Abstract
In a previous study, 50% calorie restriction in mice from days 1.5–11.5 of pregnancy resulted in reduced placental weights and areas, relatively sparing of labyrinth zone area compared to junctional zone area, and dramatic changes in global gene expression profiles. However, little lasting effect was seen on adult offspring of these pregnancies, with a slight reduction in adiposity in males, and some changes in liver gene expression in both sexes. The goals of the present study were to determine whether the placental changes induced by caloric restriction in early pregnancy had permanent, irreversible effects on the placenta, and whether the changes in liver gene expression in adult offspring were present before birth. There were no differences in placental weights or areas, or the areas of individual placental zones near term in mice that had previously been food restricted. Global gene expression profiles at d18.5 were indistinguishable in placentas from control and previously food restricted mothers. In fetuses from restricted dams at d18.5, liver expression of Gck, a key regulator of glycogen synthesis was reduced, whereas its expression was increased in livers from adult offspring of restricted dams. Ppara expression was also reduced in fetal livers from restricted dams at d18.5, but not in adult offspring livers. We conclude that alterations in the placenta caused by nutrient restriction in early pregnancy are reversible, and that alterations in gene expression in livers of adult offspring are not a result of changes initiated during pregnancy and maintained through adulthood.
Introduction
The Developmental Origins of Health and Disease field has shown that nutritional insults during pregnancy can have lasting consequences for the health of offspring. The timing of the insult is critical. In the seminal Dutch Hunger Winter studies, severe maternal food restriction during early pregnancy had the most deleterious consequences for offspring in later life, including higher rates of obesity, diabetes and cardiovascular disease (Ravelli et al. 1976; Painter et al. 2005). In contrast, those exposed in mid-gestation had greater rates of obstructive airway disease and microalbuminuria, and those exposed in late gestation had only a slight reduction in glucose tolerance (Roseboom et al. 2006). Similarly, in the sheep, nutrient restriction from days 28–78 of gestation led to heavier, fatter, and less glucose tolerant offspring (Gardner et al. 2005, Ford et al. 2007). In rats, exposure to 50% nutrient restriction during the first ten days of pregnancy also led to increased weight gain and adiposity in offspring (Jones & Friedman 1982, Jones et al. 1984, Anguita et al. 1993, Sardinha et al. 2006).
It has been proposed that food restriction during early gestation has these effects because it is a critical period for placental development, and the altered placenta could then continue to influence the fetus through the rest of gestation (Barker et al. 1990, Dandrea et al. 2001, Barker et al. 2010). Placental development is altered by maternal nutrient restriction in early gestation in ways that suggest compensatory adaptation to maintain consistent nutrient supplies to the fetus. For example, in ewes that are food-restricted during early gestation, and then returned to normal feeding in late pregnancy, overgrowth of the placenta is observed at term (Foote et al. 1959, Clarke et al. 1998, Dandrea et al. 2001). Similarly, term placental weights were increased in women who were exposed to the Dutch Hunger Winter during the first trimester of pregnancy (Lumey 1998).
We have previously shown that in the mouse, 50% food restriction from days 1.5–11.5 of pregnancy results in changes in placental morphology and gene expression. Specifically, food restriction reduced the weight of the placenta, and the area of the junctional zone, while the area of the labyrinth zone, the part of the placenta responsible for nutrient exchange, was preserved. (Schulz et al. 2012). Similar placental adaptations were observed by others in mice restricted through gestational day 16 (Sferruzzi-Perri et al. 2011). However, food restriction from d1.5–11.5 had only modest effects on mouse offspring (Pennington et al. 2012). As adults, their body weights, glucose tolerance, and response to a high fat diet were not different than those of offspring from control pregnancies. Among males only, offspring of food restricted mothers had slightly lower adiposity and greater bone strength than offspring of controls, and increases in Igf1r mRNA and 18s rRNA levels in the liver (Pennington et al. 2012).
The goal of the present study was to determine whether the adaptations observed in the placenta in response to maternal food restriction are permanent or reversible. After food restriction from pregnancy d1.5–11.5, dams were returned to ad libitum feeding until near term. We examined placental morphology and gene expression on day 18.5 in order to identify lasting effects in the placenta. We also compared expression of key metabolic genes in fetal livers to determine whether differences previously noted in livers of the adult offspring begin prenatally.
Materials and Methods
Animals
All animal procedures were approved by the University of Missouri Animal Care and Use Committee. Female Swiss Webster (ND4) mice, age 6–8 weeks, ≥ 25g, were obtained from Harlan Laboratories (Indianapolis, IN), or born at the University of Missouri to Swiss Webster mice obtained from Harlan, and individually housed at the University of Missouri. Mice were acclimated for 1 week to AIN-93G diet for pregnancy and lactation (Research Diets, New Brunswick, NJ) in a powdered, blue-dyed form presented in specialized dishes for weighing (Townsend et al. 2008). Average daily individual food consumption levels were estimated by weighing food every other day for 10–14 days. Females were introduced to male cages, and the day in which a copulatory plug was detected was considered pregnancy d 0.5. Females were returned to individual cages, and at d1.5, randomly assigned to one of two treatment groups: (1) ad libitum fed plus saline injection (control) or (2) food restricted to 50% of prior consumption levels plus saline injection (restricted). Saline injections were given to permit comparisons to our previous studies, in which we examined placentas at d11.5 and offspring during adulthood using the same two treatment groups (Pennington et al. 2012, Schulz et al. 2012). In those studies, saline injections were used to control for leptin injections given to a third treatment group, which was not included in this study. Injections were given intraperitoneally once daily at 1400h and mice were fed at 1600h. All mice were fed ad libitum without injections from d11.5–18.5, at which time they were euthanized by CO2 inhalation followed by cervical dislocation for collection of blood, placentas, and fetuses.
Placental Morphology
After weighing, half of the total number of placentas from each dam were fixed overnight in 4% paraformaldehyde, then dehydrated and paraffin embedded (IDEXX/RADIL, Columbia, MO). Each placenta was bisected mid-sagitally and embedded cut-face down. Three pairs of serial 5 μm sections were cut at 50 μm intervals and stained with hematoxylin and eosin (H&E). The largest face on each slide was chosen for morphological analysis. Overlapping images were photographed with a 4x objective lens on an Olympus Ix81 microscope, and assembled into a single, high-resolution image covering the entire placental cross-section. Total cross-sectional area, junctional zone, and labyrinth zone areas were measured by manual outlining using the freehand selection tool in ImageJ software (NIH) while blinded to treatment group. Cross sectional measurements were averaged from three placentas per dam to obtain n=1 (total n=8 control, n=8 restricted). For comparison of placental blood spaces, maternal blood sinuses and fetal blood vessels within the labyrinth were identified in H&E stained sections by the appearance of nucleated (fetal) and enucleated (maternal) red blood cells, as well as the location of fetal endothelial cells and trophoblast, and manually outlined with the freehand selection tool within ImageJ software, also while blinded to treatment group. Five frames were photographed per placenta with a 40x objective, and 2–3 placentas were analyzed per dam to obtain n=1 (total n=7 per treatment group).
Leptin and insulin measurement
Terminal blood samples from dams were collected by cardiac puncture at d18.5. Serum was obtained by centrifugation of blood samples at 1000 × g for 12 minutes. Leptin and insulin were measured by commercial sandwich ELISA (Millipore, Billerica, MA).
Statistical analysis
Dams were considered the experimental unit such that fetal weights, placental weights, and placental areas were averaged within each dam to obtain n=1. Student’s t-tests were used to compare means, and F-test to compare variances between control and restricted dams.
Microarray analysis
Half of the total number of placentas from each dam were cut in two, and one piece was frozen in TriReagent (Sigma, St. Louis, MO). Following homogenization and phase separation according to TriReagent instructions, the aqueous phase was further purified using RNEasy Mini Kit (Qiagen, Valencia CA). Microarray analysis was conducted on placentas from five dams from each of the two treatment groups. RNA from two placentas was pooled to obtain one sample for each dam. Preparation of cRNA, microarray hybridization, and scanning and cluster analysis were carried out by GenUs Biosystems (Northbrook, IL). Mouse 4×44 whole genome arrays (Agilent, Design ID 026655) containing over 39,000 probes were scanned on an Agilent G2565 Microarray Scanner. Data were analyzed with Agilent Feature Extraction, GeneSpring GX v7.3.1, and R 3.0.1 software.
The primary analysis of microarray gene expression data was conducted using the Linear Models for Microarray Data (limma) package (Gentleman R 2005)] and the Agi4×44PreProcess package available through the Bioconductor project(Gentleman et al. 2004)for use with R statistical software. Data quality was examined via an extensive battery of metrics obtained using the Agi4×44PreProcess package. Foreground mean signal was used with background median signal for background correction in order to produce positive corrected intensities via the normexp function in limma. Between-array normalization was done via quantile normalization(Bolstad et al. 2003). Because Agilent 4×44 chips contain up to 10 replicated (non-control) probes for a given set, these were collapsed into a single value by computing the median of the probes intensities belonging to the same set. Finally, independent filtering was carried out on probe sets that were not putatively expressed across the sample (as defined by log2 mean signal intensity >8); these were excluded (n= 26,881) from further statistical analyses in order to reduce false positives, as they contain very little information content relative to the analysis at hand and doing so increases power (Bourgon et al. 2010).
After this preprocessing was completed, the statistical analysis was performed using an empirical Bayesian moderated t-test (Smyth 2004) applied to normalized intensity for each gene (n=11,608 remaining after preprocessing), where the restricted group was compared to the control group. The comparisons are expressed as fold changes (res/ctl) along with adjusted p-values. Adjustment to the p-values was made to account for multiple testing using the false discovery rate (FDR) method of Benjamini and Hochberg. We chose 10% as our FDR-cutoff for declaring statistical significance.
Fetal Liver Gene Expression
Liver samples were collected from each of the d18.5 fetuses, placed in TRI Reagent (Sigma), and stored at −80°C until further use. Samples were homogenized in Trireagent with an Omni GLH Homogenizer and centrifuged to remove excess polysaccharides. After phase separation, RNA was further purified by using the RNeasy isolation procedure (Qiagen) according to manufacturer’s protocol for RNA cleanup. Genomic DNA was removed with TurboDNase (Ambion) according to manufacturer’s protocol.
RNA was extracted from the liver of one female fetus each from five dams per treatment group and from one male fetus each from four dams per treatment group, and analyzed by custom RT2 Profiler PCR arrays (Qiagen, Valencia, CA) on an Applied Biosystems 7500 real-time PCR system (Applied Biosystems, Foster City, CA) using RT2 SYBR Green ROX qPCR master mix according to manufacturer’s protocol, as previously described (Pennington et al. 2012). For the PCR arrays, 1 μg of total RNA was reverse transcribed using the RT2 first strand kit (Qiagen, Germantown, MD). Three housekeeping genes, 18s rRNA (18s ribosomal RNA), Actb (actin-β), and Hprt (hypoxanthine phosphoribosyltransferase 1) were examined; however, due to variation in 18s and Hprt, only Actb was used to calculate fold changes. Manufacturer’s external reverse transcription and positive PCR controls were also included. Fold differences in expression were calculated according to the ΔΔCt method (Livak & Schmittgen 2001).
Expression of two additional genes, Cd36 and Srebp1c was examined by real-time RT-PCR in fetal livers. There was some evidence of differentially susceptibility to non-alcoholic fatty liver disease (NAFLD) in offspring in our previous study (Pennington et al. 2012) and these genes have been implicated in NAFLD (Shimomura et al. 1999, Lemoine et al. 2006, Koonen et al. 2007, Anderson & Borlak 2008, Greco et al. 2008). Additionally, their expression in offspring liver is altered in other developmental programming paradigms, such as maternal high fat/high sugar or low-carbohydrate feeding (Zhang et al. 2005, Bruce et al. 2009, Gregorio et al. 2010). Total RNA (500ng) was reverse transcribed with SuperScript III reverse transcriptase (Invitrogen/Life Technologies, Grand Island, NY), using random hexamer primers, according to manufacturer’s protocol. Data were normalized to the average Ct of Actb and Gapdh. Expression of these control genes was first determined not to differ between control and restricted samples; for Actb, Ct values were 19.8 + 0.9 for controls and 20.2 + 0.5 for restricted; for Gapdh, Ct values were 21.3 + 0.9 for controls and 21.1 + 0.6 for restricted. Primer sequences were designed by using Primer Express software (Applied Biosystems), synthesized by Integrated DNA Technologies (Coralville, IA) and validated by performing serial dilutions of template cDNA. Primer sequences were Cd36 (5′-GGTCCTTACACTACAGAGTTCGTTA-3′ and 5′-CATTGGGCTGTACAAAAGACACA-3′) and Srebp1c (5′-TGGTGGGCACTGAAGCAAA-3′ and (5′-TGGTGGGCACTGAAGCAAA-3′ and 5′-GCAAGAAGCGGATGTAGTCGAT-3′).
Results
Maternal Characteristics
Whereas 90% of ad libitum (control) fed dams remained pregnant after initial detection of a copulatory plug, roughly one third of pregnancies were lost in the restricted dams by day 18.5, though this was not a statistically significant difference (Table 1). In addition, one mouse in the control group and two mice in the restricted group gave birth prior to d18.5, and no fetal or placental tissues could be collected. There were no differences between treatment groups in maternal serum leptin or insulin concentrations at the time of sacrifice, nor in maternal weights (Table 1). Thus, maternal characteristics recovered completely in the 1 week following return to ad libitum feeding.
Table 1.
Pregnancy Characteristics. N d0.5 = number of mice assigned to each group with copulatory plugs. N d18.5 = number of mice from which placental and fetal tissues were collected at d18.5. Number in parenthesis indicates number of mice that delivered prior to d18.5. The remaining mice were no longer pregnant at d18.5. There were no significant differences between groups.
| Treatment group | Control | Restricted |
|---|---|---|
| N d0.5 | 10 | 16 |
| N d18.5 | 8 (1) | 9 (2) |
| Pre-pregnancy weight (g) | 27.5 ± 0.3 | 26.3 ± 0.5 |
| Maternal weight at d18.5 (g) | 44.2 ±1.1 | 42.9 ± 4.2 |
| Litter size at d18.5 | 10.8± 0.7 | 9.4 ± 0.9 |
| Maternal serum leptin d18.5 | 13.9 ± 3.2 ng/ml | 16.1 ± 1.5 ng/ml |
| Maternal serum insulin d18.5 | 2.04 ± 0.39 ng/ml | 1.56 ± 0.39 ng/ml |
Placental and Fetal Weights
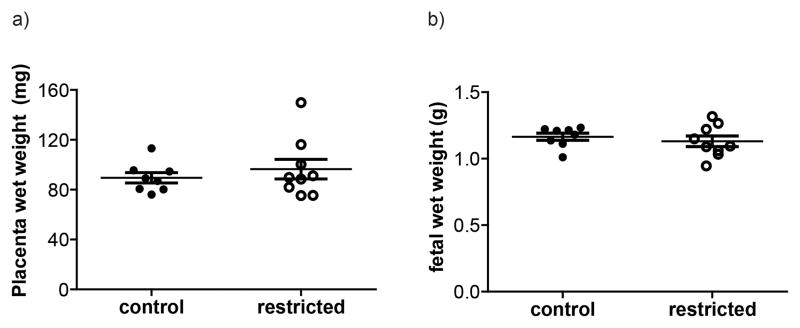
There were no differences in litter sizes between control and restricted dams at day 18.5 (Table 1). There also were no significant differences in mean fetal or placental weights (Figure 1), or the ratio of fetal:placental weights, a measure of placental efficiency (not shown). When fetal and placental weights were separated by fetal sex, there were still no differences between treatment groups (data not shown).
Figure 1.
(a) Placental wet weights at d18.5. Neither means or variances (p=0.08) were significantly different (d) Fetal weights at d18.5. There was no significant difference. Each marker represents the mean for one litter. Lines represent the treatment group mean, and bars show standard error.
Placental Morphology
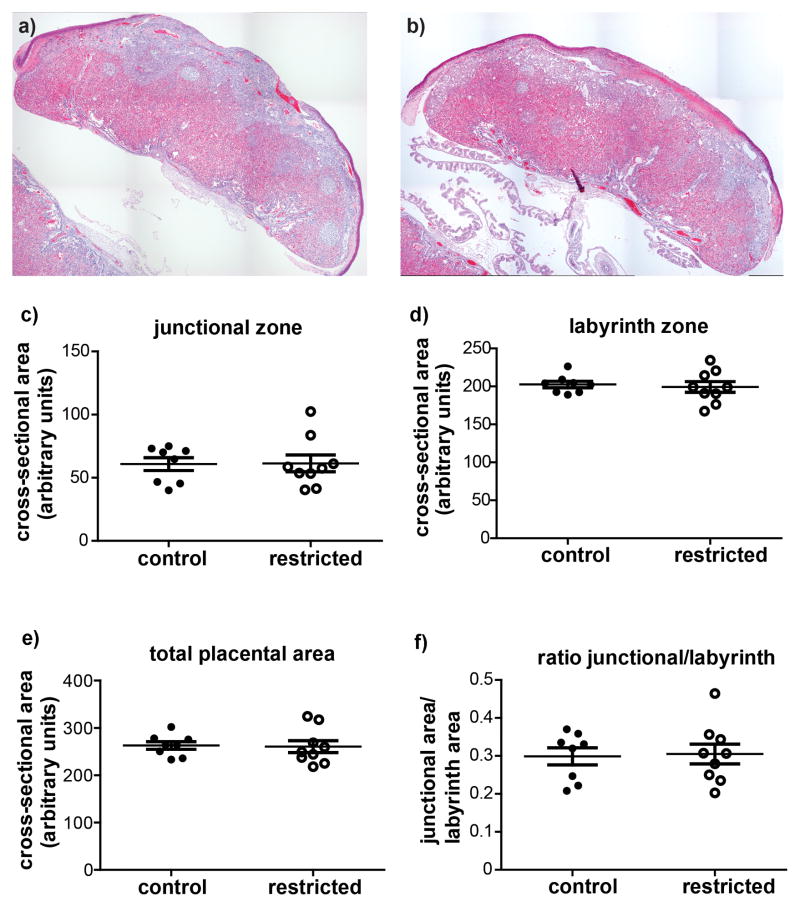
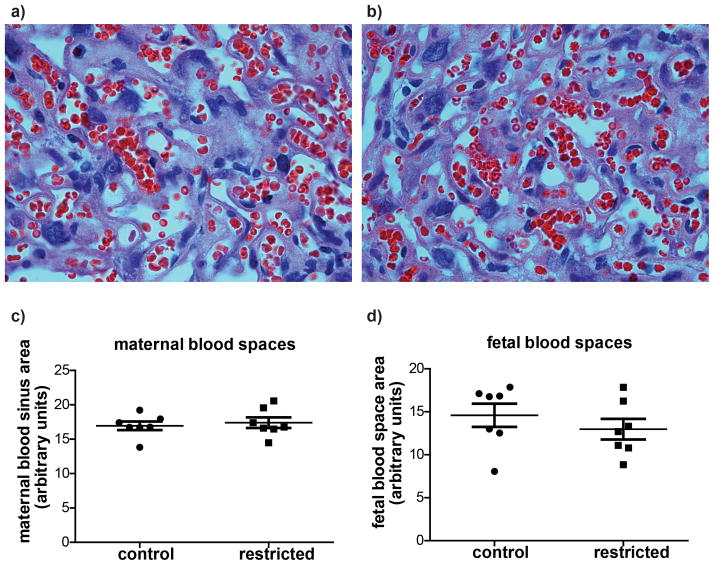
There were no differences in total cross-sectional area of the placenta or cross-sectional areas of the junctional or labyrinth zones between control and previously food restricted dams (Figure 2). The cross-sectional areas of the maternal blood sinuses and the fetal blood vessels within the labyrinth zone were also examined, and found not to differ significantly between treatment groups (Figure 3).
Figure 2.
Representative composite images of placentas from (a) control and (b) previously restricted dams at pregnancy d18.5. Cross-sectional areas of the (c) junctional zone, and (d) labyrinth zone (e) total cross-sectional area and (f) the ratio of the junctional zone/labyrinth zone areas were not different between control and restricted dams. Each marker represents the mean from one litter. Horizontal lines are the means for each maternal treatment group, and bars represent SEM.
Figure 3.
Representative images of labyrinth zone photographed with a 40x objective lens from (a) control and (b) previously food restricted dams. There were no differences in cross-sectional areas of (c) maternal or (d) fetal blood spaces. Each marker represents the mean from one litter. Horizontal lines are the means for each maternal treatment group, and bars represent SEM.
Placental Gene expression
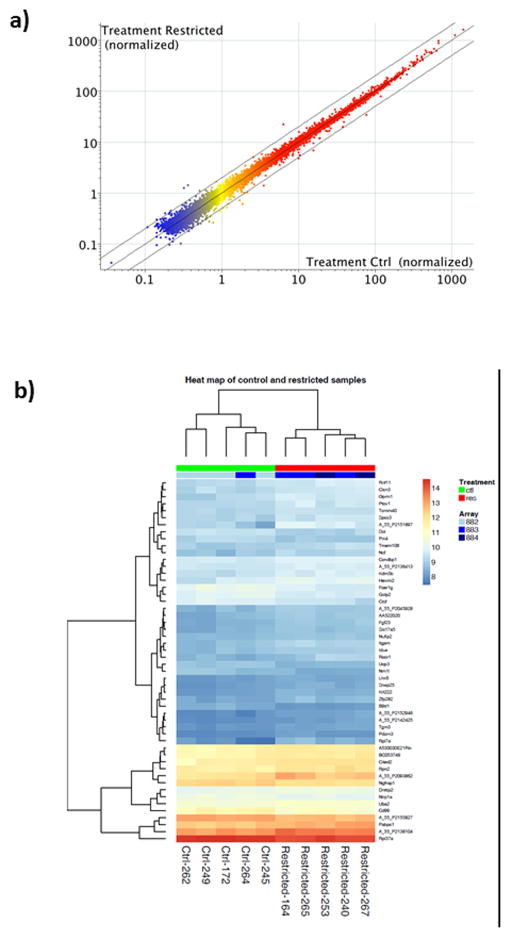
Microarray analysis was conducted on pooled RNA from two placentas each from five dams in each of the treatment groups. Both heat map and scatter plot visualization of array data reveal highly similar patterns of gene expression between control and restricted placentas at d18.5 (Figure 4). Using a 10% FDR cutoff, there were no differentially expressed genes between placentas of control and restricted dams. The 50 genes showing the highest probability of differential expression are listed in Supplemental Table 1. The smallest adjusted p-value was 0.41.
Figure 4.
Whole genome microarray analysis of five pairs of placentas from control and food restricted dams. (a) Scatter plot: genes present (above background) in all replicates of at least one treatment (15,430 probes) are displayed as normalized to the 75th percentile of each array. Red=High expression, Yellow=Medium expression, Blue=Low expression. Center line= identical expression, and outer lines = 2-fold difference. (b) Heat map based on 50 genes most likely to be differentially expressed. No significant differences in expression were found.
Fetal Liver
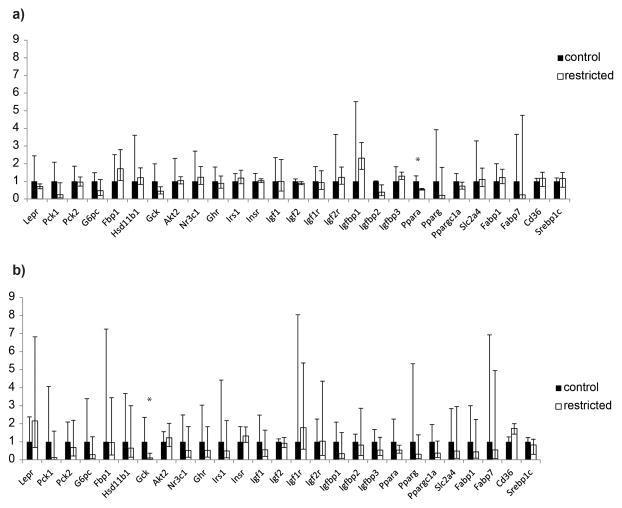
Fetal liver gene expression was compared between offspring groups by using the same real-time PCR arrays previously used to identify differences in the adult offspring from these treatments (Pennington et al. 2012). Expression of genes encoding two key fatty acid transporters, Cd36 and Srepb1c were also examined by real-time PCR in these samples. Ppara expression was significantly lower in livers of fetuses from restricted dams overall, although when the sexes were analyzed separately, the difference was only significant within male offspring (Figure 4). Expression of Gck was significantly reduced in fetal livers regardless of sex, although when the sexes were analyzed separately, the difference was only significant within females (Figure 5).
Figure 5.
Relative expression of key metabolic genes in the liver of (a) male and (b) female fetuses from control (black) and restricted (white) dams determined by real-time PCR. Each is expressed as fold-change relative to control sample mean. Error bars indicated fold-changes calculated from mean ± standard deviation of ΔΔCt * p=0.04 **p=0.01.
Discussion
When mice were placed on 50% food restriction from d1.5–11.5 of pregnancy, they had significantly reduced placental weights, placental junctional zone areas, and junctional to labyrinth zone ratios, along with somewhat reduced fetal blood space areas within the labyrinth zone (Schulz et al. 2012) at d11.5. Over two hundred gene expression differences were identified by microarray, including PCR-confirmed decreases in Ceacam12, Gbp1 and Tpbpa. However, the adult offspring of these pregnancies had largely normal metabolism, so here we asked whether the placental abnormalities observed at day 11.5 were sustained until late pregnancy. The answer is no; we detected no significant differences in placental weights, morphology or gene expression at d18.5 in mice that had been food restricted from days 1.5–11.5 when compared to controls.
This finding is consistent with the hypothesis that lasting changes in the placenta are responsible for lasting changes in offspring physiology that follow early pregnancy insults. In humans and sheep, early pregnancy nutrient restriction that led to obesity in offspring also led to oversized placentas at term. We found that in the mouse, early pregnancy nutrition did not lead to placental abnormalities at term and did not lead to offspring obesity. Thus, recovery of normal placental morphology and gene expression may underlie species differences in the effects of early pregnancy undernutrition on adult phenotype. As a caveat, only one strain of outbred mouse was used, so this conclusion may not be applicable to all mice. For example, strain differences in remodeling of the placental vasculature have been documented between inbred C57Bl/6 and outbred CD1 mice (Rennie et al. 2012).
The ability of the mice to recover normal placental function and yet not overcompensate by term following early pregnancy food restriction may be a function of timing. The timescales for development in the sheep and human are much longer than in the mouse, leaving more time for compensation to become overcompensation. On the other hand, the rat has only a 1–2 day longer gestation period than the mouse, and food restriction during early gestation can lead to offspring obesity in rats. In a series of studies, Jones and Friedman found that male rat offspring became obese following 50% maternal food restriction from day 1–10, and Palou et al. saw a similar effect in offspring of rats with just 20% food restriction (Palou et al. 2010). In contrast, Anguita et al. found that female rat offspring became obese following 50% maternal food restriction from day 1–14, but male offspring weighed less than offspring of controls (Anguita et al. 1993). All of these studies initiated food restriction on the morning following mating, whereas our food restriction began on the next day, which may also explain the differing long term consequences. The periconceptual period is critical for some developmental programming effects, particularly that of dietary methyl donors (Sinclair et al. 2007). The effects on offspring of total caloric restriction during the period prior to d1.5 mouse pregnancy remains to be tested.
There were two differences in gene expression in fetal livers at d18.5 from previously food restricted dams. In the fetal livers from restricted dams, Gck mRNA was reduced nearly 10-fold. Gck encodes glucokinase, the enzyme that converts glucose to glucose-6-phosphate in the first step of glycolysis. In both humans and mice, loss-of-function mutations in Gck are associated with mature onset diabetes of the young (MODY) and reduced birth weights, likely due to its role in glucose sensing and insulin release in the pancreas (Terauchi et al. 2000, Shields et al. 2010). In the liver, Gck regulates glycogen synthesis, but fetuses depend largely on other hexokinases for this function (Cifuentes et al. 2008), and mice with Gck knockout in liver do not develop abnormal blood glucose until after weaning (Wang et al. 2013). Thus, the significance of this dramatic expression difference for fetal physiology is not clear. Gck expression and methylation have previously been shown to be altered in adult rats following prenatal exposure to BPA (Ma et al. 2013), suggesting it is a target of fetal programming. However, the decreased expression we observed in fetal livers from restricted dams was not present in female offspring of restricted dams at 29 weeks of age (Pennington et al. 2012) and Gck expression was significantly increased in livers of adult males from restricted dams (Pennington et al. 2012). These changes in liver Gck expression suggest complex regulation by maternal undernutrition.
The other expression difference was a reduction in Ppara expression in the livers of fetuses from previously food restricted mothers. Ppara, a member of the peroxisome proliferator-activated receptor family, is a key regulator of lipid metabolism in the liver. It is hypothesized to play a protective role in rat fetuses of dams with type I diabetes (Martinez et al., 2011). A reduction in Ppara expression was also observed in the livers of adult rats that had been exposed to maternal undernutrition throughout pregnancy (Gluckman et al. 2007). However, the opposite effect, an increase in liver Ppara expression, was observed at postnatal d6 in rats born to dams fed a protein restricted diet throughout pregnancy (Lillycrop et al. 2005). The decreased Ppara expression present in fetal livers from restricted dams was not detected in the adult offspring of restricted mothers at 29 weeks of age (Pennington et al. 2012). However, Ppara expression was only measured in the adult offspring following high fat diet consumption. The high fat diet eliminated the lower adiposity that male offspring displayed on a standard diet, so it may also have erased differences in Ppara expression (Pennington et al. 2012). A caveat to interpretation of either difference in liver gene expression is that it is not known whether these changes are reflected by changes in protein production.
A strength of the liver analyses was the separation of samples by fetal sex, as our previous study had shown differing effects of maternal treatment on male and female adult offspring (Pennington et al. 2012). Similarly, it was recently shown in the baboon that food restriction induced consistent changes in gene expression in female, but not male placentas (Cox et al. 2013), and murine female placentas also respond more dramatically to high-fat diet changes (Mao et al. 2010). Thus, a drawback of the placental microarray analysis is that RNA from two placentas was pooled at random from each dam, so that results cannot be separated by sex. Treatment effects that are limited to one sex may not have been detected. However, a strength of the pooling approach is the ability to sample multiple placentas from each dam, despite the cost limitations of the array approach.
Overall, the results of this study show that the mouse placenta can recover normal morphology and gene expression by term despite early growth restriction resulting from maternal nutrient restriction in early pregnancy. While there were still differences in fetal liver gene expression at d18.5, further study is needed to identify their physiological significance. It has been proposed in the DOHAD field that the mechanism by which maternal nutrient restriction in early pregnancy influences adult health is via permanent alterations in the placenta. It was the goal of this series of studies to better understand that mechanism. By exposing dams to early pregnancy nutrient restriction, then examining the placenta at mid-and now late pregnancy, we would identify the lasting changes in gene expression and placental morphology that underlie programming of the adult. Instead, we encountered an unexpected phenomenon; early placental changes were reversible, and the offspring were normal. Additional studies are needed to uncover mechanisms by which the placenta is able to recover and protect the fetus from permanent damage.
Supplementary Material
Microarray analysis showing the fifty genes with the greatest probability of differential expression between control and restricted placentas. Average normalized expression levels are given in column e, the t-statistic in column F, unadjusted and adjusted p-values in columns G and H, and mean fold-change in restricted vs. control placentas in column J.
Acknowledgments
GRANT SUPPORT: Eunice Kennedy Shriver National Institute of Child Health and Human Development HD055231.
Thank you to Jessica Schlitt and Lisa Mao for technical assistance. This work was supported by grant number HD055231 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development.
Footnotes
Disclaimer. This is not the definitive version of record of this article. This manuscript has been accepted for publication in Reproduction, but the version presented here has not yet been copy edited, formatted or proofed. Consequently, the journal accepts no responsibility for any errors or omissions it may contain. The definitive version is available at doi: 10.1530/REP-15-0010 2015 Society for Reproduction and Fertility.”
DECLARATION OF INTERESTS
There are no conflicts of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- Anderson N, Borlak J. Molecular mechanisms and therapeutic targets in steatosis and steatohepatitis. Pharmacol Rev. 2008;60:311–357. doi: 10.1124/pr.108.00001. [DOI] [PubMed] [Google Scholar]
- Anguita RM, Sigulem DM, Sawaya AL. Intrauterine food restriction is associated with obesity in young rats. J Nutr. 1993;123:1421–1428. doi: 10.1093/jn/123.8.1421. [DOI] [PubMed] [Google Scholar]
- Barker DJ, Bull AR, Osmond C, Simmonds SJ. Fetal and placental size and risk of hypertension in adult life. Bmj. 1990;301:259–262. doi: 10.1136/bmj.301.6746.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJ, Gelow J, Thornburg K, Osmond C, Kajantie E, Eriksson JG. The early origins of chronic heart failure: impaired placental growth and initiation of insulin resistance in childhood. Eur J Heart Fail. 2010;12:819–825. doi: 10.1093/eurjhf/hfq069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Bourgon R, Gentleman R, Huber W. Independent filtering increases detection power for high-throughput experiments. Proc Natl Acad Sci U S A. 2010;107:9546–9551. doi: 10.1073/pnas.0914005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce KD, Cagampang FR, Argenton M, Zhang J, Ethirajan PL, Burdge GC, Bateman AC, Clough GF, Poston L, Hanson MA, McConnell JM, Byrne CD. Maternal high-fat feeding primes steatohepatitis in adult mice offspring, involving mitochondrial dysfunction and altered lipogenesis gene expression. Hepatology. 2009;50:1796–1808. doi: 10.1002/hep.23205. [DOI] [PubMed] [Google Scholar]
- Cifuentes D, Martinez-Pons C, Garcia-Rocha M, Galina A, Ribas de Pouplana L, Guinovart JJ. Hepatic glycogen synthesis in the absence of glucokinase: the case of embryonic liver. J Biol Chem. 2008;283:5642–5649. doi: 10.1074/jbc.M706334200. [DOI] [PubMed] [Google Scholar]
- Clarke L, Heasman L, Juniper DT, Symonds ME. Maternal nutrition in early-mid gestation and placental size in sheep. Br J Nutr. 1998;79:359–364. doi: 10.1079/bjn19980060. [DOI] [PubMed] [Google Scholar]
- Cox LA, Li C, Glenn JP, Lange K, Spradling KD, Nathanielsz PW, Jansson T. Expression of the placental transcriptome in maternal nutrient reduction in baboons is dependent on fetal sex. J Nutr. 2013;143:1698–1708. doi: 10.3945/jn.112.172148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandrea J, Wilson V, Gopalakrishnan G, Heasman L, Budge H, Stephenson T, Symonds ME. Maternal nutritional manipulation of placental growth and glucose transporter 1 (GLUT-1) abundance in sheep. Reproduction. 2001;122:793–800. [PubMed] [Google Scholar]
- Foote WC, Pope AL, Chapman AB, Casida LE. Reproduction in the Yearling Ewe as Affected by Breed and Sequence of Feeding Levels. II. Effects on Fetal Development. J Anim Sci. 1959;18:463–474. [Google Scholar]
- Ford SP, Hess BW, Schwope MM, Nijland MJ, Gilbert JS, Vonnahme KA, Means WJ, Han H, Nathanielsz PW. Maternal undernutrition during early to mid-gestation in the ewe results in altered growth, adiposity, and glucose tolerance in male offspring. J Anim Sci. 2007;85:1285–1294. doi: 10.2527/jas.2005-624. [DOI] [PubMed] [Google Scholar]
- Gardner DS, Tingey K, Van Bon BW, Ozanne SE, Wilson V, Dandrea J, Keisler DH, Stephenson T, Symonds ME. Programming of glucose-insulin metabolism in adult sheep after maternal undernutrition. Am J Physiol Regul Integr Comp Physiol. 2005;289:R947–954. doi: 10.1152/ajpregu.00120.2005. [DOI] [PubMed] [Google Scholar]
- Gentleman RHW, Carey VJ, Irizarry RA, Dudoit S. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluckman PD, Lillycrop KA, Vickers MH, Pleasants AB, Phillips ES, Beedle AS, Burdge GC, Hanson MA. Metabolic plasticity during mammalian development is directionally dependent on early nutritional status. Proc Natl Acad Sci U S A. 2007;104:12796–12800. doi: 10.1073/pnas.0705667104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco D, Kotronen A, Westerbacka J, Puig O, Arkkila P, Kiviluoto T, Laitinen S, Kolak M, Fisher RM, Hamsten A, Auvinen P, Yki-Jarvinen H. Gene expression in human NAFLD. Am J Physiol Gastrointest Liver Physiol. 2008;294:G1281–1287. doi: 10.1152/ajpgi.00074.2008. [DOI] [PubMed] [Google Scholar]
- Gregorio BM, Souza-Mello V, Carvalho JJ, Mandarim-de-Lacerda CA, Aguila MB. Maternal high-fat intake predisposes nonalcoholic fatty liver disease in C57BL/6 offspring. Am J Obstet Gynecol. 2010;203:495.e491–498. doi: 10.1016/j.ajog.2010.06.042. [DOI] [PubMed] [Google Scholar]
- Jones AP, Friedman MI. Obesity and adipocyte abnormalities in offspring of rats undernourished during pregnancy. Science. 1982;215:1518–1519. doi: 10.1126/science.7063860. [DOI] [PubMed] [Google Scholar]
- Jones AP, Simson EL, Friedman MI. Gestational undernutrition and the development of obesity in rats. J Nutr. 1984;114:1484–1492. doi: 10.1093/jn/114.8.1484. [DOI] [PubMed] [Google Scholar]
- Koonen DP, Jacobs RL, Febbraio M, Young ME, Soltys CL, Ong H, Vance DE, Dyck JR. Increased hepatic CD36 expression contributes to dyslipidemia associated with diet-induced obesity. Diabetes. 2007;56:2863–2871. doi: 10.2337/db07-0907. [DOI] [PubMed] [Google Scholar]
- Lemoine M, Barbu V, Girard PM, Kim M, Bastard JP, Wendum D, Paye F, Housset C, Capeau J, Serfaty L. Altered hepatic expression of SREBP-1 and PPARgamma is associated with liver injury in insulin-resistant lipodystrophic HIV-infected patients. AIDS. 2006;20:387–395. doi: 10.1097/01.aids.0000206503.01536.11. [DOI] [PubMed] [Google Scholar]
- Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr. 2005;135:1382–1386. doi: 10.1093/jn/135.6.1382. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lumey LH. Compensatory placental growth after restricted maternal nutrition in early pregnancy. Placenta. 1998;19:105–111. doi: 10.1016/s0143-4004(98)90105-9. [DOI] [PubMed] [Google Scholar]
- Ma Y, Xia W, Wang DQ, Wan YJ, Xu B, Chen X, Li YY, Xu SQ. Hepatic DNA methylation modifications in early development of rats resulting from perinatal BPA exposure contribute to insulin resistance in adulthood. Diabetologia. 2013;56:2059–2067. doi: 10.1007/s00125-013-2944-7. [DOI] [PubMed] [Google Scholar]
- Mao J, Zhang X, Sieli PT, Falduto MT, Torres KE, Rosenfeld CS. Contrasting effects of different maternal diets on sexually dimorphic gene expression in the murine placenta. Proc Natl Acad Sci U S A. 2010;107:5557–5562. doi: 10.1073/pnas.1000440107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez N, White V, Kurtz M, Higa R, Capobianco E, Jawerbaum A. Activation of the nuclear receptor PPARalpha regulates lipid metabolism in foetal liver from diabetic rats: implications in diabetes-induced foetal overgrowth. Diabetes Metab Res Rev. 2011;27:35–46. doi: 10.1002/dmrr.1151. [DOI] [PubMed] [Google Scholar]
- Painter RC, Roseboom TJ, Bleker OP. Prenatal exposure to the Dutch famine and disease in later life: an overview. Reprod Toxicol. 2005;20:345–352. doi: 10.1016/j.reprotox.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Palou M, Priego T, Sanchez J, Palou A, Pico C. Sexual dimorphism in the lasting effects of moderate caloric restriction during gestation on energy homeostasis in rats is related with fetal programming of insulin and leptin resistance. Nutr Metab (Lond) 2010;7:69. doi: 10.1186/1743-7075-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington KA, Harper JL, Sigafoos AN, Beffa LM, Carleton SM, Phillips CL, Schulz LC. Effect of food restriction and leptin supplementation on fetal programming in mice. Endocrinology. 2012;153:4556–4567. doi: 10.1210/en.2012-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravelli GP, Stein ZA, Susser MW. Obesity in young men after famine exposure in utero and early infancy. N Engl J Med. 1976;295:349–353. doi: 10.1056/NEJM197608122950701. [DOI] [PubMed] [Google Scholar]
- Rennie MY, Detmar J, Whiteley KJ, Jurisicova A, Adamson SL, Sled JG. Expansion of the fetoplacental vasculature in late gestation is strain dependent in mice. Am J Physiol Heart Circ Physiol. 2012;302:H1261–1273. doi: 10.1152/ajpheart.00776.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseboom T, de Rooij S, Painter R. The Dutch famine and its long-term consequences for adult health. Early Hum Dev. 2006;82:485–491. doi: 10.1016/j.earlhumdev.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Sardinha FL, Telles MM, Albuquerque KT, Oyama LM, Guimaraes PA, Santos OF, Ribeiro EB. Gender difference in the effect of intrauterine malnutrition on the central anorexigenic action of insulin in adult rats. Nutrition. 2006;22:1152–1161. doi: 10.1016/j.nut.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Schulz LC, Schlitt JM, Caesar G, Pennington KA. Leptin and the Placental Response to Maternal Food Restriction During Early Pregnancy. Biol Reprod. 2012 doi: 10.1095/biolreprod.112.103218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sferruzzi-Perri AN, Vaughan OR, Coan PM, Suciu MC, Darbyshire R, Constancia M, Burton GJ, Fowden AL. Placental-specific Igf2 deficiency alters developmental adaptations to undernutrition in mice. Endocrinology. 2011;152:3202–3212. doi: 10.1210/en.2011-0240. [DOI] [PubMed] [Google Scholar]
- Shields BM, Freathy RM, Hattersley AT. Genetic influences on the association between fetal growth and susceptibility to type 2 diabetes. J Dev Orig Health Dis. 2010;1:96–105. doi: 10.1017/S2040174410000127. [DOI] [PubMed] [Google Scholar]
- Shimomura I, Bashmakov Y, Horton JD. Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. J Biol Chem. 1999;274:30028–30032. doi: 10.1074/jbc.274.42.30028. [DOI] [PubMed] [Google Scholar]
- Sinclair KD, Allegrucci C, Singh R, Gardner DS, Sebastian S, Bispham J, Thurston A, Huntley JF, Rees WD, Maloney CA, Lea RG, Craigon J, McEvoy TG, Young LE. DNA methylation, insulin resistance, and blood pressure in offspring determined by maternal periconceptional B vitamin and methionine status. Proc Natl Acad Sci U S A. 2007;104:19351–19356. doi: 10.1073/pnas.0707258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Terauchi Y, Kubota N, Tamemoto H, Sakura H, Nagai R, Akanuma Y, Kimura S, Kadowaki T. Insulin effect during embryogenesis determines fetal growth: a possible molecular link between birth weight and susceptibility to type 2 diabetes. Diabetes. 2000;49:82–86. doi: 10.2337/diabetes.49.1.82. [DOI] [PubMed] [Google Scholar]
- Townsend KL, Lorenzi MM, Widmaier EP. High-fat diet-induced changes in body mass and hypothalamic gene expression in wild-type and leptin-deficient mice. Endocrine. 2008;33:176–188. doi: 10.1007/s12020-008-9070-1. [DOI] [PubMed] [Google Scholar]
- Wang R, Gao H, Xu W, Li H, Mao Y, Wang Y, Guo T, Wang X, Song R, Li Z, Irwin DM, Niu G, Tan H. Differential expression of genes and changes in glucose metabolism in the liver of liver-specific glucokinase gene knockout mice. Gene. 2013;516:248–254. doi: 10.1016/j.gene.2012.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wang C, Terroni PL, Cagampang FR, Hanson M, Byrne CD. High-unsaturated-fat, high-protein, and low-carbohydrate diet during pregnancy and lactation modulates hepatic lipid metabolism in female adult offspring. Am J Physiol Regul Integr Comp Physiol. 2005;288:R112–118. doi: 10.1152/ajpregu.00351.2004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Microarray analysis showing the fifty genes with the greatest probability of differential expression between control and restricted placentas. Average normalized expression levels are given in column e, the t-statistic in column F, unadjusted and adjusted p-values in columns G and H, and mean fold-change in restricted vs. control placentas in column J.