Abstract
In mammals, global DNA demethylation in vivo occurs in the pre-implantation embryo and in primordial germ cells (PGCs) where it is hypothesized to create a blank slate or “tabula rasa” upon which new DNA methylation patterns are written. However, global DNA demethylation in vivo is far from complete with a small number of loci protected from demethylation. Failure to demethylate, or overt demethylation results in compromised differentiation. Recent work has shown that reversion of primed human pluripotent stem cells to the naïve state leads to unbridled DNA demethylation which has unknown consequences on the quality differentiated cells created in vitro. Taken together understanding DNA methylation remodeling is critical for understanding the epigenetic foundations of life, and the quality of stem cells for regenerative medicine.
The kinetics of DNA demethylation in vivo
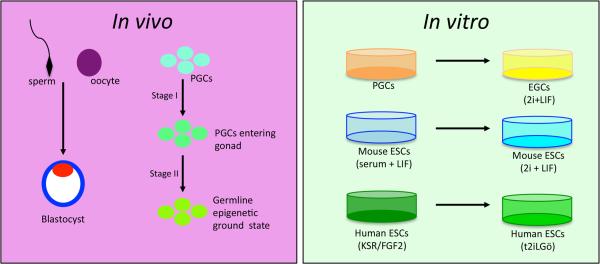
Global DNA demethylation in vivo occurs in the pre-implantation embryo and in primordial germ cells (PGCs) of mammals (Figure 1). Modifying the molecular pathways that regulate global DNA demethylation is incompatible with development and differentiation, illustrating the crucial role for global DNA demethylation in health and fertility [1,2]. Historically it was thought global DNA demethylation in vivo results in the removal of cytosine methylation from the entire genome [3]. However, using unbiased genome-wide sequencing approaches at single-base resolution it is now appreciated that DNA demethylation in vivo does not create a blank slate. Instead, persistently methylated cytosines are found at certain classes of transposons, as well as some rare intragenic regions such as exons, promoters, splice sites and CG islands [4–12]. The result is a unique hypomethylated landscape in the pre-implantation embryo and PGCs which is required to launch the next stages of development and differentiation.
Figure 1.
Global DNA methylation remodeling occurs in vivo during pre-implantation embryo development and in primordial germ cells (PGCs). In PGCs demethylation occurs in two stages as represented by two arrows. Global DNA methylation remodeling in vitro occurs during culture-induced reversion of PGCs to embryonic germ cells (EGCs) in medium containing 2i+LIF (called “2i” in the text), reversion of mouse ESCs cultured in a medium containing serum + LIF (called “serum” in the text) to 2i, and reversion of primed hESCs cultured in FGF2/Knockout Serum replacer (KSR) to the naïve state in a medium called titrated 2i + LIF + Gö6983.
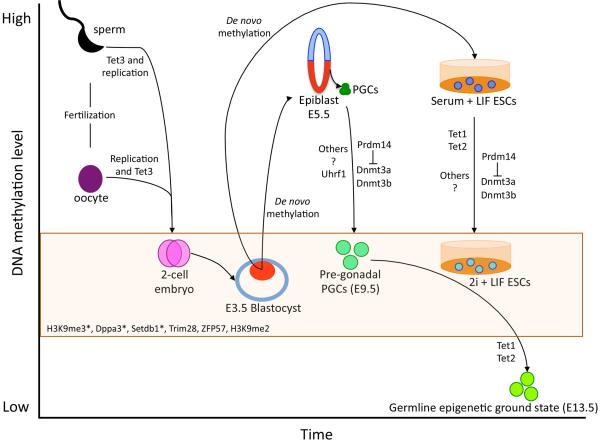
The epigenome of the pre-implantation embryo is created after fertilization of a haploid oocyte and sperm [13] (Figure 2). Removal of cytosine methylation from the maternal mouse genome after fertilization follows a slow kinetics, closely coupled to DNA replication [14–18]. In contrast the hyper-methylated paternal genome is rapidly demethylated at fertilization prior to the first cleavage to create the 2-cell embryo [14–18]. Recent work indicates that both the maternal and paternal genomes are demethylated prior to forming the 2-cell embryo, and that DNA demethylation involves oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) by Tet methylcytosine dioxygenase 3 (Tet3) [1,15–17,19]. Specifically, DNA demethylation of the maternal genome prior to the 2-cell stage involves a minor role for oxidation by Tet3 with a major role for passive replication-coupled DNA demethylation, in contrast, demethylation of the paternal genome involves a major role for Tet3 and replication-coupled DNA demethylation (Figure 2). After the 2-cell stage, the methylation landscape continues to change in the pre-implantation embryo through loss of DNA methylation during cleavage divisions ultimately generating a blastocyst composed of trophoblast and inner cell mass cells. The inner cell mass of the pre-implantation blastocyst at embryonic (E) day 3.5 exhibits the lowest average CpG methylation of all pre-implantation embryonic stages [4]. This hypomethylated state is relatively transient, with global re-methylation occurring rapidly by E5.5 in the epiblast during implantation. This restores DNA methylation to the levels observed in somatic cells of the embryo [4] (Figure 2). At this point, the methylated epiblast is poised to generate all cell types in the body, including PGCs.
Figure 2.
Kinetics of DNA demethylation in vitro and in vivo and the molecules involved in regulating this process. Orange box represents the stages in which the genome still contains methylated cytosines that are protected from global DNA demethylation. These include imprinting control centers (ICCs) and some transposons. In the box are the molecules identified to protect ICCs and transposons from precocious demethylation (see text for more details). Inner cell mass in blastocyst and epiblast in the embryo are shown in red. (*) Indicates that the protection mechanism functions both in pre-implantation embryos and PGCs.
In the post-implantation embryo PGCs are specified from the epiblast at ~E6.25, resulting in an average of forty definitive PGCs by E7.25 that exhibit global DNA methylation levels similar to the surrounding somatic cells [10,12,20,21]. Then, around 24–36 hours after specification, the PGC genome globally demethylates to around 50% of the levels quantified in the epiblast. In PGCs, this DNA demethylation event is referred to as stage I DNA methylation reprogramming (Figure 1). Next, the PGC genome demethylates further by locus-specific DNA demethylation from E9.5 to E13.5. This is referred to as stage II DNA methylation reprogramming [12,14,21]. The combined effects of stage I (global DNA demethylation) plus stage II (local DNA demethylation) creates an epigenetic landscape at E13.5 that has been termed the germline epigenetic ground state (Figure 2).
Although the E3.5 blastocyst and E13.5 PGCs are hypomethylated, the absolute levels of cytosine methylation in PGCs is lower because fewer cytosines are protected from DNA demethylation during PGC differentiation (Figure 2). For example, in the pre-implantation embryo cytosine methylation is protected at imprinting control centers (ICCs), some transposable elements [4,5,11] and transient maternally methylated CG islands (CGI) [4,5,13]. In PGCs, cytosine methylation is lost from ICCs and almost all transposons, with only the youngest transposable elements and pericentromeric heterochromatin retaining DNA methylation [9,10,12,14,21]. These persistently methylated sites in PGCs lead to interesting health-related questions regarding DNA methylation inheritance in the germline and susceptibility to disease in the next generation. For example the creation of abnormal epialleles, or the demethylation of an active transposon could cause permanent epigenetic or genetic changes to germline DNA respectively.
DNA demethylation in vitro
Uncovering mechanisms of DNA demethylation in vivo is difficult to achieve because of low cell numbers and technical challenges in working with pre-implantation embryos. An alternate model to study DNA demethylation involves the use of in vitro pluripotent cell types. Specifically, the reversion of PGCs to embryonic germ cells (EGCs), reversion of serum cultured mouse embryonic stem cells (ESCs), to the naïve ground state, and more recently, reversion of primed human ESCs (hESCs) to the naïve state (Figure 1). The advantage of studying DNA demethylation in vitro is the large number of cells that are amenable to biochemical studies. The caveat is the heterogeneity in which cells enter and exit from the process of DNA demethylation, and the different methylation landscapes that emerge at the conclusion of reversion, which may have important implications to the use of hypomethylated pluripotent stem cells in regenerative medicine.
The majority of mechanistic insights on pathways that regulate global DNA demethylation in vitro have been uncovered by reverting mouse ESCs from serum containing medium plus leukemia inhibitory factor (LIF) (herein referred to as “serum”), to a defined medium with LIF and inhibitors of mitogen activated protein kinase and glycogen synthase kinase-3 (2i). [22–24]. Global DNA demethylation following reversion from serum to 2i takes around 72 hours. This dramatic and widespread loss of DNA methylation also occurs when switching primed human ESCs to the naïve state in medium containing titrated 2i, LIF and Gö6983 [25]. However, the timing of global DNA demethylation during human reversion remains to be determined. Furthermore, in the mouse, the degree of DNA demethylation with reversion in vitro is sex-dependent, meaning that reverted female ESC lines are more hypomethylated than reverted male ESC lines [24,26]. The mechanism(s) that contribute to this sex-specific difference, and whether this also applies to human cells are unknown but warrant more investigation.
Mechanisms that remove DNA methylation globally from the genome
One of the major mechanisms for promoting DNA demethylation when reverting mouse ESCs to 2i involves up-regulation of the DNA binding protein Prdm14 (positive regulatory domain 1 binding factor 1 (PRD1-BF1) and retinoblastoma interacting zinc finger (RIZ) homology domain containing 14) [22,23,27,28]. Deleting Prdm14 in 2i-cultured ESCs causes up-regulation of the de novo DNA methyltransferases Dnmt3b, Dnmt3a and an increase in the level of DNA methylation in the genome [22,26,27]. Deleting Dnmt3a, Dnmt3b or both in serum cultured ESCs is not sufficient to cause the same degree of DNA demethylation that occurs when switching ESCs from serum to 2i [23], suggesting that additional mechanisms must be at-play. Expressing specific splice variants of Dnmt3a and Dnmt3b together with Dnmt3l in 2i-cultured wild type ESCs is sufficient to increase the levels of DNA methylation at specific loci, however the affect on the whole genome is unknown [27].
An additional mechanism that removes DNA methylation from the genome together with Prdm14 involves the pluripotency-specific dioxygenases Tet1 and Tet2 [23,26]. Seventy-two hours after switching ESCs from serum to 2i, a significant enrichment in 5hmC is observed in the genome [23]. However, deleting both Tet1 and Tet2 prior to reversion only delays DNA demethylation [23]. Instead, Tets have localized effects on demethylating the genomic landscape [26]. Therefore, it appears that additional mechanisms are yet to be uncovered that function independently or synergistically with Prdm14, Tet1 and Tet2 to demethylate mammalian genomic DNA.
Locus specific protection from DNA demethylation
During in vitro DNA demethylation in 2i, ICCs are protected from demethylation through what is speculated to be the activity of Dnmt1/Uhrf1 maintenance methylation [22,23,26]. However, our recent work suggests that reversion of primed hESCs to the naïve state causes locus-specific erosion of methylation beyond what is observed in both the pre-implantation embryo and in human PGCs [6]. Thus understanding the mechanisms that protect loci during global DNA demethylation is critical to develop strategies that prevent abnormal loss of DNA methylation in vitro.
Clues for mechanisms that protect loci from global DNA demethylation came from studying the pre-implantation the embryo, beginning with a maternally supplied protein called Developmental pluripotency associated 3 (Dppa3). This protein functions to protect some (but not all) ICCs from DNA demethylation prior to E3.5 [29]. At Dppa3 target sites, it is now appreciated that histone H3 lysine 9 dimethylation (H3K9me2) functions as a substrate for Dppa3, with loss of H3K9me2 leading to failed recruitment of Dppa3 and loss of DNA methylation [30]. Protecting sites from overt DNA demethylation in vivo is critical as an oocyte-specific deletion of Dppa3 leads to failed pre-implantation embryo development. A second maternal protein called Trim28 (Kap1) was recently shown to recruit SET domain bifurcated 1 (Setdb1) to chromatin, causing local deposition of H3K9me3 [31]. Trim28 is tethered to DNA by Krüppel-associated box domain zinc finger proteins (ZFPs), with ZFP57 being expressed by both the oocyte and embryo and enriched at the same ICCs as Trim28 [31]. Deletion of maternal and zygotic supplies of ZFP57 also results in a loss in methylation at multiple ICCs, and similar to Dppa3 leads to failed preimplantation embryo development [32]. Therefore, protection from locus-specific demethylation in the pre-implantation embryo (and possibly during reversion in 2i) involves complex cross-talk between DNA binding proteins and chromatin.
Mechanisms that promote DNA demethylation in PGCs
Similar to reversion in 2i, one of the major mechanisms responsible for promoting global DNA demethylation in mouse PGCs is Prdm14 [2]. Deleting Prdm14 in the embryo causes transcriptional derepression of the de novo methyltransferases Dnmt3a and Dnmt3b and a failure of the PGCs to undergo global DNA demethylation and the germline cells die [2]. However, unlike DNA demethylation in 2i, the maintenance methylation machinery is also handicapped in PGCs, with loss of Uhrf1 beginning soon after PGC specification [33]. These two events, repression of Dnmt3a/Dnmt3b and repression of Uhrf1 are speculated to be the major mechanisms for stage I global DNA demethylation in PGCs (Figure 1). The second stage of PGC demethylation occurs as cells enter the genital ridge and involves an orderly increase in Tet1 and Tet2 in single PGCs from E9.5-E13.5, oxidation of 5mC to 5hmC and rapid cell division [9,10,34–36].
Our recent work suggests that the initial global loss of cytosine methylation from the PGC genome in stage I occurs independently from Tet1 and Tet2, and that loss of methylation from ICCs is an intrinsic feature of demethylating germline cells that requires cell division in stage II [10,36,37]. These stage-specific events in PGC methylation remodeling combined with the critical role of Prdm14 in regulating DNA demethylation lead to an intriguing hypothesis that global DNA demethylation in vitro in pluripotent stem cells resembles the first stage of methylation remodeling in PGCs. If this is the case, what is acting to prevent 2i cultured ESCs and the germline cells from precocious demethylation of ICCs and certain transposons?
One candidate for protecting DNA methylation during stage I of PGC methylation remodeling is Dppa3, although its role appears very mild [38]. An alternate mechanism similar to the strategies used by pre-implantation embryos is H3K9me3, which is deposited primarily by Setdb1 in PGCs [39]. In ESCs cultured in serum, Setdb1 protects the long terminal repeats of retrotransposons and imprinted genes from demethylation in the absence of Dnmt3a and Dnmt3b [40]. Similarly in PGCs, loss of Setdb1 results in the derepression of some endogenous retroviruses and a heterogeneous reduction in DNA methylation particularly at sites that are normally highly enriched in H3K9me3 [39].
Concluding remarks
Global DNA methylation remodeling is an exquisite process where failure to demethylate leads to problems in embryo development and differentiation, and failure to protect loci from DNA demethylation results in the same fate. Therefore maintaining the correct balance of DNA demethylation and protection has tremendous implications when considering the use of globally hypomethylated pluripotent stem cells in regenerative medicine. Evaluating mechanisms of DNA demethylation in vitro indicate that the combined action of repressed Dnmt3b and Dnmt3a and oxidation of 5mC to 5hmC are localized in their effects, leaving open a major question of how a pluripotent stem cell genome is globally depleted of DNA methylation, and what barriers are in place to prevent unbridled DNA demethylation in vitro. Solving these questions is critical to developing strategies that prevent DNA methylation remodeling from going awry in vitro, and for understanding the epigenetic foundations of life in vivo.
Acknowledgements
ATC is supported by funds from the NIH/NICHD, HD058047, HD079546 and HD075795.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 2.Yamaji M, Seki Y, Kurimoto K, Yabuta Y, Yuasa M, Shigeta M, Yamanaka K, Ohinata Y, Saitou M. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat Genet. 2008;40:1016–1022. doi: 10.1038/ng.186. [DOI] [PubMed] [Google Scholar]
- 3.Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic and extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]; ** Classic analysis of DNA methylation in the mouse embryo and in mouse germline cells. I highly recommend that everyone who works on DNA methylation should read this original work. While reading this classic paper, it is important to keep in mind that these experiment were performed nearly thirty years ago, and these classic studies set the foundation for all future work.
- 4.Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith ZD, Chan MM, Humm KC, Karnik R, Mekhoubad S, Regev A, Eggan K, Meissner A. DNA methylation dynamics of the human preimplantation embryo. Nature. 2014;511:611–615. doi: 10.1038/nature13581. [DOI] [PMC free article] [PubMed] [Google Scholar]; * First DNA methylation map at single base resolution in the human pre-implantation embryo
- 6.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 2001;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 7.Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 8.Gkountela S, Zhang K, Shafiq T, W-W L, Hargan-Calvopina J, Chen P-Y, Clark A. DNA demethylation dynamics in the human prenatal germline. Cell. 2015;161:1425–1436. doi: 10.1016/j.cell.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamaguchi S, Hong K, Liu R, Inoue A, Shen L, Zhang K, Zhang Y. Dynamics of 5-methylcytosine and 5-hydroxymethylcytosine during germ cell reprogramming. Cell research. 2013;23:329–339. doi: 10.1038/cr.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hargan-Calvopina J, Cook H, Vincent J, Nee K, Clark A. The Aorta Gonad Mesonephros organ culture recapitulates 5hmC reorganization and replication-dependent and independent loss of DNA methylation in the germline. Stem cells and development. 2015 doi: 10.1089/scd.2014.0410. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y, Yan J, Ren Z, Lin SA, Li J, et al. The DNA methylation landscape of human early embryos. Nature. 2014;511:606–610. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]; * First DNA methylation map at single base resolution in the human pre-implantation embryo
- 12.Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Molecular cell. 2012;48:849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; * First DNA methylation map at single base resolution of mouse PGCs.
- 13.Smallwood S, Tomizawa S, Krueger F, Ruf N, Carli N, Segonds-Pichon A, Sato S, Hata K, Andrews S, Kelsey G. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat Genetics. 2011;43:811–814. doi: 10.1038/ng.864. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This work revealed the intermediate nature of oocyte genome remethylation, and introduced the concept of maternally derived transient imprints.
- 14.Wang L, Zhang J, Duan J, Gao X, Zhu W, Lu X, Yang L, Li G, Ci W, Li W, et al. Programming and inheritance of parental DNA methylomes in mammals. Cell. 2014;157:979–991. doi: 10.1016/j.cell.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; * First description of DNA demethylation from the perspective of maternal and paternal genomes at base resolution.
- 15.Guo F, Li X-L, Liang D, Li T, Zhu P, Guo H, Wu X, Wen L, Bu T-P, Hu B, et al. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell stem cell. 2014 doi: 10.1016/j.stem.2014.08.003. [DOI] [PubMed] [Google Scholar]; * Analysis of paternal and maternal DNA demethylation in zygotes at single base resolution.
- 16.Shen LIA, He J, Liu Y, Lu F, Zhang Y. Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes. Cell stem cell. 2014;15:459–470. doi: 10.1016/j.stem.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Analysis of paternal and maternal DNA methylation in zygotes at single base resolution and strong evidence that DNA replication drives the demethylation of the majority of cytosines in the paternal pronucleus.
- 17.Peat J, Dean W, Clark S, Krueger F, Smallwood S, Ficz G, Kim J, Marioni J, Hore T, Reik W. Genome-wide bisulfite sequencing in zygotes identifies demethylation targets and maps the contribution of TET3 oxidation. Cell reports. 2014;9:1990–2000. doi: 10.1016/j.celrep.2014.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]; *First whole genome bisulfite sequencing analysis of mouse zygotic genomes.
- 18.Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nature communications. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- 20.Seki Y, Hayashi K, Itoh K, Mizugaki M, Saitou M, Matsui Y. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol. 2005;278:440–458. doi: 10.1016/j.ydbio.2004.11.025. [DOI] [PubMed] [Google Scholar]; **Classic analysis of epigenetic marks, including DNA methylation at single cell resolution using immunofluorescence. The work from this paper is still the standard today in the dynamics of mouse PGC epigenetic remodeling.
- 21.Kobayashi H, Sakurai T, Miura F, Imai M, Mochiduki K, Yanagisawa E, Sakashita A, Wakai T, Suzuki Y, Ito T, et al. High-resolution DNA methylome analysis of primordial germ cells identifies gender-specific reprogramming in mice. Genome research. 2013;23:616–627. doi: 10.1101/gr.148023.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leitch H, McEwen K, Turp A, Encheva V, Carroll T, Grabole N, Mansfield W, Nashun B, Knezovich J, Smith A, et al. Naive pluripotency is associated with global DNA hypomethylation. Nat Struct Mol Biol. 2013;20:311–316. doi: 10.1038/nsmb.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ficz G, Hore T, Santos F, Lee H, Dean W, Arand J, Krueger F, Oxley D, Paul Y, Walter J, et al. FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell stem cell. 2013;13:351–359. doi: 10.1016/j.stem.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; **Comprehensive work showing describing the dynamics and mechanisms of DNA methylation remodeling in ESCs with reversion to 2i.
- 24.Habibi E, Brinkman A, Arand J, Kroeze L, Kerstens H, Matarese F, Lepikhov K, Gut M, Brun-Heath I, Hubner N, et al. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell stem cell. 2013;13:360–369. doi: 10.1016/j.stem.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, Oxley D, Santos F, Clarke J, Mansfield W, et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hackett J, Dietmann S, Murakami K, Down T, Leitch H, Surani M. Synergistic mechanisms of DNA demethylation during transition to ground-state pluripotency. Stem Cell Reports. 2013;1:518–531. doi: 10.1016/j.stemcr.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamaji M, Ueda J, Hayashi K, Ohta H, Yabuta Y, Kurimoto K, Nakato R, Yamada Y, Shirahige K, Saitou M. PRDM14 ensures naive pluripotency through dual regulation of signaling and epigenetic pathways in mouse embryonic stem cells. Cell stem cell. 2013;12:368–382. doi: 10.1016/j.stem.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 28.Grabole NTJ, Hackett JA, Kim S, Tang F, Leitch HG, Magnúsdóttir E, Surani MA. Prdm14 promotes germline fate and naive pluripotency by repressing FGF signalling and DNA methylation. EMBO J. 2013;14:629–637. doi: 10.1038/embor.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol. 2007;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura T, Liu Y, Nakashima H, Umehara H, Inoue K, Matoba S, Tachibana M, Ogura A, Shinkai Y, Nakano T. PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature. 2012;486:415–419. doi: 10.1038/nature11093. [DOI] [PubMed] [Google Scholar]
- 31.Messerschmidt D, de Vries W, Ito M, Solter D, Ferguson-Smith A, Knowles BB. Trim28 Is Required for Epigenetic Stability During Mouse Oocyte to Embryo Transition. Science. 2012;335:1499–1502. doi: 10.1126/science.1216154. [DOI] [PubMed] [Google Scholar]
- 32.Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, Ferguson-Smith A. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell. 1998;15:547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kagiwada S, Kurimoto K, Hirota T, Yamaji M, Saitou M. Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. The EMBO journal. 2013;32:340–353. doi: 10.1038/emboj.2012.331. [DOI] [PMC free article] [PubMed] [Google Scholar]; **Comprehensive analysis of PGCs revealing the close relationship between cell proliferation and loss of DNA methylation from imprinting control centers.
- 34.Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339:448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamaguchi S, Shen L, Liu Y, Sendler D, Zhang Y. Role of Tet1 in erasure of genomic imprinting. Science. 2013;504:460–464. doi: 10.1038/nature12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vincent JJ, Huang Y, Chen PY, Feng S, Calvopina JH, Nee K, Lee SA, Le T, Yoon AJ, Faull K, et al. Stage-Specific Roles for Tet1 and Tet2 in DNA Demethylation in Primordial Germ Cells. Cell stem cell. 2013;12:470–478. doi: 10.1016/j.stem.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliveros-Etter, Li Z, Nee K, Hosohama L, Hargan-Calvopina J, Lee S, Joti P, Yu J, Clark A. PGC reversion to pluripotency involves erasure of DNA methylation from imprinting control centers followed by locus-specific remethylation. Stem Cell Reports. 2015 doi: 10.1016/j.stemcr.2015.07.006. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakashima H, Kimura T, Kaga Y, Nakatani T, Seki Y, Nakamura T, Nakano T. Effects of Dppa3 on DNA Methylation Dynamics During Primordial Germ Cell Development in Mice. Biology of reproduction. 2013;88:1–9. doi: 10.1095/biolreprod.112.105932. [DOI] [PubMed] [Google Scholar]
- 39.Liu S, Brind'Amour J, Karimi M, Shirane K, Bogutz A, Lefebvre L, Sasaki H, Shinkai Y, Lorincz M. Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes Dev. 2015;28:2041–2055. doi: 10.1101/gad.244848.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leung D, Du T, Wagner U, Xie W, Lee A, Goyal P, Li Y, Szulwach K, Jin P, Lorincz M, et al. Regulation of DNA methylation turnover at LTR retrotransposons and imprinted loci by the histone methyltransferase Setdb1. PNAS. 2014;111:6690–6695. doi: 10.1073/pnas.1322273111. [DOI] [PMC free article] [PubMed] [Google Scholar]