Abstract
HLA genotyping via next generation sequencing (NGS) poses challenges for the use of HLA allele names to analyze and discuss sequence polymorphism. NGS will identify many new synonymous and non-coding HLA sequence variants. Allele names identify the types of nucleotide polymorphism that define an allele (non-synonymous, synonymous and non-coding changes), but do not describe how polymorphism is distributed among the individual features (the flanking untranslated regions, exons and introns) of a gene. Further, HLA alleles cannot be named in the absence of antigen-recognition domain (ARD) encoding exons. Here, a system for describing HLA polymorphism in terms of HLA gene features (GFs) is proposed. This system enumerates the unique nucleotide sequences for each GF in an HLA gene, and records these in a GF enumeration notation that allows both more granular dissection of allele-level HLA polymorphism and the discussion and analysis of GFs in the absence of ARD-encoding exon sequences.
Keywords: HLA, Nomenclature, Gene Feature Enumeration, Next Generation Sequencing, IHIW, 17th Workshop
1. Introduction
The human leucocyte antigen (HLA) genes are well known as the most polymorphic loci in the human genome. The extensive sequence polymorphism known for the HLA alleles is curated by the ImMunoGeneTics (IMGT)/HLA Database[1], which annotates the individual features for each gene [nucleotide sequences of each exon, intron and flanking untranslated region (UTR)] and gene product (encoded protein sequences). Here, exons, introns and UTRs are collectively referred to as gene features (GFs) to distinguish them from “sequence features” described elsewhere [2].
The World Health Organization Nomenclature Committee for factors of the HLA system (HLA Nomenclature Committee) assigns a unique allele name to each unique HLA nucleotide sequence[3]. Each HLA allele name consists of four colon-delimited fields (e.g., HLA-A*01:01:01:01). The first field identifies the allele family (for all genes but HLA-DPB1); the second field enumerates the unique protein sequences for the alleles in a given allele family, in the order in which they were identified; the third field enumerates sequences with synonymous substitutions for a given protein sequence, in the order in which they were identified; and the fourth field enumerates sequences with nucleotide substitutions in UTRs and introns for a given synonymous sequence in an exon, in the order in which they were identified. HLA-DPB1 lacks allele-families; the first field identifies unique protein sequences for all but the DPB1*02 and *04 alleles, for which two distinct protein sequences each are known [3–5].
The IMGT/HLA Database is updated every three months, and the number of named HLA gene and pseudogene sequences increases with each update. For example, 9,946 HLA alleles had been named as of December of 2013[6]; this number increased to 12,242 in December of 2014[7], and 13,412 HLA alleles have been named as of July of 2015. Increases in the number of new allele sequences included in the database have followed the adoption of new genotyping technologies by the Histocompatibility and Immunogenetics (H&I) community, often in conjunction with international HLA and immunogenetics workshops (IHIWs).
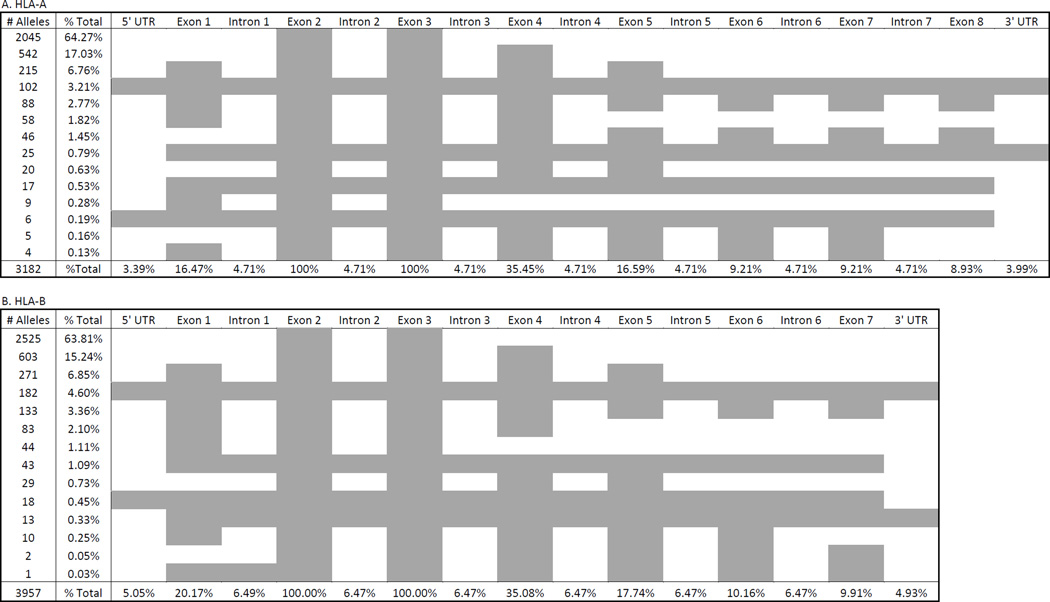
The IMGT/HLA Database annotation, based on European Molecular Biology Laboratory (EMBL) formats, is available as hla.dat and hla.xml files from ftp.ebi.ac.uk. These files identify and characterize the nucleotide sequences corresponding to specific GFs for each HLA allele. As illustrated in Table 1, each HLA gene can have a different number of GFs, but all HLA genes have a 3’ and 5’ UTR, at least four exons and at least three introns. However, for most HLA genes, full-length sequence is unavailable for the majority of alleles. As illustrated in Figure 1, nucleotide sequences for more than 60% of HLA-A, -B, -C, and –DRB1 alleles in IMGT/HLA Database version 3.21.1 are available only for exons 2 and 3 of the class I genes, and exon 2 of the class II genes, as these exons encode the antigen recognition domain (ARD). Fewer than 8% of the alleles at these loci have full-length sequences, describing nucleotide sequence for all of an allele’s GFs. Many of these full-length sequences have been generated using next generation sequencing (NGS) technologies, and the number of HLA alleles included in the database seems poised to increase dramatically as NGS technologies become widely used for HLA genotyping by the H&I and genomics communities, and as part of the 17th IHIW.
Table 1.
Maximum Lengths of Gene Features in 11 HLA Genes in IMGT/HLA Database Release 3.21.1
| Locus | 5' UTR | Exon 1 | Intron 1 | Exon 2 | Intron 2 | Exon 3 | Intron 3 | Exon 4 | Intron 4 | Exon 5 | Intron 5 | Exon 6 | Intron 6 | Exon 7 | Intron 7 | Exon 8 | 3' UTR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HLA-A | 300 | 73 | 130 | 293 | 242 | 314 | 600 | 280 | 102 | 117 | 442 | 33 | 142 | 48 | 169 | 5 | 300 |
| HLA-B | 284 | 73 | 129 | 272 | 250 | 281 | 575 | 277 | 104 | 120 | 441 | 33 | 107 | 44 | 364 | ||
| HLA-C | 283 | 73 | 130 | 277 | 250 | 297 | 587 | 277 | 124 | 138 | 440 | 33 | 107 | 48 | 164 | 5 | 171 |
| HLA-DPA1 | 523 | 100 | 3584 | 246 | 340 | 282 | 214 | 155 | 4331 | ||||||||
| HLA-DPB1 | 366 | 100 | 4536 | 264 | 4014 | 282 | 547 | 111 | 329 | 30 | 963 | ||||||
| HLA-DQA1 | 746 | 82 | 3858 | 249 | 445 | 282 | 429 | 155 | 436 | ||||||||
| HLA-DQB1 | 530 | 109 | 1458 | 270 | 2889 | 291 | 517 | 111 | 485 | 24 | 611 | 14 | 206 | ||||
| HLA-DRB1 | 607 | 100 | 10306 | 272 | 3464 | 282 | 702 | 111 | 487 | 24 | 1142 | 14 | 325 | ||||
| HLA-DRB3 | 327 | 100 | 7681 | 270 | 2302 | 282 | 684 | 111 | 473 | 24 | 799 | 14 | 579 | ||||
| HLA-DRB4 | 313 | 100 | 9563 | 270 | 2741 | 282 | 704 | 111 | 474 | 24 | 302 | 14 | 570 | ||||
| HLA-DRB5 | 0 | 100 | 0 | 270 | 0 | 282 | 0 | 111 | 0 | 24 | 0 | 14 | 0 |
The maximum length of the nucleotide sequences for each gene feature (GF) [untranslated region (UTR), exon or intron] for each HLA gene in 12,332 HLA alleles in IMGT/HLA Database release 3.21.1 is shown.
Blank cells indicate that no GF exists for that gene. Values of 0 indicate that no sequences for that GF have been included in available IMGT/HLA Database annotations.
Figure 1.
Percentages of HLA-A, -B, -C and -DRB1 Alleles with Nucleotide Sequences for Sets of Gene Features in IMGT/HLA Database Release Version 3.21.1
Each of the four panels details the percentage of alleles for which the nucleotide sequence of sets of gene features (GFs) is known at the HLA-A, -B, -C, or -DRB1 locus. Grey boxes represent GFs for which nucleotide sequence is known for a given percentage of alleles that locus.
The % Total value at the bottom of each column represents the percentage of alleles for which nucleotide sequence for each individual GF is known. Each % Total value in the second column represents the percentage of alleles for which nucleotide sequence for the GFs shown in grey in that row are known.
The total number of alleles at each locus with available nucleotide sequences is shown at the bottom of the first column.
1.1 Application of NGS Technology Highlights Current Nomenclature Limitations
The four colon-delimited field nomenclature for HLA alleles developed in step with genotyping technologies, as greater insights into the nature and scope of HLA polymorphism became available[4, 8–12]. While it provides insight into the types of polymorphism that distinguish alleles, this nomenclature does not identify the patterns and location of polymorphism across GFs at a given locus; the extent of the nucleotide sequence represented by an HLA allele name cannot be inferred from that name. The former issue has been partially addressed by extending allele names to identify those alleles that share identical ARD-encoding exon sequences (G groups of alleles, e.g., HLA-A*01:01:01G), as well as those alleles that encode identical ARD protein sequences (P groups of alleles, e.g., A*01:01P)[3], as these GFs constitute the largest fraction of the database. However, outside of the G group extension, alleles that share nucleotide sequences for other GFs cannot easily be identified. For example, class I alleles that share identical sequences for one of the ARD-encoding exons, but not the other, cannot be identified using G groups.
The sequences of ARD-encoding exons are required for all nucleotide sequence submissions to the HLA Nomenclature Committee via the IMGT/HLA Database, and novel nucleotide sequences for non-coding GFs must be submitted as part of full-length sequences. As a result, an HLA allele name cannot be assigned to a novel nucleotide sequence for an individual GF of interest (e.g., the 3’ UTR of HLA-C [13, 14]) in the absence of nucleotide sequences for ARD-encoding GFs.
Klitz and Hedrick[15] have estimated that millions of alleles persist in the human population for each HLA gene. As NGS technologies extend sequence knowledge into non-ARD encoding GFs, the number of alleles distinguished by synonymous and non-coding variants can be expected to increase dramatically; for example, as illustrated in Table 1, introns 1 and 2 of class II genes can be several thousand nucleotides long, and are likely to have accumulated many nucleotide variants. These variants will be noted in the third and fourth fields of allele names, and it does not seem out of the case to imagine allele names like HLA-DRB1*01:01:100:1004 in the near future. As the number of full-length HLA gene sequences generated increases, it seems likely that a large fraction of them will be unique.
Given the inability to determine which GFs are represented in an HLA allele name, the inability to assign allele names to individual non-ARD-encoding GFs, and the impending likelihood of a large number of unique full-length gene sequences, the utility of the HLA nomenclature is limited for managing, exchanging, discussing and analyzing nucleotide sequences for HLA GFs without the context of ARD-encoding GFs.
Here, a gene feature enumeration (GFE) notation is proposed as a supplement to the current HLA nomenclature for the purposes of cataloging nucleotide sequence polymorphisms for non-ARD-encoding GFs, discussing and analyzing HLA alleles in the context of polymorphism distributed between GFs, and capturing novel nucleotide sequences for non-ARD-encoding GFs generated via NGS technologies. This GFE approach is being developed as part of the 17th IHIWS Informatics Component.
2. Gene Feature Enumeration
HLA allele name nomenclature enumerates non-synonymous, synonymous and non-coding nucleotide variants in the second through fourth fields of an allele name. To supplement this approach, the unique sequences in each GF of a given HLA gene can be sequentially numbered, and applied to construct a second name for that allele consisting of one field for each GF, containing the unique number for that GF nucleotide sequence and delimited by dashes, prefaced with the allele name followed by a ‘w’ (for Workshop) to identify the provisional nature of this notation[16, 17]. This GFE notation is illustrated in Table 2.
Table 2.
Gene Feature Enumerations for Three HLA-A Alleles in IMGT/HLA Database Release 3.21.1
| IMGT/HLA Allele | Gene Feature Enumeration Notation | 5' UTR | Exon 1 | Intron 1 | Exon 2 | Intron 2 | Exon 3 | Intron 3 | Exon 4 | Intron 4 | Exon 5 | Intron 5 | Exon 6 | Intron 6 | Exon 7 | Intron 7 | Exon 8 | 3' UTR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HLA-A*01:01:01:01 | HLA-Aw1-1-1-1-1-1-1-1-1-1-1-1-1-1-1-1-1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| HLA-A*01:01:01:02N | HLA-Awp7-1-1-1-p1-1-1-1-1-1-1-1-1-1-1-1-p1 | p7 | 1 | 1 | 1 | p1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | p1 |
| HLA-A*01:01:02 | HLA-Awp0-p0-p0-1-p0-102-p0-p0-p0-p0-p0-p0-p0-p0-p0-p0-p0 | p0 | p0 | p0 | 1 | p0 | 102 | p0 | p0 | p0 | p0 | p0 | p0 | p0 | p0 | p0 | p0 | p0 |
For each HLA-A allele, the number assigned to each gene feature (GF) [flanking untranslated region (UTR), exon, or intron] represents a unique sequence for that GF, enumerated as described in section 2.1. GFs for which no sequence is available are assigned a value of p0. The GF enumeration (GFE) notation is a compilation of the enumerations for each GF, delimited with dashes, and prefixed with the HLA gene name and the letter ‘w’. Values that include a p and a non-zero number identify partial sequences for a given GF that are found in longer, full-length sequences for that GF, as described in section 2.1.
For example, the HLA-A gene includes 17 GFs (Table 1); any HLA-A allele can be represented as a unique haplotype of 17 GFs. As illustrated in Table 2, and in Supplementary Table S1, HLA-A*01:01:01:01 can be described in GFE notation as HLA-Aw1-1-1-1-1-1-1-1-1-1-1-1-1-1-1-1-1, identifying the constituent sequences of its GFs. In this case, the sequences for each HLA-A*01:01:01:01 GF are the first sequences numbered for those HLA-A GFs. The approach applied to assign these GF numbers is described in section 2.1.
Not all nucleotide sequences for a GF of an HLA gene are the same length. In some cases, these length differences are due to incomplete sequence of the GF in question, and in other cases they are due to insertion-deletion mutations. Using GFE notation, GF nucleotide sequences that exactly match longer nucleotide sequences for that GF are numbered separately; these sequences are prefaced with a ‘p’ to indicate that they are partial sequences, and may represent multiple “full-length” GF sequences. The first partial sequence identified at a given GF is numbered as p1, and the next unique partial sequence at that GF is numbered as p2, etc.
For example, HLA-A*01:01:01:02N is a null allele that results from a four nucleotide deletion in HLA-A intron 2. The GFE for this allele is HLA-Awp7-1-1-1-p1-1-1-1-1-1-1-1-1-1-1-1-p1. Comparing this GFE to that for HLA-A*01:01:01:01, it becomes clear that intron 2 and the 5’ and 3’ UTRs of A*01:01:01:02N are shorter than those for A*01:01:01:01. The issue of distinguishing length variants of GFs (which should be numbered as “full-length” GF sequences) from partial sequences resulting from methodological variation is discussed in section 2.3.
In instances where the sequence for a GF is not known, p0 is used to denote unknown sequence. For example, HLA-A*01:01:02 is one of the 2045 HLA-A alleles for which only exon 2 and 3 sequence is known; this allele can be represented as HLA-Awp0-p0-p0-1-p0-102-p0-p0-p0-p0-p0-p0-p0-p0-p0-p0-p0. By comparing the GFEs for HLA-A*01:01:01:01 and *01:01:02, it is immediately clear that these alleles share identical exon 2 sequences, that *01:01:02 differs from *01:01:01:01 only in its exon 3 sequence, and that the sequences of most *01:01:02 GFs are unknown.
2.1 Assigning Gene Feature Numbers
The example GFEs in Tables 2 and 3 and Supplementary Table 1 were generated using the hla.xml database export available from ftp.ebi.ac.uk. Nucleotide sequence information for each GF in each HLA gene was isolated and enumerated in order of allele accession numbers, so that older alleles are assigned the lowest numbers for a given GF.
Table 3.
Enumerated Gene Features for HLA-DQA1 in IMGT/HLA Database Release 3.21.1
| IMGT/HLA Allele Name | Gene Feature Enumeration Notation | 5' UTR | Exon 1 | Intron 1 | Exon 2 | Intron 2 | Exon 3 | Intron 3 | Exon 4 | 3' UTR |
|---|---|---|---|---|---|---|---|---|---|---|
| HLA-DQA1*01:01:01 | HLA-DQA1wp0-1-p0-1-p0-1-p0-1-p0 | p0 | 1 | p0 | 1 | p0 | 1 | p0 | 1 | p0 |
| HLA-DQA1*01:01:02 | HLA-DQA1w10-1-12-1-3-1-9-9-9 | 10 | 1 | 12 | 1 | 3 | 1 | 9 | 9 | 9 |
| HLA-DQA1*01:01:03 | HLA-DQA1wp0-1-p0-18-p0-1-p0-9-p0 | p0 | 1 | p0 | 18 | p0 | 1 | p0 | 9 | p0 |
| HLA-DQA1*01:02:01:01 | HLA-DQA1w1-1-1-2-1-1-1-2-1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 |
| HLA-DQA1*01:02:01:02 | HLA-DQA1w1-1-17-2-1-1-1-2-1 | 1 | 1 | 17 | 2 | 1 | 1 | 1 | 2 | 1 |
| HLA-DQA1*01:02:01:03 | HLA-DQA1w1-1-16-2-1-1-1-2-1 | 1 | 1 | 16 | 2 | 1 | 1 | 1 | 2 | 1 |
| HLA-DQA1*01:02:01:04 | HLA-DQA1w12-1-18-2-12-1-1-2-10 | 12 | 1 | 18 | 2 | 12 | 1 | 1 | 2 | 10 |
| HLA-DQA1*01:02:02 | HLA-DQA1wp0-1-p0-2-p0-2-p0-2-p0 | p0 | 1 | p0 | 2 | p0 | 2 | p0 | 2 | p0 |
| HLA-DQA1*01:02:03 | HLA-DQA1wp0-1-p0-2-p0-10-p0-2-p0 | p0 | 1 | p0 | 2 | p0 | 10 | p0 | 2 | p0 |
| HLA-DQA1*01:02:04 | HLA-DQA1wp0-9-p0-2-p0-2-p0-2-p0 | p0 | 9 | p0 | 2 | p0 | 2 | p0 | 2 | p0 |
| HLA-DQA1*01:03:01:01 | HLA-DQA1w2-1-2-3-2-3-2-3-2 | 2 | 1 | 2 | 3 | 2 | 3 | 2 | 3 | 2 |
| HLA-DQA1*01:03:01:02 | HLA-DQA1w13-1-19-3-2-3-2-3-2 | 13 | 1 | 19 | 3 | 2 | 3 | 2 | 3 | 2 |
| HLA-DQA1*01:04:01:01 | HLA-DQA1w3-2-3-1-3-1-3-4-3 | 3 | 2 | 3 | 1 | 3 | 1 | 3 | 4 | 3 |
| HLA-DQA1*01:04:01:02 | HLA-DQA1w3-2-15-1-3-1-3-4-3 | 3 | 2 | 15 | 1 | 3 | 1 | 3 | 4 | 3 |
| HLA-DQA1*01:04:02 | HLA-DQA1wp0-2-p0-1-p0-10-p0-4-p0 | p0 | 2 | p0 | 1 | p0 | 10 | p0 | 4 | p0 |
| HLA-DQA1*01:05:01 | HLA-DQA1w3-3-4-1-3-1-3-1-3 | 3 | 3 | 4 | 1 | 3 | 1 | 3 | 1 | 3 |
| HLA-DQA1*01:05:02 | HLA-DQA1wp0-2-p0-1-p0-1-p0-1-p0 | p0 | 2 | p0 | 1 | p0 | 1 | p0 | 1 | p0 |
| HLA-DQA1*01:06 | HLA-DQA1wp0-p0-p0-13-p0-p0-p0-p0-p0 | p0 | p0 | p0 | 13 | p0 | p0 | p0 | p0 | p0 |
| HLA-DQA1*01:07Q | HLA-DQA1wp3-2-15-15-3-1-3-4-p3 | p3 | 2 | 15 | 15 | 3 | 1 | 3 | 4 | p3 |
| HLA-DQA1*01:08 | HLA-DQA1wp0-p0-p0-2-p0-17-p0-p0-p0 | p0 | p0 | p0 | 2 | p0 | 17 | p0 | p0 | p0 |
| HLA-DQA1*01:09 | HLA-DQA1wp0-p0-p0-2-p0-18-p0-p0-p0 | p0 | p0 | p0 | 2 | p0 | 18 | p0 | p0 | p0 |
| HLA-DQA1*01:10 | HLA-DQA1wp3-1-19-17-2-3-2-3-p5 | p3 | 1 | 19 | 17 | 2 | 3 | 2 | 3 | p5 |
| HLA-DQA1*01:11 | HLA-DQA1wp4-1-22-2-1-1-1-12-p6 | p4 | 1 | 22 | 2 | 1 | 1 | 1 | 12 | p6 |
| HLA-DQA1*01:12 | HLA-DQA1wp0-p0-p0-1-p0-20-p0-p0-p0 | p0 | p0 | p0 | 1 | p0 | 20 | p0 | p0 | p0 |
| HLA-DQA1*02:01 | HLA-DQA1w4-4-5-4-4-4-4-5-4 | 4 | 4 | 5 | 4 | 4 | 4 | 4 | 5 | 4 |
| HLA-DQA1*03:01:01 | HLA-DQA1w5-4-6-5-5-4-5-6-5 | 5 | 4 | 6 | 5 | 5 | 4 | 5 | 6 | 5 |
| HLA-DQA1*03:01:03 | HLA-DQA1wp0-p0-p0-19-p0-4-p0-p0-p0 | p0 | p0 | p0 | 19 | p0 | 4 | p0 | p0 | p0 |
| HLA-DQA1*03:02 | HLA-DQA1w6-5-7-5-6-5-5-6-5 | 6 | 5 | 7 | 5 | 6 | 5 | 5 | 6 | 5 |
| HLA-DQA1*03:03:01 | HLA-DQA1w5-4-6-5-5-5-5-6-5 | 5 | 4 | 6 | 5 | 5 | 5 | 5 | 6 | 5 |
| HLA-DQA1*03:03:02 | HLA-DQA1wp0-4-p0-5-p0-19-p0-6-p0 | p0 | 4 | p0 | 5 | p0 | 19 | p0 | 6 | p0 |
| HLA-DQA1*04:01:01 | HLA-DQA1wp0-6-p0-6-p0-6-p0-7-p0 | p0 | 6 | p0 | 6 | p0 | 6 | p0 | 7 | p0 |
| HLA-DQA1*04:01:02:01 | HLA-DQA1wp2-6-13-6-11-11-10-7-p1 | p2 | 6 | 13 | 6 | 11 | 11 | 10 | 7 | p1 |
| HLA-DQA1*04:01:02:02 | HLA-DQA1wp5-6-23-6-11-11-10-7-p7 | p5 | 6 | 23 | 6 | 11 | 11 | 10 | 7 | p7 |
| HLA-DQA1*04:02 | HLA-DQA1w11-6-14-6-11-12-10-7-p2 | 11 | 6 | 14 | 6 | 11 | 12 | 10 | 7 | p2 |
| HLA-DQA1*04:03N | HLA-DQA1wp0-p0-p0-14-p0-p0-p0-p0-p0 | p0 | p0 | p0 | 14 | p0 | p0 | p0 | p0 | p0 |
| HLA-DQA1*04:04 | HLA-DQA1wp0-p0-p0-6-p0-14-p0-p0-p0 | p0 | p0 | p0 | 6 | p0 | 14 | p0 | p0 | p0 |
| HLA-DQA1*05:01:01:01 | HLA-DQA1wp1-7-8-7-7-7-6-7-6 | p1 | 7 | 8 | 7 | 7 | 7 | 6 | 7 | 6 |
| HLA-DQA1*05:01:01:02 | HLA-DQA1w14-7-21-7-8-7-6-7-6 | 14 | 7 | 21 | 7 | 8 | 7 | 6 | 7 | 6 |
| HLA-DQA1*05:01:02 | HLA-DQA1wp0-p1-p0-8-p0-p0-p0-p0-p0 | p0 | p1 | p0 | 8 | p0 | p0 | p0 | p0 | p0 |
| HLA-DQA1*05:02 | HLA-DQA1wp0-p0-p0-9-p0-p0-p0-p0-p0 | p0 | p0 | p0 | 9 | p0 | p0 | p0 | p0 | p0 |
| HLA-DQA1*05:03 | HLA-DQA1w7-7-9-7-8-8-6-7-6 | 7 | 7 | 9 | 7 | 8 | 8 | 6 | 7 | 6 |
| HLA-DQA1*05:04 | HLA-DQA1wp0-p0-p0-10-p0-p0-p0-p0-p0 | p0 | p0 | p0 | 10 | p0 | p0 | p0 | p0 | p0 |
| HLA-DQA1*05:05:01:01 | HLA-DQA1w8-8-10-7-9-9-7-8-7 | 8 | 8 | 10 | 7 | 9 | 9 | 7 | 8 | 7 |
| HLA-DQA1*05:05:01:02 | HLA-DQA1w8-8-10-7-9-9-12-8-7 | 8 | 8 | 10 | 7 | 9 | 9 | 12 | 8 | 7 |
| HLA-DQA1*05:05:01:03 | HLA-DQA1w8-8-20-7-9-9-11-8-p4 | 8 | 8 | 20 | 7 | 9 | 9 | 11 | 8 | p4 |
| HLA-DQA1*05:06 | HLA-DQA1wp0-7-p0-7-p0-15-p0-7-p0 | p0 | 7 | p0 | 7 | p0 | 15 | p0 | 7 | p0 |
| HLA-DQA1*05:07 | HLA-DQA1wp0-7-p0-7-p0-8-p0-10-p0 | p0 | 7 | p0 | 7 | p0 | 8 | p0 | 10 | p0 |
| HLA-DQA1*05:08 | HLA-DQA1wp0-8-p0-7-p0-16-p0-8-p0 | p0 | 8 | p0 | 7 | p0 | 16 | p0 | 8 | p0 |
| HLA-DQA1*05:09 | HLA-DQA1wp0-10-p0-7-p0-9-p0-8-p0 | p0 | 10 | p0 | 7 | p0 | 9 | p0 | 8 | p0 |
| HLA-DQA1*05:10 | HLA-DQA1wp0-p0-p0-16-p0-9-p0-p0-p0 | p0 | p0 | p0 | 16 | p0 | 9 | p0 | p0 | p0 |
| HLA-DQA1*05:11 | HLA-DQA1w8-8-10-7-9-9-13-11-7 | 8 | 8 | 10 | 7 | 9 | 9 | 13 | 11 | 7 |
| HLA-DQA1*06:01:01 | HLA-DQA1w9-6-11-11-10-6-8-7-8 | 9 | 6 | 11 | 11 | 10 | 6 | 8 | 7 | 8 |
| HLA-DQA1*06:01:02 | HLA-DQA1wp0-p0-p0-12-p0-p1-p0-p0-p0 | p0 | p0 | p0 | 12 | p0 | p1 | p0 | p0 | p0 |
| HLA-DQA1*06:02 | HLA-DQA1wp0-p0-p0-11-p0-13-p0-p0-p0 | p0 | p0 | p0 | 11 | p0 | 13 | p0 | p0 | p0 |
A p0 value identifies gene features (GFs) for which no sequence is available in the IMGT/HLA Database.
Values that consist of a p followed by a non-zero number are described in Table 4.
When the available nucleotide sequence for a GF is very short, it may be found in all full length nucleotide sequences for that GF. Similarly, large partial sequences may be found in multiple full length sequences. For example, as shown in Table 3, the exon 1 sequence for DQA1*05:01:02 is identified as p1. As shown in Table 4, DQA1 exon 1 sequence p1 is 13 nucleotides long and is a partial match to full length DQA1 exon 1 sequences 1, 4, 5, 6, 7, 8 and 9. Exon 1 sequence p1 is found in 79% of DQA1 alleles with full length sequences for this GF.
Table 4.
Enumeration of Partial Sequences for DQA1 Sequence Features in IMGT/HLA Database Release 3.21.1
| Locus | Gene Feature |
Partial Sequence |
Partial Sequence Length |
Matching Longer Sequences |
|---|---|---|---|---|
| HLA-DQA1 | 3' UTR | p1 | 328 | 8 |
| HLA-DQA1 | 3' UTR | p2 | 80 | 8 |
| HLA-DQA1 | 3' UTR | p3 | 323 | 3 |
| HLA-DQA1 | 3' UTR | p4 | 237 | 6/7 |
| HLA-DQA1 | 3' UTR | p5 | 147 | 2 |
| HLA-DQA1 | 3' UTR | p6 | 315 | 1/10 |
| HLA-DQA1 | 3' UTR | p7 | 143 | 8 |
| HLA-DQA1 | Exon 1 | p1 | 13 | 1/4/5/7/8/6/9 |
| HLA-DQA1 | Exon 3 | p1 | 1 | 1/3/4/5/7/8/9/6/11/12/2/10/15/16/19/13/14/17/18/20 |
| HLA-DQA1 | 5' UTR | p1 | 19 | 1/2/3/5/6/7/8/9/10/11/12/13/14 |
| HLA-DQA1 | 5' UTR | p2 | 132 | 9/11 |
| HLA-DQA1 | 5' UTR | p3 | 329 | 3/13 |
| HLA-DQA1 | 5' UTR | p4 | 304 | 1/2 |
| HLA-DQA1 | 5' UTR | p5 | 125 | 9/11 |
Partial Sequence: Short sequences that are partial matches to full-length sequences of a given feature are enumerated separately; the enumeration of these sequences is prefaced with the letter ‘p’.
Partial Sequence Length: The number of nucleotides in the partial sequence.
Matching Longer Sequences: A slash delimited list of enumerations of full-length sequences that contain the partial sequence in question.
2.2 Applications of Enumerated Gene Features
GFE notation allows the rapid identification of GFs that are shared by alleles with apparently unrelated allele names. For example, in Table 3, it is clear that several DQA1*04 and *05 alleles (e.g., *04:02 and *05:01:01:01) share identical exon 4 sequences (sequence number 7); that *02:01 and *03:01:01 share identical exon 3 sequences (sequence 4), as do *04:01:01 and *06:01:01 (sequence 6); and that *06:01:01 shares identical exon 1 sequence (sequence 6) with four *04 alleles. In this respect, GFE is similar to the G group approach for identifying alleles with identical ARD-encoding GFs, but allows any combination of GFs to be compared. By identifying the relationship between alleles across all GFs, GFE notation facilitates the investigation of alleles that share, or are distinct at, non-ARD-encoding GFs.
In addition, GFE notation allows alleles that share particular GFs to be combined by converting the enumeration for other GFs to zeros. For example, DQA1*04:01:01, *04:01:02:01, *04:01:02:02, *04:02 and *06:01:01 can be identified as HLA-DQA1w0-6-0-0-0-0-0-0-0, as these alleles all share the same exon 1 nucleotide sequence. Here, a 0 is used to denote GF sequence that is being ignored, whereas a p0 denotes an unknown GF sequence. In this manner, alleles can be grouped for analysis by the variation at individual GFs, or selected GF sets, without having to parse the sequence information represented by the allele name. In this respect, GFE notation offers a solution to the potential problem of evaluating the significance of hypothetical alleles such as “HLA-DRB1*01:01:100:1004” and “HLADRB1* 01:03:76:408”; using these allele names, the simple option for analysis would be to truncate them to two-field names (HLA-DRB1*01:01 and *01:03), ignoring any synonymous or non-coding variation. Using GFE notation, it may be possible to identify GFs that distinguish these alleles in terms of analytical significance.
Finally, GFE allows the consideration of nucleotide sequence variants in non-ARD-encoding GFs, and specifically in the absence of any non-ARD-encoding GF sequences. For example, if 3' UTR sequences are being studied, those 3' UTR sequences can be considered in the context of known polymorphism at a locus by using GFE notation that pertains to the 3' UTR only (e.g., HLA-DQA1w0-0-0-0-0-0-0-0-7 vs HLA-DQA1w0-0-0-0-0-0-0-0-10). GFE notation also allows for novel GF sequence variants to be compared with sequence variants that are already in the IMGT/HLA Database by assigning a new number to the novel variant. For example, the database includes 10 unique HLA-DQA1 3’ UTR sequences; a novel HLADQA1 3’ UTR can be named with GFE notation as HLA-DQA1wp0-p0-p0-p0-p0-p0-p0-p0-11 in the absence of exon 2 nucleotide sequences.
2.3 A Service for Managing Gene Feature Enumeration
GFE notation is not intended to replace HLA allele names; the long histories of HLA nomenclature and the H&I field, and the notoriety of specific HLA allele names preclude these names from ever being retired. GFE notation is proposed as a means for managing and discussing HLA polymorphism by acknowledging the underlying structure of the genes. As such, it is best managed in an automated fashion, with new GFE notations added as new HLA alleles are named, and updated as the nucleotide sequences of extant HLA allele names are extended to new GFs, with each IMGT/HLA Database release.
An internet-based service would make GFE notations publically accessible for online or offline use, and would permit the automated inter-conversion of allele names and GFE notations. This service is under development as part of a 17th IHIW Informatics Component project (ihiws.org/informatics-of-genomic-data/), and will be made available as an open-source product. Development of the service is ongoing, with code available online at github.com/nmdp-bioinformatics/service-feature. Interested investigators are encouraged participate in the development of the service by enrolling in this IHIW project.
This GFE service will persist a database of GFE’s based on a specific future IMGT/HLA Database release version and update that database to incorporate future IMGT/HLA Database releases. Alleles added to the IMGT/HLA Database with new GF sequences will receive GFE notations that incorporate new numbers reflecting the novel GF sequences. However, GF-level knowledge of the elements that distinguish alleles will be insufficient for many use cases. The GFE service will also maintain annotations describing more granular relationships between GF sequences within and between loci (e.g., characterizing the nucleotide polymorphisms that distinguish unique sequences for a given GF, or for GFs within and between loci), fostering more granular investigations of HLA polymorphism.
In addition, the GFE service will distinguish apparent partial sequences that result from length variations (indels) from those that result from technological or methodological limitations (e.g., primer positions) that result in a portion of a GF going unsequenced. GF sequences that reflect insertion/deletion mutations will be treated as full-length GF sequences, while sequences resulting from technological constraints and limitations, will be treated as partial sequences. Precise characterizations of partial sequences, and full-length and sequence length variations will be included in the GF sequence annotations.
Finally, the GFE service will also register novel, uncurated HLA gene sequences generated by 17th IHIW NGS projects, and eventually by any HLA sequencing effort, prior to submission to the IMGT/HLA Database, or in instances when ARD-encoding GF sequences are not available. Undoubtedly, many of the sequences so registered will result from sequencing errors, and should not be included in the IMGT/HLA Database. However, genuine novel nucleotide sequences may presumably be reported by multiple sequencing efforts, and the GFE service would serve as a clearing house for such sequences.
A service such as described above could be applied to other highly polymorphic genetic systems that make use of EMBL-formatted sequence annotation. In addition, the application of a coordinate-system that divides nucleotide sequence into other genetic features (e.g., individual codons, or user-defined features) than GFs could allow the same enumerations and operations to be performed at other levels of structural and biological organization.
3. Conclusions
Gene feature enumeration is a novel approach to describing HLA polymorphism that takes the structural elements of an HLA gene into account. This approach should be relatively easy to adopt, because it relies on information resources already provided by the IMGT/HLA Database. GFE notation will not supplant HLA allele names, but can complement them as the database accumulates sequences for non-ARD-encoding GFs generated via NGS methods. As knowledge of the regulatory roles of non-coding nucleotide sequences and their functional impacts on the HLA genes grows, it seems possible that the definition of an HLA allele may someday include promoters, enhancers and other intergenic sequences. GFE enumeration can accommodate this kind of growth in our understanding of immunogenetics in ways that allele names cannot. Given the inevitable changes that NGS methods will have on the H&I field, now is the time to discuss the best means of adapting to them.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health (NIH) grants U01AI067068, awarded by the National Institute of Allergy and Infectious Disease (NIAID), and R01GM109030, awarded by the National Institute of General Medical Sciences (NIGMS). The content presented is solely the responsibility of the author and does not necessarily represent the official views of the NIH, NIAID, NIGMS or United States Government. The gene feature enumeration approach was workshopped as part of the second Data Standards Hackathon for Next Generation Sequencing in February of 2015. The input of Henry Erlich, Marcelo Fernandez-Viña, Damian Goodridge, Martin Maiers, Nezih Cereb, Christina Chaivorapol, Hans-Peter Eberhard, Gottfried Fischer, Mathjis Groeneweg, Jan Hoffman, Jill A. Hollenbach, Tzvetana Kerelska, Shana McDevitt, Bob Milius, Carlheinz Müller, Jürgen Sauter, Gregory Turenchalk and Jennifer Zhang is very much appreciated in the development of this work.
Abbreviations
- ARD
Antigen Recognition Domain
- EMBL
European Molecular Biology Laboratory
- GF
Gene Feature
- GFE
Gene Feature Enumeration
- HLA
Human Leucocyte Antigen
- IHIW
International HLA and Immunogenetics Workshop
- IMGT
ImMunoGeneTics
- NGS
Next Generation Sequencing
- UTR
Untranslated Region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Robinson J, et al. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 2015;43(Database issue):D423–D431. doi: 10.1093/nar/gku1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomson G, et al. Sequence feature variant type (SFVT) analysis of the HLA genetic association in juvenile idiopathic arthritis. Pac Symp Biocomput. 2010:359–370. doi: 10.1142/9789814295291_0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marsh SG, et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens. 2010;75(4):291–455. doi: 10.1111/j.1399-0039.2010.01466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marsh SG, et al. Nomenclature for factors of the HLA system, 2002. Hum Immunol. 2002;63(12):1213–1268. doi: 10.1016/s0198-8859(02)00769-3. [DOI] [PubMed] [Google Scholar]
- 5.Bodmer JG, et al. Nomenclature for factors of the HLA system, 1989. Tissue Antigens. 1990;35(1):1–8. doi: 10.1111/j.1399-0039.1990.tb01749.x. [DOI] [PubMed] [Google Scholar]
- 6.Marsh SG. Nomenclature for factors of the HLA system, update December 2013. Tissue Antigens. 2014;83(3):229–235. doi: 10.1111/tan.12310. [DOI] [PubMed] [Google Scholar]
- 7.Marsh SG. Nomenclature for factors of the HLA system, update December 2014. Hum Immunol. 2015;24(15):00007-5. doi: 10.1016/j.humimm.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 8.Bodmer WF, et al. Nomenclature for factors of the HLA system 1984. Immunogenetics. 1984;20(6):593–601. doi: 10.1007/BF00430318. [DOI] [PubMed] [Google Scholar]
- 9.Nomenclature for factors of the HLA system, 1987. Immunogenetics. 1988;28(6):391–398. doi: 10.1007/BF00355369. [DOI] [PubMed] [Google Scholar]
- 10.Bodmer JG, et al. Nomenclature for factors of the HLA system, 1990. Hum Immunol. 1991;31(3):186–194. doi: 10.1016/0198-8859(91)90025-5. [DOI] [PubMed] [Google Scholar]
- 11.Bodmer JG, et al. Nomeclature for factors of the HLA system, 1995. Human Immunology. 1995;41:149–164. doi: 10.1016/0198-8859(95)00071-b. [DOI] [PubMed] [Google Scholar]
- 12.Bodmer JG, et al. Nomenclature for factors of the HLA System, 1996. Hum Immunol. 1997;53(1):98–128. doi: 10.1016/S0198-8859(97)00031-1. [DOI] [PubMed] [Google Scholar]
- 13.McCutcheon JA, et al. Low HLA-C expression at cell surfaces correlates with increased turnover of heavy chain mRNA. J Exp Med. 1995;181(6):2085–2095. doi: 10.1084/jem.181.6.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulkarni S, et al. Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature. 2011;472(7344):495–498. doi: 10.1038/nature09914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klitz W, Hedrick P, Louis EJ. New reservoirs of HLA alleles: pools of rare variants enhance immune defense. Trends Genet. 2012;28(10):480–486. doi: 10.1016/j.tig.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 16.Nomenclature for factors of the HL-A system. Bull World Health Organ. 1972;47(5):659–662. [PMC free article] [PubMed] [Google Scholar]
- 17.Nomenclature for factors of the HLA system. Bull World Health Organ. 1975;52(3):261–265. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.