Abstract
This study aims to systematically determine the cytochrome P450s (CYPs) activities and expressions in hepatocellular carcinoma (HCC) patients to support their optimal use in personalized treatment of HCC. Activities of seven major drug-metabolizing CYP enzymes (CYP1A2, 2A6, 2C8, 2C9, 2D6, 2E1 and 3A4) were determined in tumors and pericarcinomatous tissues harvested from 26 patients with HBV-positive HCC using probe substrates. Protein and mRNA levels of these CYPs were also measured using isotope label-free LC-MS/MS method and real-time PCR, respectively. Maximal metabolic velocity (Vmax) of CYP probe substrates were decreased by 2.5- to 30-fold in tumor microsomes, accompanied by a corresponding decrease in their protein and mRNA expression levels. However, Km values and turnover numbers of substrates in tumor microsomes were not changed. High correlations between activities and CYP protein levels were also observed, but the correlation between activities and mRNA levels were often poor. There was a major decrease in the degree of correlation in CYP expression in tumor tissues, suggesting that CYP expression levels are greatly disrupted by the tumorigenic process. Our unprecedented systemic study of the effects of HCC on CYPs demonstrated that activities of CYPs were seriously impaired and their expression patterns were severely altered by HCC. We proposed that determination of the CYP protein expression profile by LC-MS/MS in each patient is a promising approach that can be clinically used for individualized treatment of HCC.
Keywords: cytochrome P450, hepatocellular carcinoma, personalized treatment, metabolizing activity, turnover number
1. INTRODUCTION
Liver cancer is the fifth most common cancer and the second most frequent cause of death from cancer worldwide [1]. Hepatocellular carcinoma (HCC) represents more than 90% of primary liver cancers and is a major global health problem. Chronic infection with hepatitis B virus (HBV) is the most clearly established risk factor for HCC, and approximately 54% of cases can be attributed to HBV infection [2]. In China where 50% of the world’s liver cancer cases and deaths occurred [1], more than 90% of the HCC patients are also diagnosed as having HBV infection [3–4]. Surgical resection remains the gold standard for HCC eradication. However, more than 80% of patients with HCC are not surgical candidates and other options such as chemotherapy and chemoembolization are often considered [5].
The efficacy of anticancer therapy is limited by our inability to predict patient outcomes such as tumor response and toxicity. Drug-metabolizing enzymes in tumors or their pericarcinomatous tissues play key roles in the activation or inactivation of numerous cytotoxic drugs, and influence the susceptibility of host and neoplasm to their effects. Cytochrome P450 (CYPs) play critically important roles in the biotransformation of many endogenous and exogenous compounds including drugs and dietary components [6]. In typical human livers, five subfamilies, including CYP3A, CYP2C, CYP2D, CYP1A, and CYP2E are mainly expressed, and responsible for 70–80% of phase I metabolism of 90% of marketed drugs [7–9]. However, CYPs not only function in the detoxification of xenobiotics but may also be involved in the activation of potential (pro-) carcinogens and mutagens [10]. Furthermore, it has been suggested that the local expression of CYPs in tumors is very important for the management of cancer since these functionally associated enzymes might be involved in both the development of HCC and in determining the anti-cancer drug sensitivity of such tumor [11–12].
Rates and extents of drug metabolism by tumor CYPs can be used as a marker for potential mechanism of drug resistance or a means to achieve optimal chemotherapy. This is particularly true in HCC, where CYPs are normally most abundantly expressed. This is because drugs that are rapidly and extensively metabolized in the tumor but slowly metabolized in the normal tissues surrounding the tumor are unlikely to have a good benefit/risk ratio for a patient [13–14]. Therefore, a detailed understanding of the differential expression and activity of CYPs within HCC tumors and their pericarcinomatous tissues may provide opportunities for improved therapeutic outcome. However, knowledge on activities of CYPs in HCC tumors is limited, and few studies have directly assessed protein extracts of human tumor tissues for specific enzyme kinetics, the capability to metabolize cytotoxic substrates. Differential gene expression levels of some CYP isoforms in HCC and pericarcinomatous tissues have been reported previously [15]. But changes in mRNA levels may not necessarily reflect that in activities of CYPs, which might limit the clinical implementation of mRNA determination. Additionally, CYPs are also subject to significant inter- and intra-individual variability, displaying genetic polymorphism and differences in activity inducibility [12]. So even if the expressions and activities of CYPs in average tissues are relatively well described, it is still difficult to predict the tumor response and toxicity in each patient. Therefore, an approach to monitoring the expression and activities of CYPs in HCC tumors in individual patients is necessary for individualized treatment of HCC.
MS-based protein quantification analysis is a novel approach developed recently and widely used in proteomic studies. This method normally relies on analysis of proteotypic peptides in trypsinized protein samples using chemically synthesized isotope labeled or label-free peptides as calibration standards. Therefore MS-based quantifications are totally different from the traditional immunometric methods such as western blots and ELISA that are generally restricted by the limited availability of specific antibodies. This new approach makes it possible to sensitively (detecting peptides down to the femtomole range) and easily (simultaneously quantifying 50 or more proteins) determine the absolute amounts of proteins with high degree of sequence homology (allowing peptides differing in molecular weight by less than 1 Da to be distinguished) in complex biological matrices, such as quantifying CYPs and UGTs superfamilies in biological tissues [16–17] or cultured cell lines [18]. And these MS-based protein quantification methods were highlighted as a promising approach for basic research of drug development such as drug-drug interaction study and PK/PD modeling. We recently also developed an isotope label-free LC-MS/MS method to determine the absolute amounts of CYPs and UGTs isoforms in human liver tissues and the results showed that this method was accurate and reproducible, and could lead to a better understanding of hepatic disposition and metabolism of drugs [19].
In this study, we systematically expounded activities and expression levels of seven major CYPs enzymes in liver tissues of HCC patients. The differential expression of CYPs enzymes within the tumor microenvironment compared with the surround normal tissues were monitored using the recently published LC-MS/MS method that can simultaneously measure these CYPs in one run using small sample size. We believed that our data showed why it would be beneficial for us to use LC-MS/MS for the determination of CYP levels that can be used for personalized treatment of HCC.
2. MATERIALS AND METHODS
2.1. Chemicals and Reagents
Substrates and metabolite standards were obtained from the following sources: phenacetin, acetaminophen, coumarin, 7-hydroxycoumarin, tolbutamide, 4-hydroxytolbutamide, dextromethorphan, dextrorphan, chlorzoxazone, 6-hydroxychlorzoxazone and 6β-hydroxytestonsterone were purchased from Sigma-Aldrich (St. Louis, MO). Testosterone, Taxol and 6-hydroxytaxol were from Melone-Pharmaceutical (Dalian, China). NADPH generating system was purchased from BD Biosciences (Woburn, MA). Sequencing grade trypsin was purchased from Promega (Madison, WI). PureLink RNA Mini Kit was purchased from Life Technologies (Gaithersburg, MD). PrimeScript™ RT Reagent Kit and SYPR Premix Ex Taq™ II (TliRnaseH Plus) were purchased from Takara Bio (Dalian, China). All other chemical were used as received.
2.2. Human Liver tissues
Approvals for tissue collection and studies were obtained from the NanFang Hospital Research Ethics Committee. All subjects, who also tested positive with HBV, had undergone anatomic or limited hepatectomy for HCC resection at Affiliated NangFang Hospital of Southern Medical University, Guangzhou, China. HCC tissues and matched pericarcinomatous tissues (tissue macroscopically dissected from the tumor, which was 1 cm away from the tumor lesions [20]) were obtained from 26 Chinese subjects (aged between 33 and 74 years; 51±11 years, mean±SD). The surgical specimens were confirmed by pathological examination and clinicopathological parameters. HCC tissues were classified into five grades (grade cannot be assessed, well differentiated, moderately differentiated, poorly differentiated and undifferentiated) according to the AJCC Cancer Staging Manual [21]. Only the moderately and/or poorly differentiated cases were selected in the present study. All the pericarcinomatous tissues were free of visible tumor nodules or tumor cell invasion, and most of them suffered from hepatitis B infection and/or cirrhosis (Supplementary Table S1). HCC and pericarcinomatous tissues were kept in ice-cold saline immediately after resection and used in preparation of liver microsomes right away (30 min usually) or stored in liquid nitrogen before RNA extraction.
2.3. Preparation of Human Liver Microsomes
Liver microsomes of tumors (tHLMs-individual) and matched pericarcinomatous tissues (nHLMs-individual) from 25 donors were processed using standard differential centrifugation procedures, which is essentially the same as those described previously in our publication [19]. A portion of 15 tHLMs-individual or matched nHLMs-individual were pooled together, named as nHLMs-pooled or tHLMs-pooled, based on equivalent protein amount to ensure the mixture represent the average of 15 individuals. Reference pooled human liver microsomes (rHLMs-pooled, derived from a pool of 22 Caucasian, 1 Hispanic and 1 African American, average age: 48±14 years) purchased from BD Biosciences were used as a control.
2.4. Total RNA extraction
Total RNA of liver tissues was isolated and purified using the PureLink RNA Mini Kit according to the manufacturer’s instructions. The concentration and purity of the total RNA was spectrometrically assessed using a Biospec-nano spectrophotometer (Shimadzu, Japan). The quality of RNA was determined based on A260/A280 ratio, which was 1.7–2.0 for all RNA preparations [22].
2.5. Measurement of CYP Activities in Human Liver Microsomes
Activities of the seven most important CYP isoforms, including CYP1A2, 2A6, 2C8, 2C9, 2D6, 2E1 and 3A4, were measured using human liver microsomes and probe substrates [23]. The enzyme kinetic profiles of the target CYP isoforms in tHLMs-pooled, nHLMs-pooled and rHLMs-pooled were measured by incubation with a range concentration of probe substrates in vitro. And the activities of these isoforms in each patient were determined by incubation of microsomes with specific substrate concentrations, at which best specificity was obtained. Assay details are provided in Supplementary Table S2. The assay conditions, such as protein concentration and incubation time, were validated to provide linear enzyme kinetics. In general, microsomes were mixed on ice with buffer (pH 7.4; 100 mM Tris-HCl buffer for CYP2A6, and 50mM potassium phosphate buffer for the other isoforms), NADPH generating system and substrate to a final incubation volume of 250μl (500μl for CYP3A4). Incubations were carried out in a shaking water bath (150 rpm) at 37°C for an optimized period. The final solvent concentration (acetonitrile or methanol) was ≤ 1%. Incubations were typically terminated by addition of 200μl of ice-cold methanol containing internal standard (except for the CYP3A4 assay; see below). The terminated incubation mixtures, as well as standard curve and quality control samples that composed of the same matrix materials but without NADPH generating system, were centrifuged for 30 min at 18, 000×g, and the supernatants was subjected to UHPLC for analysis. Incubations for the CYP3A4 assay were terminated by addition of 4 ml ice-cold dichloromethane containing internal standard. After centrifugation, the supernatant was transferred to another glass tube and evaporated to dryness. The residue was reconstituted with 200 μl methanol-water (50:50, v/v) for injection. All incubations were performed in triplicate.
Formation of 7-hydroxycoumarin (CYP2A6), 6-hydroxytaxol (CYP2C8), dextrorphan (CYP2D6) was determined using an Agilent 1290 Infinite UHPLC system coupled with Agilent 6460 triple quadruple mass spectrometer equipped with an ESI source (Agilent Technologies Inc., USA) in positive ionization mode. The formation of acetaminophen (CYP1A2), 4-hydroxytolbutamide (CYP2C9), 6-hydroxychlorzoxazone (CYP2E1), or 6β-hydroxytestonsterone (CYP3A4) was monitored using Agilent 1290 Infinite UHPLC or Waters ACQUITY UPLC on an Agilent Zorbax SB-C18 column (1.8 μm, 3.0×100 mm), with an UV detector. All the analytical methods were validated and the optimal instrument-dependent and compound-dependent parameters were summarized in Supplementary Table S3. MassHunter Acquisition Software Rev. B.6.00 (Agilent Technologies) or Empower Software (Waters) was used for data acquisition.
2.6. Absolute Quantification of CYP Proteins in Human Liver Microsomes by Using an Isotope Label-free LC-MS/MS Method
Protein expression levels of the seven CYP isoforms were determined simultaneously by using an isotope label-free LC-MS/MS method as described previously, with some minor modifications [19]. Protein expression levels were determined by quantifying proteotypic peptides produced by (high purity) trypsin digestion, using synthetic standard peptides (APeptide Co., Shanghai, China). All samples were analyzed by using an Agilent 6460 triple quadruple mass spectrometer coupled with 1290 Infinite UHPLC system. The liquid chromatography separation was carried out on an Agilent Zorbax SB-C18 column (1.8 μm, 3.0×100 mm). Sample volume of 10 μl was injected to the column and flow rate was 0.3ml/min. Sample rack and column temperatures were 10°C and 40°C, respectively. Mobile phase A was high purity HPLC-grade water with 0.1% (v/v) formic acid, whereas mobile phase B was 100% acetonitrile with 0.1% (v/v) formic acid. The gradient profile used was as follows (time: mobile phase A: mobile phase B): 0 min: 95:5, 10 min: 55:45, 12–14 min: 20:80, 15 min: 95:5; post time, 3min. Quantification was performed in positive ion mode and the mass spectrometer was set up to run a Dynamic MRM experiment for peptides. The instrument settings were as follows: capillary voltage, 4000 V; nebulizer gas, 30 psi; gas temperature and sheath gas heater were 250 and 300°C, respectively; gas flow and sheath gas flow were 15 and 11 L/min; MRM detection window, 60 s and target scan time, 600 ms. The MRM transitions selected for each peptide were the same as described previously [19] and their compound-dependent parameters were listed in Supplementary Table S4.
2.7. Real-time PCR Gene Expression Analysis
Full-length sequences of the target genes were obtained from GeneBank, and the primers (forward and reverse) were designed using Primer Premier 6.0 (Premier Biosoft, CA) software based on the criteria described previously [24]. The uniqueness of the primer sequences designed was assessed with the BLAST search (National Center for Biotechnology Information). Details of primers and inventory used in this study are provided in Supplementary Table S5. All these oligonucleotide primers were custom-synthesized by BGI Tech. (Shenzhen, China).
For first-strand cDNA synthesis, 1600 ng of total RNA was reverse-transcribed in a final volume of 40 μl following the protocol for the PrimeScript™ RT Reagent Kit. Reactions were diluted twenty times with water for all real time PCR experiments. Real time PCR was performed in an ABI 7500 Fast Real-Time PCR System (Applied Biosystems) using SYPR Premix Ex Taq™ II. Thermocycling was carried out in a final volume of 20 μl containing 2.0 μl of the diluted cDNA samples, 40 nm each of the primers (forward and reverse), and 10.4 ul of SYBR Premix Ex Taq II (including Taq DNA polymerase, reaction buffer, and deoxynucleotide triphosphate mixture). The PCR amplification consisted of 40 cycles (95°C for 5 s, 60°C for 34 s) after an initial denaturation step (95°C for 30 s). All measurements were performed in triplicate. Relative expression differences were calculated using the comparative ΔΔCt method [25], and Ct values were normalized to GAPDH expression levels. The relative expression levels of the target genes were presented as 2−ΔΔCt×102, and the LLOQ was defined as 2−ΔΔCt=1×10−5.
2.8. Data Analysis
Shapiro-Wilk test of normality was used to check the distribution shape of the data, using the SPSS Statistics 20.0 software. Paired-samples T-test was employed to analyze normally distributed data. For non-normally distributed data, Wilcoxon’s sign rank test was used. Correlation analyses were performed using Pearson product-moment correlation for normally distributed data, and Spearman’s rank correlation for non-normally distributed data. A p-value of < 0.05 was considered to be the minimum level of statistical significance (two-tailed) for all the statistical analyses.
Kinetic parameters were estimated fitting the proper models (Michaelis-Menten, autoactivation, substrate inhibition or biphasic kinetic) to the substrate concentrations and initial rates using GraphPad Prism 6.0 software, aided by profiles of the Eadie-Hofstee plots as previously described [26].
Turnover number (TON), which indicates the catalytic capacity of enzyme, is defined as the number of molecules of substrate that an enzyme protein can convert to product per unit of time (a turnover rate), and can be calculated as follows:
where V is the reaction rate at a specific substrate concentration, and Et is the total enzyme concentration. Since the absolute amount of each CYP isoform was quantified in human liver microsomes of patients with HCC using LC-MS/MS method, we were able to obtain the TONs of probe substrates metabolized by CYPs in tumors or pericarcinomatous tissues.
3. RESULTS
3.1. Activities of CYP Enzymes were Decreased Significantly in Tumor Tissues of Patients with HCC
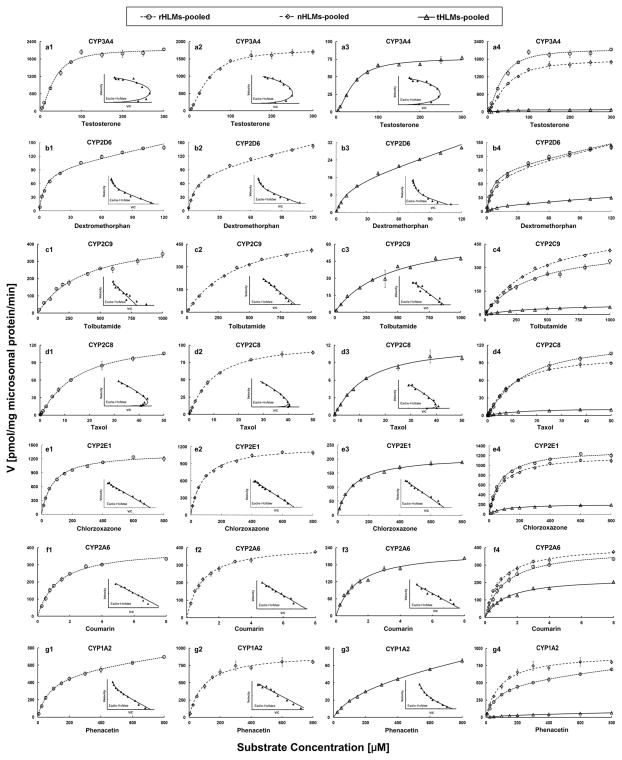
Kinetic profiles of seven CYP enzymes were determined in tHLMs-pooled, nHLMs-pooled, and rHLMs-pooled. As shown in Fig. 1 and Supplementary Table S6, activities of all these CYPs were remarkably decreased in the tHLMs-pooled compared with those in the nHLMs-pooled and rHLMs-pooled. The intrinsic clearance (CL) of testosterone (calculated by 6β-hydroxytestosterone) metabolized by CYP3A4 was more than 20-fold lower in tHLMs-pooled (1.95 μl/mg/min) than in nHLMs-pooled (40.05 μl/mg/min) as a consequence of a decrease of its Vmax of metabolism (76.71±2.22 versus 1796±34.67 pmol/mg/min). The CL of probe substrates metabolized by CYP2C9 and CYP2D6, which were active in the metabolism of ~40% marketed drugs [27], were decreased over 6-fold in HCC tumors. The activity of CYP1A2 involved in precarcinogen activation was also drastically declined (35-fold) in tHLMs-pooled (0.28μl/mg/min) compared with that in nHLMs-pooled (9.68 μl/mg/min). Additionally, the activities of other major CYP isoforms expressed in human liver, including CYP2C8, 2A6 and 2E1, were also assessed, and remarkable decreases (ranging from 2.5- to 11-fold) in the CL were observed in HCC tumors.
Figure 1.
Enzyme kinetic analysis for measuring activities of seven CYPs in rHLMs-pooled (Fig. a1–g1, circles and dotted line), nHLMs-pooled (Fig. a2–g2, diamonds and dashed line) and tHLMs-pooled (Fig. a3–g3, triangles and solid line) by using specific probe substrates. In each figure, the inset shows Eadie-Hofstee plot. (Fig. a4–g4) comparison of activity of each CYP isoform in rHLMs-, tHLMs- and nHLMs-pooled. Each data point represents an average of three determinations and the error bar represents standard deviation of the mean (n=3).
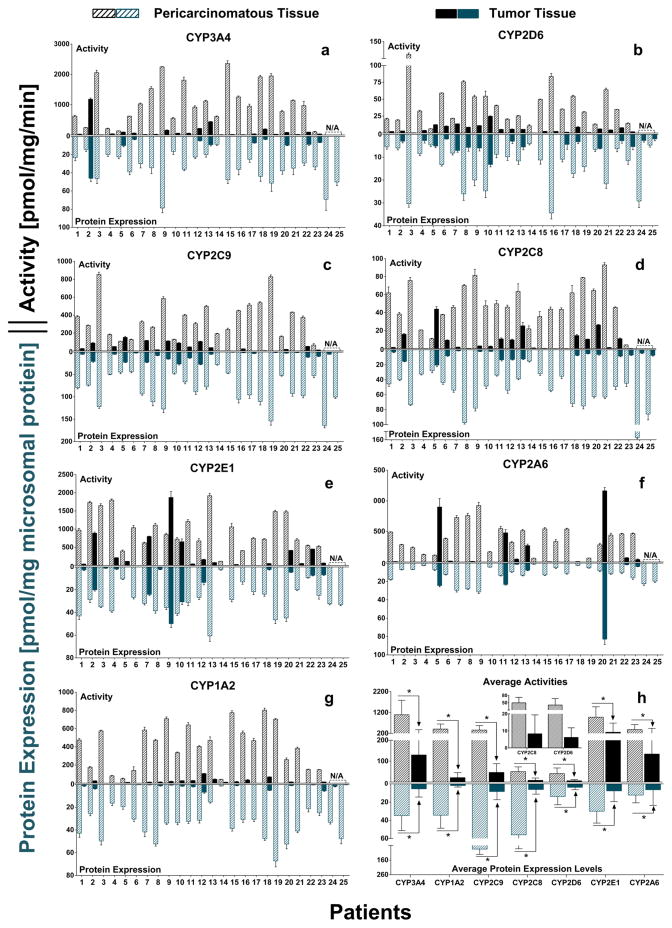
CYPs activities in individuals with HCC were also measured by using selected concentrations of probe substrates. As shown in Fig. 2 and Supplementary Table S7, the activities of CYP3A4, the most important drug metabolizing CYP isoform, were significantly decreased (ranging from 2 to 95-fold) in tumors of 96% patients compared with their corresponding pericarcinomatous tissues. The average formation rate of 6β-hydroxytestosterone metabolized by CYP3A4 in nHLMs-individual was 1092.2±693.9 pmol/mg protein/min, while that in tHLMs-individual was 129.6±248.0 pmol/mg protein/min. Amoung the CYP isoforms most attributable to the carcinogenesis processes including CYP1A2, 2A6 and 2E1, activity of CYP1A2 was drastically decreased in tumors by more than 10 folds in 83% patients. The average activity of CYP1A2 in 23 tHLM-individual was 19-fold lower than that in nHLMs-individual. In contrast, activity of CYP2A6 was decreased by more modest 2.8 fold, and 2 in 23 subjects showed an increased level (p<0.05). This smaller decrease was similar to that of CYP2E1 (3.8 fold decrease).
Figure 2.
Activities and protein expression levels of seven CYP enzymes in human liver microsomes prepared from HCC tumors (tHLMs-individual) and matched pericarcinomatous tissues (nHLMs-individual) (Fig. 2a–2g). Activities of CYPs were measured using probe substrates, and absolute protein amounts were quantified using an isotope label-free LC-MS/MS method. All the experiments were performed in triplicate, and data are presented as mean±SD. Paired-samples T-test was used for data analysis. (h) Average activity and protein expressions of each CYP isoform in 25 tumors and pericarcinomatous tissues. Wilcoxon’s sign rank test was used for data analysis. “*” denotes statistical significance (p<0.05).
3.2. Protein and mRNA Expression Levels of CYP Enzymes were Significantly Down-regulated in Tumor Tissues of Patients with HCC
In order to clarify the reason for the significantly reduced activities of CYPs in HCC tumor microsomes, CYPs protein and mRNA expression levels were also determined in tumors and matched pericarcinomatous tissues of each patient using isotope label-free LC-MS/MS method and real-time PCR, respectively. The protein expression profiles of the measured CYPs were depicted in Fig. 2 and Supplementary Table S8. Group comparison of CYP3A4 protein expression in tumors and paired pericarcinomatous tissues showed a mean amount of 5.5±8.9 and 34.8±16.5 pmol/mg microsomal protein, respectively, corresponding to a 6.3-fold reduction in tumors. Among the most abundantly expressed CYP isoforms (i.e., top 3) which are CYP2C9, CYP2C8 and CYP1A2, all of them were expressed at remarkably lower levels in tumors than in matched pericarcinomatous tissues of all 25 patients, and the difference in average CYP amount was 10.1-, 9.1-, and 19.3-fold, respectively (Fig. 2h).
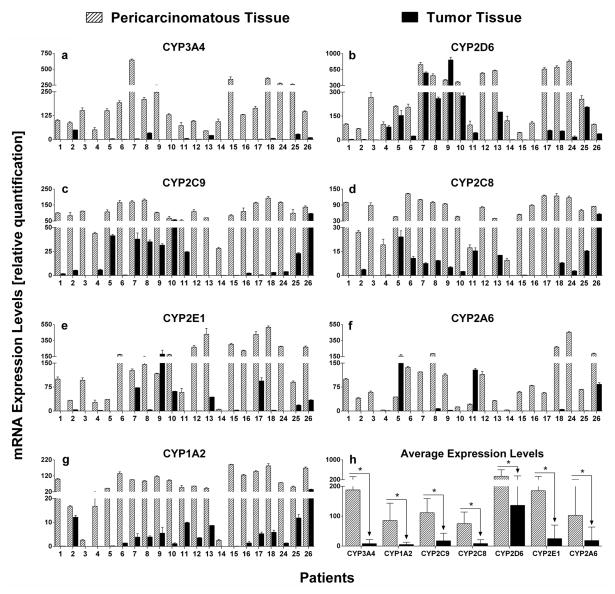
The mRNA expression levels of CYPs in tumors and pericarcinomatous tissues were also measured. The results showed that CYP3A4, 2D6, 1A2, 2C8, and 2E1 were unequivocally expressed in all samples of HCC tumors and paired non-tumor tissues (Fig. 3 and Supplementary Table S9). In contrast, CYP2C9 and CYP2A6 were undetectable in 1 (No. 13) and 3 samples (No. 7, 12, and 13) of tumors, respectively. We found that in pairwise comparisons, aloss of CYP expression in tumors was evident in most patients. The mRNA levels of CYP3A4 and CYP1A2 were drastically decreased (by 23- and 16-fold on average, respectively) in tumors of all subjects, and greater than 10-fold decrease were observed in over 75% of subjects. On the other hand, mRNA expression levels of CYP2D6 were decreased less than 3 fold in 42% of subjects by only a more modest extent of 2.5-fold. The average mRNA expression levels of CYP2C9, 2C8, 2E1, and 2A6 were decreased significantly by 6.3-, 9.0, -7.4, and 5.6-fold, respectively.
Figure 3.
mRNA expressions of seven CYP enzymes in tumors and matched pericarcinomatous tissues of 21 human livers with HCC(Fig. 3a–3g). Relative expression differences were calculated using the comparative ΔΔCt method, and Ct values were normalized to GAPDH expression levels. All measurements were performed in triplicate and the relative expression levels of the target genes were presented as 2−ΔΔCt×102. Data are presented as mean±SD, and paired-samples T-test was used for data analysis. (h) Average mRNA expression levels of each CYP isoform in 21 tumors and pericarcinomatous tissues. Wilcoxon’s sign rank test was used for data analysis. “*” denotes statistical significance (p<0.05).
3.3. Correlation of Protein and mRNA Expression Levels with Activities of CYP Enzymes
The activities of seven CYPs were compared with their protein and mRNA expression levels. As shown in Table 1, there was a much better correlation of enzyme activity to protein expression than to mRNA expression. For all the measured CYPs in pericarcinomatous tissues, high correlations between activities and protein expressions were observed with correlation coefficient values of more than 0.69 (in particular, correlation coefficient for CYP2D6 and 2A6 were 0.93 and 0.92, respectively). In HCC tumors, CYPs showed a similar correlation between activities and protein levels with correlation coefficient of better than 0.54. Moreover, slopes of correlation between protein and activity of CYPs in tumors was comparable to that in pericarcinomatous tissues with maximal difference of 1.7 fold, suggesting the similar catalytic efficiencies for CYP enzymes present in tumors and pericarcinomatous tissues.
Table 1.
Correlation of Enzyme Activity to Protein Level and mRNA Expression Level of CYP enzymes.
| Isoforms | Activity vs Protein
|
Activity vs mRNA
|
Protein vs mRNA
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pericarcinomatous
|
Tumor
|
Pericarcinomatous
|
Tumor
|
Pericarcinomatous
|
Tumor
|
|||||||
| r2a | slopeb | r2 | slope | r2 | slope | r2 | slope | r2 | slope | r2 | slope | |
| CYP3A4 | 0.804* | 35.6 | 0.728* | 25.5 | 0.492* | 4.5 | 0.426 | 23.1 | 0.690* | 0.12 | 0.590* | 0.85 |
| CYP2D6 | 0.928* | 3.0 | 0.767* | 1.7 | < 0.100 | - | 0.670* | 0.02 | 0.259 | 0.03 | 0.827* | 0.01 |
| CYP2C9 | 0.799* | 4.6 | 0.851* | 4.6 | 0.204 | 2.9 | 0.565* | 2.6 | 0.543* | 0.66 | 0.593* | 0.49 |
| CYP2C8 | 0.792* | 1.0 | 0.907* | 1.5 | 0.458 | 0.6 | 0.712* | 1.4 | 0.584* | 0.59 | 0.578* | 0.88 |
| CYP2E1 | 0.727* | 32.2 | 0.687* | 33.1 | < 0.100 | - | 0.491* | 10.2 | < 0.100 | 0.11 | 0.564* | 0.31 |
| CYP2A6 | 0.917* | 30.3 | 0.828* | 29.2 | 0.337 | 3.2 | 0.269 | 4.9 | 0.412 | 0.11 | 0.138 | 0.17 |
| CYP1A2 | 0.688* | 13.4 | 0.544* | 13.7 | 0.590* | 4.3 | 0.577* | 4.8 | 0.477* | 0.29 | 0.482 | 0.40 |
r2 is Pearson’s or Spearman’s correlation coefficient, and “*” denotes statistical significance of association (p < 0.05).
slope of regression line.
Protein expression levels of CYPs were also compared with mRNA levels. As shown in Table 1, a moderate correlation was observed between protein and mRNA levels of CYP3A4, 2C9 and 2C8 in both tumors and pericarcinomatous tissues with correlation coefficient of 0.6. For CYP2D6 and CYP2E1, better correlations were observed in tumors (0.83 and 0.56, respectively) compared with that in pericarcinomatous tissues (no correlation).
3.4. Catalytic Capacity of CYP Enzyme was not Seriously Impaired in HCC Tumors
As shown in Table 2, with the exception of CYP3A4, TONs of CYP enzymes in HCC tumors were nearly identical to the values observed in pericarcinomatous tissues with similar ranges of 95% confidence interval (p>0.05), suggesting that the catalytic efficiency of CYPs was not seriously impaired in HCC tumors. Catalytic efficiency of CYP2D6 in tumors was significantly lower than that in matched pericarcinomatous tissues, while catalytic efficiency of CYP2C9 was significantly higher in tumors but the maximal difference was only 1.6-fold. There is no significant difference between TONs of CYP3A4 tHLMs- and nHLMs-pooled. However, both of them are more than 2-fold lower than that in rHLMs-pooled. Moreover, a large interindividual variability of TON of CYPs was observed in human livers (Table 2). TONs of CYP3A4 and CYP2C8 in pericarcinomatous tissues exhibited the greatest interindividual differences, with about 19-fold difference between the donors with the highest and lowest levels (57.0 versus 3.0 pmol 6β-hydroxytestosterone/min/pmol CYP3A4, and 1.7 versus 0.1 pmol 6-hydroxytaxol/min/pmol CYP2C8). The other isoforms, including CYP2C9, 2D6, 1A2, 2E1, and 2A6 varied from modest 2.7 to 4.5 fold. This may be caused by the genetic polymorphisms of CYPs, by which people were divided into groups of poor, intermediate, extensive and ultrarapid metabolizers [28].
Table 2.
Summary of Turnover Numbers of Probe Substrate Metabolized by CYP Enzymes.
| Isoforms | TON (HLM-pooled)a
|
TON (nHLMs-individual)b
|
TON (tHLMs-individual)c
|
nd | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | t | r | Mean | SD | Max | Min | Max/Min | 95% CIe | Mean | SD | Max | Min | Max/Min | 95% CI | ||
| pmol metabolite formed/min/pmol CYP enzyme | ||||||||||||||||
| CYP3A4 | 24.7 | 17.4 | 50.3 | 27.5 | 17.6 | 57.0 | 3.0 | 19.0 | 14.9~40.1 | 24.2 | 20.7 | 62.1 | 5.1 | 12.2 | 9.4~39.0 | 10 |
| CYP2D6 | 6.5 | 4.8 | 6.4 | 2.9* | 1.1 | 5.6 | 1.3 | 4.5 | 2.2~3.5 | 1.8* | 1.2 | 5.1 | 0.3 | 16.8 | 1.2~2.5 | 15 |
| CYP2C9 | 5.3 | 5.1 | 6.7 | 4.0# | 1.2 | 6.4 | 2.4 | 2.7 | 3.3~4.8 | 6.4# | 3.5 | 16.0 | 3.3 | 4.9 | 4.3~8.5 | 13 |
| CYP2C8 | 1.5 | 1.5 | 3.0 | 1.0 | 0.4 | 1.7 | 0.1 | 18.3 | 0.7~1.2 | 1.5 | 1.0 | 4.3 | 0.04 | 107.5 | 0.9~2.1 | 14 |
| CYP2E1 | 13.3 | 13.3 | 14.0 | 31.2 | 15.7 | 60.4 | 17.7 | 3.4 | 20.0~42.4 | 35.2 | 25.5 | 91.6 | 9.9 | 9.2 | 17.0~53.4 | 10 |
| CYP2A6 | 20.4 | 19.0 | 12.9 | 34.0 | 9.6 | 45.8 | 16.0 | 2.9 | 25.1~42.8 | 27.8 | 12.2 | 41.3 | 11.2 | 3.7 | 16.5~39.0 | 7 |
| CYP1A2 | 30.0 | 41.3 | 37.7 | 11.6 | 5.1 | 19.8 | 5.9 | 3.3 | 6.9~16.4 | 12.2 | 5.8 | 19.8 | 2.4 | 8.1 | 6.8~17.6 | 7 |
turnover numbers of probe substrates metabolized by CYPs enzymes in nHLMs-, tHLMs- and rHLMs-pooled, which was calculated as the ratio of maximal metabolic rate (Vmax) to the total amount of CYPs enzyme reported previously [19].
turnover numbers of probe substrates metabolized by CYPs enzymes in nHLMs- and tHLMs-individuals. Each value was calculated at a specific concentration of probe substrate that was showed in Supplementary Table S2.
numbers of donors were selected for calculating turnover numbers. CYP activities and protein amounts could be accurately quantified (higher than LLOQ) in both tumor tissues and matched pericarcinomatous tissues of these patients.
95% CI represents the 95% confidence interval. Paired-samples T-test or Wilcoxon’s sign rank test was used for data analysis.
“*” or “#” denotes statistical significance (p<0.05).
The results of kinetic analysis showed that the Km values in different groups (nHLMs-, tHLMs- and rHLMs-pooled) were comparable (with maximal difference of <1.9-fold) for all the measured CYPs, indicating that liver disease did not alter the affinity of CYPs (Fig. 1 and Supplementary Table S6). Additionally, Eadie-Hofstee plots in Fig. 1 showed that, with exception of CYP1A2, the kinetic mechanisms of CYPs were not changed in human livers with cirrhosis or HCC compared with healthy livers.
3.5. Activities of CYP Enzymes were not Seriously Impaired in Pericarcinomatous Tissues of HCC Tumors
Although suffering from hepatitis B infection and/or cirrhosis, the activities of all the determined CYPs in nHLMs-pooled were comparable to that in rHLMs-pooled. As shown in Fig. 1 and Supplementary Table S6, The CL and Vmax values of probe substrates metabolized by the seven measured CYPs enzymes in nHLMs-pooled were similar to that in rHLMs-pooled with a maximal difference of 1.6 fold, which were consistent with the alterations of protein and mRNA expressions reported previously [19, 29]. Additionally, further studies with large sample size of healthy human livers were necessary for reaffirming this initial observation.
3.6. Comparison of Activity or Expression Levels between Seven CYP Isoforms
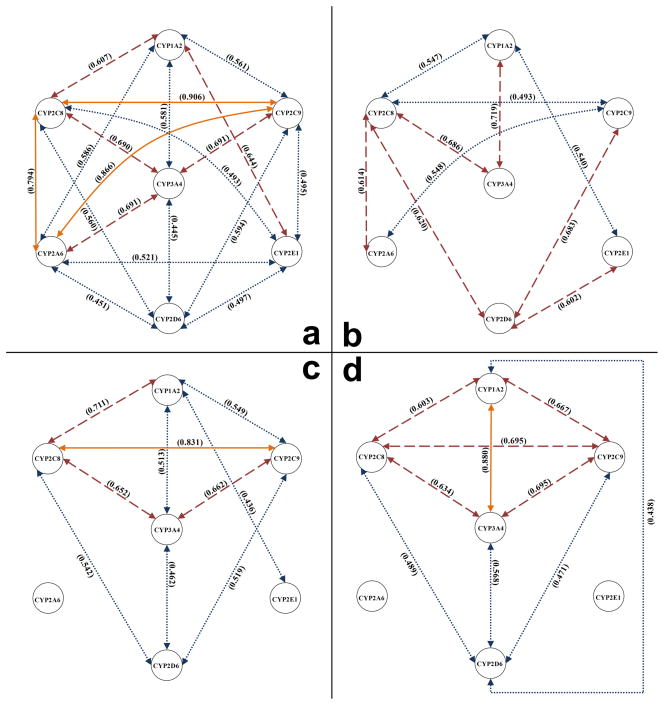
The activities, mRNA or protein expression levels of the seven measured CYPs were compared. As shown in Fig. 4a, mRNA expressions of CYPs were generally correlated to each other in pericarcinomatous tissues of HCCs. A high degree of correlation was observed between mRNA expressions of CYP2C8 and CYP2C9, which are both transcribed from a cluster of CYP genes located on chromosome 10q24, with a correlation coefficient of 0.906. mRNA expression of CYP2A6, which is located on chromosome 19q13, was also found to be highly correlated to that of CYP2C9 and CYP2C8 (r2=0.866 and 0.794, respectively). Medium correlations were observed between CYP3A4 and the other isoforms except for CYP2D6 and 2E1. mRNA expressions of CYP2D6 and CYP2E1 showed only weak to medium correlations to the other CYPs expressions. In HCC tumors, correlations of the mRNA expressions of CYPs were generally weakened or even lost, although correlations between CYP2D6 and CYP2C9/2C8/2E1 got slightly better (Fig. 4b).
Figure 4.
Correlation Analysis of Various mRNA and Protein Levels. (a) Correlations between mRNA expression levels of seven CYP isoforms in pericarcinomatous tissues of patients with HCC (n=21). (b) Correlations between mRNA expression levels of seven CYPs in HCC tumors (n=21). (c) Correlations between protein expression levels of seven CYPs in pericarcinomatous tissues (n=25). (d) Correlations between activities of seven CYPs in pericarcinomatous tissues (n=23). Correlation analyses were performed using Pearson product-moment correlation for normally distributed data, and Spearman’s rank correlation for non-normally distributed data. Two of the seven isoforms were linked with each other using yellow solid lines (r2 ≥ 0.8), red dashed lines (0.8> r2 ≥ 0.6), or blue dotted lines (r2 < 0.6), when significant correlation (p<0.05) was observed.
As shown in Fig. 4c, protein expressions of CYP3A4, 1A2, 2C8, 2C9 and 2D6 were significantly correlated to each other with a similar degree of correlation between their mRNA expression levels in pericarcinomatous tissues of HCC. In contrast, no significant correlation was observed when comparing the protein expression of CYP2A6 and CYP2E1 to the other CYP isoforms (except for correlation between CYP2E1 and 1A2), even though their mRNA expressions showed a medium to high correlation to each other. This indicated that CYP2A6 and 2E1 might be regulated in a unique posttranscriptional mode such as alteration of the translational efficiency and/or stability of mRNA and protein [30]. A similar pattern of correlations observed for protein expression and enzyme activity (Fig. 4c and d). The correlations between activities of CYP1A2 and CYP2D6 (r2=0.438, p<0.05) were better than that between their protein expression levels (r2=0.329, p>0.05) and mRNA expression levels (r2=0.105, p>0.05). This may be caused by the lack of high degree of specificity of CYP1A2 probe substrate phenacetin, which was also found to be a substrate of CYP2D6 [31].
3.7. Clinicopathological relevance of CYP expression levels in HCC
Activities, protein and mRNA expression levels were statistically compared with histology and clinicopathological parameters of HCC patients. However, no correlation was observed, and we suspected that a larger number of samples are probably required to establish such relationships.
4. DISCUSSION
Our unprecedented systemic study of the effects of HCC on CYPs has provided strong evidence to argue strongly for the development of individualized therapy for each HCC patient. This is because all the major drug-metabolizing CYPs are severely dysregulated by liver cancer carcinogenesis such that the different drugs will be affected differently in different patients (i.e., there is no general pattern to follow for all HCC patients). Because liver biopsies are available, our results also support the use of highly sensitive LC-MS/MS method that requires small sample volume as a means to quantify the protein and activity levels of CYPs in both HCC and pericarcinomatous tissues. We believe that these levels are indicative of relevant CYP activities and can be used to guide clinical selection of drugs for developing an individualized treatment plan for the HCC patients.
Our systemic studies of the impact of HCC on liver CYPs are unprecedented. First, we derived our results using human HCC tumors removed from patients, whereas most previous publications have focused on the effect of other chronic liver diseases (e.g., cirrhosis) on CYPs [32–33]. Second, we employed three complementary sets of measurements: activities, protein determination by LC-MS and quantitative PCR whereas in a few earlier studies on HCC, the authors employed primarily quantitative PCR [15, 29]. We and others have shown that the PCR-derived gene expression levels are often not correlated with CYP activities, which were commonly viewed as a consequence of post-transcriptional regulations (e.g. mRNA and protein stabilization/degradation) [34]. Third, our studies provided the first sets of evidence that are enriched with correlations between activities, mRNA and protein levels to support the theory that HCC tissues generally express much less as well as more variable (or less predictable) levels of seven CYPs that are important for drug metabolism. Therefore, for patients suffering from inoperable HCC, liver biopsy followed by LC-MS/MS CYP quantitation may be a good starting point for the development of an individualized chemotherapy or chemoablation therapy. Ideally, selection of drugs based on HCC CYP levels could identify drugs that are highly active in the tumor but less or non-toxic in normal liver cells. Because we can use a small amount of tissues to rapidly quantitate these CYPs, liver biopsy samples should provide sufficient amount of tissues for determining the CYP levels.
Our results may also explain why sorafenib, a first-line therapeutic drug for liver cancer, was quite effective against HCC and yet have only manageable side effects. The compound was recently shown by us to have significantly decreased metabolism in HCC tumor microsomes [35], probably as a consequence of lost of CYP3A4 and UGT1A9 activities. The results of our studies, which showed a large decrease in CYP3A4 activities in HCC tissues, suggest that sorafenib could serve as an ideal drug for both chemoembolization and hepatic drug infusion. Indeed, a PubMed search conducted in March 19, 2015 using chemoembolization and sorafenib produced more than 300 hits, most of which are relevant for its use in chemoembolization.
We could apply this approach of individualized therapy to other anticancer drugs that are substrates of these seven CYPs since we found that the activities of seven major CYPs in human livers were significantly decreased in HCC tumors. Interestingly, enzyme activities in adjacent non-tumor (or pericarcinomatous) tissues were similar to that in healthy human livers (rHLM-pooled) (Fig. 1). This suggests that the decreases in CYPs activities were localized mainly to the malignant regions or tumor tissues. The latter is similar to what Hong et al. observed previously [29], where mRNA expressions of CYP1A2, 2C9, 2E1 and 3A4 were drastically decreased in HCC tumors but not in the matched pericarcinomatous tissues or liver tissues with HBV cirrhosis.
We have stated that changes in protein levels as measured by LC-MS/MS could be particularly suitable for determining the corresponding CYP activity in HCC patients. However, metabolic rates of a substrate are impacted by the expression levels of CYPs or the structures of the enzymes (via mutation for example). How could we solve this concern? First of all, we found high degree of correlations between activities and CYP protein amounts in both tumors and matched pericarcinomatous tissues (Fig. 2 and Table 1). We then determined the catalytic efficiency of an enzyme, where changes in enzyme structures due to post-translational modifications and/or noncovalent binding of allosteric effectors could be measured [36]. In this study, similar turnover numbers (with maximal difference of ~2 fold) of probe substrates metabolized by CYPs (except for CYP3A4) were observed (Table 2), suggesting that the catalytic efficiencies of CYPs were not seriously impaired in HCC tumors. Moreover, kinetic analysis also showed same kinetic mechanisms with similar Km values (Fig. 1 and Supplementary Table S6). This indicated that the functional properties of CYPs were not impaired seriously by HCC. Therefore, the drastic decrease in activities of CYPs in HCC tumors was likely to be caused by lower protein expression levels, not lower catalytic efficiency of the expressed enzymes. The latter suggest that there were no significant amount of CYP mutants present in the HCC or they would have been measured and counted as normal CYPs, unless the mutation occurred in our proteotypic peptide region (which is highly unlikely).
We have also said that it would be especially important to derive individualized treatment plan for HCC because each HCC patient has a unique CYP expression profile. The unique expression profile was the result of CYP dysregulation in HCC as shown in Fig. 4. The dysregulation could be demonstrated in multiple levels. First, the mRNA expressions of CYPs were generally correlated to each other in pericarcinomatous tissues, but those correlations worsened considerably in HCC samples. Good correlation is expected in absence of major diseases since CYPs gene expressions are tightly regulated by overlapping nuclear receptors. For example, CYP 2C9, 2C8 and 2A6 could be regulated by the same nuclear receptors including PXR (NR1I2), CAR (NR1I3), HNF4A, and PPARA, which might explain the high correlation between their mRNA expression levels (Fig. 4a) [37–38]. The correlations between CYPs mRNA expressions were often drastically reduced (albeit not always) in HCC samples, which indicates less co-regulation of these CYPs in HCC tumors (Fig. 4b). Second, there is significant alteration of expression of nuclear receptors governing CYP expression. It was reported that expression levels of nuclear receptors such as PXR, CAR, and AHR were significant decreased in HCC tumors compared with matched pericarcinomatous tissue [29]. These changes in nuclear receptors expression should impact the expression of CYPs, as shown in the current study. Lastly, changes in other expression modulators or the epigenetic alterations such as hypermethylation may also result in decreased expression of CYP genes [39–40]. Changes in inflammation signaling through the Toll-like receptor (TLR) pathway may also affect the expression of CYPs in these patients who are all HBV-positive [41–42]. For example, Raunio et al. [43] reported that CYP2A6 protein could be overexpressed selectively in a subpopulation of HCC tumor cells. They also found that chronic inflammation associated with HCC could lead to an elevation of CYP2A6 content in areas adjacent to inflammation. Taken together, there is strong evidence to conclude that CYP expression in HCC is severely dysregulated compared to the matching pericarcinomatous tissues, and multiple factors may be involved to cause this dysregulation.
The efficacy of anticancer therapy is usually limited by an inability to predict tumor response and toxicity of chemotherapy drugs [12]. Therefore, an important question for HCC therapy is: would the large differences (mostly down-regulation and sometimes drastic) in the activities of seven major CYPs (Fig. 2) observed here make a significant difference in outcome if they were integrated into design of chemotherapy or chemoablation therapy? We hypothesize that an improvement is highly likely although a perspective clinical would be needed. There are several evidences to support our hypothesis. First of all, some anticancer drugs need activation via CYPs. For example, doxorubicin and tegafur that are activated by CYP3A4 and CYP2A6 respectively, are the most commonly described agents for systemic or chemoembolization HCC therapy [13, 44–45]. For patient NO.20, in which CYP2A6 was overexpressed and CYP3A4 was decreased in tumors (Fig. 2a and f), tegafur (activated by CYP2A6) would be a better choice. In contrast, doxorubicin (activated by CYP3A4) could be a better choice for the treatment of patient NO.2, in which CYP3A4 was overexpressed in tumors. On the other hand, many anticancer drugs are inactivated by CYPs. For example, etoposide, which was inactivated by CYP3A4, 1A2 and 2E1 [13, 46], could be more widely used in patients with HCC, since these CYP isoforms were poorly decreased in most HCC tumors. Taxol, another important anticancer drug used in HCC, was employed as a probe substrate of CYP2C8 (responsible for 85% of taxol metabolism [47]) in the current study and was found with drastically decreased clearance rate in most HCC tumors (Fig. 1 and 2). Hence, we believe that the CYP expression levels should be applied to HCC therapy, and together with evidence of susceptibility of HCC to particular drugs derived from other pharmacodynamic or pharmacogenetic studies, they can be integrated to design the best individualized therapy for HCC patients.
It is expected to predict the effect reliably for drugs mainly metabolized by a single isoform (e.g. taxol). However, it will be complicated to predict the effects of drugs disposed by multiple isoforms such as etoposid (inactivated by CYP3A4, 1A2 and 2E1), and contributions of each enzyme to drug metabolism should be taken into account, together with the alterations of enzyme activities. Additionally, significant inter-individual variations of CYP expressions were observed among the patients, which was probably due to genetic polymorphisms and/or other confounding factors such as diet and alcohol consumption. These factors should also be considered for personalized therapy. For example, expression of CYP3A4 in pericarcinomatous tissue of patient NO.3 was approximately 10-fold higher than in NO.4 (Fig. 2a). Therefore, sorafenib was expected to inactivated by CYP3A4 more rapidly and have less toxicity in the non-tumor tissues of NO.3 than in NO.4, while high degree of cytotoxicity were expected to be observed in tumors of both patients.
Lastly, we argue strongly that LC-MS/MS derived CYP levels are the most relevant readout for clinical action. The primary reason is that high correlations between enzymes activities and protein expressions of CYPs were observed in both HCC tumors and pericarcinomatous tissues. This result suggested that activities of CYPs are better predicted by their protein levels than by mRNA expressions levels. This conclusion is supported by earlier investigation reported by this and other laboratories [34]. Although our current method are somewhat demanding technically, the use of isotope labeled signature peptides could substantially decrease this burden [16–17].
In conclusion, the activities and protein amounts of seven major CYP enzymes were found to be significantly and sometimes drastically decreased in HCC tumors, probably as a result of either decreased transcriptions or decreased mRNA stabilities. However, the catalytic capacities of these enzymes were not changed with similar turnover numbers of substrates in tumors and pericarcinomatous tissues. High correlations between enzymes activities and protein expression levels of CYPs were observed in both tumors and pericarcinomatous tissues of HCC, suggesting that activities of CYP enzymes could be well predicted by their protein amounts. Therefore, we proposed that determination of the CYP protein expression profile by LC-MS/MS in each patient is a promising approach that can be clinically used for individualized treatment of HCC.
Supplementary Material
Acknowledgments
Financial Support: This work was supported by grants from the Key International Joint Research Project of National Natural Science Foundation of China (81120108025), Science and Technology Project of Guangzhou City (2015 09010004), and Guangdong Natural Science Foundation (2015 AD030312012) to Z. Liu, and M. Hu was also supported by a grant from the National Institute of Health Grant, GM070737.
Abbreviations
- HCC
hepatocellular carcinoma
- rHLMs-pooled
reference human liver microsomes prepared from healthy human livers
- nHLMs-individual
human liver microsomes prepared from pericarcinomatous tissue of a single donor with HCC
- nHLMs-pooled
pooled human liver microsomes prepared from pericarcinomatous tissues of 15 donors, which contain same amount of donor microsomes from each subject
- tHLMs-individual
human liver microsomes prepared from tumor tissue of a single donor with HCC
- tHLMs-pooled
pooled human liver microsomes prepared from tumor tissues of 15 donors, which contain same amount of donor microsomes from each subject
- CL
intrinsic clearance
- TON
turnover number
- PXR
pregnane X receptor
- CAR
constitutive and rostane receptor
- HNF4A
hepatocyte nuclear factor 4 alpha
- PPAR
peroxisome proliferator-activated receptor alpha
- AHR
aryl hydrocarbon receptor
- RXR
retinoid X receptor
Footnotes
Conflict of Interest Statement: The authors confirm that there are no conflicts of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- 1.IARC. Estimated cancer incidence, mortality and prevalence worldwide in 2012. Globocan 2012: Liver cancer. 2012 Available from: http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx.
- 2.European Association For The Study Of The Liver. European Organisation For Research And Treatment Of Cancer. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56:908–943. doi: 10.1016/j.jhep.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Bisceglie AMD. Hepatitis B and hepatocellular carcinoma. Hepatology. 2009;49:S56–S60. doi: 10.1002/hep.22962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hiyashi N, Manns MP. Prevention of progression in chronic liver disease. Netherlands: Springer; 2004. [Google Scholar]
- 5.Brown DB, Geschwind JFH, Soulen MC, Millward SF, Sacks D. Society of interventional radiology position statement on chemoembolization of hepatic malignancies. Journal of Vascular and Interventional Radiology. 2006;17(2):217–223. doi: 10.1097/01.RVI.0000196277.76812.A3. [DOI] [PubMed] [Google Scholar]
- 6.Sheweita SA. Drug-metabolizing enzymes: mechanisms and functions. Curr Drug Metab. 2000;1:107–132. doi: 10.2174/1389200003339117. [DOI] [PubMed] [Google Scholar]
- 7.Sharma RR. Enzyme inhibition and bioapplications. In: Badal S, Shields M, Delgoda R, editors. Cytochrome P450 Enzyme Inhibitors Nature. In Tech; 2012. pp. 39–56. [Google Scholar]
- 8.Ruano G, Villagra D, Szarek B, Windemuth A, Kocherla M, Gorowski K, et al. Physiogenomic analysis of CYP450 drug metabolism correlates dyslipidemia with pharmacogenetic functional status in psychiatric patients. Biomark Med. 2011;5:439–49. doi: 10.2217/bmm.11.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, Tawa GJ, Wallqvist A. Identifying cytochrome P450 functional networks and their allosteric regulatory elements. PloS one. 2013;8:e81980. doi: 10.1371/journal.pone.0081980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez-Antona C, Ingelman-Sundberg M. Cytochrome P450 parmacogenetics and cancer. Oncogene. 2006;25(11):1679–91. doi: 10.1038/sj.onc.1209377. [DOI] [PubMed] [Google Scholar]
- 11.Oyama T, Kagawa N, Kunugita N, Kitagawa K, Ogawa M, Yamaguchi T, et al. Expression of cytochrome P450 in tumor tissues and its association with cancer development. Front Biosci. 2004;9:1967–1976. doi: 10.2741/1378. [DOI] [PubMed] [Google Scholar]
- 12.Michael M, Doherty MM. Drug metabolism by tumours: its nature, relevance and therapeutic implications. Expert Opinion on Drug Metabolism & Toxicology. 2007;3(6):783–803. doi: 10.1517/17425255.3.6.783. [DOI] [PubMed] [Google Scholar]
- 13.McFadyen MC, Melvin WT, Murray GI. Cytochrome P450 enzymes: novel options for cancer therapeutics. Molecular Cancer Therapeutics. 2004;3(3):363–371. [PubMed] [Google Scholar]
- 14.Miyoshi Y, Taguchi T, Kim SJ, Tamaki Y, Noguchi S. Prediction of response to docetaxel by immunohistochemical analysis of CYP3A4 expression in human breast cancers. Breast Cancer. 2005;12(1):11–15. doi: 10.2325/jbcs.12.11. [DOI] [PubMed] [Google Scholar]
- 15.Iizuka N, Oka M, Hamamoto Y, Mori N, Tamesa T, Tangoku A, et al. Altered levels of cytochrome P450 genes in hepatitis B or C virus-infected Liver Identified by oligonucleotide microarray. Cancer Genomics & Proteomics. 2004;1:53–58. [PubMed] [Google Scholar]
- 16.Kawakami H, Ohtsuki S, Kamiie J, Suzuki T, Abe T, Terasaki T. Simultaneous absolute quantification of 11 cytochrome P450 isoforms in human liver microsomes by liquid chromatography tandem mass spectrometry with in silico target peptide selection. Journal of Pharmaceutical Sciences. 2011;100(1):341–352. doi: 10.1002/jps.22255. [DOI] [PubMed] [Google Scholar]
- 17.Fallon JK, Neubert H, Hyland R, Goosen TC, Smith PC. Targeted Quantitative Proteomics for the Analysis of 14 UGT1As and-2Bs in Human Liver Using NanoUPLC–MS/MS with Selected Reaction Monitoring. Journal of proteome research. 2013;12(10):4402–4413. doi: 10.1021/pr4004213. [DOI] [PubMed] [Google Scholar]
- 18.Schaefer O, Ohtsuki S, Kawakami H, Inoue T, Liehner S, Saito A, Ebner T. Absolute quantification and differential expression of drug transporters, cytochrome P450 enzymes, and UDP-glucuronosyl transferases in cultured primary human hepatocytes. Drug Metabolism and Disposition. 2012;40(1):93–103. doi: 10.1124/dmd.111.042275. [DOI] [PubMed] [Google Scholar]
- 19.Yan T, Gao S, Peng X, Shi J, Xie C, Li Q, et al. Significantly decreased and more variable expression of major CYPs and UGTs in liver microsomes prepared from HBV-positive human hepatocellular carcinoma and matched pericarcinomatous tissues determined using an isotope label-free UPLC-MS/MS method. Pharm Res. 2015;32(3):1141–57. doi: 10.1007/s11095-014-1525-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W, Yang LY, Huang GW, Lu WQ, Yang ZL, Yang JQ, et al. Genomic analysis reveals RhoC as a potential marker in hepatocellular carcinoma with poor prognosis. Br J Cancer. 2004;90(12):2349–55. doi: 10.1038/sj.bjc.6601749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.American Joint Committee on Cancer. AJCC Cancer Staging Manual. 7. New York: Springer; 2010. [Google Scholar]
- 22.Franca A, Melo LD, Cerca N. Comparison of RNA extraction methods from biofilm samples of Staphylococcus epidermidis. BMC Research Notes. 2011;4(1):572. doi: 10.1186/1756-0500-4-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.US Food and Drug Administration. Drug development and drug interactions: table of substrates, inhibitors and inducers. (Internet) Available from: http://www.fda.gov/drugs/developmentapprovalprocess/developmentresources/druginteractionslabeling/ucm093664.htm.
- 24.Abd-Elsalam KA. Bioinformatic tools and guideline for PCR primer design. African Journal of Biotechnology. 2003;2(5):91–95. [Google Scholar]
- 25.Hilario E, Mackay J. Protocols for nucleic acid analysis by nonradioactive probes. Humana Press; 2007. [Google Scholar]
- 26.Houston JB, Kathryn EK. In vitro-in vivo scaling of CYP kinetic data not consistent with the classical Michaelis-Menten model. Drug Metabolism and Disposition. 2000;28(3):246–254. [PubMed] [Google Scholar]
- 27.Guengerich FP. Cytochromes P450, drugs, and diseases. Molecular Interventions. 2003;3(4):194. doi: 10.1124/mi.3.4.194. [DOI] [PubMed] [Google Scholar]
- 28.Bozina N, Bradamante V, Lovric M. Genetic polymorphism of metabolic enzymes P450 (CYP) as a susceptibility factor for drug response, toxicity, and cancer risk. Archives of Industrial Hygiene and Toxicology. 2009;60(2):217–242. doi: 10.2478/10004-1254-60-2009-1885. [DOI] [PubMed] [Google Scholar]
- 29.Hong C, Shen ZY, Xu W, Fan YT, Li J, Lu YF, et al. Expression of P450 and nuclear receptors in normal and end-stage Chinese livers. World Journal of Gastroenterology. 2014;20(26):8681. doi: 10.3748/wjg.v20.i26.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kocarek TA, Zangar RC, Novak RF. Post-transcriptional regulation of rat CYP2E1 expression: role of CYP2E1 mRNA untranslated regions in control of translational efficiency and message stability. Archives of biochemistry and biophysics. 2000;376(1):180–190. doi: 10.1006/abbi.2000.1704. [DOI] [PubMed] [Google Scholar]
- 31.Venkatakrishnan K, Moltke LLV, Greenblatt DJ. Human cytochromes P450 mediating phenacetin O-deethylation in vitro: Validation of the high affinity component as an index of CYP1A2 activity. Journal of Pharmaceutical Sciences. 1998;87(12):1502–1507. doi: 10.1021/js980255z. [DOI] [PubMed] [Google Scholar]
- 32.Pasanen M, Rannala Z, Tooming A, Sotaniemi EA, Pelkonen O, Rautio A. Hepatitis A impairs the function of human hepatic CYP2A6 in vivo. Toxicology. 1997;123(3):177–184. doi: 10.1016/s0300-483x(97)00119-4. [DOI] [PubMed] [Google Scholar]
- 33.Furlan V, Demirdjian S, Bourdon O, Magdalou J, Taburet AM. Glucuronidation of drugs by hepatic microsomes derived from healthy and cirrhotic human livers. Journal of Pharmacology and Experimental Therapeutics. 1999;289(2):1169–1175. [PubMed] [Google Scholar]
- 34.Glubb DM, Etheridge AS, Seiser E, Innocenti F. Liver expression quantitative trait loci (eQTL) and related approaches in pharmacogenomic studies. In: Padmanabhan S, editor. Handbook of pharmacogenomics and stratified medicine. Waltham: Elsevier; 2014. pp. 120–121. [Google Scholar]
- 35.Ye L, Yang X, Guo E, Chen W, Lu L, Wang Y, et al. Sorafenib metabolism is significantly altered in the liver tumor tissue of hepatocellular carcinoma patient. PLoS One. 2014;9(5):e96664. doi: 10.1371/journal.pone.0096664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oliveira AP, Sauer U. The importance of post-translational modifications in regulating Saccharomyces cerevisiae metabolism. FEMS Yeast Research. 2012;12(2):104–117. doi: 10.1111/j.1567-1364.2011.00765.x. [DOI] [PubMed] [Google Scholar]
- 37.Wen X. Nuclear Receptors in Drug Metabolism. New Jersey: John Wiley & Sons, Inc; 2009. [Google Scholar]
- 38.Chai XJ, Zeng S, Xie W. Nuclear receptors PXR and CAR: implications for drug metabolism regulation, pharmacogenomics and beyond. Expert Opinion on Drug Metabolism & Toxicology. 2013;9(3):253–266. doi: 10.1517/17425255.2013.754010. [DOI] [PubMed] [Google Scholar]
- 39.Shen J, Wang S, Zhang YJ, Kappil M, Wu HC, Kibriya MG, et al. Genome-wide DNA methylation profiles in hepatocellular carcinoma. Hepatology. 2012;55:1799–808. doi: 10.1002/hep.25569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hector HV, Lambert MP, Calvez-Kelm FL, Gouysse G, Sandrine MC, Tavtigian SV, et al. Hepatocellular carcinoma displays distinct DNA methylation signatures with potential as clinical predictors. PloS One. 2010;5:e9749. doi: 10.1371/journal.pone.0009749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghose R, Guo T, Vallejo JG, Gandhi A. Differential role of Toll-interleukin 1 receptor domain-containing adaptor protein in Toll-like receptor 2-mediated regulation of gene expression of hepatic cytokines and drug-metabolizing enzymes. Drug Metabolism and Disposition. 2011;39(5):874–81. doi: 10.1124/dmd.110.037382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Machida K. TLRs, alcohol, HCV, and tumorigenesis. Gastroenterology Research and Practice, 2011. 2010:article ID 518674. doi: 10.1155/2010/518674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raunio H, Juvonen R, Pasanen M, Pelkonen O, Paakko P, Soini Y. Cytochrome P4502A6 (CYP2A6) expression in human hepatocellular carcinoma. Hepatology. 1998;27(2):427–432. doi: 10.1002/hep.510270217. [DOI] [PubMed] [Google Scholar]
- 44.Ishikawa T, Ichida T, Sugitani S, Tsuboi Y, Genda T, Sugahara S, et al. Improved survival with oral administration of enteric-coated tegafur/uracil for advanced stage IV-A hepatocellular carcinoma. Journal of Gastroenterology and Hepatology. 2001;16(4):452–459. doi: 10.1046/j.1440-1746.2001.02352.x. [DOI] [PubMed] [Google Scholar]
- 45.Dhanasekaran R, Kooby DA, Staley CA, Kauh JS, Khanna V, Kim HS. Comparison of conventional transarterial chemoembolization (TACE) and chemoembolization with doxorubicin drug eluting beads (DEB) for unresectable hepatocelluar carcinoma (HCC) Journal of Surgical Oncology. 2010;101(6):476–480. doi: 10.1002/jso.21522. [DOI] [PubMed] [Google Scholar]
- 46.Aita P, Robieux I, Sorio R, Tumolo S, Corona G, Cannizzaro R, et al. Pharmacokinetics of oral etoposide in patients with hepatocellular carcinoma. Cancer Chemotherapy and Pharmacology. 1999;43(4):287–294. doi: 10.1007/s002800050897. [DOI] [PubMed] [Google Scholar]
- 47.van Schaik RH. Cancer treatment and pharmacogenetics of cytochrome P450 enzymes. Investigational new drugs. 2005;23(6):513–522. doi: 10.1007/s10637-005-4019-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.