Abstract
The development of next-generation sequencing (NGS) technologies for HLA and KIR genotyping is rapidly advancing knowledge of genetic variation of these highly polymorphic loci. NGS genotyping is poised to replace older methods for clinical use, but standard methods for reporting and exchanging these new, high quality genotype data are needed. The Immunogenomic NGS Consortium, a broad collaboration of histocompatibility and immunogenetics clinicians, researchers, instrument manufacturers and software developers, has developed the Minimum Information for Reporting Immunogenomic NGS Genotyping (MIRING) reporting guidelines. MIRING is a checklist that specifies the content of NGS genotyping results as well as a set of messaging guidelines for reporting the results. A MIRING message includes five categories of structured information – message annotation, reference context, full genotype, consensus sequence and novel polymorphism – and references to three categories of accessory information – NGS platform documentation, read processing documentation and primary data. These eight categories of information ensure the long-term portability and broad application of this NGS data for all current histocompatibility and immunogenetics use cases. In addition, MIRING can be extended to allow the reporting of genotype data generated using pre-NGS technologies. Because genotyping results reported using MIRING are easily updated in accordance with reference and nomenclature databases, MIRING represents a bold departure from previous methods of reporting HLA and KIR genotyping results, which have provided static and less-portable data. More information about MIRING can be found online at miring.immunogenomics.org.
Keywords: NGS, HLA, KIR, MIRING, genotyping, data standards
1. Introduction
Next-generation sequencing (NGS) offers high-throughput generation of phased sequences for the highly polymorphic human leucocyte antigen (HLA) and killer-cell immunoglobulin-like receptor (KIR) genes, allowing their rapid, high-resolution genotyping. NGS methods may be more generally described as single-molecule sequencing methods[1]. In some cases, these methods offer full-gene sequence results[1–4]. In general, all NGS methods offer higher resolution and lower ambiguity genotypes than standard methods such as “Sanger” sequencing based typing (SBT), and sequence-specific oligonucleotide probe (SSOP) or primer (SSP) methods[3–6], and do not require the use of secondary genotyping methods to resolve ambiguities.
Any method for genotyping HLA and KIR using genomic DNA requires at least three components: the genotyping instrument, reference sequences, and analysis software. The genotyping instrument generates primary sequence data, which is interpreted by the analysis software, using the reference sequences to identify the subject’s genotype. A wide variety of instruments, reference sequence resources, and data analysis programs are available for both NGS and pre-NGS genotyping approaches, and are used in different combinations.
In some cases, the different methods may not generate the same results for a given subject. Such discrepancies may derive from the instrumentation, reference sequences, software, or a combination of these components. However, as Hollenbach et al.[7] have described, there is no standard format for reporting a genotyping result or for documenting the components that were applied to generate that result. In the absence of such documentation, the source of discrepancies in genotyping results is rarely identifiable. In addition, it becomes impossible to directly relate the HLA and KIR genotypes of subjects genotyped using different methods, as genetic differences between individuals may not be distinguishable from methodological differences between genotyping approaches. This lack of clarity has important implications for meta-analytical approaches to population or disease association studies that seek to combine and compare data across different studies. In general, ambiguity regarding the source of genotyping discrepancies impedes technical advances and optimization, and frustrates reproducible research.
Guidelines for reporting and documenting genotyping results are essential for evaluating HLA and KIR genotypes generated using different instruments, reference sequences or data-analysis programs. The active and ongoing development of NGS methods requires the adoption of a single extensible and adaptable standard for reporting and documenting NGS genotyping results.
Here we describe the Minimum Information for Reporting NGS Genotyping (MIRING) checklist, a set of Minimum Information for Biological and Biomedical Investigations (MIBBI)[8, 9] reporting guidelines developed by a consortium of immunogenomic researchers and clinicians, NGS instrument manufacturers and software developers, HLA and KIR sequence database developers and administrators, bone marrow donor registries and donor centers.
2. Description of MIRING
2.1 MIRING Development
The standard reporting of HLA and KIR genotypes is a long unmet need of the histocompatibility and immunogenetics community [7, 10–14]. The specific need for NGS genotype reporting guidelines emerged from a survey of Immunogenomic data management and analysis practices[15], carried out by the Immunogenomic Data Analysis Working Group (IDAWG) as part of the 16th International HLA and Immunogenetics Workshop (IHIW)[16]. The survey uncovered a lack of consistency between laboratories and the resulting impact on downstream analytical results. The development of MIRING began with the formation of the Immunogenomic Next Generation Sequencing Data Consortium (INGSDC) (ngs.immunogenomics.org) by the IDAWG and the HLA Information Exchange Data Format Standards (HIEDFS) group. The INGSDC met several times between 2012 and 2014, and identified the minimum information needed to accurately report NGS genotyping results for the HLA and KIR genes for clinical and research applications. Further MIRING development took place as part of the BeTheMatch Foundation’s Data Standards ‘Hackathons’ for NGS-based typing held in September of 2014 and February of 2015 (dash.immunogenomics.org). Implementations of MIRING are being evaluated as part of a 17th IHIW Informatics Component (ihiws.org/informatics-of-genomicdata/) project; bioinformatic tools for generating, exchanging and consuming MIRING messages are being developed as part of this project as well. The participation of interested investigators in this IHIWS project is welcome.
2.2 MIRING Goals
To meet the current needs of the histocompatibility and immunogenetics community for reporting and exchanging NGS genotype data, the elements of a MIRING message were designed with the following goals:
To facilitate downstream analyses and data management for current research and clinical use cases for molecular genotyping data in the histocompatibility and immunogenetics field.
To permit the re-analysis of NGS HLA or KIR genotyping results in the context of past, present and (foreseeable) future molecular nomenclatures and methods of describing HLA and KIR allele diversity.
To permit the comparison and evaluation of genotyping performance between different NGS platforms and analysis methods.
To enable molecular genotyping results generated using SBT, SSOP and SSP genotyping technologies to be incorporated if required.
That the MIRING elements be sufficient to permit the accurate reporting of NGS data generated for other highly-polymorphic regions of the human genome.
2.3. MIRING Elements
MIRING is both a checklist of elements that constitute a NGS HLA or KIR genotyping result, and a set of messaging guidelines for transmitting that NGS HLA or KIR genotyping result. Genotyping reports can be generated from a MIRING message. The MIRING guidelines include semantic definitions for a MIRING message, but are not intended to impose syntactic constraints on the message; they are principles that must be met, regardless of the structure of the message.
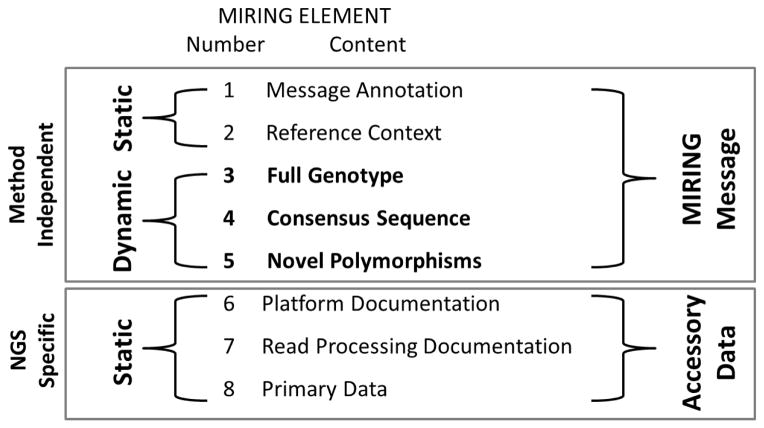
MIRING comprises eight primary elements, and their constituents (Table 1). Elements 1–5 constitute the MIRING message, suitable for reporting a genotyping result. Elements 6–8 constitute the contextual resource for MIRING messages, but are not included in MIRING messages; instead, these elements are referenced in MIRING messages. Where possible, MIRING elements are consistent with established formats for describing genetic and genomic data (e.g., FASTA[17–19], FASTQ[20], Variant Call Format (VCF)[21] and Genotype List (GL) String formats[22]), and leverage existing genetic and genomic data-resources (e.g., the IMGT/HLA and IPD-KIR Databases[23], the NCBI Genetic Testing Registry (GTR)[24] and International Nucleotide Sequence Database Collaboration (INSDC)[25]).
Table 1.
Definition of MIRING Elements and Formats
| Number | Element | Components | Messaging Instructions and Notes | |
|---|---|---|---|---|
| 1 | Message Annotation | |||
| 1.1 | Unique MIRING Message Identifier | Identifies the MIRING message generator (e.g., an International Organization for Standardization (ISO)/International Electrotechnical Commission (IEC) standard 6523 organization identifier (OID)[29, 30]) and the specific MIRING message. | ||
| 1.2 | Message Generator Contact Information | Email, mailing address, website, phone number, etc. | ||
| 1.3 | Platform Documentation (MIRING element 6) Reference | e.g., citation of a peer-reviewed publication or an entry in the National Center for Biotechnology Information (NCBI) Genetic Testing Registry (GTR)[24)] (ncbi.nlm.nih.gov/gtr/) | ||
| 1.4 | Read Processing Documentation (MIRING element 7) Reference | A reference to the location of a structured report documenting the use of programs/scripts (including parameters and order of use) to process the primary read data in order to make allele calls. | ||
| 1.5 | Primary Data Availability | A: Public, and available as defined in MIRING element 1.6 B: Private, and potentially available by contacting the message generator as defined in MIRING element 1.2 |
||
| 1.6 | Primary Data (MIRING element 8) Reference | Provided when permitted. e.g., referenced to data in the NCBI Sequence Read Archive (SRA) (ncbi.nlm.nih.gov/sra/) | ||
| 2 | Reference Context | |||
| 2.1 | Reference Sequence Database Version for Allele Calling | Identify for each locus included in the message | ||
| 2.2 | Individual Reference Sequences Applied | Identify the source database and accession number of each individual sequence applied in the message. | ||
| 2.2.1 | Reference Sequence Identifier | A unique identifier ranging from 0 to n-1, where n is the number of reference sequences in MIRING element 2.2. | ||
| 2.3 | Reference Sequence Source Type | Specified for MIRING elements 2.1 and 2.2 A: Public and curated B: Public and uncurated C: Not public D: No reference | ||
| 3 | Full Genotype | Defined in MIRING elements 3.1 and 3.2 | ||
| 3.1 | Pertinent Locus/Loci | Genetic loci as defined in an International Nucleotide Sequence Database Collaboration (INSDC) resource[25]. All of the loci tested as part of the work reported in the MIRING message should be included. | ||
| 3.2 | Formatted Genotype | If a genotype is detected for a given locus, report that genotype in Genotype List (GL) String format[22], or an equivalent format. If a locus is identified in MIRING element 3.1, but no sequence data are generated for that locus, report that genotype as ‘Absent’ in the GL String. | ||
| 3.3 | Genotype Uniform Resource Identifier (URI) | e.g., derived from the GL Service (gl.nmdp.org) | ||
| 4 | Consensus Sequence | The use of MIRING elements 4.1 and 4.2 to describe the consensus sequence allows the sequences to be reported in FASTA format[17–19]. FASTA format is not a required component of a MIRING message, but a FASTA formatted component of a MIRING-derived genotype report should use the pipe-delimited header format described in 4.2. | ||
| 4.1 | Consensus Sequence Block (CSB) | A contiguous nucleotide sequence organized in the 5′ to 3′ direction and written using IUPAC and IUBMB nucleotide base symbols[36]. Multiple sequence blocks may be included in a MIRING message. | ||
| 4.2 | Consensus Sequence Descriptor | For FASTA representations of consensus sequence, assign each CSB a pipe-delimited descriptor comprised by MIRING elements 4.2.1 – 4.2.7. | ||
| 4.2.1 | Consensus Sequence Block Identifier | Ranges from 0 to n-1, where n is the number of CSBs included in the message. CSB identifier numbers must increase in the 5′ to 3′ order of CSBs. | ||
| 4.2.2 | Reference Sequence Identifier | MIRING Element 2.2.1 pertinent to each CSB. If the reference sequence is identified as being of type D (no Reference; MIRING element 2.3) the entire CSB is considered to be a novel polymorphism, but does not need to be independently documented as part of MIRING element 5. | ||
| 4.2.3 | Reference Sequence Coordinate | The position in the reference sequence (MIRING element 2.2.1) (indexed from 0) corresponding to the 1st position of the CSB. | ||
| 4.2.4 | Phase Set | When phase information is available, identify the lowest numbered CSB (using MIRING element 4.2.1) sharing phase with a given CSB; assign the lowest numbered CSB in a phase set its own CSB identifier. If no phase information is available for a given CSB, assign that CSB its own CSB identifier | ||
| 4.2.5 | Copy Number | 1 to n, where n is the number of distinct sequences represented by the CSB (e.g., haploid = 1, diploid = 2, etc.). | ||
| 4.2.6 | Reference Sequence Match | 1: CSB exactly matches the sequence range (MIRING element 4.2.3) of the reference sequence (MIRING Element 2.2.1). 0: CSB does not exactly match the sequence range of the reference sequence. When reference sequence match = 0, a description of novel polymorphisms (MIRING element 5) is expected unless value for MIRING element 2.3 = D. |
||
| 4.2.7 | Sequence Continuity | 1: no sequence gaps occur between a given CSB and the preceding CSB in the same phase set. 0: there is no phase information for this consensus sequence block, or there is a sequence gap between this CSB and the preceding CSB. |
||
| 5 | Novel Polymorphisms | Define novel polymorphisms (identified in MIRING element 4.2.6) using MIRING elements 5.1 – 5.8. These elements allow novel polymorphisms to be reported using variant call format (VCF)[21] or an equivalent. VCF is not a required component of a MIRING message, but a VCF component of a MIRING-derived genotype report should use the format described in elements 5.1–5.8. | ||
| 5.1 | Reference | MIRING element 2.2.1 | ||
| 5.2 | Position | The position in the reference sequence (MIRING element 2.2.1) (indexed from 1) corresponding to the position of the reported variant sequence. | ||
| 5.3 | Variant Identifier | A composite value comprised by the CSB identifier including the variant, and a number ranging from 0 to n-1, where n is the number of sequence variants reported, separated by a pipe (e.g. 0|12). | ||
| 5.4 | Reference Sequence | The sequence in the reference (MIRING element 2.2.1) at the position (MIRING element 5.2). | ||
| 5.5 | Variant Sequence | The variant sequence identified at the position (MIRING element 5.2). This is the equivalent of the VCF ALT column[21]. | ||
| 5.6 | Quality Score | Quality score for the sequence variant reported in MIRING element 5.5. This is the equivalent of the VCF QUAL column[21]. | ||
| 5.7 | Quality Filter Status | PASS or FAIL value for MIRING element 5.6 | ||
| 5.8 | INSDC Accession Number | When possible, provide a GenBank or EMBL-ENA accession number for the novel sequence | ||
| 6 | Platform Documentation | A peer-reviewed publication, or the identifier of a record deposited in the NCBI GTR or an equivalent resource, documenting the specific details of the methodology and pertinent versions of the platform and instrument-dependent analysis software applied to obtain the unmapped reads and quality scores (MIRING element 8). Relevant platform-dependent information must include: instrument version, instrument-dependent software version identifier(s), reagent versions and lot number, sequence read lengths, expected amplicon/insert length, reference sequences applied, and sequence feature/region targeted. Inclusion of primer target locations is optional. |
||
| 7 | Read Processing Documentation | The specific details of the instrument-independent processing of the primary data (MIRING element 8), documented using the SRA Analysis XSD XML Schema[38] or an equivalent; e.g., instrument-independent analysis software version identifier(s), analysis software parameters used, details of the cutoff values and reference sequences (defined in MIRING element 2) used to filter the data for read quality and/or mapping quality, along with the final read depth obtained and a confidence score of the zygosity for the SNPs used to infer the final genotype. This information is not included in the MIRING message, but must be associated with the primary data (MIRING element 8) by the message generator, and can be accessed using MIRING element 1. | ||
| 8 | Primary Data | When permitted, unmapped reads with quality scores (e.g., Sanger FASTQ[20] or standard flowgram format (SFF)[40] formatted files), as generated by the instrument (defined in MIRING element 6), must be retained and should be made available as the primary NGS data. Adapter sequences may be excluded from the primary data. These primary data are not included in the MIRING message, but are accessed using MIRING element 1. | ||
MIRING Element 1: Message Annotation
Each MIRING message must include a unique identifier that links the message contents to external information excluded from the MIRING message. For example, individual subject identifiers protected by the U.S. Health Insurance Portability and Accountability Act of 1996 (HIPAA)[26], the Canadian Personal Information Protection and Electronic Documents Act (PIPEDA)[27], and EU Directive 95/46/EC[28] are excluded from MIRING messages, and should be reported and transmitted outside the scope of the message. MIRING message annotation must allow unambiguous identification of the organization that generated the message, as well as the unambiguous identification of any MIRING message generated by that organization. For example, organizations can be identified unambiguously using an International Organization for Standardization (ISO)/International Electrotechnical Commission (IEC) standard 6523 organization identifier (OID)[29, 30]. MIRING message annotation must also include contact information for the organization that generated the message, along with references to the location and availability of platform (MIRING element 6) and read processing (MIRING element 7) documentation, and the primary data (MIRING element 8) from which the MIRING message was generated.
MIRING Element 2: Reference Context
Comparison to specific well-characterized and annotated reference sequences is crucial for NGS genotyping of HLA and KIR genes. Reference sequences may be used for the mapping and processing of reads, as well as for the determination of a genotype based on a consensus sequence. These sequences are stored in a variety of databases, including the Genome Reference Consortium (GRC)[31] and IMGT/HLA and IPD-KIR Databases[23]. Clear identification both of the database versions and of the individual sequences applied in an NGS genotyping must be included in each MIRING message. For example, GRCh38.p4 or IMGT/HLA Database release 3.21.1 describe the current versions of the GRC human genome and IMGT/HLA Databases, respectively; GL000251.2. and HLA00001 are the accession numbers for the GRCh38.p4 alternate reference locus number 2, (derived from the COX cell line) and IMGT/HLA Database HLA-A*01:01:01:01 allele, respectively.
Any reference database or individual reference sequence used to generate a NGS genotype should be documented as part of MIRING element 2. To allow the assessment of the confidence in the genotyping, this documentation should indicate whether or not the database is public, and if a public database is curated. If no reference database is used, this should also be indicated. For instances when either previously unexplored gene features (e.g., HLA-DRB5 introns) or a genomic region that is unrepresented in any genomic alignment (e.g., DR1 or DR8 haplotypes of the HLA-DRB region[32]) is being sequenced, the absence of available reference sequence at the time of the genotyping should be noted.
MIRING Element 3: Full Genotype
Some NGS methods provide phased, full-gene resolution data. However, many NGS genotyping approaches do not return such results, and genotyping ambiguity [7] is not resolved. In order to evaluate genotyping results across specimens, NGS instruments, genotyping and analysis methods, the complete set of alleles and genotype combinations that are possible for a given set of sequence data using a given reference sequence database must be provided.
GL String[22] format can be used to describe the genotype at each locus, including genotyping ambiguity and known allelic phase between loci. A “best guess,” estimate or imputation of an unambiguous genotype should not be included in a MIRING message. When available, a reference to an external uniform resource identifier (URI) for the GL String should be included as well.
In addition, genes that were specifically targeted, but yielded no sequence data should be explicitly identified. Due to structural variation among the HLA[32] and KIR loci[33–35], genes that are present in some individuals may be completely absent in others. For example, individuals homozygous for HLA-DRB1*01 alleles have no HLA-DRB3, HLA-DRB4 or HLA-DRB5 genes[32], and individuals homozygous for the KIR A haplotype have no KIR2DS1, KIR2DS3/5 or KIR2DL5A genes[33]. When a gene could have been detected by a given NGS instrument, but no sequence for that gene is generated for a subject, the locus in question should be identified in the MIRING message, with the genotype reported as “Absent” for that locus.
MIRING Element 4: Consensus Sequence
Depending on the NGS approach applied, consensus sequence for an individual reported allele may be generated as a single, gene-length consensus sequence block (CSB) (e.g., resulting from de novo assembly), or as shorter phased or unphased CSBs (e.g., corresponding to individual exons). These sequences should be written using the single-letter symbols for nucleotide bases and incompletely specified bases defined by the International Union of Pure and Applied Chemistry (IUPAC) and International Union of Biochemistry and Molecular Biology (IUBMB)[36]. Two CSBs would be reported for heterozygous individuals where complete phase was known for a given gene.
Each CSB should be accompanied by MIRING elements 4.2.1–4.2.7, which identify the CSB, the reference sequence (as defined in MIRING element 2) to which it has been aligned along with its position and identity to that reference sequence, or the absence of a reference sequence, any phase and continuity between CSBs, and the inferred copy number for each CSB. For diploid loci, the copy number values for homologous CSBs should sum to 2; however, due to copy number variation of some HLA and KIR genes[32, 33], some individuals are truly haploid for a given gene (copy number of 1), while others may have more than two copies of a given gene (e.g., copy number of 3 or 4). For example, some individuals have four copies of the KIR2DS3 and KIR2DL5 genes[33].
A CSB can be described in FASTA format by including a header line that comprises MIRING elements 4.2.1–4.2.7, formatted as defined in Table 1 and illustrated in Figure 1. Although this header format should be used for FASTA presentation of CSBs in a genotyping report, MIRING elements 4.2.1 – 4.2.7 can be recorded separately and differently within a MIRING message.
Figure 1. FASTA Consensus Sequence Blocks with MIRING Headers.
FASTA Consensus Sequence Blocks with MIRING Headers Consensus sequences in MIRING messages are arranged in consensus sequence blocks (CSBs). CSBs are equivalent to FASTA formatted sequences that use a MIRING-specific descriptor as the header line. CSBs representing phased exon 2 and exon 3 sequences for an HLA-A allele are shown in Figure 1A and 1B. A MIRING CSB descriptor consists of seven fields of information, delimited by pipes (|) as shown in Figure 1C.
Each CSB is identified with a unique index, beginning from 0, in the Consensus Sequence Block Identifier field. CSB identifiers must increase in the 5′ to 3′ direction of CSBs.
Each reference sequence is identified with a unique index, beginning from 0, in the Reference Sequence Identifier field. Each index is defined in MIRING element 2.
The Reference Sequence Coordinate field identifies the first position of the pertinent CSB in the pertinent reference sequence.
The Phasing Group field identifies CSBs between which phase is known. Each set of phased CSBs will be identified with the same index, beginning from 0.
The Copy Number field identifies the number of distinct sequences represented by each CSB (e.g., 1 = haploid, 2 = diploid, etc.).
The Reference Sequence Match field identifies CSBs that exactly match the sequence range of the pertinent reference sequence (value = 1) or that do not match the sequence range of the reference sequence (value = 0).
When phase is indicated for CSBs, the Sequence Continuity field indicates if there are gaps (of any size) between those phased CSBs. A value of 1 in this field indicates that there are no gaps between the pertinent CSB and the most immediately 5′ CSB in phase with that CSB. A value of 0 in this field indicates a sequence gap between the pertinent CSB and the most immediately 5′ CSB in phase, or that no phase information is available for the pertinent CSB.
(A) CSB 0 and header, (B) CSB 1 and header, (C) Guide to interpreting the MIRING headers.
1: Reference sequence for CSB 0 is IMGT/HLA Database release version 3.21.1 A_nuc.fasta HLA00005 HLA-A*02:01:01:01. Reference sequence for CSB 1 is IMGT/HLA Database release version 3.21.1 A_nuc.fasta HLA10254 HLA-A*66:01:02.
MIRING Element 5: Novel Sequence Polymorphisms
The HLA and KIR genes are highly polymorphic, and the number of alleles reported to public databases is expected to increase dramatically with the use of NGS genotyping[37]. In the context of a MIRING message, novel polymorphism includes nucleotide sequence variants not yet present in a curated, public reference sequence database (e.g., the IMGT/HLA Database). The explicit identification of novel polymorphisms is an important element of both clinical and research genotyping, and must be documented in a MIRING message. Depending on how a MIRING message is generated, a CSB (as defined in MIRING element 4) representing novel sequence variants may have been submitted to a non-curated public reference sequence database [e.g., GenBank or the European Molecular Biology Laboratory (EMBL) European Nucleotide Archive (ENA)] as a novel sequence.
Novel sequence variants can be described through the use of MIRING elements 5.1–5.8, which define the reference sequence, variant position, variant sequence, quality score and quality filter status, and (if available) a GenBank or EMBL-ENA accession number for the novel sequence (e.g. L28096). This accession number can be included in the GL String (MIRING element 4.2.2, e.g. HLA-DRB1*L28096).
Use of MIRING elements 5.1–5.8 is sufficient to describe novel sequence variants in VCF as part of a genotyping report. A GenBank or EMBL-ENA accession number can be linked to the variant identifier (MIRING element 5.3) in the VCF meta-data. However, actual VCF is not a required component of a MIRING message, and MIRING elements 5.1–5.8 can be recorded separately and differently within a MIRING message.
MIRING Element 6: Platform Documentation
The specifics of each instrument, methodological approach (e.g., whole-genome sequencing, target enrichment, targeted amplicon sequencing) and reagent set applied to generate the primary read data (MIRING element 8) upon which the genotyping result is based should be documented. This documentation can take the form of a citation to a peer-reviewed publication or a reference to a structured documentation of the instrument and methodology in a publically accessible resource (e.g., the NCBI’s Genetic Testing Registry (GTR)). This documentation is not included in the MIRING message, but the reference to the resource must be included in MIRING element 1.
MIRING Element 7: Read Processing Documentation
After the primary read data (MIRING element 8) have been generated by the NGS instrument, they may be scrutinized for quality and length, modified or subjected to various bioinformatics filters before allele calls are made and a genotype result is generated. To enable the replication of the genotyping result and the evaluation of the bioinformatics process itself, the software and version used for both the genotyping and the processing steps applied must be documented. While these read processing steps are idiosyncratic to the combination of NGS components that have been applied in the genotyping effort, they can be accommodated in the NCBI’s Sequence Read Archive (SRA) Analysis XSD XML Schema[38, 39]. Each MIRING message should reference a report, based on the Analysis XSD or an equivalent, describing each program or script applied in the processing of reads, the order in which they were applied, the software versions and the pertinent parameters used. Where possible, this report should be associated with the primary read data (MIRING element 8), or made available by the MIRING message generator. This report should not be included in the MIRING message, but a reference to the location of the read processing documentation, or instructions for obtaining access to that documentation, should be included in MIRING element 1.
MIRING Element 8: Primary Data
The reads generated by the NGS instrument applied for the typing should be made available for re-analysis, either via deposition in a public database [e.g., the SRA, NCBI’s Genotype and Phenotype database (dbGAP) or an equivalent] or directly from the data generators. Primary data should take the form of unmapped reads with quality scores (e.g. Sanger FASTQ, SFF[40], or unmapped BAM including quality scores). The primary data are not included in the MIRING message, but a reference to the location of the primary data, or instructions for obtaining access to those data, should be included in MIRING element 1.
Because each MIRING message is assigned a unique identifier (MIRING element 1), it is possible for a MIRING message generator to produce multiple distinct messages for a single specimen from one set of primary data. Depending on read processing parameters applied and references used, each message may use a different subset of reads. Therefore, the MIRING message generator must maintain an archive for a given set of primary data, which identifies the reads pertinent to each MIRING message. When the primary data are publically available (as defined as part of MIRING element 1), the MIRING message generator must make this information available as well.
3. Strengths and Limitations of MIRING
MIRING represents a bold departure from previous methods of reporting HLA and KIR genotyping results. Previously, genotyping results have been maintained as static entities constrained by the existing references and nomenclature, with insufficient reference context and sequence information provided to foster genotype reassessment. By contrast, MIRING elements 3–5 are dynamic in that they can change with the reference context (MIRING element 2) - for instance when the database is updated. By providing access to the primary read data, and by including the consensus sequence in the MIRING message, the genotype and novel polymorphism information in a MIRING message can be updated with each reference allele sequence database revision.
The MIRING checklist identifies the minimum information needed to provide adequate documentation of an NGS HLA and KIR genotype. This documentation should be sufficient to reproduce or permit the reinterpretation of the reported genotype from the primary read data, and to allow informed comparisons of genotyping results for the same subject generated using different NGS genotyping instruments, reference sequences and analytical software. In cases when such genotyping results differ, use of MIRING messages to report those genotypes should facilitate the rapid identification of the sources of such discrepancies.
Many pieces of information pertinent to a genotyping experiment are not included in MIRING messages. Protected subject identifiers, phenotypic and demographic subject details, specimen details (e.g., preparation, quantification), specifics of the activity for which the genotyping effort was undertaken, descriptions and selection criteria for the loci genotyped, interpretations of the genotyping result (e.g., a ‘best call’ for ambiguous genotypes, or identifying a donor as a match to a patient), and funding sources for the genotyping are excluded from MIRING messages. Although this information can be associated with MIRING messages as part of a larger message if required, MIRING’s main purpose is to document those elements of a genotyping experiment that foster the archival utility of the genotyping result.
The MIRING checklist has been developed via community consensus, in order to meet the data management and exchange needs of the histocompatibility and immunogenetics community. Much of the information included in a MIRING message will not be pertinent to all current HLA and KIR genotyping use cases (e.g., clinical care, basic research, instrument validation, software development), but all such use cases can be met using the same MIRING message. Applications that parse MIRING messages and provide information tailored to each use case will make use of the same message. Given the rapid exploration of NGS technologies and methodologies, use of MIRING messages will allow the transparent evaluation of different instruments and genotyping approaches, encouraging improvement and standardization of all NGS methods.
The elements of the MIRING checklist allow MIRING messages to pertain to a single locus or multiple loci, but not to multiple subjects. Data for discrete subjects are reported in distinct MIRING messages, and multiple messages can be generated for each subject. Proper management of MIRING message identifiers then becomes an essential part of the MIRING messaging system. A central repository for MIRING messages will greatly facilitate and simplify the exchange of genotyping results among centers and researchers.
As illustrated in Figure 2, the elements of a MIRING message can be divided into distinct categories. MIRING elements 6–8 are specific to NGS methodologies, but are not included in the MIRING message. Therefore, MIRING messaging could be extended to include other genotyping methodologies (e.g., SBT, SSOP or SSP) by broadening the scope of information reported in elements 6–8 to document the methodologies, software, and raw data pertinent to these other methodologies. However, the structure or content of MIRING elements 1–5 would not change. Only the details of the MIRING message would change; for example, CSBs would decrease in length for SSOP- and SSP-based MIRING messages, to accommodate the shorter length of the hybridized sequences[41], and ambiguity in the full genotypes reported would increase. CSB copy number values would change from 1 (haploid) to 2 (diploid) for most Sanger sequencing-based MIRING messages (unless sequencing was applied to isolated chromosomes, or on the basis of group-specific amplification[42]).
Figure 2. MIRING Checklist Categories.
The eight elements of the MIRING checklist are either included in a MIRING message or exist as accessory data that are referenced in the MIRING message. MIRING message elements 1 and 2 and 6–8 are static in that they pertain to events that occurred when the genotyping instrument was applied to generate the primary read data. MIRING elements 3–5 are dynamic in that they can change if MIRING element 2, changes (e.g., a future release of the IPD-KIR database may result in a change to the ambiguity level of a genotype). MIRING elements 6–8 are also specific to NGS platforms. Valid MIRING messages could be generated on the basis of Sanger sequence-based typing (SBT), sequence-specific oligonucleotide probe (SSOP) and priming (SSP) methods, requiring changes to the content of MIRING elements 6–8 alone.
Although, the MIRING checklist was developed with HLA and KIR genotypes in mind, it can be applied to report genotype data for any highly-polymorphic genetic system. In addition, MIRING is sufficiently flexible that it can accommodate future developments in sequencing technology. As the cost of generating high-quality genomic information decreases, the need to report and exchange these data in a straightforward and reproducible manner will increase. As genomic data accumulate, specific genes, haplotype-blocks and chromosomal regions will be revealed as medically relevant, and their polymorphism can be documented and reported via a MIRING message.
Examples of Histoimmunogenetics Markup Language version 1.0 messages that comply with MIRING standards and principles are included in the paper by Milius et al. [43] included in this issue.
4. Conclusions
MIRING messages foster the portability of HLA and KIR genotype data in a standard format, allowing the dynamic re-analysis of these medically important results in the context of continual genomic discovery. The data recorded in a MIRING message are essential for the systematic traceability of a NGS genotyping result; this traceability is critical for reproducible research and the meaningful archiving of modern genotyping results. The MIRING checklist is sufficiently broad in scope that genotyping results generated using NGS technologies, older genotyping technologies and future methods can be accommodated in a MIRING message. Reporting of NGS genotype data as MIRING messages promotes transparency across varying applications of NGS components, facilitating comparison, and therefore the improvement and ongoing development of NGS technology. Finally, the widespread application of the MIRING checklist for exchanging NGS genotyping results will allow the leveraging of public data resources for meta-analysis, study replication and new discovery. More information about MIRING can be found online at miring.immunogenomics.org.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grants U01AI067068 (SJM and JAH) and U01AI090905 (PRP and PJN), awarded by the National Institute of Allergy and Infectious Disease (NIAID), and R01GM109030 (SJM and JAH), awarded by the National Institute of General Medical Sciences (NIGMS), and by Office of Naval Research (ONR) grant N00014-08-1-1207 (MM and RPM). The content presented is solely the responsibility of the authors and does not necessarily represent the official views of the NIH, NIAID, NIGMS, ONR, Department of Defense or United States Government. We thank all of the participants in the INGSDC for their input and useful discussions regarding the development of MIRING.
Abbreviations
- BAM
Binary Alignment/Map
- CSB
Consensus Sequence Block
- dbGAP
Genotype and Phenotype Database
- EMBL
European Molecular Biology Laboratory
- ENA
European Nucleotide Archive
- GL
Genotype List
- GRC
Genome Reference Consortium
- GTR
Genetic Testing Registry
- HLA
Human Leukocyte Antigen
- HIEDFS
HLA Information Exchange Data Format Standards
- HIPAA
Health Insurance Portability and Accountability Act
- IEC
International Electrotechnical Commission
- IDAWG
Immunogenomic Data Analysis Working Group
- IHIW
International HLA and Immunogenetics Workshop
- IMGT
ImMunoGeneTics
- INGSDC
Immunogenomic Next Generation Sequencing Data Consortium
- INSDC
International Nucleotide Sequence Database Collaboration
- IPD
Immuno Polymorphism Database
- ISO
International Organization for Standardization
- IUBMB
International Union of Biochemistry and Molecular Biology
- IUPAC
International Union of Pure and Applied Chemistry
- KIR
Killer-cell Immunoglobulin-like Receptor
- MIBBI
Minimum Information for Biological and Biomedical Investigations
- MIRING
Minimum Information for Reporting Immunogenomic NGS Genotyping
- NCBI
National Center for Biotechnology Information
- NGS
Next Generation Sequencing
- OID
Organization Identifier
- PIPEDA
Personal Information Protection and Electronic Documents Act
- SBT
Sanger sequencing Based Typing
- SFF
Standard Flowgram Format
- SRA
Sequence Read Archive
- SSOP
Sequence-Specific Oligonucleotide Probe
- SSP
Sequence-Specific Priming
- URI
Uniform Resource Identifier
- VCF
Variant Call Format
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Erlich HA. HLA typing using next generation sequencing: An overview. Hum Immunol. 2015 doi: 10.1016/j.humimm.2015.03.001. In Press. [DOI] [PubMed] [Google Scholar]
- 2.Erlich H. HLA DNA typing: past, present, and future. Tissue Antigens. 2012;80(1):1–11. doi: 10.1111/j.1399-0039.2012.01881.x. [DOI] [PubMed] [Google Scholar]
- 3.De Santis D, et al. 16(th) IHIW : review of HLA typing by NGS. Int J Immunogenet. 2013;40(1):72–6. doi: 10.1111/iji.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ozaki Y, et al. HLA-DRB1, -DRB3, -DRB4 and -DRB5 genotyping at a super-high resolution level by long range PCR and high-throughput sequencing. Tissue Antigens. 2014;83(1):10–6. doi: 10.1111/tan.12258. [DOI] [PubMed] [Google Scholar]
- 5.Sanchez-Mazas A, Meyer D. The relevance of HLA sequencing in population genetics studies. J Immunol Res. 2014;2014:971818. doi: 10.1155/2014/971818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lange V, et al. Cost-efficient high-throughput HLA typing by MiSeq amplicon sequencing. BMC Genomics. 2014;15:63. doi: 10.1186/1471-2164-15-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollenbach JA, et al. A community standard for immunogenomic data reporting and analysis: proposal for a STrengthening the REporting of Immunogenomic Studies statement. Tissue Antigens. 2011;78(5):333–44. doi: 10.1111/j.1399-0039.2011.01777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kettner C, et al. Meeting Report from the Second "Minimum Information for Biological and Biomedical Investigations" (MIBBI) workshop. Stand Genomic Sci. 2010;3(3):259–66. doi: 10.4056/sigs.147362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor CF, et al. Promoting coherent minimum reporting guidelines for biological and biomedical investigations: the MIBBI project. Nat Biotechnol. 2008;26(8):889–96. doi: 10.1038/nbt.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mack SJ, et al. Population samples and genotyping technology. Tissue Antigens. 2007;69(Suppl 1):188–91. doi: 10.1111/j.1399-0039.2006.00768.x. [DOI] [PubMed] [Google Scholar]
- 11.Single RM, et al. 14th International HLA and Immunogenetics Workshop: report of progress in methodology, data collection, and analyses. Tissue Antigens. 2007;69(Suppl 1):185–7. doi: 10.1111/j.1399-0039.2006.00767.x. [DOI] [PubMed] [Google Scholar]
- 12.Mack SJ, et al. Proposal for HLA data validation. 2008 Jun;2008 [cited 2014 October 26]; Available from: https://immport.niaid.nih.gov/docs/standards/Proposal_For_HLA_Data_Validation_Version_2.doc. [Google Scholar]
- 13.Bochtler W, et al. An update to the HLA Nomenclature Guidelines of the World Marrow Donor Association, 2012. Bone Marrow Transplant. 2013;48(11):1387–8. doi: 10.1038/bmt.2013.93. [DOI] [PubMed] [Google Scholar]
- 14.Helmberg W, et al. Virtual DNA analysis as a platform for interlaboratory data exchange of HLA DNA typing results. Tissue Antigens. 1999;54(4):379–85. doi: 10.1034/j.1399-0039.1999.540407.x. [DOI] [PubMed] [Google Scholar]
- 15.Hollenbach JA, et al. 16(th) IHIW: immunogenomic data-management methods. report from the immunogenomic data analysis working group (IDAWG) Int J Immunogenet. 2013;40(1):46–53. doi: 10.1111/iji.12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Middleton D, Marsh SGE. 16th International HLA and Immunogenetics Workshop (IHIW) Introduction. Int J Immunogenet. 2013;40(1):1. doi: 10.1111/iji.12034. [DOI] [PubMed] [Google Scholar]
- 17.Lipman DJ, Pearson WR. Rapid and sensitive protein similarity searches. Science. 1985;227(4693):1435–41. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- 18.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988;85(8):2444–8. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neuwald AF, et al. Extracting protein alignment models from the sequence database. Nucleic Acids Research. 1997;25:1665–1677. doi: 10.1093/nar/25.9.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cock PJ, et al. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010;38(6):1767–71. doi: 10.1093/nar/gkp1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Danecek P, et al. The variant call format and VCF tools. Bioinformatics. 2011;27(15):2156–8. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milius RP, et al. Genotype List String: a grammar for describing HLA and KIR genotyping results in a text string. Tissue Antigens. 2013;82(2):106–12. doi: 10.1111/tan.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson J, Halliwell JA, Marsh SGE. IMGT/HLA and the Immuno Polymorphism Database. Methods Mol Biol. 2014;1184:109–21. doi: 10.1007/978-1-4939-1115-8_5. [DOI] [PubMed] [Google Scholar]
- 24.Rubinstein WS, et al. The NIH genetic testing registry: a new, centralized database of genetic tests to enable access to comprehensive information and improve transparency. Nucleic Acids Res. 2013;41(Database issue):D925–35. doi: 10.1093/nar/gks1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakamura Y, Cochrane G, Karsch-Mizrachi I. The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2013;41(Database issue):D21–4. doi: 10.1093/nar/gks1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.An Act To amend the Internal Revenue Code of 1986 to improve portability and continuity of health insurance coverage in the group and individual markets, to combat waste, fraud, and abuse in health insurance and health care delivery, to promote the use of medical savings accounts, to improve access to long-term care services and coverage, to simplify the administration of health insurance, and for other purposes. (Brief title: Health Insurance Portability and Accountability Act of 1996). (PL 104-191, 21 Aug. 1996). 1996, http://www.constitution.org/uslaw/sal/110_statutes_at_large.pdf: United States Statutes at Large 110. p. 1936–2103.
- 27.An Act to support and promote electronic commerce by protecting personal information that is collected, used or disclosed in certain circumstances, by providing for the use of electronic means to communicate or record information or transactions and by amending the Canada Evidence Act, the Statutory Instruments Act and the Statute Revision Act (Brief title: Personal Information Protection and Electronic Documents Act), April 13, 2000, http://laws-lois.justice.gc.ca/PDF/P-8.6.pdf: Statutes of Canada 2000, Chapter 5.
- 28.Directive 95/46/EC on the protection of individuals with regard to the processing of personal data and on the free movement of such data, E.P.a.t. Council, Editor October 24, 1995: Official Journal L281, 23.11.1995, p31–50. p. 20.
- 29.32, I.I.J.S. Information technology — Structure for the identification of organizations and organization parts — Part 1: Identification of organization identification schemes. 1998 [cited 2014 October 26]; Available from: https://www.iso.org/obp/ui/#iso:std:iso-iec:6523:-1:ed-1:v1:en.
- 30.32, I.I.J.S. Information technology — Structure for the identification of organizations and organization parts — Part 2: Registration of organization identification schemes. 1998 [cited 2014 October 26]; Available from: https://www.iso.org/obp/ui/#iso:std:iso-iec:6523:-2:ed-1:v1:en.
- 31.Church DM, et al. Modernizing reference genome assemblies. PLoS Biol. 2011;9(7):e1001091. doi: 10.1371/journal.pbio.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andersson G. Evolution of the human HLA-DR region. Front Biosci. 1998;27(3):d739–45. doi: 10.2741/a317. [DOI] [PubMed] [Google Scholar]
- 33.Pyo CW, et al. Different patterns of evolution in the centromeric and telomeric regions of group A and B haplotypes of the human killer cell Ig-like receptor locus. PLoS One. 2010;5(12):e15115. doi: 10.1371/journal.pone.0015115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uhrberg M, et al. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7(6):753–63. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 35.Norman PJ, et al. Co-evolution of human leukocyte antigen (HLA) class I ligands with killer-cell immunoglobulin-like receptors (KIR) in a genetically diverse population of sub-Saharan Africans. PLoS Genet. 2013;9(10):e1003938. doi: 10.1371/journal.pgen.1003938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cornish-Bowden A. Nomenclature for incompletely specified bases in nucleic acid sequences: recommendations 1984. Nucleic Acids Res. 1985;13(9):3021–30. doi: 10.1093/nar/13.9.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klitz W, Hedrick P, Louis EJ. New reservoirs of HLA alleles: pools of rare variants enhance immune defense. Trends Genet. 2012;28(10):480–6. doi: 10.1016/j.tig.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 38.National Center for Biotechnology Information (US), editor Analusis Submission Guide. Bethesda (MD): 2010. SRA Handbook. http://trace.ncbi.nlm.nih.gov/Traces/sra/?view=xml_schemas. [Google Scholar]
- 39.Kodama Y, Shumway M, Leinonen R. The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res. 2012;40(Database issue):D54–6. doi: 10.1093/nar/gkr854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malde K. Flower: extracting information from pyrosequencing data. Bioinformatics. 2011;27(7):1041–2. doi: 10.1093/bioinformatics/btr063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Helmberg W. Storage and utilization of HLA genomic data--new approaches to HLA typing. Rev Immunogenet. 2000;2(4):468–76. [PubMed] [Google Scholar]
- 42.Witter K, et al. Group-specific amplification of HLA-DQA1 revealed a number of genomic full-length sequences including the novel HLA alleles DQA1*01:10 and DQA1*01:11. Tissue Antigens. 2014;83(1):49–51. doi: 10.1111/tan.12270. [DOI] [PubMed] [Google Scholar]
- 43.Milius RP, et al. Histoimmunogenetics Markup Language 1.0: Reporting Next Generation Sequencing-based HLA and KIR Genotyping. Hum Immunol. 2015 doi: 10.1016/j.humimm.2015.08.001. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]