Abstract
CD5+ B cell origins and their pre-disposition to lymphoma are long-standing issues. Transfer of fetal and adult liver bone marrow Pro-B cells generates B cells with distinct phenotypes: fetal cells generate IgMhighIgDlowCD5+, whereas adult cells IgMlowIgDhighCD5−. This suggests a developmental switch in B lymphopoiesis, similar to the switch in erythropoiesis. Comparison of mRNA and miRNA expression in fetal and adult Pro-B cells revealed differential expression of Lin28b mRNA and Let-7 miRNA, providing evidence that this regulatory axis functions in the switch. Recent work has shown that Arid3a is a key transcription factor mediating fetal-type B-cell development. Lin28b-promoted fetal development generates CD5+ B cells as a consequence of positively selected self-reactivity. CD5+ B cells play important roles in clearance of apoptotic cells and in protective immune responses, but also pose a risk of progression to leukemia/lymphoma. Differential Lin28b expression in fetal and adult human B-cell precursors showed that human B-cell development may resemble mouse, with self-reactive “innate-like” B cells generated early in life. It remains to be determined whether such human B cells have a higher propensity to leukemic progression. This review describes our recent research with CD5+ B cells and presents our perspective on their role in disease.
Keywords: B-cell development, stem cells, B1a cells, Transgenic mouse models, B-cell leukemia, CD5+ B cells
Introduction
CD5+ B cells were originally identified in the autoimmune mouse strain, NZB, and were shown to produce certain autoantibodies in mice [1, 2]. Later, we found that CD5+ B-cell numbers were enriched in the peritoneal cavity [3] and this enabled monitoring of CD5+ B-cell generation in mice after precursors cell transfer. Unexpectedly, we found inefficient generation of CD5+ B cells by precursors in bone marrow from adult mice (>2 months old), in contrast with CD5+ B-cell generation by hematopoietic precursors in neonatal liver [3]. Such biased production, along with the fact that CD5+ B cells self-renew and persist throughout life [4], prompted to the hypothesis that a large fraction of CD5+ B cells present in adult mice are generated early in life [5].
Later, we subdivide B lineage cells in bone marrow B-cell development, identifying a stage where cells have immunoglobulin (Ig) heavy chain DJ rearrangements on both chromosomes, but lack productively rearranged VDJs [6]. We called these Pro-B cells, to distinguish them from Pre-B cells that express heavy chain protein in their cytoplasm. When Pro-B cells were transferred into immunodeficient SCID mice that lack lymphoid populations, they generated mature B cells, but not other lymphoid cells and not a self-renewing precursor pool [6]. Pro-B cells with a phenotype similar to those in bone marrow have been identified in fetal liver, and they possessed partial IgH rearrangements, similar to bone marrow [7].
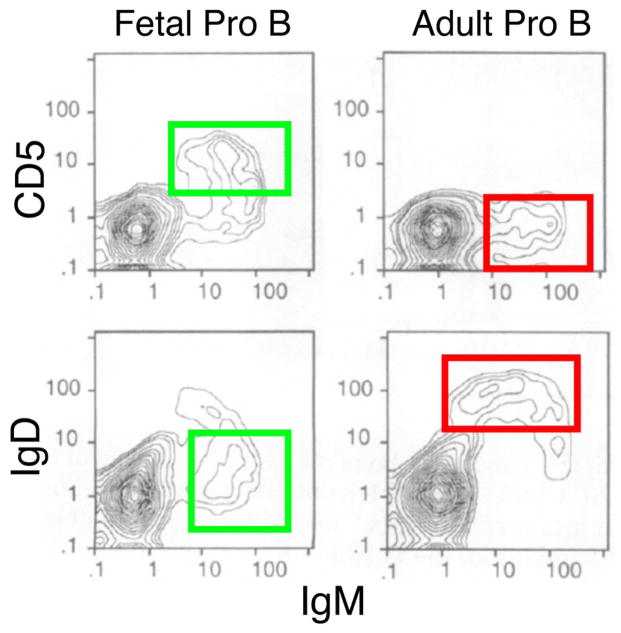
We performed cell transfer experiments of these Pro-B cells into SCID mouse recipients, comparing the B cells generated from these committed B-cell precursors isolated from fetal and adult sources [7]. Flow cytometry analysis showed that the B cells generated by these Pro-B cells at two different stages in the animal’s life are very different, with fetal precursors generating cells with a B1 phenotype (IgMhighIgDlow, many CD5+) and adult precursors generating cells with a B2 phenotype (IgMlowIgDhigh, predominantly CD5−) (Fig. 1). Thus, we proposed that a “developmental switch” may occur in B lymphopoiesis, similar to that long-recognized in erythropoiesis [8], with fetal (B-1) development generating mostly B1 B cells and adult development (B-2) generating mostly B2 B cells.
Figure 1. A developmental switch in B lymphopoiesis.
Fetal and adult Pro-B cells were transferred to SCID mice (n=4 of each type). Three weeks after, CD5/IgM and IgD/IgM profiles of spleen B cells were analyzed. Fetal phenotype (B1) marked by green box, adult phenotype (B2) marked by red box. Data shown are representative of 4 independent experiments.
A word on nomenclature
Many years ago, shortly after the first meeting devoted to CD5+ B cells in Miami in 1991, the terms B-1 and B-2 were proposed to describe these phenotypically distinct B cells [9]. This was a compromise among scientists with two different views of these cells: 1) CD5+ B cells arise from distinctive B-cell development (“lineage hypothesis”); and 2) CD5+ B cells arise by distinctive activation (“activation hypothesis”). Recent work provides the underlying mechanism for some of the distinctions between fetal and adult B-cell development, providing strong support for the lineage hypothesis [10, 11]. Furthermore, it is clear that CD5+ B cells are selected by antigen [12] and that BCR signaling upregulates CD5 expression [13], so in a sense, both views are correct. We propose the nomenclature suggested above, using B-1 and B-2 to describe distinctive fetal and adult development, and use B1a (B220lowerCD5+), B1b (B220lowerCD5−CD11b+) and B2 (B220higherCD5−) to describe distinctive B-cell phenotypes. In general, most CD5+ B cells are generated by B-1 development and most B2/follicular B cells by B-2 development [7, 11]. We think that this terminology is important, since some B-1 development occurs in adults, particularly giving rise to CD5− B1b cells [14–16]. Such B-1 development in adult bone marrow may represent the progeny of infrequent stem cells that continue to express low levels of the fetal-type regulator, Lin28b [10]. Moreover, it is clear that certain types of activation and tolerance can generate cells from B-2 development with a CD5+ phenotype [17, 18], even if such cells do not persist for an extended period.
Aspects of what we discuss below have also been previously reviewed as part of two presentations at a recent meeting devoted to B1 B cells [19, 20].
Distinctions between fetal and adult B lymphopoiesis
Obvious questions raised by the Pro-B transfer experiments were for example how such a switch is mediated and what it consists of. We first attempted to analyze gene expression differences in fetal and adult Pro-B cells by performing global mRNA analysis [17]. Microarray analysis revealed a striking difference in the expression of terminal deoxynucleotidyl transferase (TdT), a gene that encodes the enzyme mediating non-germline nucleotide addition (N-addition) at the Ig heavy chain junctions [21]. A paucity of such N-addition limits CDR3 diversity, a critical component of Ig antigen recognition [22]. Furthermore, low TdT favors rearrangement of gene segments that share short regions of homology, limiting diversity [23]. Thus differential expression of TdT is one reason for the difference between B-1 and B-2 development, since the IgH repertoires generated in their progeny will differ. However, this is not the complete explanation, since a TdT null mouse generated by gene targeting developed a mature pool of B cells that resembles B2/follicular B cells [24].
Development of B-cell pools with distinctive heavy chains could also occur by altered dependence on pre-BCR signaling [25]. Bone marrow B-cell development depends critically on assembly of a newly-rearranged VDJ-μ heavy chain with pre-existing B-lineage specific proteins encoded by the genes λ5 and VpreB that together make up the surrogate light chain (SLC) [26, 27]. Assembly of the IgH chain with SLC produces the pre-BCR, a complex that mediates a tonic signal indicating that a productive Ig heavy chain has been generated [28]. The pre-BCR also induces a number of changes in developing pre-B cells, including down-regulation of the Rag proteins [29] and rapid proliferation of pre-B cells. An IgH rearrangement that is frequent in B1 B cells, but rare in B2, utilizes a gene from the VH11 family [30, 31]. These heavy chains are nearly always paired with a specific light chain, a member of the Vκ9 family; this BCR recognizes the haptenic group phosphatidyl choline, exposed on aged erythrocytes [32]. Analysis of B-cell development in transgenic mice bearing this heavy chain revealed poor expression of the VH11-μ transgene and extensive endogenous IgH expression in spleen B cells of adult mice [25].
Failure of effective pre-BCR signaling results in continued rearrangement, explaining why an IgH-μ transgene may not block endogenous IgH locus rearrangement [33]. Analysis of assembly of the VH11-μ transgene with SLC, by immunoprecipitation of SLC components followed by western blotting with IgH-μ in transfected Pro-B cells, revealed weak assembly of the Pre-BCR [25]. This was unexpected, since this heavy chain was isolated from the CD5+ B-cell pool, whereVH11 rearrangements constitute 5–15% of the BCR repertoire [25]. This finding suggested a potential difference in dependence on pre-BCR signaling between fetal B-1 and adult B-2 development. Analysis of pre-B-cell proliferation in several IgH-μ transgenic mice, comparing cells isolated from fetal liver with those isolated from adult bone marrow (all on a Rag-1 null background to block endogenous VH gene expression) revealed a clear result: a human IgH-μ and the high-copy VH11 Tg lines inhibited fetal proliferation, whereas a low-copy VH11 Tg line allowed ongoing proliferation to continue [25]. This means that optimal fetal B-1 development occurs with lower pre-BCR expression compared to bone marrow B-2 development, so different sets of VDJ segments will be expanded in the pre-B pool from fetal liver and bone marrow.
Mechanism underlying distinctions between fetal and adult B-cell development
Analysis using the anti-Thymocyte/Thy-1 autoreactive (ATA) transgenic system (see BCR signaling, CD5+ B cells, and chronic leukemia) showed that BCR signaling is critical for B-1 development and is less important for B-2 development [13]; we investigated expression of microRNAs, since these tune TCR responses during T cell development [34]. A clear result emerged, with expression of Let-7 family members higher in bone marrow and lower in fetal liver [34]. This prompted us to examine the RNA-binding protein Lin28b, since it functions in a kind of binary switch with Let-7: Lin28b sequesters precursor Let-7, preventing its processing into functional microRNA and, in turn, Let-7 targets Lin28b mRNA for degradation [35].
Recently the Muljo laboratory showed that retroviral expression of human Lin28b in mouse bone marrow hematopoietic stem cells could induce the generation of B1a B cells in cell transfer experiments [10]. They also found that innate-like γδ T cells were induced by Lin28b. This was not completely unexpected since Ikuta and Weissman showed many years ago that certain innate-like γδ T cells were only generated from fetal stem cells [36]. Thus the fetal/adult switch in B (and T) cell development is regulated by the Lin28b/Let-7 axis. The Lin28b/Let-7 axis functions to regulate a variety of metabolic, proliferation, and differentiation pathways [37, 38]. For example, the Lin28b blocking of Let-7 function releases repression of a network of proto-oncogenes, including the insulin-PI3K-mTOR pathway, Ras, Myc, Hmga2, and the Igf2bps, resulting in increased metabolism and growth [39, 40]. Such reprogramming of metabolism also enhances tissue repair [41]. Let-7 targets genes involved in cellular proliferation, so expression of the Lin28b protein can result in increased proliferation and even malignancy [42–44].
In order to understand how Lin28b was regulating fetal B-1 development, we determined the genes reciprocally perturbed by mis-expression of Lin28b in bone marrow and Let-7 in fetal liver B lineage progenitors using microarray analysis [37]. We identified a short list of genes that included a transcription factor, Arid3a, also known as BRIGHT. The 3′ untranslated region of this gene includes target sites for miRNAs that are found at higher levels in bone marrow relative to fetal precursors, Let-7 and miR-125b. Following this lead, we cloned Arid3a into a retrovirus and expressed it in BM Pro-B stage cells. We found that Arid3a over-expression in BM Pro-B was sufficient to generate large numbers of B1a B cells, similar to that obtained by transduction of Lin28b. Next we generated Arid3a knockdown constructs and transduced fetal Pro-B cells, finding that we could reciprocally decrease the production of B1a cells. Therefore we concluded that Arid3a is a key mediator of the developmental switch in B lymphopoiesis [11].
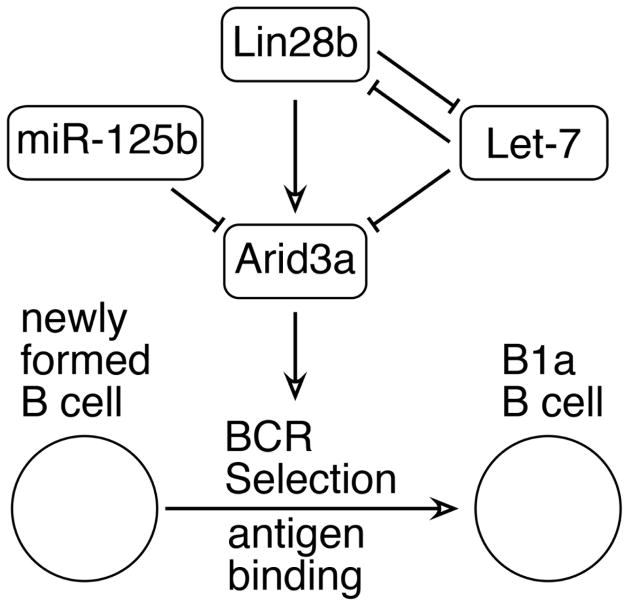
Arid3a was originally identified by its capacity to bind to IgH V segments, increasing their expression level in a cell line treated with antigen and IL-5 [45]. Later work showed that Arid3a was not restricted to activated B cells, but was also expressed in B-cell progenitors [46]. Further analysis showed that Arid3a binding site motifs were common in the promoter regions of both human and mouse VH genes [47]. Thus Arid3a may be altering accessibility of VH genes to rearrangement in fetal development. Furthermore, Arid3a has been found to shuttle between the cytoplasm and the nucleus [48]. In the cytoplasm, Arid3a has been shown to interact with the Tec kinase Btk [49] and, in fact, functional Btk is required for Arid3a activity [50], by phosphorylation of TFII-I which then forms a tripartite complex with Arid3a and Btk. Importantly, Arid3a has also been found to alter BCR signaling, due to its association with BCR-containing lipid rafts [51]. Thus, altered expression of Arid3a may change the initial VH repertoire and also impact selection at the newly-formed B-cell stage. A model for Lin28b/Let-7/Arid3a regulation of the developmental switch in B lymphopoiesis is shown in Figure 2.
Figure 2. Model for regulation of fetal-type (B-1) development.
B-1 development is regulated by the Lin28b/Let-7 axis and is promoted by the Arid3a transcription factor. Lin28b activates transcription of Arid3a, whereas Let-7 targets this mRNA for degradation. Arid3a modulates BCR signaling, altering antigen selection during the transition from newly-formed B to mature B.
BCR signaling, CD5+ B cells, and chronic leukemia
The ATA BCR system enabled us to test the role of BCR signaling in B1a generation, and discover differences in B-cell fate arising from different levels of BCR signaling in B-1 and B-2 development [12, 52, 53]. This unmutated ATA IgM (VH3609/Vk21) was identified as a B1a BCR in the SM/J mice [54] that are non-autoimmune but that have an elevated level of ATA IgM autoantibody in serum [55]. The BCR binds thymocyte membrane and was later shown to recognize highly glycosylated Thy-1 (CD90), abundantly expressed on immature thymocyte membranes [56], including dying thymocytes. We generated VH3609μTg mice, and also VDJ knock-in mice, finding that mature B1a cells with this BCR accumulated in the presence of Thy-1 self-antigen, but not in the absence of Thy-1, demonstrating for the first time that positive selection occurs in mouse B cells [12, 53], as has long been known for T cells.
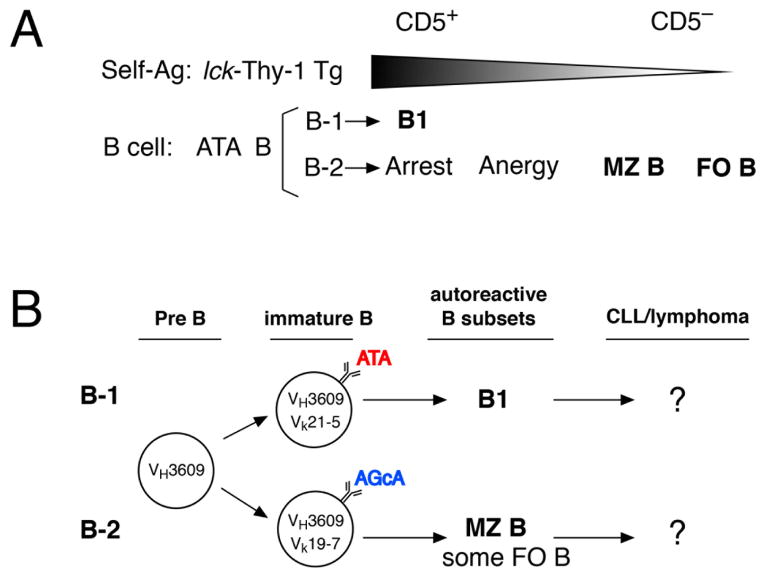
Analysis of “VHVL double-transgenic” ATAμκ mice on mouse backgrounds where lck-Thy-1 transgenes resulted in different levels of Thy-1, resulting in different intensities of BCR signaling, revealed that high BCR signaling during early/neonatal B-1 development led to positive selection, whereas comparable signaling led to maturation arrest during B-2 development in BM [52]. However, during B-2 development a weaker BCR signal (lower Thy-1 background) promoted MZ B-cell generation and very low or absent BCR signaling allowed FO B-cell generation [13], as diagrammed in Figure 3A. Thus, positive selection occurs in B-2 cell development, generating different mature subsets depending on differences in BCR signaling, although strong signaling in adult BM did not yield mature B1a cells. Instead some CD5+ B cells were generated that failed to mature, revealed by continued expression of CD93/AA4.1[52]. This indicates that the threshold for BCR tolerance and requirement for BCR signaling differ between fetal B-1 and adult B-2 development.
Figure 3. BCR signaling strength governs B-cell subset fate and progression to leukemia.
A. Generation of different B-cell subsets governed by BCR signal strength. The ATA μκTg mouse line on a Thy-1 knockout background (ATAμκTg.ThyKO) was crossed with lck-Thy-1 transgenic mouse lines expressing different levels of Thy-1 (from higher than normal level, wildtype level, lower, lowest, and completely absent). High levels promote the B1 cell fate from B-1 development, while very low levels generate MZ B cells and FO B-cell maturation occurs in the absence of Thy-1. B. In mice expressing VH3609μ at the pre-B-cell stage, expression of different light chains results in the generation of different B-cell subsets, either B1 from B-1 development or MZ B (and some FO B cells) from B-2 development. These BCRs have different autoreactivity but both can produce autoantibody, as natural autoreactive B cells: ATA, anti-thymocyte autoantibody, and AGcA, anti-intestinal goblet cell/mucin 2 autoreactivity. We are now asking whether B cells in these different autoreactive subsets carry a similar risk of progression to CLL.
B-cell chronic lymphocytic leukemia (B-CLL) is the most common form of adult leukemia in Western countries, with an incidence that increases with advancing age [57]. In the early 1980s, expression of the CD5 pan-T cell surface glycoprotein on B-CLL was noted as a unique feature of human B-CLL, in contrast with other B-cell lymphomas [58, 59]. This was a major impetus prompting us to test for Ly-1/CD5 expression on normal mouse B cells. Importantly, a year after transfer of purified peritoneal B1 cells into young irradiated mice, together with bone marrow stem cells, B1a of peritoneal B1 origin (IgMb allele+) were clearly detectable in both spleen and peritoneal cavity, together with BM derived CD5− B cells (IgMa allele+) constituting the predominant B-cell pool. Using B1a B cells from spleen of 1–2 week old neonatal mice for transfer also revealed persistence of B1a [60]. Thus B1a B cells persist for life and potentially could originate B-CLL. Importantly, BCRs on B-CLLs have been shown to encode self-reactivity [61].
In a physiologically normal self-antigen environment, we found that during B-2 development in bone marrow, autoreactive marginal zone (MZ) B cells can be generated from VH3609μ+ immature B cells, by pairing this heavy chain with a Vk19-17 light chain (Figure 3B). This MZ B associated VH3609/Vk19-17 BCR IgM has autoreactivity to intestinal goblet cell granules (AGcA), predominantly recognizing the highly glycosylated polymatrix form of intact mucin 2 glycoprotein [53]. In such mice, CD5+ B1a B cells are generated by pairing the transgenic heavy chain with a Vk21-5 light chain, yielding ATA B cells from B-1 development [12]. Thus, autoreactive B cells can be generated in different B-cell subsets with different autoreactivities and/or different affinities [13, 53]. By crossing μκTg mice with these autoreactive BCRs [52, 53] with the CLL-promoting Eμ-hTCL1 transgenic mouse [62], we are now asking whether these different autoreactive B-cell subsets (and FO B cells generated in the same mice) pose similar risks for progression to CLL, or whether only certain B cells behave in this fashion.
Human CD5+ B cells
An obvious question arises whether these findings apply to human B-cell development. Many years ago we investigated the frequency of human B cells expressing CD5 and found that normal donors could be classified into CD5+ B low and CD5+ B high, with rheumatoid arthritis patients enriched for the CD5+ B high pattern[63]. Furthermore, sorted CD5+ B cells were enriched for production of rheumatoid factor, an autoantibody. Finally, examination of B-cell in cord blood [63], the progeny of fetal development, showed a very high level of CD5+ B cells. Considering the similarities to mouse B1 B cells, it is reasonable to ask whether there is differential expression of Lin28b in human B lineage progenitors. Intriguingly, there are recent reports showing that Lin28b mRNA expression is high in fetal and low in adult hematopoietic stem cells [10, 64]. Thus it is reasonable to hypothesize that this regulatory axis is functioning in human lymphoid development, similar to mouse.
Following the differentiation of Lin28b+ progenitor cells to mature B cells in humanized mice may give insights into the selection, phenotype, and specificities of human B1 B cells. We raise this issue because CD5 can be induced by BCR signaling and humans are exposed to a wide array of pathogens that mice maintained in clean animal facilities are not. Furthermore, there is a report that CD5 is transiently expressed on most immature human B cells [65]. Recently there has been considerable controversy over the identification and characterization of human B1 B cells [66–69]. Additional analysis by the same group has described a CD11b+ subset of this population that is increased in patients with systemic lupus erythematosus and stimulates T cells [70]; this analysis has generated more controversy [71]. The analysis of B cells differentiated from Lin28b+ progenitors, either natural or transduced, may help to settle this issue, at least in the context of a human equivalent of a pool of fetal-generated B cells that persist into adults, where the phenotype of such cells may not precisely mirror that identified in mice.
Conclusions
What is the significance of fetal-generated B cells in mouse or man? Distinctive features of this cell pool in mouse are autoreactive BCRs and self-renewal throughout life. As mice age, B1 B cells can generate clonal expansions, as detected by distinctive bands on a southern blot with a JH probe [72]. Such clones can also progress to a frank leukemia or lymphoma stage [73]. This process is accelerated by B-lineage targeted transgenic expression of the human TCL1 gene [62]. Thus B1 B cells clearly pose a risk in aged mice of dysregulated growth and even leukemia.
We wonder if a similar process takes place in humans, where a self-reactive population of B cells, possibly generated during distinctive B-1 development, may generate an expanded clone. This may be the origin of monoclonal B-cell lymphocytosis (MBL), detected in relatives of chronic lymphocytic leukemia patients [74]. Such clonal expansions likely have a low probability of progressing to B-CLL depending on antigen stimulation and second hits, either genetic or epigenetic. Significantly, in B-CLL stereotyped (recurrent) VH or VL segments have been described, suggesting antigen selection [75]. Approximately 50% of B-CLLs have BCRs that are unmutated and these cases have a poorer prognosis compared to those with hypermutated BCRs [76]. BCRs from B-CLLs have been shown to include several different types of self-reactivity [77] and recently there has been a report that many B-CLLs may be self-binding [78]. Possibly such unmutated B-CLL cases arise from a germline encoded self-reactive B cell, similar to B1 B cells in mice. This issue clearly merits further investigation and examination of the role of the Lin28b/Let-7 axis (and Arid3a) in human B-cell development may help to shed light on the origin of a B-cell pool with a potential risk for progression to B-CLL.
Acknowledgments
This work presented here was supported by NIH grants RO1 AI026782 (R.R.H), RC1 CA145445 (R.R.H & K.H.), RO1 CA129330 (K.H.), R01 AI049335 (K.H.), and the FCCC Blood Cell Development and Cancer Keystone initiative.
Footnotes
Conflict of interest
The authors declare no commercial or financial conflict of interest.
References
- 1.Hayakawa K, Hardy RR, Parks DR, Herzenberg LA. The “Ly-1 B” cell subpopulation in normal, immunodefective, and autoimmune mice. J Exp Med. 1983;157:202–218. doi: 10.1084/jem.157.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayakawa K, Hardy RR, Honda M, Herzenberg LA, Steinberg AD, Herzenberg LA. Ly-1 B cells: functionally distinct lymphocytes that secrete IgM autoantibodies. Proc Natl Acad Sci U S A. 1984;81:2494–2498. doi: 10.1073/pnas.81.8.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayakawa K, Hardy RR, Herzenberg LA, Herzenberg LA. Progenitors for Ly-1 B cells are distinct from progenitors for other B cells. J Exp Med. 1985;161:1554–1568. doi: 10.1084/jem.161.6.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayakawa K, Hardy RR, Stall AM, Herzenberg LA, Herzenberg LA. Immunoglobulin-bearing B cells reconstitute and maintain the murine Ly-1 B cell lineage. Eur J Immunol. 1986;16:1313–1316. doi: 10.1002/eji.1830161021. [DOI] [PubMed] [Google Scholar]
- 5.Hayakawa K, Hardy RR. Normal, autoimmune, and malignant CD5+ B cells: the Ly-1 B lineage? Annu Rev Immunol. 1988;6:197–218. doi: 10.1146/annurev.iy.06.040188.001213. [DOI] [PubMed] [Google Scholar]
- 6.Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardy RR, Hayakawa K. A developmental switch in B lymphopoiesis. Proc Natl Acad Sci U S A. 1991;88:11550–11554. doi: 10.1073/pnas.88.24.11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papayannopoulou T, Nakamoto B, Agostinelli F, Manna M, Lucarelli G, Stamatoyannopoulos G. Fetal to adult hemopoietic cell transplantation in humans: insights into hemoglobin switching. Blood. 1986;67:99–104. [PubMed] [Google Scholar]
- 9.Kantor A. A new nomenclature for B cells. Immunol Today. 1991;12:388. doi: 10.1016/0167-5699(91)90135-G. [DOI] [PubMed] [Google Scholar]
- 10.Yuan J, Nguyen CK, Liu X, Kanellopoulou C, Muljo SA. Lin28b reprograms adult bone marrow hematopoietic progenitors to mediate fetal-like lymphopoiesis. Science. 2012;335:1195–1200. doi: 10.1126/science.1216557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Y, Li YS, Bandi S, Tang L, Shinton SA, Hayakawa K, Hardy RR. Lin28b promotes fetal B lymphopoiesis through the transcription factor Arid3a. J Exp Med. 2015;212:569–580. doi: 10.1084/jem.20141510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayakawa K, Asano M, Shinton SA, Gui M, Allman D, Stewart CL, Silver J, Hardy RR. Positive selection of natural autoreactive B cells. Science. 1999;285:113–116. doi: 10.1126/science.285.5424.113. [DOI] [PubMed] [Google Scholar]
- 13.Wen L, Brill-Dashoff J, Shinton SA, Asano M, Hardy RR, Hayakawa K. Evidence of marginal-zone B cell-positive selection in spleen. Immunity. 2005;23:297–308. doi: 10.1016/j.immuni.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 14.Kantor AB, Stall AM, Adams S, Herzenberg LA, Herzenberg LA. Differential development of progenitor activity for three B-cell lineages. Proc Natl Acad Sci U S A. 1992;89:3320–3324. doi: 10.1073/pnas.89.8.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang CA, Henry C, Iacomini J, Imanishi-Kari T, Wortis HH. Adult bone marrow contains precursors for CD5+ B cells. Eur J Immunol. 1996;26:2537–2540. doi: 10.1002/eji.1830261039. [DOI] [PubMed] [Google Scholar]
- 16.Montecino-Rodriguez E, Leathers H, Dorshkind K. Identification of a B-1 B cell-specified progenitor. Nature immunology. 2006;7:293–301. doi: 10.1038/ni1301. [DOI] [PubMed] [Google Scholar]
- 17.Cong YZ, Rabin E, Wortis HH. Treatment of murine CD5- B cells with anti-Ig, but not LPS, induces surface CD5: two B-cell activation pathways. Int Immunol. 1991;3:467–476. doi: 10.1093/intimm/3.5.467. [DOI] [PubMed] [Google Scholar]
- 18.Hippen KL, Tze LE, Behrens TW. CD5 maintains tolerance in anergic B cells. J Exp Med. 2000;191:883–890. doi: 10.1084/jem.191.5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li YS, Zhou Y, Tang L, Shinton SA, Hayakawa K, Hardy RR. A developmental switch between fetal and adult B lymphopoiesis. Ann N Y Acad Sci. 2015 doi: 10.1111/nyas.12769. [DOI] [PubMed] [Google Scholar]
- 20.Hayakawa K, Formica AM, Colombo MJ, Ichikawa D, Shinton SA, Brill-Dashoff J, Hardy RR. B cells generated by B-1 development can progress to chronic lymphocytic leukemia. Ann N Y Acad Sci. 2015 doi: 10.1111/nyas.12768. [DOI] [PubMed] [Google Scholar]
- 21.Landau NR, Schatz DG, Rosa M, Baltimore D. Increased frequency of N-region insertion in a murine pre-B-cell line infected with a terminal deoxynucleotidyl transferase retroviral expression vector. Mol Cell Biol. 1987;7:3237–3243. doi: 10.1128/mcb.7.9.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu JL, Davis MM. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity. 2000;13:37–45. doi: 10.1016/s1074-7613(00)00006-6. [DOI] [PubMed] [Google Scholar]
- 23.Feeney AJ. Predominance of VH-D-JH junctions occurring at sites of short sequence homology results in limited junctional diversity in neonatal antibodies. J Immunol. 1992;149:222–229. [PubMed] [Google Scholar]
- 24.Gilfillan S, Dierich A, Lemeur M, Benoist C, Mathis D. Mice lacking TdT: mature animals with an immature lymphocyte repertoire. Science. 1993;261:1175–1178. doi: 10.1126/science.8356452. [DOI] [PubMed] [Google Scholar]
- 25.Wasserman R, Li YS, Shinton SA, Carmack CE, Manser T, Wiest DL, Hayakawa K, Hardy RR. A novel mechanism for B cell repertoire maturation based on response by B cell precursors to pre-B receptor assembly. The Journal of experimental medicine. 1998;187:259–264. doi: 10.1084/jem.187.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vettermann C, Herrmann K, Jack HM. Powered by pairing: the surrogate light chain amplifies immunoglobulin heavy chain signaling and pre-selects the antibody repertoire. Semin Immunol. 2006;18:44–55. doi: 10.1016/j.smim.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Melchers F, ten Boekel E, Seidl T, Kong XC, Yamagami T, Onishi K, Shimizu T, Rolink AG, Andersson J. Repertoire selection by pre-B-cell receptors and B-cell receptors, and genetic control of B-cell development from immature to mature B cells. Immunol Rev. 2000;175:33–46. [PubMed] [Google Scholar]
- 28.Melchers F. The pre-B-cell receptor: selector of fitting immunoglobulin heavy chains for the B-cell repertoire. Nat Rev Immunol. 2005;5:578–584. doi: 10.1038/nri1649. [DOI] [PubMed] [Google Scholar]
- 29.Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 30.Hardy RR, Carmack CE, Shinton SA, Riblet RJ, Hayakawa K. A single VH gene is utilized predominantly in anti-BrMRBC hybridomas derived from purified Ly-1 B cells. Definition of the VH11 family. Journal of immunology. 1989;142:3643–3651. [PubMed] [Google Scholar]
- 31.Carmack CE, Shinton SA, Hayakawa K, Hardy RR. Rearrangement and selection of VH11 in the Ly-1 B cell lineage. The Journal of experimental medicine. 1990;172:371–374. doi: 10.1084/jem.172.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mercolino TJ, Arnold LW, Hawkins LA, Haughton G. Normal mouse peritoneum contains a large population of Ly-1+ (CD5) B cells that recognize phosphatidyl choline. Relationship to cells that secrete hemolytic antibody specific for autologous erythrocytes. J Exp Med. 1988;168:687–698. doi: 10.1084/jem.168.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.ten Boekel E, Melchers F, Rolink AG. Changes in the V(H) gene repertoire of developing precursor B lymphocytes in mouse bone marrow mediated by the pre-B cell receptor. Immunity. 1997;7:357–368. doi: 10.1016/s1074-7613(00)80357-x. [DOI] [PubMed] [Google Scholar]
- 34.Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, Klein LO, Davis MM, Chen CZ. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 35.Mayr F, Schutz A, Doge N, Heinemann U. The Lin28 cold-shock domain remodels pre-let-7 microRNA. Nucleic Acids Res. 2012;40:7492–7506. doi: 10.1093/nar/gks355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ikuta K, Kina T, MacNeil I, Uchida N, Peault B, Chien YH, Weissman IL. A developmental switch in thymic lymphocyte maturation potential occurs at the level of hematopoietic stem cells. Cell. 1990;62:863–874. doi: 10.1016/0092-8674(90)90262-d. [DOI] [PubMed] [Google Scholar]
- 37.Viswanathan SR, Daley GQ. Lin28: A microRNA regulator with a macro role. Cell. 2010;140:445–449. doi: 10.1016/j.cell.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 38.Zhu H, Shyh-Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, Urbach A, Thornton JE, Triboulet R, Gregory RI, Altshuler D, Daley GQ. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shinoda G, Shyh-Chang N, Soysa TY, Zhu H, Seligson MT, Shah SP, Abo-Sido N, Yabuuchi A, Hagan JP, Gregory RI, Asara JM, Cantley LC, Moss EG, Daley GQ. Fetal deficiency of lin28 programs life-long aberrations in growth and glucose metabolism. Stem Cells. 2013;31:1563–1573. doi: 10.1002/stem.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shyh-Chang N, Daley GQ. Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell. 2013;12:395–406. doi: 10.1016/j.stem.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shyh-Chang N, Zhu H, Yvanka de Soysa T, Shinoda G, Seligson MT, Tsanov KM, Nguyen L, Asara JM, Cantley LC, Daley GQ. Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell. 2013;155:778–792. doi: 10.1016/j.cell.2013.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, Chin L, Brown D, Slack FJ. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 43.Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O’Sullivan M, Lu J, Phillips LA, Lockhart VL, Shah SP, Tanwar PS, Mermel CH, Beroukhim R, Azam M, Teixeira J, Meyerson M, Hughes TP, Llovet JM, Radich J, Mullighan CG, Golub TR, Sorensen PH, Daley GQ. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Madison BB, Liu Q, Zhong X, Hahn CM, Lin N, Emmett MJ, Stanger BZ, Lee JS, Rustgi AK. LIN28B promotes growth and tumorigenesis of the intestinal epithelium via Let-7. Genes Dev. 2013;27:2233–2245. doi: 10.1101/gad.224659.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herrscher RF, Kaplan MH, Lelsz DL, Das C, Scheuermann R, Tucker PW. The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: a B cell-specific trans-activator that describes a new DNA-binding protein family. Genes Dev. 1995;9:3067–3082. doi: 10.1101/gad.9.24.3067. [DOI] [PubMed] [Google Scholar]
- 46.Webb CF, Smith EA, Medina KL, Buchanan KL, Smithson G, Dou S. Expression of bright at two distinct stages of B lymphocyte development. J Immunol. 1998;160:4747–4754. [PubMed] [Google Scholar]
- 47.Goebel P, Montalbano A, Ayers N, Kompfner E, Dickinson L, Webb CF, Feeney AJ. High frequency of matrix attachment regions and cut-like protein x/CCAAT-displacement protein and B cell regulator of IgH transcription binding sites flanking Ig V region genes. J Immunol. 2002;169:2477–2487. doi: 10.4049/jimmunol.169.5.2477. [DOI] [PubMed] [Google Scholar]
- 48.Kim D, Tucker PW. A regulated nucleocytoplasmic shuttle contributes to Bright’s function as a transcriptional activator of immunoglobulin genes. Mol Cell Biol. 2006;26:2187–2201. doi: 10.1128/MCB.26.6.2187-2201.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Webb CF, Yamashita Y, Ayers N, Evetts S, Paulin Y, Conley ME, Smith EA. The transcription factor Bright associates with Bruton’s tyrosine kinase, the defective protein in immunodeficiency disease. J Immunol. 2000;165:6956–6965. doi: 10.4049/jimmunol.165.12.6956. [DOI] [PubMed] [Google Scholar]
- 50.Rajaiya J, Hatfield M, Nixon JC, Rawlings DJ, Webb CF. Bruton’s tyrosine kinase regulates immunoglobulin promoter activation in association with the transcription factor Bright. Mol Cell Biol. 2005;25:2073–2084. doi: 10.1128/MCB.25.6.2073-2084.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt C, Kim D, Ippolito GC, Naqvi HR, Probst L, Mathur S, Rosas-Acosta G, Wilson VG, Oldham AL, Poenie M, Webb CF, Tucker PW. Signalling of the BCR is regulated by a lipid rafts-localised transcription factor, Bright. EMBO J. 2009;28:711–724. doi: 10.1038/emboj.2009.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hayakawa K, Asano M, Shinton SA, Gui M, Wen LJ, Dashoff J, Hardy RR. Positive selection of anti-thy-1 autoreactive B-1 cells and natural serum autoantibody production independent from bone marrow B cell development. J Exp Med. 2003;197:87–99. doi: 10.1084/jem.20021459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ichikawa D, Asano M, Shinton SA, Brill-Dashoff J, Formica AM, Velcich A, Hardy RR, Hayakawa K. Natural Anti-Intestinal Goblet Cell Autoantibody Production from Marginal Zone B Cells. J Immunol. 2015;194:606–614. doi: 10.4049/jimmunol.1402383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hayakawa K, Carmack CE, Hyman R, Hardy RR. Natural autoantibodies to thymocytes: origin, VH genes, fine specificities, and the role of Thy-1 glycoprotein. J Exp Med. 1990;172:869–878. doi: 10.1084/jem.172.3.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eisenberg RA, Theofilopoulos AN, Andrews BS, Peters CJ, Thor L, Dixon FJ. Natural thymocytotoxic autoantibodies in autoimmune and normal mice. J Immunol. 1979;122:2272–2278. [PubMed] [Google Scholar]
- 56.Gui M, Wiest DL, Li J, Kappes D, Hardy RR, Hayakawa K. Peripheral CD4+ T cell maturation recognized by increased expression of Thy-1/CD90 bearing the 6C10 carbohydrate epitope. J Immunol. 1999;163:4796–4804. [PubMed] [Google Scholar]
- 57.Morton LM, Wang SS, Devesa SS, Hartge P, Weisenburger DD, Linet MS. Lymphoma incidence patterns by WHO subtype in the United States, 1992–2001. Blood. 2006;107:265–276. doi: 10.1182/blood-2005-06-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Royston I, Majda JA, Baird SM, Meserve BL, Griffiths JC. Human T cell antigens defined by monoclonal antibodies: the 65,000-dalton antigen of T cells (T65) is also found on chronic lymphocytic leukemia cells bearing surface immunoglobulin. J Immunol. 1980;125:725–731. [PubMed] [Google Scholar]
- 59.Wang CY, Good RA, Ammirati P, Dymbort G, Evans RL. Identification of a p69,71 complex expressed on human T cells sharing determinants with B-type chronic lymphatic leukemic cells. J Exp Med. 1980;151:1539–1544. doi: 10.1084/jem.151.6.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hayakawa K, Hardy RR, Stall AM, Herzenberg LA. Immunoglobulin-bearing B cells reconstitute and maintain the murine Ly-1 B cell lineage. Eur J Immunol. 1986;16:1313–1316. doi: 10.1002/eji.1830161021. [DOI] [PubMed] [Google Scholar]
- 61.Herve M, Xu K, Ng YS, Wardemann H, Albesiano E, Messmer BT, Chiorazzi N, Meffre E. Unmutated and mutated chronic lymphocytic leukemias derive from self-reactive B cell precursors despite expressing different antibody reactivity. J Clin Invest. 2005;115:1636–1643. doi: 10.1172/JCI24387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bichi R, Shinton SA, Martin ES, Koval A, Calin GA, Cesari R, Russo G, Hardy RR, Croce CM. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:6955–6960. doi: 10.1073/pnas.102181599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hardy RR, Hayakawa K, Shimizu M, Yamasaki K, Kishimoto T. Rheumatoid factor secretion from human Leu-1+ B cells. Science. 1987;236:81–83. doi: 10.1126/science.3105057. [DOI] [PubMed] [Google Scholar]
- 64.McWilliams L, Su KY, Liang X, Liao D, Floyd S, Amos J, Moody MA, Kelsoe G, Kuraoka M. The human fetal lymphocyte lineage: identification by CD27 and LIN28B expression in B cell progenitors. J Leukoc Biol. 2013;94:991–1001. doi: 10.1189/jlb.0113048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee J, Kuchen S, Fischer R, Chang S, Lipsky PE. Identification and characterization of a human CD5+ pre-naive B cell population. J Immunol. 2009;182:4116–4126. doi: 10.4049/jimmunol.0803391. [DOI] [PubMed] [Google Scholar]
- 66.Griffin DO, Holodick NE, Rothstein TL. Human B1 cells in umbilical cord and adult peripheral blood express the novel phenotype CD20+ CD27+ CD43+ CD70. J Exp Med. 2011;208:67–80. doi: 10.1084/jem.20101499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perez-Andres M, Grosserichter-Wagener C, Teodosio C, van Dongen JJ, Orfao A, van Zelm MC. The nature of circulating CD27+CD43+ B cells. J Exp Med. 2011;208:2565–2566. doi: 10.1084/jem.20112203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Descatoire M, Weill JC, Reynaud CA, Weller S. A human equivalent of mouse B-1 cells? J Exp Med. 2011;208:2563–2564. doi: 10.1084/jem.20112232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Griffin DO, Holodick NE, Rothstein TL. Human B1 cells are CD3-: A reply to “A human equivalent of mouse B-1 cells?” and “The nature of circulating CD27+CD43+ B cells”. J Exp Med. 2011;208:2566–2569. doi: 10.1084/jem.20111761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Griffin DO, Rothstein TL. A small CD11b(+) human B1 cell subpopulation stimulates T cells and is expanded in lupus. J Exp Med. 2011;208:2591–2598. doi: 10.1084/jem.20110978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reynaud CA, Weill JC. Gene profiling of CD11b(+) and CD11b(−) B1 cell subsets reveals potential cell sorting artifacts. J Exp Med. 2012;209:433–434. doi: 10.1084/jem.20120402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stall AM, Farinas MC, Tarlinton DM, Lalor PA, Herzenberg LA, Strober S, Herzenberg LA. Ly-1 B-cell clones similar to human chronic lymphocytic leukemias routinely develop in older normal mice and young autoimmune (New Zealand Black-related) animals. Proc Natl Acad Sci U S A. 1988;85:7312–7316. doi: 10.1073/pnas.85.19.7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Forster I, Gu H, Rajewsky K. Germline antibody V regions as determinants of clonal persistence and malignant growth in the B cell compartment. EMBO J. 1988;7:3693–3703. doi: 10.1002/j.1460-2075.1988.tb03251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fazi C, Scarfo L, Pecciarini L, Cottini F, Dagklis A, Janus A, Talarico A, Scielzo C, Sala C, Toniolo D, Caligaris-Cappio F, Ghia P. General population low-count CLL-like MBL persists over time without clinical progression, although carrying the same cytogenetic abnormalities of CLL. Blood. 2011;118:6618–6625. doi: 10.1182/blood-2011-05-357251. [DOI] [PubMed] [Google Scholar]
- 75.Widhopf GF, 2nd, Rassenti LZ, Toy TL, Gribben JG, Wierda WG, Kipps TJ. Chronic lymphocytic leukemia B cells of more than 1% of patients express virtually identical immunoglobulins. Blood. 2004;104:2499–2504. doi: 10.1182/blood-2004-03-0818. [DOI] [PubMed] [Google Scholar]
- 76.Klein U, Dalla-Favera R. New insights into the pathogenesis of chronic lymphocytic leukemia. Semin Cancer Biol. 2010;20:377–383. doi: 10.1016/j.semcancer.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 77.Chu CC, Catera R, Zhang L, Didier S, Agagnina BM, Damle RN, Kaufman MS, Kolitz JE, Allen SL, Rai KR, Chiorazzi N. Many chronic lymphocytic leukemia antibodies recognize apoptotic cells with exposed nonmuscle myosin heavy chain IIA: implications for patient outcome and cell of origin. Blood. 2010;115:3907–3915. doi: 10.1182/blood-2009-09-244251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Duhren-von Minden M, Ubelhart R, Schneider D, Wossning T, Bach MP, Buchner M, Hofmann D, Surova E, Follo M, Kohler F, Wardemann H, Zirlik K, Veelken H, Jumaa H. Chronic lymphocytic leukaemia is driven by antigen-independent cell-autonomous signalling. Nature. 2012;489:309–312. doi: 10.1038/nature11309. [DOI] [PubMed] [Google Scholar]