pooled data). *P < 0.05. (n=20 per cell type). mmu, mouse; rno, rat.
In order to determine if certain miRNAs are uniquely expressed in stem cells, miRNA in stem cells (Lgr5high) and daughter stem cells (Lgr5low) were compared. miR-125a-5p, miR-190b and miR-191 were enriched in stem cells compared to daughter cells (Figure 8B). Interestingly, miR-125a-5p, miR-190b and miR-191 were also significantly (P <0.05) up-regulated in stem cells compared to differentiated cells (see Table 2 in (99)). This suggests that miR-125a-5p, miR-190b and miR-191 may be required by stem cells and their expression decreases as cells differentiate.
Carcinogen modulates stem cell miRNAs
To determine the effect of carcinogen on colonic stem cell miRNA expression patterns, both Lgr5high and Lgr5negative sorted cells in AOM and saline treated groups were compared. Interestingly, distinct effects of carcinogen on miRNA expression in Lgr5high vs Lgr5negative cells were observed (see Table 3 in (99)). This suggests that stem cell miRNAs are uniquely modulated by carcinogen.
Diet modulates stem cell miRNAs following exposure to carcinogen
To determine the effects of dietary bioactives on miRNA, e.g., miR-19b, miR-26b and miR-203, expression in stem cells and differentiated cells, select colonocyte populations from carcinogen injected Lgr5-cre-EGFP mice fed the CCA or FPA experimental diets were examined. Analysis of FACS sorted stem cells revealed that twelve miRNAs were differentially expressed (P <0.05), ten of which were up-regulated and two miRNAs were down-regulated in Lgr5high sorted cells in CCA vs FPA treatment (see Table 4A in (99)). These miRNAs were not altered in Lgr5high cells in presence of saline (CCS vs FPS) (see Table 4B in (99)). To our surprise, miR-19b, miR-26b and miR-203 were significantly up-regulated (P ≤0.05) in Lgr5high stem cells obtained from CCA vs FPA. This was unexpected based on previous observations using a mixed population of colonic mucosal cells from rats fed a similar diet (34). In addition, we noted that 5 miRNAs - miR-10a, miR-21, miR-200a, miR-203 and miR-218 that were altered in Lgr5high cells as compared to Lgr5negative cells (see Table 2 in (99)) were also modulated by diet.
With respect to the effects of diet (CCA vs FPA) in presence of carcinogen on differentiated cells (Lgr5neg), 6 miRNAs were significantly (P ≤0.05) altered (see Table 5 in (99)). Out of these, miR-19a and miR-19b were significantly (P ≤0.05) down-regulated in CCA compared to FPA treatment groups.
Diet modulates PTK2B expression in differentiated cells
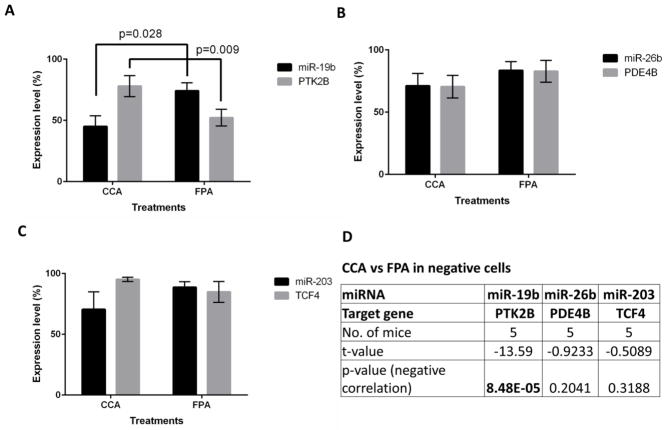
To determine if the validated targets of miR-19b, miR-26b and miR-203, i.e., PTK2B, PDE4B and TCF4 (Figures 1, 3 & 5), respectively, were altered in stem cells and differentiated cells in CCA and FPA groups, RNA sequencing was carried out on the same samples for which miRNA profiling was performed. As part of the initial characterization, we assessed the expression of several stem cell makers and self-renewal genes in Lgr5high, Lgr5low and Lgr5negative cells, including Lgr5 expression. The expression level of fast cycling stem cell markers (e.g., Lgr5, Ascl2, Olfm4, Lrig1) was up-regulated in Lgr5high cells as compared to Lgr5negative cells (Supplemental Figure 10) similar to a previous report (45). The expression of PTK2B and miR-19b were reciprocally modulated in the bulk population of differentiated cells (Lgr5negative) from CCA vs FPA fed animals (Figure 9A). A significant (P <0.002) negative correlation between miR-19 and PTK2B in differentiated cells was detected using a linear regression model (Figure 9D). This observation is consistent with the data reported in Figures 1&2. In contrast, the expression of PDE4B and TCF4, direct targets of miR-26b and miR-203, respectively, was not altered in the bulk population of differentiated cells from CCA vs FPA fed animals (Figures 9B & C). Furthermore, to understand the mechanism of action of these miRNAs and their targets at the stem cell level, we examined at the expression of PTK2B, PDE4B and TCF4, specifically in Lgr5high cells. To our surprise, the changes in the expression of the targets were not as expected (i.e., decreased in CCA vs FPA treatment). In contrast, there was no change in expression of PTK2B and TCF4 in CCA group as compared to FPA group in Lgr5high cells (fold change = 0.93, p-value = 0.320 and fold change = 1.08, p-value= 0.137, respectively). The expression of PDE4B was significantly downregulated in CCA vs FPA group in Lgr5high cells (fold change = 1.48, p – value = 0.010). These findings suggest that stem cell responses to diet may be distinct from the bulk cell population in the colon.
Figure 9. Expression of miRNA and their targets in Lgr5negative cells sorted cells from CCA and FPA samples.
miRNA profiling and RNA sequencing was performed using Lgr5negative cells sorted from CCA and FPA treated mice and expression of (A) miR-19b and PTK2B, (B) miR-26b and PDE4B and (C) miR-203 and TCF4 was determined, n=5 per group. (D) Linear regression analysis was performed to determine the significant negative correlation between miRNAs and their putative targets.
Fish oil and pectin alter the number of apoptotic stem cells in mouse distal colon
Reduced apoptosis is a significant risk factor for development of colon cancer (46). Hence, we determined the effects of fish oil and pectin feeding on the number of apoptotic stem cells in the distal colon. As shown in Figure 10, a higher number of apoptotic stem cells per crypt column was observed in saline-injected fish oil/pectin (FPS) fed animals as determined by TUNEL assay. Interestingly, AOM exposure suppressed the proapoptotic effect (FPA vs FPS) in Lgr5-positive stem cells.
Figure 10. Effect of diet on stem cell apoptosis.
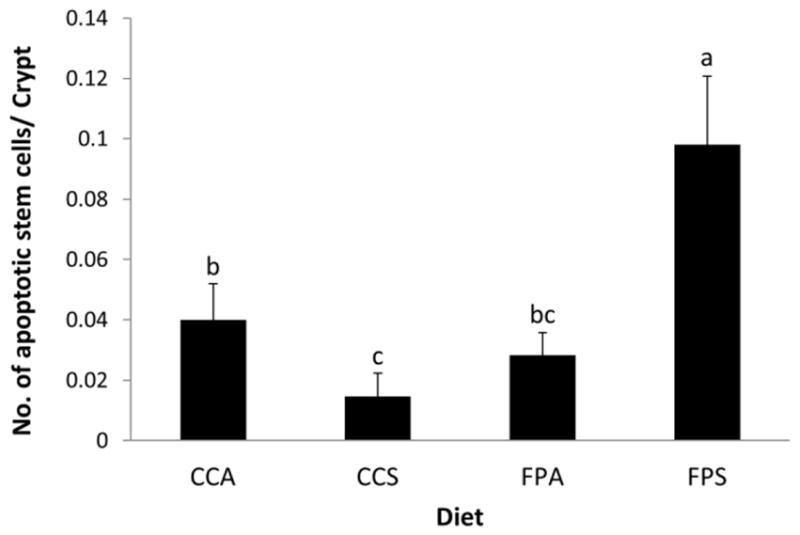
Saline injected fish oil/pectin fed mice exhibited higher levels of apoptosis in stem cells. Bars with different letters are significantly different (P <0.05 by one tail test with Mixed Procedure and GENMOD Procedure).
Discussion
We have recently demonstrated that n-3 PUFA uniquely modulate carcinogen-directed miRNA signatures in the colon (33). In addition, previous studies have shown that the bioactive compounds associated with dietary fish oil (DHA and EPA) and fermentable fiber (butyrate) synergize to protect against colon carcinogenesis, primarily by enhancing apoptosis (29, 31, 46–48). With regard to dietary combination, i.e., fat x fiber interaction in the presence of carcinogen (CCA vs FPA), we observed a coherent relationship between miR-19b, miR-26b, miR-203 and PTK2B, PDE4B and TCF4, respectively (34). These analyses were performed using a heterogeneous population of crypt cells. Since these diet/carcinogen modulated-miRNAs may exhibit distinct functions in different cell types, their role in colonic epithelial stem cells warrants investigation. Therefore, to further elucidate the biological effects of diet and carcinogen on miRNAs and their targets in stem versus differentiated cell populations in the intestinal crypt, an integrated targeted approach was used.
We initially investigated the role of three miRNAs previously shown to be selectively modulated by bioactive n-3 PUFA in combination with fermentable fiber and carcinogen. By manipulating miRNA levels and performing luciferase reporter assays, we demonstrated that PDE4B and TCF4 are direct targets of miR-26b and miR-203, respectively. Even though PTK2B protein levels were modulated by miR-19b, there was no change in luciferase reporter activity, indicating the absence of direct binding between miR19b and PTK2B. These data suggest that miR-19b modulates PTK2B upstream target(s).
We have previously documented the ability of dietary fish oil/pectin combination to increase miR-26b expression in the colon (34). This is noteworthy, because the suppression of miR-26b expression has been reported in various cancers including colon cancer, hepatocellular carcinoma, breast cancer and prostate cancer (49–51). In addition, miR-26b status has been associated with the invasiveness and metastasis of colorectal cancer cells (49). PDE4B (cAMP-specific 3′,5′-cyclic phosphodiesterase 4B) acts as an oncogene in colorectal and other cancers (52) by limiting cAMP-associated apoptosis (53, 54). In the present study, we observed that apoptosis was induced with increasing levels of miR-26b (Figure 7), which is consistent with published reports (49–51). In addition, previous studies from our laboratory have shown that fish oil - pectin combination act synergistically to induce apoptosis, both in vivo and in vitro, by recruiting a p53 independent, oxidation sensitive, Ca2+ -mediated intrinsic mitochondrial pathway (29, 55). Based on this evidence, we propose a putative mechanism describing miR-26 action in the colon (Supplemental Figure 11). By increasing mucosal levels of miR-26b, fish oil-pectin combination decreases PDE4B expression, which promotes cyclic adenosine monophosphate (cAMP) formation (56). This can lead to reduction of phosphorylation at threonine 308 and serine 473 of Akt (i.e. pAKT) (57) and activation of protein kinase A (PKA) (56). From a chemoprotective standpoint, PKA can promote apoptosis (58). Moreover, decreased pAKT activity can result in a reduction in PIP3 levels and decrease the lipid kinase activity of PI3K (57). Interestingly, we have observed that other dietary chemoprotective agents, e.g., curcumin, are capable of blocking intestinal dextran sodium sulphate-induced increase in PDE4B (59). In addition, EZH2, one of the targets of miR-26b, is modulated by curcumin in pancreatic cancer cells (50, 60), suggesting that miR-26b and its targets may be modulated by chemoprotective bioactive dietary agents. Collectively, these findings suggest that miR-26b acts as a “tumor suppressor” by targeting PDE4B and other genes.
The increased expression of miR-203 in HCT116 cells was associated with decreased levels of one of its predicted targets, TCF4 (Figure 5). miR-203 is down-regulated in a number of cancers and was up-regulated in the colonic mucosa of rats fed a fish oil - pectin combination diet (34, 61–66). miR-203 down-regulation appears to be mediated by epigenetic silencing. For example, studies in breast cancer and multiple myeloma cells have shown that the miR-203 promoter is hypermethylated (61, 67). Moreover, Lena et al. demonstrated that miR-203 is down-regulated during epithelial commitment of embryonic stem cells (63). Through overexpression or knockdown of miR-203 levels in HCT116 cells, we showed that TCF4 expression was inversely correlated to the level of miR-203. Also, luciferase reporter assays showed that miR-203 directly binds to TCF4 and mutation of the miR-203 binding site in the TCF4 3′ UTR abolished the effect of miR-203 (Figure 4). These findings are noteworthy because TCF4 is a well-known transcription factor expressed in the intervillus pocket of the developing intestine and is required to establish proliferative progenitor cells of prospective crypts in the embryonic intestine (68). TCF4 is regulated by the Wnt signaling pathway and controls stem cell fate in the intestine (69). Indeed, it is well appreciated that an increase in TCF4 levels promotes TCF4-β-catenin interaction, which can lead to the inappropriate activation of a TCF4 target gene program (70). Previously, we have demonstrated that colonic-β-catenin signaling, an upstream mediator of TCF4, is suppressed in carcinogen injected rats fed a fish oil + pectin (FPA) as compared to control diet (CCA) (42). Based on our findings, we propose a pathway by which CCA combination enhances hypermethylation of the miR-203 promoter, resulting in the reduced expression of mature miR-203 (Supplemental Figure 12). Hence, TCF4 protein levels would increase, thus promoting cell proliferation. Further work is needed in order to determine how FPA modulates the expression of mature miR-203 in the colon.
Recent findings lend support to the concept that in some circumstances, carcinogen exposure can perturb homeostasis within the stem cell compartment and promote malignant transformation (71). “Adult” somatic stem cells of the colon are of particular interest because they sustain self-renewal capacity and are targets for cancer initiating mutations (72–74). Lgr5 is regarded as a specific marker for identification of intestinal stem cells (21). Therefore, it is important to know precisely how intestinal stem cell populations respond to environmental factors, specifically bioactive dietary agents. To date, the effect of a chemoprotective n-3 PUFA/pectin diet on miRNAs including miR-19b, miR-26b and miR-203 in stem cells during initiation of colon cancer has not been documented. Hence, in this study, we used the Lgr5-EGFP-IRES-creERT2 knock in mouse model to examine miRNA expression profiles in stem and differentiated cell populations. Moreover, since transformation of adult stem cells is an extremely efficient route towards initiation of intestinal cancer (74), Lgr5-EGFP-IRES-creERT2 knock in mice were injected with AOM, a colon carcinogen. The AOM-induced colon tumor model was selected because it provides a clear distinction between tumor initiation and promotion (75). We hypothesized that the miRNA expression profiles would differ between stem cells and differentiated cells. Of the 384 rodent miRNAs identified, 26 miRNAs in stem cells were significantly altered compared to differentiated cells (see Table 2 in (99)). We noted that select miRNAs, e.g., miR-203, miR-145, miR-27a, miR-215, had a reduced expression in Lgr5high cells as compared to Lgr5negative cells. These miRNAs were also reported to be necessary for differentiation of other tissue stem cells (76–79). Notch signaling has been shown to be important for Lgr5high stem cells (80) and in our data, Hes-1, a component of Notch signaling was upregulated in Lgr5high cells (Supplemental Figure 10). miR-200b, a target of Notch signaling (81), was also downregulated in stem cells. In addition, several members of miR-17-92 clusters such as miR-17, miR-92a and miR-20b were upregulated in stem cells as compared to differentiated cells. It has been shown that this miRNA cluster is also highly expressed in embryonic stem cells and plays a role in cell proliferation (82, 83). Also, several miRNAs such as miR-10a, miR-155 and miR-191 have been shown to be important for self-renewal of hematopoietic stem cells (84–87).
To determine stem cell specific miRNAs, we compared Lgr5high vs Lgr5low daughter cells. Interestingly, three miRNAs, miR-125a-5p, miR-190 and miR-191 exhibited significantly (P <0.05) higher levels in the stem cells compared to daughter cells and differentiated cells (Figure 8B). These miRNAs were not differentially expressed in daughter stem cells compared with differentiated cells, suggesting that they might serve a specific role in intestinal stem cells. Specifically, miR-125 has been shown to be reported to play an important role in normal cell homeostasis in different tissues (88). Higher levels of miR-125 have been associated with hematopoietic and embryonic stem cells (89). Concurrently, some of the validated targets of miR-125 such as Mcl-1, TNFA1P3, Tp53inp1 and Orp9 were also significantly downregulated in Lgr5high stem cells (Supplemental Table III–A).
Next, we wanted to identify miRNAs that were altered by AOM in Lgr5high and Lgr5negative cells (see Table 3 in (99)). We observed that miR-92a, a well-characterized oncomiR in colorectal cancer (90–94) was upregulated in stem cells, while miR-124 a tumor suppressor in colorectal cancer was downregulated in stem cells as compared to differentiated cells (95–97). Interestingly, other members of miR-17-92, e.g., miR-18a and miR-20b, were also upregulated in AOM-Lgr5negative cells. This suggests that the miR-17-92 family is modulated by carcinogen in both stem cells and differentiated cells. In addition, expression of miR-193b and miR-224 were also altered in differentiated cells as shown previously in colorectal cancer (13, 98).
Another interesting outcome of the study was that dietary bioactive agents differentially modulated miRNAs based on their location in the colonic crypt. In stem cells, miR-19b, miR-27b, miR-26b and miR-203 were up-regulated by CCA compared to FPA. However, in differentiated cells, let-7a, let-7e and miR-19b were significantly down-regulated (P <0.05) in CCA fed mice, similar to our previous study in which scraped mucosa containing a heterogeneous population of cells from the rat colon was assayed (34). In order to elucidate the role of miRNAs in stem cells, it is necessary to determine the function of these miRNAs by identifying their targets. This was determined by performing global mRNA profiling by RNA sequencing from the same samples that were used to assess miRNA expression patterns. Importantly, we observed a down-regulation of miR-19b and up-regulation of PTK2B in differentiated cells, which is consistent with our previous study (34), in which scraped mucosa predominantly containing differentiated cells was used.
We also observed that the increase in apoptosis in FPA compared to CCA treatment in our previous studies (31) was not seen in stem cells. This along with the fact that no change was observed in PDE4B or PTK2B in the stem cells suggests that the function of miR-19b, miR-26b and miR-203 may be distinct from differentiated cells. Another interesting observation was that corn oil/cellulose vs fish oil/pectin feeding reduced miR-125a-5p expression (see Table 4B in (99)) and increased its putative gene targets, Mcl-1, Bcl-w, ARID3B, IGF2, and IL6R, in Lgr5high cells (Supplemental Table III–B). This is noteworthy, because these genes are generally considered to drive an anti-apoptotic phenotype (88). Collectively, these data suggest that diet-modulated miR-125a-5p may be necessary for stem cell maintenance. Hence, a comprehensive analysis and identification of additional downstream targets of miR-125a-5p warrants further investigation.
In summary, our data indicate for the first time that colonic stem cells exhibit a unique miRNA signature. In addition, the chemoprotective effect of combined fish oil and pectin feeding, which antagonizes the oncogenic effects of carcinogen, modulated the expression of select miRNAs. To our knowledge, this is the first study to utilize Lgr5+ reporter mice to determine the impact of diet and carcinogen on miRNA expression in intestinal stem cells. The effect of the highly chemoprotective fish oil + pectin combination on miRNA expression and their targets on mucosal physiology is summarized in Supplemental Figure 13.
Supplementary Material
Highlights.
Carcinogen modulates stem cell miRNAs
n-3 PUFA and butyrate modulates stem cell miRNAs following exposure to carcinogen
miR-125a-5p, miR-190 and miR-191 are colonic stem cell (Lgr5+) specific miRNAs.
miR-125a-5p and their targets are modulated by chemoprotective diet.
PDE4B and TCF4 -directly target miR-26b and 203; miR-19b indirectly targets PTK2B.
Acknowledgments
Funding
We gratefully acknowledge grant support from the National Institutes of Health grants (CA168312, CA129444, P30ES023512 and F32DK107108) and the American Institute for Cancer Research.
Footnotes
Conflict of interest Statement: None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 2.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 3.Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2746–51. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, et al. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- 6.Piepoli A, Tavano F, Copetti M, Mazza T, Palumbo O, Panza A, et al. Mirna expression profiles identify drivers in colorectal and pancreatic cancers. PLoS One. 2012;7:e33663. doi: 10.1371/journal.pone.0033663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michael MZ, SMOC, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–91. [PubMed] [Google Scholar]
- 8.Bandres E, Cubedo E, Agirre X, Malumbres R, Zarate R, Ramirez N, et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903–6. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 10.Ng EK, Tsang WP, Ng SS, Jin HC, Yu J, Li JJ, et al. MicroRNA-143 targets DNA methyltransferases 3A in colorectal cancer. Br J Cancer. 2009;101:699–706. doi: 10.1038/sj.bjc.6605195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arndt GM, Dossey L, Cullen LM, Lai A, Druker R, Eisbacher M, et al. Characterization of global microRNA expression reveals oncogenic potential of miR-145 in metastatic colorectal cancer. BMC Cancer. 2009;9:374. doi: 10.1186/1471-2407-9-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO molecular medicine. 2012;4:143–59. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caruso S, Bazan V, Rolfo C, Insalaco L, Fanale D, Bronte G, et al. MicroRNAs in colorectal cancer stem cells: new regulators of cancer stemness? Oncogenesis. 2012;1:e32. doi: 10.1038/oncsis.2012.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iliopoulos D, Rotem A, Struhl K. Inhibition of miR-193a expression by Max and RXRalpha activates K-Ras and PLAU to mediate distinct aspects of cellular transformation. Cancer research. 2011;71:5144–53. doi: 10.1158/0008-5472.CAN-11-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Runtsch MC, Round JL, O’Connell RM. MicroRNAs and the regulation of intestinal homeostasis. Frontiers in genetics. 2014;5:347. doi: 10.3389/fgene.2014.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez-Garcia C, Klein AM, Simons BD, Winton DJ. Intestinal stem cell replacement follows a pattern of neutral drift. Science. 2010;330:822–5. doi: 10.1126/science.1196236. [DOI] [PubMed] [Google Scholar]
- 17.Arnold CP, Tan R, Zhou B, Yue SB, Schaffert S, Biggs JR, et al. MicroRNA programs in normal and aberrant stem and progenitor cells. Genome Res. 2011;21:798–810. doi: 10.1101/gr.111385.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shackleton M, Quintana E, Fearon ER, Morrison SJ. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell. 2009;138:822–9. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi RU, Miyazaki H, Ochiya T. The role of microRNAs in the regulation of cancer stem cells. Frontiers in genetics. 2014;4:295. doi: 10.3389/fgene.2013.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, Li W, Nan F, Ren F, Wang H, Xu Y, et al. MicroRNA expression profile of colon cancer stem-like cells in HT29 adenocarcinoma cell line. Biochem Biophys Res Commun. 2011;404:273–8. doi: 10.1016/j.bbrc.2010.11.106. [DOI] [PubMed] [Google Scholar]
- 21.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–7. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 22.Chapkin RS, Davidson LA, Ly L, Weeks BR, Lupton JR, McMurray DN. Immunomodulatory effects of (n-3) fatty acids: putative link to inflammation and colon cancer. The Journal of nutrition. 2007;137:200S–4S. doi: 10.1093/jn/137.1.200S. [DOI] [PubMed] [Google Scholar]
- 23.Chapkin RS, McMurray DN, Lupton JR. Colon cancer, fatty acids and anti-inflammatory compounds. Curr Opin Gastroenterol. 2007;23:48–54. doi: 10.1097/MOG.0b013e32801145d7. [DOI] [PubMed] [Google Scholar]
- 24.Tavani A, Pelucchi C, Parpinel M, Negri E, Franceschi S, Levi F, et al. n-3 polyunsaturated fatty acid intake and cancer risk in Italy and Switzerland. Int J Cancer. 2003;105:113–6. doi: 10.1002/ijc.11018. [DOI] [PubMed] [Google Scholar]
- 25.Reddy BS, Patlolla JM, Simi B, Wang SH, Rao CV. Prevention of colon cancer by low doses of celecoxib, a cyclooxygenase inhibitor, administered in diet rich in omega-3 polyunsaturated fatty acids. Cancer research. 2005;65:8022–7. doi: 10.1158/0008-5472.CAN-05-0212. [DOI] [PubMed] [Google Scholar]
- 26.Fan YY, Zhan Y, Aukema HM, Davidson LA, Zhou L, Callaway E, et al. Proapoptotic effects of dietary (n-3) fatty acids are enhanced in colonocytes of manganese-dependent superoxide dismutase knockout mice. The Journal of nutrition. 2009;139:1328–32. doi: 10.3945/jn.109.106203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan YY, Ran Q, Toyokuni S, Okazaki Y, Callaway ES, Lupton JR, et al. Dietary fish oil promotes colonic apoptosis and mitochondrial proton leak in oxidatively stressed mice. Cancer Prev Res (Phila) 2011;4:1267–74. doi: 10.1158/1940-6207.CAPR-10-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davidson LA, Nguyen DV, Hokanson RM, Callaway ES, Isett RB, Turner ND, et al. Chemopreventive n-3 polyunsaturated fatty acids reprogram genetic signatures during colon cancer initiation and progression in the rat. Cancer Res. 2004;64:6797–804. doi: 10.1158/0008-5472.CAN-04-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crim KC, Sanders LM, Hong MY, Taddeo SS, Turner ND, Chapkin RS, et al. Upregulation of p21Waf1/Cip1 expression in vivo by butyrate administration can be chemoprotective or chemopromotive depending on the lipid component of the diet. Carcinogenesis. 2008;29:1415–20. doi: 10.1093/carcin/bgn144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho Y, Kim H, Turner ND, Mann JC, Wei J, Taddeo SS, et al. A chemoprotective fish oil- and pectin-containing diet temporally alters gene expression profiles in exfoliated rat colonocytes throughout oncogenesis. The Journal of nutrition. 2011;141:1029–35. doi: 10.3945/jn.110.134973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong MY, Lupton JR, Morris JS, Wang N, Carroll RJ, Davidson LA, et al. Dietary fish oil reduces O6-methylguanine DNA adduct levels in rat colon in part by increasing apoptosis during tumor initiation. Cancer Epidemiol Biomarkers Prev. 2000;9:819–26. [PubMed] [Google Scholar]
- 32.Chang WC, Chapkin RS, Lupton JR. Predictive value of proliferation, differentiation and apoptosis as intermediate markers for colon tumorigenesis. Carcinogenesis. 1997;18:721–30. doi: 10.1093/carcin/18.4.721. [DOI] [PubMed] [Google Scholar]
- 33.Davidson LA, Wang N, Shah MS, Lupton JR, Ivanov I, Chapkin RS. n-3 Polyunsaturated fatty acids modulate carcinogen-directed non-coding microRNA signatures in rat colon. Carcinogenesis. 2009;30:2077–84. doi: 10.1093/carcin/bgp245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shah MS, Schwartz SL, Zhao C, Davidson LA, Zhou B, Lupton JR, et al. Integrated microRNA and mRNA expression profiling in a rat colon carcinogenesis model: effect of a chemo-protective diet. Physiol Genomics. 2011;43:640–54. doi: 10.1152/physiolgenomics.00213.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiang Y, Song Y, Wang Z, Chen Y, Yue Z, Xu H, et al. Aberrant expression of miR-203 and its clinical significance in gastric and colorectal cancers. J Gastrointest Surg. 2011;15:63–70. doi: 10.1007/s11605-010-1367-8. [DOI] [PubMed] [Google Scholar]
- 36.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 37.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 38.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–5. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 39.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan YY, Zhang J, Barhoumi R, Burghardt RC, Turner ND, Lupton JR, et al. Antagonism of CD95 signaling blocks butyrate induction of apoptosis in young adult mouse colonic cells. Am J Physiol. 1999;277:C310–9. doi: 10.1152/ajpcell.1999.277.2.C310. [DOI] [PubMed] [Google Scholar]
- 41.Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O’Leary JJ, Ruan Q, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2010;11:141–7. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 42.Vanamala J, Glagolenko A, Yang P, Carroll RJ, Murphy ME, Newman RA, et al. Dietary fish oil and pectin enhance colonocyte apoptosis in part through suppression of PPARdelta/PGE2 and elevation of PGE3. Carcinogenesis. 2008;29:790–6. doi: 10.1093/carcin/bgm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, et al. miR-19 is a key oncogenic component of mir-17-92. Genes & development. 2009;23:2839–49. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu N, Zhao X, Liu M, Liu H, Yao W, Zhang Y, et al. Role of microRNA-26b in glioma development and its mediated regulation on EphA2. PLoS One. 2011;6:e16264. doi: 10.1371/journal.pone.0016264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Munoz J, Stange DE, Schepers AG, van de Wetering M, Koo BK, Itzkovitz S, et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. The EMBO journal. 2012;31:3079–91. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang WL, Chapkin RS, Lupton JR. Fish oil blocks azoxymethane-induced rat colon tumorigenesis by increasing cell differentiation and apoptosis rather than decreasing cell proliferation. The Journal of nutrition. 1998;128:491–7. doi: 10.1093/jn/128.3.491. [DOI] [PubMed] [Google Scholar]
- 47.Davidson LA, Brown RE, Chang WC, Morris JS, Wang N, Carroll RJ, et al. Morphodensitometric analysis of protein kinase C beta(II) expression in rat colon: modulation by diet and relation to in situ cell proliferation and apoptosis. Carcinogenesis. 2000;21:1513–9. [PubMed] [Google Scholar]
- 48.Sanders LM, Henderson CE, Hong MY, Barhoumi R, Burghardt RC, Wang N, et al. An increase in reactive oxygen species by dietary fish oil coupled with the attenuation of antioxidant defenses by dietary pectin enhances rat colonocyte apoptosis. The Journal of nutrition. 2004;134:3233–8. doi: 10.1093/jn/134.12.3233. [DOI] [PubMed] [Google Scholar]
- 49.Ma YL, Zhang P, Wang F, Moyer MP, Yang JJ, Liu ZH, et al. Human embryonic stem cells and metastatic colorectal cancer cells shared the common endogenous human microRNA-26b. J Cell Mol Med. 2011;15:1941–54. doi: 10.1111/j.1582-4934.2010.01170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koh CM, Iwata T, Zheng Q, Bethel C, Yegnasubramanian S, De Marzo AM. Myc enforces overexpression of EZH2 in early prostatic neoplasia via transcriptional and post-transcriptional mechanisms. Oncotarget. 2011;2:669–83. doi: 10.18632/oncotarget.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu XX, Li XJ, Zhang B, Liang YJ, Zhou CX, Cao DX, et al. MicroRNA-26b is underexpressed in human breast cancer and induces cell apoptosis by targeting SLC7A11. FEBS Lett. 2011;585:1363–7. doi: 10.1016/j.febslet.2011.04.018. [DOI] [PubMed] [Google Scholar]
- 52.Tsunoda T, Ota T, Fujimoto T, Doi K, Tanaka Y, Yoshida Y, et al. Inhibition of Phosphodiesterase-4 (PDE4) activity triggers luminal apoptosis and AKT dephosphorylation in a 3-D colonic-crypt model. Mol Cancer. 2012;11:46. doi: 10.1186/1476-4598-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Narita M, Murata T, Shimizu K, Nakagawa T, Sugiyama T, Inui M, et al. A role for cyclic nucleotide phosphodiesterase 4 in regulation of the growth of human malignant melanoma cells. Oncol Rep. 2007;17:1133–9. [PubMed] [Google Scholar]
- 54.Smith PG, Wang F, Wilkinson KN, Savage KJ, Klein U, Neuberg DS, et al. The phosphodiesterase PDE4B limits cAMP-associated PI3K/AKT-dependent apoptosis in diffuse large B-cell lymphoma. Blood. 2005;105:308–16. doi: 10.1182/blood-2004-01-0240. [DOI] [PubMed] [Google Scholar]
- 55.Turk HF, Kolar SS, Fan YY, Cozby CA, Lupton JR, Chapkin RS. Linoleic acid and butyrate synergize to increase Bcl-2 levels in colonocytes. Int J Cancer. 2010 doi: 10.1002/ijc.25323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Palladino MA, Bahjat FR, Theodorakis EA, Moldawer LL. Anti-TNF-alpha therapies: the next generation. Nat Rev Drug Discov. 2003;2:736–46. doi: 10.1038/nrd1175. [DOI] [PubMed] [Google Scholar]
- 57.Kim S, Jee K, Kim D, Koh H, Chung J. Cyclic AMP inhibits Akt activity by blocking the membrane localization of PDK1. The Journal of biological chemistry. 2001;276:12864–70. doi: 10.1074/jbc.M001492200. [DOI] [PubMed] [Google Scholar]
- 58.Robinson-White AJ, Hsiao HP, Leitner WW, Greene E, Bauer A, Krett NL, et al. Protein kinase A-independent inhibition of proliferation and induction of apoptosis in human thyroid cancer cells by 8-Cl-adenosine. J Clin Endocrinol Metab. 2008;93:1020–9. doi: 10.1210/jc.2007-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jia Q, Ivanov I, Zlatev ZZ, Alaniz RC, Weeks BR, Callaway ES, et al. Dietary fish oil and curcumin combine to modulate colonic cytokinetics and gene expression in dextran sodium sulphate-treated mice. Br J Nutr. 2011;106:519–29. doi: 10.1017/S0007114511000390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bao B, Ali S, Banerjee S, Wang Z, Logna F, Azmi AS, et al. Curcumin analogue CDF inhibits pancreatic tumor growth by switching on suppressor microRNAs and attenuating EZH2 expression. Cancer research. 2012;72:335–45. doi: 10.1158/0008-5472.CAN-11-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Z, Zhang B, Li W, Fu L, Fu L, Zhu Z, et al. Epigenetic Silencing of miR-203 Upregulates SNAI2 and Contributes to the Invasiveness of Malignant Breast Cancer Cells. Genes Cancer. 2011;2:782–91. doi: 10.1177/1947601911429743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bueno MJ, Perez de Castro I, Gomez de Cedron M, Santos J, Calin GA, Cigudosa JC, et al. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 2008;13:496–506. doi: 10.1016/j.ccr.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 63.Lena AM, Shalom-Feuerstein R, Rivetti di Val Cervo P, Aberdam D, Knight RA, Melino G, et al. miR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ. 2008;15:1187–95. doi: 10.1038/cdd.2008.69. [DOI] [PubMed] [Google Scholar]
- 64.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–36. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feber A, Xi L, Luketich JD, Pennathur A, Landreneau RJ, Wu M, et al. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg. 2008;135:255–60. doi: 10.1016/j.jtcvs.2007.08.055. discussion 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mathe EA, Nguyen GH, Bowman ED, Zhao Y, Budhu A, Schetter AJ, et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res. 2009;15:6192–200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wong KY, Liang R, So CC, Jin DY, Costello JF, Chim CS. Epigenetic silencing of MIR203 in multiple myeloma. Br J Haematol. 2011;154:569–78. doi: 10.1111/j.1365-2141.2011.08782.x. [DOI] [PubMed] [Google Scholar]
- 68.Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, et al. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–83. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 69.van Es JH, Jay P, Gregorieff A, van Gijn ME, Jonkheer S, Hatzis P, et al. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat Cell Biol. 2005;7:381–6. doi: 10.1038/ncb1240. [DOI] [PubMed] [Google Scholar]
- 70.Hatzis P, van der Flier LG, van Driel MA, Guryev V, Nielsen F, Denissov S, et al. Genome-wide pattern of TCF7L2/TCF4 chromatin occupancy in colorectal cancer cells. Mol Cell Biol. 2008;28:2732–44. doi: 10.1128/MCB.02175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324:1670–3. doi: 10.1126/science.1171837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Willis ND, Przyborski SA, Hutchison CJ, Wilson RG. Colonic and colorectal cancer stem cells: progress in the search for putative biomarkers. J Anat. 2008;213:59–65. doi: 10.1111/j.1469-7580.2008.00917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McDonald SA, Preston SL, Lovell MJ, Wright NA, Jankowski JA. Mechanisms of disease: from stem cells to colorectal cancer. Nat Clin Pract Gastroenterol Hepatol. 2006;3:267–7. doi: 10.1038/ncpgasthep0473. [DOI] [PubMed] [Google Scholar]
- 74.Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–11. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 75.Reddy BS, Burill C, Rigotty J. Effect of diets high in omega-3 and omega-6 fatty acids on initiation and postinitiation stages of colon carcinogenesis. Cancer research. 1991;51:487–91. [PubMed] [Google Scholar]
- 76.Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 2008;452:225–9. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–58. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 78.Ma Y, Yao N, Liu G, Dong L, Liu Y, Zhang M, et al. Functional screen reveals essential roles of miR-27a/24 in differentiation of embryonic stem cells. The EMBO journal. 2015;34:361–78. doi: 10.15252/embj.201489957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jones MF, Hara T, Francis P, Li XL, Bilke S, Zhu Y, et al. The CDX1-microRNA-215 axis regulates colorectal cancer stem cell differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E1550–8. doi: 10.1073/pnas.1503370112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.VanDussen KL, Carulli AJ, Keeley TM, Patel SR, Puthoff BJ, Magness ST, et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012;139:488–97. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gao XJ, Liu JW, Zhang QG, Zhang JJ, Xu HT, Liu HJ. Nobiletin inhibited hypoxia-induced epithelial-mesenchymal transition of lung cancer cells by inactivating of Notch-1 signaling and switching on miR-200b. Die Pharmazie. 2015;70:256–62. [PubMed] [Google Scholar]
- 82.Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nature methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 83.Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Developmental biology. 2007;310:442–53. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vasilatou D, Papageorgiou S, Pappa V, Papageorgiou E, Dervenoulas J. The role of microRNAs in normal and malignant hematopoiesis. European journal of haematology. 2010;84:1–16. doi: 10.1111/j.1600-0609.2009.01348.x. [DOI] [PubMed] [Google Scholar]
- 85.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–6. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 86.Guo S, Lu J, Schlanger R, Zhang H, Wang JY, Fox MC, et al. MicroRNA miR-125a controls hematopoietic stem cell number. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14229–34. doi: 10.1073/pnas.0913574107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang L, Flygare J, Wong P, Lim B, Lodish HF. miR-191 regulates mouse erythroblast enucleation by down-regulating Riok3 and Mxi1. Genes & development. 2011;25:119–24. doi: 10.1101/gad.1998711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sun YM, Lin KY, Chen YQ. Diverse functions of miR-125 family in different cell contexts. Journal of hematology & oncology. 2013;6:6. doi: 10.1186/1756-8722-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shaham L, Binder V, Gefen N, Borkhardt A, Izraeli S. MiR-125 in normal and malignant hematopoiesis. Leukemia. 2012;26:2011–8. doi: 10.1038/leu.2012.90. [DOI] [PubMed] [Google Scholar]
- 90.Lao IW, Cui F, Zhu H. Quantitation of microRNA-92a in colorectal adenocarcinoma and its precancerous lesions: Co-utilization of hybridization and spectral imaging. Oncology letters. 2015;9:1109–15. doi: 10.3892/ol.2014.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Clancy C, Joyce MR, Kerin MJ. The use of circulating microRNAs as diagnostic biomarkers in colorectal cancer. Cancer biomarkers: section A of Disease markers. 2015;15:103. doi: 10.3233/CBM-140456. [DOI] [PubMed] [Google Scholar]
- 92.Abdul Majid AS, Kwag JH, Jung SH, Yim HB, Kim YD, Kang KD. Correlation between disc damage likelihood scale and optical coherence tomography in the diagnosis of glaucoma. Ophthalmologica. 2010;224:274–82. doi: 10.1159/000287350. [DOI] [PubMed] [Google Scholar]
- 93.Ke TW, Wei PL, Yeh KT, Chen WT, Cheng YW. MiR-92a Promotes Cell Metastasis of Colorectal Cancer Through PTEN-Mediated PI3K/AKT Pathway. Annals of surgical oncology. 2014 doi: 10.1245/s10434-014-4305-2. [DOI] [PubMed] [Google Scholar]
- 94.Yamada N, Nakagawa Y, Tsujimura N, Kumazaki M, Noguchi S, Mori T, et al. Role of Intracellular and Extracellular MicroRNA-92a in Colorectal Cancer. Translational oncology. 2013;6:482–92. doi: 10.1593/tlo.13280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Taniguchi K, Sugito N, Kumazaki M, Shinohara H, Yamada N, Nakagawa Y, et al. MicroRNA-124 inhibits cancer cell growth through PTB1/PKM1/PKM2 feedback cascade in colorectal cancer. Cancer letters. 2015;363:17–27. doi: 10.1016/j.canlet.2015.03.026. [DOI] [PubMed] [Google Scholar]
- 96.Koukos G, Polytarchou C, Kaplan JL, Morley-Fletcher A, Gras-Miralles B, Kokkotou E, et al. MicroRNA-124 regulates STAT3 expression and is down-regulated in colon tissues of pediatric patients with ulcerative colitis. Gastroenterology. 2013;145:842–52. e2. doi: 10.1053/j.gastro.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Park SY, Kim H, Yoon S, Bae JA, Choi SY, Jung YD, et al. KITENIN-targeting microRNA-124 suppresses colorectal cancer cell motility and tumorigenesis. Molecular therapy: the journal of the American Society of Gene Therapy. 2014;22:1653–64. doi: 10.1038/mt.2014.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kara M, Yumrutas O, Ozcan O, Celik OI, Bozgeyik E, Bozgeyik I, et al. Differential expressions of cancer-associated genes and their regulatory miRNAs in colorectal carcinoma. Gene. 2015;567:81–6. doi: 10.1016/j.gene.2015.04.065. [DOI] [PubMed] [Google Scholar]
- 99.Shah MS, Kim E, Davidson LA, Knight JM, Zoh RS, Golsdby JS, et al. Modulation of colonic stem cell microRNA and mRNA expression by dietary bioactive agents. Data in brief. doi: 10.1016/j.dib.2015.12.026. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.