Abstract
Background & Aims
Hepatocellular carcinoma (HCC) is the second most lethal cancer due to lack of effective therapies. Although promising, HCC molecular classification, which enriches potential responders to specific therapies, has not yet been assessed in clinical trials of anti-HCC drugs. We aimed to overcome these challenges by developing clinicopathological surrogate indices of HCC molecular classification.
Methods
HCC classification defined in our previous transcriptome meta-analysis (S1, S2, and S3 subclasses) was implemented in an FDA-approved diagnostic platform (Elements assay, NanoString). Ninety-six HCC tumors (training set) were assayed to develop molecular subclass-predictive indices based on clinicopathological features, which were independently validated in 99 HCC tumors (validation set). Molecular deregulations associated with the histopathological features were determined by pathway analysis. Sample sizes for HCC clinical trials enriched with specific molecular subclasses were determined.
Results
HCC subclass-predictive indices were: steatohepatitic (SH)-HCC variant and immune cell infiltrate for S1 subclass, macrotrabecular/compact pattern, lack of pseudoglandular pattern, and high serum alpha-fetoprotein (>400 ng/mL) for S2 subclass, and microtrabecular pattern, lack of SH-HCC and clear cell variants, and lower histological grade for S3 subclass. Macrotrabecular/compact pattern, a predictor of S2 subclass, was associated with activation of therapeutically targetable oncogene YAP and stemness markers EPCAM/KRT19. BMP4 was associated with pseudoglandular pattern. Subclass-predictive indices-based patient enrichment reduced clinical trial sample sizes from 121, 184, and 53 to 30, 43, and 22 for S1, S2, and S3 subclass-targeting therapies, respectively.
Conclusions
HCC molecular subclasses can be enriched by clinicopathological indices tightly associated with deregulation of therapeutically targetable molecular pathways.
Keywords: Histopathology, molecular subclass, predictive index, gene expression, clinical diagnostic
INTRODUCTION
Liver cancer, mainly hepatocellular carcinoma (HCC), is the second leading cause of cancer death worldwide, and its prognosis is still dismal (5-year survival rate generally below 15%) (GLOBOCAN 2012, globocan.iarc.fr). Underlying chronic liver disease, namely cirrhosis, serves as a fertile soil for de novo carcinogenesis, therefore early detection and complete removal or ablation of the tumors rarely prevents tumor recurrence [1]. Once the tumors reach advanced stage as a consequence of multiple rounds of de novo carcinogenesis, only one approved medical therapy is available, sorafenib, which extends patient survival by only 3 months [2, 3]. The development of improved HCC therapies has been challenging as evidenced by the series of failed phase 3 trials of various molecular targeted agents [4]. It is increasingly emerging that this is attributable to the lack of predictive biomarker of response to enrich potential responders to detect therapeutic benefit in clinical trials [4].
Genomics studies in the past decade have elucidated numerous therapeutic targets, which can be mapped onto the framework of HCC molecular classification defined in our previous transcriptome meta-analysis of global HCC populations from Asia, Europe, and the U.S., including more than 600 patients (named S1, S2, and S3 subclasses) and independently validated [5–8]. The molecular hallmarks of the HCC subclasses have become increasingly targetable by newly developed therapies. For example, inhibitors of TGF-beta pathway (a hallmark of S1), glypican-3 (a marker of S2), and MET pathway (S1/S2) have been evaluated in recent early-stage clinical trials in HCC [9–11]. Alpha-fetoprotein (AFP), a marker of S2 tumors, was targeted by AFP genetic vaccine [12]. A Src/Abl inhibitor, dasatinib, showed preferential effect in S1-like hepatoma cell lines [13, 14]. We recently found that siRNA-based silencing of YAP oncogene (activated in S2 subclass) induced HCC tumor regression [15]. These studies collectively suggest that determination of HCC molecular subclass may serve as a broadly applicable predictive biomarker of response to these therapies.
Given that the latest clinical practice guidelines recommend tumor tissue biopsy in the setting of therapeutic clinical trials [16], tools to inexpensively and robustly determine the molecular subclasses and aberrations in clinical specimens are urgently needed. However, such molecular subclass/biomarker-enriched clinical trials have been rarely conducted because of financial constraints in the biomarker component of clinical trials. In addition, although such molecular classification could refine clinical patient management by informing patient prognosis, it is still challenging to disseminate the information widely because the vast majority of HCC patients are diagnosed in developing, resource-poor countries, where the implementation of molecular biomarker assays is practically infeasible [8].
To overcome these challenges, here we aimed to develop and validate clinically readily available surrogates of HCC molecular classification based on robust clinicopathological information, which enables clinical trial enrichment for molecular targeted anti-HCC drugs. Furthermore, we implemented and evaluated a gene signature-based HCC molecular classification assay in an FDA-approved diagnostic platform applicable to clinical archived formalin-fixed, paraffin-embedded (FFPE) tissues [17]. The assay may be used to retrospectively corroborate the clinicopathological indices-based subclass prediction whenever funding and/or access to the assay facility become available in a real-world clinical setting. Of note, the assay provides objective readout unlike immunohistochemistry, which relies on molecular pathology expertise and somewhat subjective interpretation, using exactly the same material, i.e., FFPE tissue sections. Collectively, the information of HCC molecular classification will become globally accessible to facilitate therapeutic development and refined patient care.
PATIENTS AND METHODS
Patient cohorts
Archived FFPE tissues from 96 tumor foci from 88 HCC patients who underwent surgical resection between 1992 and 2012 at Toranomon Hospital were used for development of clinicopathological indices predictive of HCC molecular subclasses (training set) (Figure S1). The predictive indices were validated in an independent set of 99 HCC patients, for which the molecular subclasses were determined based on genome-wide transcriptome profiling in our previous study [5], and analyzable hematoxylin and eosin (H&E)-stained FFPE tissue sections were available (validation set). Hepatitis C virus (HCV) infection was determined by positivity of serum HCV antibody or RNA. Hepatitis B virus (HBV) infection was determined by positivity of the hepatitis B surface antigen (HBsAg). Alcohol abuse was defined as lifetime alcohol intake greater than 500kg. Non-alcoholic fatty liver disease (NAFLD) and non-alcoholic steatohepatitis (NASH) were diagnosed according to current practice guidelines [18]. The study, retrospectively analyzing archived tissues from previous treatment performed as routine clinical care, was approved and acquisition of written informed consent was waived by the institutional review board granted on the condition that all samples were anonymized. The study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a priori approval by the institution's human research committee (Program for the Protection of Human Subjects [PPHS]).
Histopathological analysis
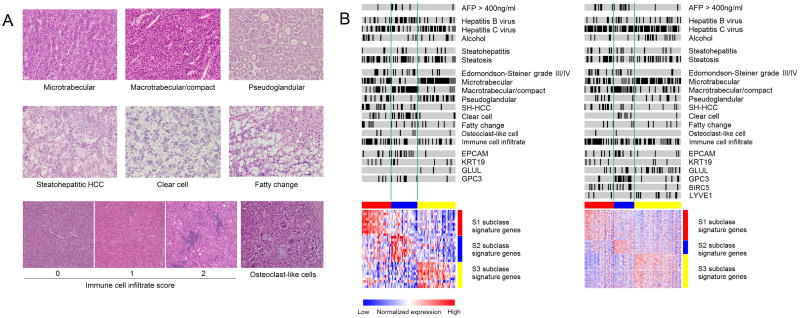
Histopathological evaluation of representative H&E stained slides was independently performed by three pathologists (T.H., S.C.W. and M.I.F.), three hepatologists (P.T., N.G., and Y.H.) and one liver surgeon (S.N.) with liver pathology training. All evaluators were blinded to information of the molecular subclass. Discrepant readings (observed in 17 tumors, 9%) were reconciled by discussion of the six evaluators. Foci within the tumors were classified based on architecture and cytological features according to the World Health Organization Classification of Tumors of the Digestive System (4th edition) and related publications [19–21]. The following histological patterns and cytological variant were determined: microtrabecular pattern, macrotrabecular pattern, compact pattern, pseudoglandular pattern, clear cell variant, steatohepatitic HCC (SH-HCC) variant, and fatty change (Figure 1A). The trabecular pattern was characterized by cords of tumor cells separated by sinusoidal-like vascular spaces, which was further classified into the microtrabecular pattern if the cords were composed of up to 10 cells (mostly 3 to 5 cells) and the macrotrabecular pattern if the cords were thicker than 10 cells without endothelial cells between the cords. The compact pattern was characterized by extensive compression of the sinusoidal spaces resulting in its solid appearance. The pseudoglandular pattern was characterized by the presence of gland-like structures that represent dilated, abnormal bile canaliculi surrounded by a ring of tumor cells. The clear cell variant was characterized by uniform tumor cells with centrally located nuclei and prominent optically clear cytoplasm, attributed to glycogen accumulation, without obvious vesicles or other cytoplasmic structures [22–24]. The steatohepatitic HCC (SH-HCC) variant is a recently described HCC variant that is characterized by tumor cells showing ballooning degeneration-like appearance often accompanied by cytoplasmic lipid droplets, Mallory-Denk bodies, pericellular fibrosis, and/or inflammatory infiltrates [25–27]. Fatty change was characterized by tumor cells with large cytoplasmic lipid-containing vacuoles that displace the nucleus to the periphery and without the features of SH-HCC variant. Foci of tumor cells forming clear subnodules/subcomponents with distinct histological patterns from other components within a tumor nodule were dissected and separately analyzed for transcriptome profiling. The histological features present in more than 30% of each profiled area were recorded and correlated with the molecular subclasses. Histological grading was determined according to the Edmondson-Steiner grading system on an entire HCC nodule [28]. Steatosis and steatohepatitis in non-tumor liver were determined by greater than 5% steatosis and presence of ballooning degeneration, respectively [29]. Immune cell infiltrate within the tumor was scored 0 to 2 according to the mean number of infiltrating lymphocytes from 5 high-powered fields (0: 0 to 5 cells, 1: 5 to 20 cells, 2: greater than 20 cells) [30]. A recently described HCC variant, chromophobe HCC as well as osteoclast-like cell were also assessed [31, 32].
Figure 1.
A. Histopathological features of HCC tumors. Architectural patterns and cytological variants analyzed for correlation with HCC molecular subclasses. B. HCC molecular subclasses and clinicopathological features in the training (left) and validation (right) sets. Black bars indicate positivity of the feature. HCC tumors with high immune cell infiltrate (score of 2) is shown by black bars. Red, blue and yellow colors in the horizontal bars above the heatmaps indicate S1, S2 and S3 tumors, respectively. Red and blue colors in the heatmaps indicate high and low gene expression, respectively.
Transcriptome-based determination of HCC molecular subclasses
To determine the S1, S2, and S3 subclasses, we implemented a classifier gene signature, originally comprised of 619 genes, in the digital transcript counting technology (Elements assay, NanoString), an FDA-approved clinical diagnostic platform capable of analyzing severely degraded FFPE RNA [17]. Because the number of gene probes is the major determinant of the assay cost, we first bioinformatically evaluated performance of the top informative HCC molecular subclass signature genes that fit the minimum unit of Elements assay (30 signature genes and 6 normalization genes). From among the 619 full HCC subclass signature genes already verified for their subclass predictive performance in our previously reported random resampling-based transcriptome meta-analysis of 9 independent HCC datasets [5], top 30 genes (10 genes for each subclass) with the largest Fisher’s inverse chi-square were chosen for the assay development (30-gene signature, Table S1). The subclass prediction was repeated with the 30-gene signature in the 9 independent HCC datasets (Table S2) using nearest template prediction algorithm as previously described [5], and concordance with the original prediction made by the 619-gene signature was evaluated. We observed an overall concordance of 85% (IQR: 80%–90%, range: 72%–94%) (Figure S2). Prediction concordance for S1, S2, and S3 subclasses were 79% (IQR: 74%–88%), 85% (IQR: 82%–93%), and 87% (IQR: 84%–89%), respectively. Based on these results, indicating that the 30-gene signature enables reasonably reproducible determination of HCC molecular subclass, we implemented the signature in the Elements assay platform.
Total RNA was isolated from three 10-micron-thick FFPE tissue sections by using High Pure RNA Paraffin Kit (Roche). Multiple histopathological foci co-existing in a tumor nodule were macro-dissected based on serial H&E stained section. Expression profiling was performed using 100ng to 500ng total RNA samples with nCounter Analysis System (NanoString). Raw scan data were extracted by nSolver software ver.1, and normalized by scaling with geometric mean of normalization gene probes by using NanoString normalizer module of GenePattern genomic analysis toolkit (www.broadinstitute.org/genepattern). The datasets are available at NCBI Gene Expression Omnibus (GEO) database (GSE59548, GSE10186).
Expression of HCC marker genes
Expression of previously documented stemness marker genes in HCC, KRT19 and EPCAM, as well as a marker gene of CTNNB1 exon 3-mutated HCC tumors, GLUL [33], were determined by reverse transcription quantitative polymerase chain reaction RT-qPCR in the training set. Total RNA was converted into cDNA using EcoDry Premix (Clontech Laboratories) on Matercycler Nexus Gradient (Eppendorf), and qPCR was performed using iQ SYBR Green Supermix (BioRad) on CFX384 Touch Real-Time PCR Detection System (Bio-Rad) following manufacturer’s instruction. Used primer sequences are summarized in Table S3. Gene expression level was calculated using delta-delta Ct method based on a housekeeping gene, RPL13A. Tumors were classified into high- or low-expression group based on one standard deviation above mean.
Bioinformatics analysis
Molecular pathway deregulations associated with the histopathological features were determined in the genome-wide transcriptome dataset of the validation set by surveying a comprehensive collection of 10,295 annotated pathway gene sets in Molecular Signature Database (MSigDB, www.broadinstitute.org/mdigdb) and a collection of liver cancer-related gene signatures in literature (Table S4) using Gene Set Enrichment Analysis (GSEA) [34]. Functionally co-regulated gene networks in association with the histopathological features were determined by using Planar Filtered Network Analysis (PFNA) algorithm [35]. Briefly, global gene co-expression network was first constructed by using the genome-wide profiles of the validation set based on Pearson correlation coefficient. Co-regulated gene modules (subnetworks) with significant and robust biological/functional links were subsequently determined on a hyperbolic surface [36, 37]. Gene modules associated with each of the histopathological features were determined by using GSEA of the module member genes on gene list in the validation transcriptome dataset rank-ordered according to differential expression between tumors with the feature and the rest by using random permutation t-test implemented in GenePattern. Key regulatory genes in the gene modules associated with each of the histopathological features were determined by Key Driver Analysis (KDA), which prioritizes driver genes by measuring the impact on the down-stream genes such that the down-stream genes were defined by n-layer neighborhood in a co-expression network with optimal n that maximizes the enrichment statistic [38]. When adjustment for high-dimensional multiple hypothesis testing was needed, Benjamini-Hochberg false discovery rate (FDR) <0.05 was regarded as statistically significant. All analyses were performed using GenePattern and R statistical language (www.r-project.org).
Statistical analysis
Categorical and unpaired continuous data were tested by Fisher’s exact test and Wilcoxon rank-sum test, respectively. Logistic regression was used to evaluate correlation between each HCC molecular subclass and the clinicopathological features (architectural patterns, cytological variants, tumor grade, microvascular invasion, presence of satellite lesions, tumor size, multiplicity, serum AFP level, AFP lens culinaris agglutinin-reactive fraction 3 [L3], and des-gamma-carboxy prothrombin [DCP]), the HCC molecular markers (KRT19, EPCAM, and GLUL), and disease etiologies (HBV, HCV, alcohol, and NAFLD). Variables with p<0.05 in univariable analysis were further evaluated in multivariable logistic regression modeling with stepwise variable selection for the development of HCC molecular subclass-predictive indices using the regression coefficients. Given that the patency of sinusoidal-like spaces within a tumor could be artificially modified during the process of tissue fixation and processing, we combined the macrotrabecular and compact patterns into a single composite variable for the correlation analysis. Two sets of cut-off values for the HCC molecular subclass-predictive indices were determined based on receiver operating characteristic (ROC) curves in the training set to maximize either (i) positive predictive value (PPV) to examine the scenarios of clinical trial enrichment or (ii) sensitivity/specificity to determine diagnostic performance of the indices in general, and applied to the validation set. Sample size required to detect a therapeutic response in clinical trials of anti-HCC drugs targeting each of the HCC molecular subclasses was calculated based on alpha error of 0.05, power of 0.90, and treatment/control ratio of 1. Objective response rate (ORR) in molecularly determined targeted subclass was assumed to range from 50% to 70% based on published lung cancer trials enriched with EGFR gene mutations- or ALK gene fusion-positive patients [39]. Given the scenario that HCC patients predicted to have the target subclass using the clinicopathological indices are enrolled into clinical trials, PPVs derived from the validation set were used to calculate proportion of patients with true targeted molecular subclass. Prevalence of S1, S2, and S3 subclasses were assumed to be 30%, 25%, and 45%, respectively, from the prevalence in the current training and validation sets and our previous study [5]. Number needed to treat (NNT) was calculated based on the same assumption on ORR and PPV. Two-tailed p-value <0.05 was considered as statistically significant. All analyses were performed using R statistical language.
RESULTS
Patients
Patients in the validation set were slightly younger and more frequently affected with HCV infection compared to the training set (Table 1). Three patients in the training set had a definite diagnosis of NAFLD/NASH as a sole etiology of HCC. Serum bilirubin was marginally higher in the validation set but still within the normal reference range. There was no significant difference in the proportion of tumors with AFP level greater than 400 ng/mL. More than 90% of the tumors were stage 0/A in both the training and validation sets.
Table 1.
Clinical characteristics of HCC patients in the training and validation sets.
| Characteristic | Training Set (88 patients) | Validation Set (99 patients) | P value |
|---|---|---|---|
| Age (y), median (IQR) | 61 (56–66) | 59 (52–64) | 0.04 |
| Male, n (%) | 70 (80%) | 78 (79%) | 1.00 |
| Etiology of chronic liver disease, n (%) | |||
| - Hepatitis C virus | 43 (49%) | 70 (71%) | <0.001 |
| - Hepatitis B virus | 25 (28%) | 23 (23%) | 0.50 |
| - Alcohol | 17 (19%) | 18 (18%) | 0.85 |
| - Non-alcoholic fatty liver diseases | 3 (3%) | 0 (0%) | 0.10 |
| Albumin (g/dL), median (IQR) | 3.7 (3.5–3.9) | 3.8 (3.5–4.1) | 0.09 |
| Bilirubin (mg/dL), median (IQR) | 0.8 (0.7–1.2) | 1.0 (0.8–1.4) | 0.01 |
| Platelet count x103 /mm3, median (IQR) | 136 (97–173) | 134 (84–204) | 0.64 |
| Child Pugh score*, median (IQR) | 5 (5–6) | 5 (5–6) | 0.68 |
| Child Pugh class, n (%) | 0.11 | ||
| A | 84 (95%) | 87 (89%) | |
| B | 4 (5%) | 11 (11%) | |
| Tumor size (cm), median (IQR) | 2.3 (2.0–2.9) | 2.2 (1.8–3.0) | 0.63 |
| Tumor number, n (%) | 0.09 | ||
| Single | 74 (84%) | 73 (84%) | |
| Multiple | 14 (16%) | 14 (14%) | |
| Presence of satellite lesions | 12 (13%) | 16 (16%) | 0.54 |
| α-fetoprotein (ng/mL), median (IQR) | 16 (4.0–88.8) | 37 (10–197) | 0.009 |
| > 400 ng/mL, n (%) | 7 (8%) | 15 (15%) | 0.17 |
| AJCC stage, n (%) | |||
| I/II | 65/23 (74%/26%) | 72/14 (82%/16%) | 0.62 |
| IIIA | 0 (0%) | 2 (2%) | |
| BCLC stage, n (%) | |||
| 0/A | 12/75 (14%/85%) | 23/59 (26%/67%) | 0.12 |
| B | 1 (1%) | 6 (7%) | |
Child-Pugh score was unavailable for 1 patient in the validation set.
AJCC: American Joint Committee on Cancer, BCLC: Barcelona Clinic Liver Cancer.
Histopathological features of HCC tumors
The micro/macrotrabecular, compact, and pseudoglandular patterns, SH-HCC, clear cell, immune cell infiltrates, and fatty change were present in more than 5% of the tumors in both training and validation sets, ascertaining general applicability of the analyzed features (Table 2). Diagnostic concordance among the three pathologists was greater than 70% for all the histopathological features (Table S5). Discordant determination was resolved by discussion among all evaluators and a consensus diagnosis was reached. There was no significant difference in their prevalence between the training and validation sets except for the pseudoglandular pattern and the clear cell variant, which were more frequently observed in the training set. The microtrabecular pattern was the most prevalent feature observed in more than half of the tumors, followed by the macrotrabecular/compact pattern observed in approximately 40% of the tumors. SH-HCC was observed in approximately 20% in both training and validation sets. We confirmed correlation of the SH-HCC variant with presence of steatosis and steatohepatitis in background liver as reported in previous studies (Table S6) [40]. Presence of cirrhosis was not associated with the SH-HCC variant (p=0.52). Relatively higher immune cell infiltrates (score of 2) were observed in approximately 40% to 50% of the tumors. Osteoclast-like cells were observed in 6 and 2 tumors in the training and validation sets, respectively, and accumulated in S1 and S2 subclasses, representing higher grade tumors. Chromophobe HCC was not observed in the current patient series, which may be due to the earlier tumor stage (median tumor size 2.3cm and 2.2cm in the training and validation sets, respectively) compared to the tumors in the original report of the variant (median tumor size 5.7cm) [31]. Fatty change had similar occurrence in the two sets, whereas the clear cell variant was more prevalent in the training set. More than 80% of the tumors were of Edmondson-Steiner grade I or II.
Table 2.
Histopathological features and HCC molecular subclasses in the training and validation sets.
| Training Set (96 HCC tumors) | Validation Set (99 HCC tumors) | P value | |
|---|---|---|---|
| Histopathological feature | |||
| Architectural pattern | |||
| Microtrabecular | 53 (55%) | 53 (54%) | 0.89 |
| Macrotrabecular/compact | 38 (40%) | 39 (39%) | 1.00 |
| Pseudoglandular | 31 (32%) | 14 (14%) | 0.004 |
| Cytological variant | |||
| SH-HCC | 19 (20%) | 17 (17%) | 0.71 |
| Clear cell | 23 (24%) | 7 (7%) | 0.001 |
| Fatty change | 17 (18%) | 10 (10%) | 0.15 |
| Edmondson-Steiner grade | |||
| I | 12 (13%) | 21 (21%) | 0.29 |
| II | 66 (69%) | 62 (63%) | |
| III | 17 (18%) | 16 (16%) | |
| IV | 1 (1%) | 0 (0%) | |
| Immune cell infiltrate score* | |||
| 0 | 15 (16%) | 18 (18%) | 0.08 |
| 1 | 42 (44%) | 28 (28%) | |
| 2 | 39 (41%) | 53 (54%) | |
| Microvascular invasion | 9 (9%) | 16 (16%) | 0.20 |
| Molecular subclass | |||
| S1 | 30 (31%) | 30 (30%) | 0.46 |
| S2 | 27 (28%) | 21 (21%) | |
| S3 | 39 (41%) | 48 (48%) | |
Immune cell infiltrate score, 0: 0 to 5 cells, 1: 5 to 20 cells, 2: greater than 20 cells.
HCC: hepatocellular carcinoma, OR: odds ratio, CI: confidence interval, SH-HCC: steatohepatitic HCC, AFP: alpha-fetoprotein
Molecular deregulations associated with histopathological features of HCC tumors
The distinct histopathological features with significant correlation with the molecular subclasses suggest that molecular pathway deregulations are related to formation of the morphological features. The microtrabecular pattern is characterized by retained normal hepatocyte functions involved in a variety of metabolic pathways and enrichment of gene signatures implicated in better HCC prognosis and biologically less aggressive molecular subclasses, including the S3 subclass [41–43] (Table S4; Table S7). The macrotrabecular/compact pattern was associated with accelerated cell cycle progression and cell proliferation, activation of an oncogene YAP recently implicated in HCC [44] as determined by induction of pathway target genes. The Yap target genes were similarly activated in the S2 subclass (Figure S3), and induction of genes regulated by transcription factors, E2F1 and MYC. The S2 subclass signature together with published liver cancer signatures of biologically aggressive subclasses [41–43], stemness markers EPCAM/KRT19, MET activation, vascular invasion, and poor prognosis were strikingly induced. The S1 subclass signature and TGF-beta signature were also enriched to lesser extent. Two gene regulatory modules (no. 15 and no. 22) were associated with the macrotrabecular/compact pattern with key driver genes, PCNA and BIRC5 (also known as survivin), implicated in cellular proliferation in HCC (Table S8; Table S9; Figure S4A, B) [45]. The pseudoglandular pattern was associated with a gene signature of HCC tumor harboring CTNNB1 exon 3 mutations accompanied with overexpression of liver-specific WNT target genes such as GLUL [42, 43]. BMP4, which is known to be involved in prostate gland formation under regulation by CTNNB1 pathway [46, 47], was identified as a key driver gene in a gene regulatory module (no. 56) associated with the pseudoglandular pattern (Table S8; Table S9; Figure S5). The SH-HCC variant was associated with activation of YAP oncogene and several kinases, including STK33, KRAS, and RAF, and pathways involved in collagen formation and extracellular matrix organization. In fact, CXCL12, ligand of a chemokine receptor CXCR4 involved in hepatic fibrosis [48, 49], was identified as a key driver gene for the SH-HCC variant together with genes encoding collagens (COL1A2, COL3A1, and COL6A2) (Table S8; Table S9; Figure S6). A CXCR4 inhibitor, AMD3100, was recently reported to inhibit intra-tumoral fibrogenesis induced by a multikinase inhibitor, sorafenib [50]. The S1 subclass signature was significantly induced in the SH-HCC variant together with signature of Chiang08 subclass Proliferation [43], further supporting our correlation analysis with logistic regression (Table 3). Clear cell variant was significantly associated with signatures of the S2 subclass, biologically more aggressive HCC tumors [41, 42], EPCAM/KRT19, and cholangiocarcinoma stem cell (Table S4; Table S7; Table 3). These findings collectively provide comprehensive overview of molecular deregulations, biomarkers, and/or potential therapeutic targets underlying the distinct histopathological features that can be further pursued in subsequent studies.
Table 3.
Tumor-related clinicopathological features associated with HCC molecular subclasses in the training set (logistic regression).
| Variable | Univariable analysis
|
Multivariable analysis
|
|||
|---|---|---|---|---|---|
| No. of HCC tumors (%) | OR (95% Cl) | P value | OR (95% Cl) | P value | |
|
|
|
||||
| S1 subclass | S1: n=30 / Rest: n=66 | ||||
| SH-HCC | 11 (37%) / 8 (12%) | 4.20 (1.47–11.97) | 0.007 | 4.25 (1.44–13.20) | 0.01 |
| Immune cell infiltrate ≥ 2 | 18 (60%) / 21 (32%) | 3.21 (1.31–7.87) | 0.01 | 3.25 (1.29–8.53) | 0.01 |
| S2 subclass | S2: n=27 / Rest: n=69 | ||||
| Microtrabecular | 4 (15%) / 49 (71%) | 0.07 (0.02–0.23) | <0.001 | ||
| Macrotrabecular/compact | 22 (81%) / 16 (23%) | 14.58 (4.75–44.69) | <0.001 | 11.99 (3.48–41.24) | <0.001 |
| Pseudoglandular | 2 (7%) / 29 (42%) | 0.11 (0.02–0.50) | 0.004 | 0.22 (0.04–1.16) | 0.07 |
| Clear cell | 14 (52%) / 9 (13%) | 7.18 (2.56–20.11) | <0.001 | ||
| Serum AFP > 400 ng/mL | 6 (22%) / 2 (3%) | 9.57 (1.80–51.03) | 0.008 | 10.81 (1.27–91.63) | 0.03 |
| S3 subclass | S3: n=39 / Rest: n=57 | ||||
| Microtrabecular | 32 (82%) / 21 (37%) | 7.84 (2.94–20.86) | <0.001 | 3.94 (1.23–12.56) | 0.02 |
| Macrotrabecular/compact | 6 (15%) / 32 (56%) | 0.14 (0.05–0.39) | <0.001 | ||
| Pseudoglandular | 19 (49%) / 11 (19%) | 3.56 (1.46–8.71) | 0.005 | ||
| SH-HCC | 1 (3%) / 18 (32%) | 0.06 (0.01–0.45) | 0.006 | 0.05 (0.01–0.44) | 0.007 |
| Clear cell | 3 (8%) / 20 (35%) | 0.15 (0.04–0.56) | 0.005 | 0.20 (0.05–0.91) | 0.04 |
| Edmondson-Steiner I or II | 36 (92%) / 42 (74%) | 4.29 (1.15–16.00) | 0.03 | 3.08 (0.65–14.58) | 0.16 |
HCC: hepatocellular carcinoma, OR: odds ratio, CI: confidence interval, SH-HCC: steatohepatitic HCC, AFP: alpha-fetoprotein
Determination of HCC molecular subclasses in the training set
We next determined the HCC molecular subclasses in the training set using a clinically applicable and inexpensive assay. From among 103 HCC tumor samples/foci from 95 patients in the training set, 96 foci (93%) from 88 patients yielded good quality RNA for the expression profiling (Figure S1). By using the molecular subclass prediction model and algorithm from our previous study without any modification, 30 (31%), 27 (28%) and 39 (41%) samples in the training set were classified into S1, S2, S3 subclasses, respectively (Table 2; Figure 1B). Prevalence of the predicted molecular subclasses was highly comparable to the results in previous studies by us and others [5, 6], indicating robust performance of the 30-gene signature assay. The highest serum AFP level in the S2 subclass, which was identified in our previous study [5], was replicated in the training set of the current study (p<0.001) (Figure S7), further supporting validity of the assay.
Correlation of clinicopathological features with HCC molecular subclasses
We next sought to determine tumor-related clinicopathological variables associated with the molecular subclasses determined by the 30-gene signature assay. Univariable logistic regression revealed several striking correlations between the clinicopathological features and the HCC molecular subclasses (Figure S8A, B, Table 3). The positive correlations of HBV infection and EPCAM overexpression with the S2 subclass identified in our previous study were confirmed [5]. Presence of the SH-HCC variant and higher immune cell infiltrates were significantly associated with the S1 subclass. Lack of the microtrabecular and the pseudoglandular patterns, presence of the macrotrabecular/compact pattern, the clear cell variant, and high serum AFP level were associated with the S2 subclass. Presence of the microtrabecular or the pseudoglandular pattern, absence of the macrotrabecular/compact pattern and the clear cell and the SH-HCC variants, and lower Edmondson-Steiner grade were associated with the S3 subclass. In multivariable modeling, both SH-HCC variant and immune cell infiltrates remained significant for association with the S1 subclass (Table 3). Association of the macrotrabecular/compact pattern and high serum AFP with the S2 subclass remained significant. The microtrabecular pattern, absence of the SH-HCC and the clear cell variants were significantly associated with the S3 subclass. These results suggest that HCC molecular subclasses can be estimated reasonably well based on assessment of the clinicopathological features. We next assessed whether molecular subclass was associated with previously published markers of hepatocarcinogenesis [51, 52] (GLUL, GPC3, LYVE1 and BIRC5) as implemented in the EASL guidelines [53]. We identified a positive association between GPC3 high expression and S2 subclass in both the training and validation sets. In the validation set, high expression of BIRC5 was associated with S1 subclass, and high expression of LYVE1 and GLUL were associated with S3 subclass (Figure S8A, B). Additionally, we assessed the association between HCC molecular subclass and patient prognosis, but there was no significant association with survival or recurrence both in training and validation sets (Table S10). This is consistent with our previous observation, in which tumor-derived molecular information had no prognostic association, where the majority of the tumors are in early stage (BCLC 0/A) and recurrent tumors were clonally unrelated with the resected primary tumors [54].
Clinicopathological indices predictive of HCC molecular subclasses
Based on the significant correlations of the clinicopathological features with the molecular subclasses, we constructed a prediction index for each subclass using the regression coefficients from the multivariable logistic regression models (see footnote of Table 4). In the training set, the area under ROC curve (AUROC) for the S1, S2 and S3 subclasses were 0.69, 0.87, and 0.85, respectively, based on which cut-off value for each predictive index was determined (Table 4; Figure S9). Each prediction index and corresponding cut-off value was evaluated in the validation set without making any modification. The validation set yielded comparable AUROC of 0.71, 0.79, and 0.73 for the S1, S2, and S3 subclasses, respectively. With the cut-off values optimized for PPV in the training set, S1, S2, and S3 subclasses in the validation set were predicted with PPVs of 60%, 50%, and 71%, respectively. Based on the cut-off values for sensitivity/specificity, all subclasses were predicted with sensitivity greater than 80% and negative predictive value nearly equal to or greater than 80% (Table S11). Based on the PPVs in the validation set and ORRs ranging from 50% to 70% assumed from previously conducted phase 2 and 3 trials of molecular targeted agents in molecular subtype-enriched lung cancer patients [39], sample sizes required to detect therapeutic benefit were reduced by 59% to 79%, and NNTs were reduced by 37% to 50% compared to a strategy enrolling “all comers” without enrichment for a specific molecular subclass (Figure S10, Table S12).
Table 4.
Performance of the clinicopathological indices predictive of HCC molecular subclasses (optimized for PPV).
| Molecular subclass | Training set
|
Validation set
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AUROC | PPV | NPV | Sensitivity | Specificity | AUROC | PPV | NPV | Sensitivity | Specificity | |
|
|
|
|||||||||
| S1 | 0.69 | 89% | 75% | 27% | 98% | 0.71 | 60% | 73% | 20% | 94% |
| S2 | 0.86 | 80% | 75% | 15% | 99% | 0.77 | 50% | 82% | 24% | 94% |
| S3 | 0.83 | 71% | 79% | 69% | 81% | 0.73 | 71% | 69% | 65% | 75% |
- S1 = 1.45 x SH-HCC (0: no, 1: yes) + 1.18 x Immune cell infiltrate (0: 0–1, 1: 2); > 2.04 was predicted as S1 subclass.
- S2 = 2.48 x Macrotrabecular/compact (0: no, 1: yes) + 2.38 x serum AFP (0: ≤ 400 ng/mL; 1: > 400 ng/mL) - 1.5 x pseudoglandular (0: no, 1: yes); > 3.67 was predicted as S2 subclass.
- S3 = 1.37 x Microtrabecular (0: no, 1: yes) - 2.97 x SH-HCC (0: no, 1: yes) - 1.59 x clear cell (0: no, 1: yes) + 1.13 x tumor grade (0: Edmondson-Steiner III or IV, 1: Edmondson-Steiner I or II); > 1.94 was predicted as S3 subclass.
HCC: hepatocellular carcinoma, AUROC: area under receiver operating characteristic curve, PPV: positive predictive value, NPV: negative predictive value, SH-HCC: steatohepatitic HCC, AFP: alpha-fetoprotein.
DISCUSSION
It is increasingly recognized that molecular biomarkers will play the key role in design and conduct of more cost-effective and expedited drug development and evaluation in clinical trials. Despite the urgent unmet need, predictive biomarker-enriched clinical trials are rarely conducted because pharmaceutical companies rarely fund the biomarker component of the trials. This issue could be resolved if clinically meaningful enrichment of specific molecular subtype is feasible by using clinically readily available predictive indices, which enable better powered clinical trial not to miss therapeutic benefit. This financial constraint is more relevant in resource-poor countries, where more than 80% of HCC patients are diagnosed. Our current study showed encouraging results as a proof of concept, demonstrating that clinical variable-based estimation of molecular subtype is a viable approach to meet this need. This strategy will also benefit other pathogen-induced cancers prevalent in developing regions of the world.
A byproduct of this study is a clinically applicable HCC molecular classification assay. Transcriptome-based clinical diagnostic development has been a challenging task due to poor reproducibility of the assay measurements due to artifacts introduced during the process of target gene amplification, which have required centralized reference laboratories to perform the assay for rigorous quality control [55]. The digital transcript counting technology without target gene amplification adapted in the current study is expected to overcome the issue. It is also worth noting that the assay is applicable even for severely degraded RNA isolated from real-world archived FFPE tissue sections [17]. The cost for clinical assay development is a prohibitive factor in developing companion biomarkers for each molecular targeted therapy, which will easily overtax currently available biomedical and financial resources. Broadly applicable biomarkers, such as our proposed molecular classification of HCC, could eliminate such efforts and enable more cost-effective drug development and biomarker-guided clinical trials.
Prognostic implication of histopathological features has been studied over the past decades in numerous studies [56]. However, no histopathological feature of HCC has been incorporated in the prognostic staging systems due to unsatisfactory reproducibility [57]. For example, prognostic association of the clear cell variant has been controversial despite its unambiguous pathological diagnostic criteria (Table S13). Similarly, we observed that prognostic association of HCC molecular subclasses in literature hugely varies across studies despite obvious presence of the subclasses [33]. Recent studies suggest that this is likely due to diversity of the disease stage represented in each study i.e. even if a tumor harbors a more aggressive molecular phenotype, the tumor has less chance to disseminate and impact patient survival if it is diagnosed at an earlier stage and successfully treated, whereas persisting liver cirrhosis and de novo HCC arisen from the cirrhotic liver have more influence on the prognosis independent of the successfully treated initial primary tumor [33]. The current study revealed surprisingly close correlation between histopathological features and molecular subclasses that links these independent observations on prognostic association of histopathological or molecular features. This finding highlights the importance to assess prognostic implication of histopathological and molecular HCC tumor characteristics according to the disease stage to elucidate their clinical utility.
The incidence and prevalence of NAFLD/NASH are increasing worldwide [58]. Epidemiological studies suggest that there are carcinogenic mechanisms unique to NASH [59]. The SH-HCC variant is a unique feature tightly linked to the presence of steatosis and steatohepatitis in the background liver [40]. Formation of this variant may provide mechanistic insights into NASH-induced HCC development that involves specific molecular pathways such as CXCL12/CXCR4-related extracellular matrix production, due to the presence of and/or genetic susceptibility to NASH. Also, it will be of interest to evaluate if SH-HCC is associated with more disseminative phenotype as seen in the S1 tumors [5] when the patients are longitudinally followed up.
In conclusion, we have successfully developed clinically readily applicable clinicopathological predictive indices of HCC molecular classification. Histopathological features of HCC tumor are accompanied with distinct molecular pathway deregulations and could also serve as surrogate markers of HCC molecular subclasses. This observation demonstrates a proof of concept that can be further pursued with refined molecular classification or other types of molecular characterization in future studies. The clinicopathological indices as well as the clinically applicable assay will serve as tools that enable wider access to the molecular information for the clinical and translational research communities to further explore therapeutic implication of the molecular characteristics of HCC, and could potentially contribute to a substantial improvement of the dismal prognosis of HCC.
Supplementary Material
Key points.
An HCC molecular classification test was implemented in a clinically applicable, FDA-approved assay platform, and successfully validated in an independent patient cohort.
Clinicopathological indices predictive of HCC molecular classification were developed and validated.
Deregulated molecular pathways tightly associated with specific histopathological features of HCC tumor were comprehensively cataloged, e.g., activation of oncogene YAP and stemness markers EPCAM/KRT19 in macrotrabecular/compact pattern and BMP4 overexpression, known to be involved in gland formation in kidney, in pseudoglandular pattern.
The clinically applicable HCC molecular classification assay and predictive indices will facilitate further evaluation of clinical utility of HCC molecular subclasses and personalized therapeutic development.
Acknowledgments
Funding
This research was supported by Uehara Memorial Foundation (to SN), FLAGS foundation, Nuovo-Soldati Cancer Research Foundation, and advanced training grant from Geneva University Hospital (to NG), and National Institute of Health (DK099558) and the Irma T. Hirschl Trust (to YH).
The nCounter Elements assay was performed at Mount Sinai qPCR Shared Resource Facility. Bioinformatics analysis was performed by using High Power Computing facility at Mount Sinai Genomics Core and Department of Scientific Computing.
Abbreviations
- AFP
alpha-fetoprotein
- AUROC
area under receiver operating characteristic
- BCLC
Barcelona Clinic Liver Cancer
- FDR
false discovery rate
- FFPE
formalin-fixed paraffin-embedded
- GSEA
Gene Set Enrichment Analysis
- H&E
hematoxylin and eosin
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- KDA
Key Driver Analysis
- NAFLD
non-alcoholic fatty liver disease
- NASH
non-alcoholic steatohepatitis
- NNT
number needed to treat
- ORR
objective response rate
- NPV
negative predictive value
- OR
odds ratio
- PFNA
Planar Filtered Network Analysis
- PPV
positive predictive value
- SH-HCC
steatohepatitic hepatocellular carcinoma
Footnotes
Conflict of Interest
Nothing to disclose.
References
- 1.Hoshida Y, Fuchs BC, Bardeesy N, Baumert TF, Chung RT. Pathogenesis and prevention of hepatitis C virus-induced hepatocellular carcinoma. J Hepatol. 2014 doi: 10.1016/j.jhep.2014.07.010. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359(4):378–90. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 3.Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S, Kim JS, et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10(1):25–34. doi: 10.1016/S1470-2045(08)70285-7. [DOI] [PubMed] [Google Scholar]
- 4.Llovet JM, Hernandez-Gea V. Hepatocellular carcinoma: reasons for phase III failure and novel perspectives on trial design. Clin Cancer Res. 2014;20(8):2072–9. doi: 10.1158/1078-0432.CCR-13-0547. [DOI] [PubMed] [Google Scholar]
- 5.Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69(18):7385–92. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kan Z, Zheng H, Liu X, Li S, Barber TD, Gong Z, et al. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome research. 2013;23(9):1422–33. doi: 10.1101/gr.154492.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turato C, Vitale A, Fasolato S, Ruvoletto M, Terrin L, Quarta S, et al. SERPINB3 is associated with TGF-beta1 and cytoplasmic beta-catenin expression in hepatocellular carcinomas with poor prognosis. Br J Cancer. 2014;110(11):2708–15. doi: 10.1038/bjc.2014.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goossens N, Sun X, Hoshida Y. Molecular classification of hepatocellular carcinoma: potential therapeutic implications. Hepat Oncol. 2015 doi: 10.2217/hep.15.26. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giannelli G, Villa E, Lahn M. Transforming growth factor-beta as a therapeutic target in hepatocellular carcinoma. Cancer Res. 2014;74(7):1890–4. doi: 10.1158/0008-5472.CAN-14-0243. [DOI] [PubMed] [Google Scholar]
- 10.Zhu AX, Gold PJ, El-Khoueiry AB, Abrams TA, Morikawa H, Ohishi N, et al. First-in-man phase I study of GC33, a novel recombinant humanized antibody against glypican-3, in patients with advanced hepatocellular carcinoma. Clin Cancer Res. 2013;19(4):920–8. doi: 10.1158/1078-0432.CCR-12-2616. [DOI] [PubMed] [Google Scholar]
- 11.Santoro A, Rimassa L, Borbath I, Daniele B, Salvagni S, Van Laethem JL, et al. Tivantinib for second-line treatment of advanced hepatocellular carcinoma: a randomised, placebo-controlled phase 2 study. Lancet Oncol. 2013;14(1):55–63. doi: 10.1016/S1470-2045(12)70490-4. [DOI] [PubMed] [Google Scholar]
- 12.Hong Y, Peng Y, Guo ZS, Guevara-Patino J, Pang J, Butterfield LH, et al. Epitope-optimized alpha-fetoprotein genetic vaccines prevent carcinogen-induced murine autochthonous hepatocellular carcinoma. Hepatology. 2014;59(4):1448–58. doi: 10.1002/hep.26893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finn RS, Aleshin A, Dering J, Yang P, Ginther C, Desai A, et al. Molecular subtype and response to dasatinib, an Src/Abl small molecule kinase inhibitor, in hepatocellular carcinoma cell lines in vitro. Hepatology. 2013;57(5):1838–46. doi: 10.1002/hep.26223. [DOI] [PubMed] [Google Scholar]
- 14.Deshmukh M, Hoshida Y. Genomic profiling of cell lines for personalized targeted therapy for hepatocellular carcinoma. Hepatology. 2013;58(6):2207. doi: 10.1002/hep.26407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitamant J, Kottakis F, Benhamouche S, Tian H, Chuvin N, Parachoniak NA, et al. YAP inhibition restores hepatocyte differentiation in advanced HCC leading to tumor regression. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.02.027. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Easl-Eortc. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56(4):908–43. doi: 10.1016/j.jhep.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Kojima K, April C, Canasto-Chibuque C, Chen X, Deshmukh M, Venkatesh A, et al. Transcriptome profiling of archived sectioned formalin-fixed paraffin-embedded (AS-FFPE) tissue for disease classification. PLoS One. 2014;9(1):e86961. doi: 10.1371/journal.pone.0086961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K, et al. The diagnosis and management of non-alcoholic fatty liver disease: practice guideline by the American Gastroenterological Association, American Association for the Study of Liver Diseases, and American College of Gastroenterology. Gastroenterology. 2012;142(7):1592–609. doi: 10.1053/j.gastro.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Goodman ZD. Neoplasms of the liver. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2007;20(Suppl 1):S49–60. doi: 10.1038/modpathol.3800682. [DOI] [PubMed] [Google Scholar]
- 20.Macsween RNM, Burt AD, Portmann B, Ferrell LD Sciencedirect (Online Service) MacSween's pathology of the liver. 6. Edinburgh: Churchill Livingstone; 2011. p. 1. online resource (1 v.) [Google Scholar]
- 21.Bosman FT World Health Organization., International Agency for Research on Cancer. WHO classification of tumours of the digestive system. 4. Lyon: International Agency for Research on Cancer; 2010. [Google Scholar]
- 22.Wu PC, Lai CL, Lam KC, Lok AS, Lin HJ. Clear cell carcinoma of liver. An ultrastructural study. Cancer. 1983;52(3):504–7. doi: 10.1002/1097-0142(19830801)52:3<504::aid-cncr2820520321>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 23.Audisio RA, Bombelli L, Lombardi L, Andreola S. A clinico-pathologic study of clear-cell hepatocellular carcinoma. Tumori. 1987;73(4):389–95. doi: 10.1177/030089168707300412. [DOI] [PubMed] [Google Scholar]
- 24.Yang SH, Watanabe J, Nakashima O, Kojiro M. Clinicopathologic study on clear cell hepatocellular carcinoma. Pathol Int. 1996;46(7):503–9. doi: 10.1111/j.1440-1827.1996.tb03645.x. [DOI] [PubMed] [Google Scholar]
- 25.Salomao M, Yu WM, Brown RS, Jr, Emond JC, Lefkowitch JH. Steatohepatitic hepatocellular carcinoma (SH-HCC): a distinctive histological variant of HCC in hepatitis C virus-related cirrhosis with associated NAFLD/NASH. The American journal of surgical pathology. 2010;34(11):1630–6. doi: 10.1097/PAS.0b013e3181f31caa. [DOI] [PubMed] [Google Scholar]
- 26.Jain D, Nayak NC, Kumaran V, Saigal S. Steatohepatitic hepatocellular carcinoma, a morphologic indicator of associated metabolic risk factors: a study from India. Archives of pathology & laboratory medicine. 2013;137(7):961–6. doi: 10.5858/arpa.2012-0048-OA. [DOI] [PubMed] [Google Scholar]
- 27.Shibahara J, Ando S, Sakamoto Y, Kokudo N, Fukayama M. Hepatocellular carcinoma with steatohepatitic features: a clinicopathological study of Japanese patients. Histopathology. 2014;64(7):951–62. doi: 10.1111/his.12343. [DOI] [PubMed] [Google Scholar]
- 28.Edmondson HA, Steiner PE. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer. 1954;7(3):462–503. doi: 10.1002/1097-0142(195405)7:3<462::aid-cncr2820070308>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 29.Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41(6):1313–21. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 30.Unitt E, Marshall A, Gelson W, Rushbrook SM, Davies S, Vowler SL, et al. Tumour lymphocytic infiltrate and recurrence of hepatocellular carcinoma following liver transplantation. J Hepatol. 2006;45(2):246–53. doi: 10.1016/j.jhep.2005.12.027. [DOI] [PubMed] [Google Scholar]
- 31.Wood LD, Heaphy CM, Daniel HD, Naini BV, Lassman CR, Arroyo MR, et al. Chromophobe hepatocellular carcinoma with abrupt anaplasia: a proposal for a new subtype of hepatocellular carcinoma with unique morphological and molecular features. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2013;26(12):1586–93. doi: 10.1038/modpathol.2013.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikeda T, Seki S, Maki M, Noguchi N, Kawamura T, Arii S, et al. Hepatocellular carcinoma with osteoclast-like giant cells: possibility of osteoclastogenesis by hepatocyte-derived cells. Pathol Int. 2003;53(7):450–6. doi: 10.1046/j.1440-1827.2003.01503.x. [DOI] [PubMed] [Google Scholar]
- 33.Hoshida Y, Toffanin S, Lachenmayer A, Villanueva A, Minguez B, Llovet JM. Molecular Classification and Novel Targets in Hepatocellular Carcinoma: Recent Advancements. Semin Liver Dis. 2010;30(1):35–51. doi: 10.1055/s-0030-1247131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang B, Gaiteri C, Bodea LG, Wang Z, Mcelwee J, Podtelezhnikov AA, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell. 2013;153(3):707–20. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song WM, Di Matteo T, Aste T. Nested hierarchies in planar graphs. Discrete Appl Math. 2011;159(17):2135–46. [Google Scholar]
- 37.Song WM, Di Matteo T, Aste T. Building complex networks with Platonic solids. Physical review E, Statistical, nonlinear, and soft matter physics. 2012;85(4 Pt 2):046115. doi: 10.1103/PhysRevE.85.046115. [DOI] [PubMed] [Google Scholar]
- 38.Tran LM, Zhang B, Zhang Z, Zhang C, Xie T, Lamb JR, et al. Inferring causal genomic alterations in breast cancer using gene expression data. BMC systems biology. 2011;5:121. doi: 10.1186/1752-0509-5-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stinchcombe TE. Recent advances in the treatment of non-small cell and small cell lung cancer. F1000prime reports. 2014;6:117. doi: 10.12703/P6-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salomao M, Remotti H, Vaughan R, Siegel AB, Lefkowitch JH, Moreira RK. The steatohepatitic variant of hepatocellular carcinoma and its association with underlying steatohepatitis. Human pathology. 2012;43(5):737–46. doi: 10.1016/j.humpath.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 41.Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40(3):667–76. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 42.Boyault S, Rickman DS, De Reynies A, Balabaud C, Rebouissou S, Jeannot E, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45(1):42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 43.Chiang DY, Villanueva A, Hoshida Y, Peix J, Newell P, Minguez B, et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer research. 2008;68(16):6779–88. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perra A, Kowalik MA, Ghiso E, Ledda-Columbano GM, Di Tommaso L, Angioni MM, et al. YAP activation is an early event and a potential therapeutic target in liver cancer development. J Hepatol. 2014 doi: 10.1016/j.jhep.2014.06.033. [DOI] [PubMed] [Google Scholar]
- 45.Ito T, Shiraki K, Sugimoto K, Yamanaka T, Fujikawa K, Ito M, et al. Survivin promotes cell proliferation in human hepatocellular carcinoma. Hepatology. 2000;31(5):1080–5. doi: 10.1053/he.2000.6496. [DOI] [PubMed] [Google Scholar]
- 46.Prins GS, Putz O. Molecular signaling pathways that regulate prostate gland development. Differentiation; research in biological diversity. 2008;76(6):641–59. doi: 10.1111/j.1432-0436.2008.00277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mehta V, Schmitz CT, Keil KP, Joshi PS, Abler LL, Lin TM, et al. Beta-catenin (CTNNB1) induces Bmp expression in urogenital sinus epithelium and participates in prostatic bud initiation and patterning. Developmental biology. 2013;376(2):125–35. doi: 10.1016/j.ydbio.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ding BS, Cao Z, Lis R, Nolan DJ, Guo P, Simons M, et al. Divergent angiocrine signals from vascular niche balance liver regeneration and fibrosis. Nature. 2014;505(7481):97–102. doi: 10.1038/nature12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hong F, Tuyama A, Lee TF, Loke J, Agarwal R, Cheng X, et al. Hepatic stellate cells express functional CXCR4: role in stromal cell-derived factor-1alpha-mediated stellate cell activation. Hepatology. 2009;49(6):2055–67. doi: 10.1002/hep.22890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Y, Huang Y, Reiberger T, Duyverman AM, Huang P, Samuel R, et al. Differential effects of sorafenib on liver versus tumor fibrosis mediated by stromal-derived factor 1 alpha/C-X-C receptor type 4 axis and myeloid differentiation antigen-positive myeloid cell infiltration in mice. Hepatology. 2014;59(4):1435–47. doi: 10.1002/hep.26790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Llovet JM, Chen Y, Wurmbach E, Roayaie S, Fiel MI, Schwartz M, et al. A molecular signature to discriminate dysplastic nodules from early hepatocellular carcinoma in HCV cirrhosis. Gastroenterology. 2006;131(6):1758–67. doi: 10.1053/j.gastro.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 52.Di Tommaso L, Franchi G, Park YN, Fiamengo B, Destro A, Morenghi E, et al. Diagnostic value of HSP70, glypican 3, and glutamine synthetase in hepatocellular nodules in cirrhosis. Hepatology. 2007;45(3):725–34. doi: 10.1002/hep.21531. [DOI] [PubMed] [Google Scholar]
- 53.European Association for the Study of The L, European Organisation For R, Treatment Of C. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56(4):908–43. doi: 10.1016/j.jhep.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 54.Hoshida Y, Villanueva A, Kobayashi M, Peix J, Chiang DY, Camargo A, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. The New England journal of medicine. 2008;359(19):1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koscielny S. Why most gene expression signatures of tumors have not been useful in the clinic. Sci Transl Med. 2010;2(14):14ps2. doi: 10.1126/scitranslmed.3000313. [DOI] [PubMed] [Google Scholar]
- 56.Tandon P, Garcia-Tsao G. Prognostic indicators in hepatocellular carcinoma: a systematic review of 72 studies. Liver Int. 2009;29(4):502–10. doi: 10.1111/j.1478-3231.2008.01957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53(3):1020–2. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lazo M, Hernaez R, Eberhardt MS, Bonekamp S, Kamel I, Guallar E, et al. Prevalence of nonalcoholic fatty liver disease in the United States: the Third National Health and Nutrition Examination Survey, 1988–1994. Am J Epidemiol. 2013;178(1):38–45. doi: 10.1093/aje/kws448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Michelotti GA, Machado MV, Diehl AM. NAFLD, NASH and liver cancer. Nat Rev Gastroenterol Hepatol. 2013;10(11):656–65. doi: 10.1038/nrgastro.2013.183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.