Abstract
Lung adenocarcinoma is caused by the combination of genetic and environmental effects, and smoking plays an important role in the disease development. Exploring the gene expression profile and identifying genes that are shared or vary between smokers and nonsmokers with lung adenocarcinoma will provide insights into the etiology of this complex cancer. We obtained RNA-seq data from paired normal and tumor tissues from 34 nonsmoking and 34 smoking patients with lung adenocarcinoma (GEO: GSE40419). R Bioconductor, edgeR, was adopted to conduct differential gene expression analysis between paired normal and tumor tissues. A generalized linear model was applied to identify genes that were differentially expressed in nonsmoker and smoker patients as well as genes that varied between these two groups. We identified 2273 genes that showed differential expression with FDR<0.05 and |logFC| >1 in nonsmoker tumor versus normal tissues; 3030 genes in the smoking group; and 1967 genes were common to both groups. Sixty-eight and 70 % of the identified genes were downregulated in nonsmoking and smoking groups, respectively. The 20 genes such as SPP1, SPINK1, and FAM83A with largest fold changes in smokers also showed similar large and highly significant fold changes in nonsmokers and vice versa, showing commonalities in expression changes for adenocarcinomas in both smokers and nonsmokers for these genes. We also identified 175 genes that were significantly differently expressed between tumor samples from nonsmoker and smoker patients. Gene expression profile varied substantially between smoker and nonsmoker patients with lung adenocarcinoma. Smoking patients overall showed far more complicated disease mechanism and have more dysregulation in their gene expression profiles. Our study reveals pathogenetic differences in smoking and nonsmoking patients with lung adenocarcinoma from tran-scriptome analysis. We provided a list of candidate genes for further study for disease detection and treatment in both smoking and nonsmoking patients with lung adenocarcinoma.
Keywords: RNA-seq, Expression analysis, Smoking, Lung cancer, Lung adenocarcinoma
Introduction
Non-small cell lung cancer (NSCLC) is the most common type of lung cancer which accounts for about 85 % of lung cancer cases in the USA [1, 2]. Adenocarcinoma is currently the most common type of NSCLC in Asian and North American patients and accounts for 30 and 40 % of primary lung cancer in male and female smokers, respectively [3] in the USA. Smoking is the most prevalent risk factor for lung cancer, and the risk increases with the length of time and number of cigarettes people smoked [4]. Research has revealed differences in molecular characteristics of lung cancers in smokers versus nonsmokers. In 2008, Massion et al. found striking differences in the genomic architecture of nonsmokers versus smokers with adenocarcinomas which suggests new molecular pathways associated with smoking in cancer development [5]. The study of epidermal growth factor receptor (EGFR), tumor protein P53 (TP53), and Kirsten rat sarcoma viral on-cogene homolog (KRAS) genes all demonstrate distinct mutation patterns and frequencies that vary between smoking and nonsmoking patients with lung cancer [6]. All these findings suggest distinct pathogenic mechanism in lung cancer between smokers and nonsmokers.
RNA-seq technology provides a revolutionary tool for transcriptome analysis. Compared with microarray platform, RNA-seq has less background noise due to image analysis and is more sensitive in detection of transcripts with low-abundance or higher fold change in expression [7]. RNA-seq analysis has provided researchers insights about lung cancer development. Cheng et al. compared the gene expression data from human airway epithelial cells between a smoker with lung cancer and a smoker without lung cancer [8]. Although there were no replicates in their study, they still detected 27 genes that were differentially expressed between the two samples. They also found that overall gene expression levels were lower in smoker with lung cancer compared with healthy smokers in general. Beane and her colleagues identified about 200 genes differentially expressed between eight smokers with lung cancer and five smoking healthy controls using RNA-seq data from airway epithelial cells [9]. In 2014, Han et al. sequenced RNA samples from paired normal and tumor tissues from 88 smokers with non-small cell lung cancer, including 54 patients with adenocarcinoma and 34 with squa-mous cell carcinoma. They identified 1063 genes that were differentially expressed between tumor and normal tissues in smoking patients including SPP1, TOP2A, and CRABP2, etc. [10]. These studies are valuable in understanding the complex tumorigenesis and providing candidate genes in lung cancer study. However, none of the earlier studies focused on identifying differences in gene expression profile between smokers and nonsmokers with lung cancer.
In 2012, Korean researchers conducted transcriptome analysis of lung adenocarcinoma aiming to identify the somatic mutations and transcriptional variations associated with lung cancer. The RNA-seq data in their analysis originated from paired normal and tumor tissues from about 70 patients including both smokers and nonsmokers. A hierarchical clustering method was used to identify cancer-up, cancer-down, and mixed regulated genes for each patient. Extremely overexpressed genes, including GNAS, CDK4, KRAS, SPP1, etc., were detected in their study [11, supplementary table 11, GEO: GSE40419].
In this study, we downloaded the RNA-seq data from GEO: GSE40419 and conducted a comprehensive pair-wise comparison analysis combining the information from all the samples. We aimed to perform a large scale transcriptome analysis and identify the genes with altered expression in lung cancer development. In addition, we looked for the differences in gene expression profiles between smoking and nonsmoking patients, which will better define the impact of smoking in lung cancer development and provide insights that might better individualize treatment strategies for the patients.
Materials and methods
RNA sequencing data and phenotype data
The transcriptome sequencing data from 68 lung adenocarcinoma patients with validated smoking status were downloaded from Gene Expression Omnibus (GEO) with accession number GSE40419. For each patient, the tumor and paired normal tissue had been sequenced. Altogether, there were about 14 billion paired-end sequence reads with average 101 bp in length. The phenotype including age at diagnosis, gender, and smoking history are also available from the public website (http://genome.cshlp.org/content/22/11/2109/suppl/DC1). The 68 cancer patients including 34 never smokers and 34 smokers, and the smokers include ever smoker and current smokers. We restricted analysis of the ever smokers to patients under 75 to ensure comparability in age ranges between the two groups. For validation of genes identified in nonsmoker group, independent RNA-seq data from six non-smoker patients were downloaded from GEO with accession number GSE37765 [12]. The age onset for these six patients varied from 44 to 70.
Differential gene expression analysis
Genotype calling of single nucleotide variants (SNVs) was conducted on 136 RNA-seq bam files from 68 individuals to confirm the paired nature of RNA-seq data from tumor and normal tissue. Pair-wise concordance rates were computed for all possible 9180 pairs including between individuals and among individual pairs. The concordance rate of genotypic values between individuals varied from 0.69 to 0.77 with mean 0.73, and for the within individuals analysis, the concordance rates varied from 0.85 to 0.97 with mean 0.94. This study confirmed the paired nature of RNA-seq data from tumor and normal tissue, since there was no overlap in the distributions of the within and among pair concordance rates (Supplementary 1 Figure S1). Pair-end RNA-seq reads were aligned to human genome assembly Ensembl GRCh37 by Tophat. HTSeq was used to count the reads by genes (http://www-huber.embl.de/users/anders/HTSeq/doc/tour.html#counting-reads-by-genes). We used R Bioconductor edgeR to perform the differential expression analysis, and we applied a general linear model: lung tissue expression~smoking+smoking:patient+smoking:tissue to accommodate the multifactor design of the experiment. This model incorporated the main effect for smoking plus interactions with patients and tissues, thus allowing us to identify genes differentially expressed in tumor versus normal tissue in nonsmoker or smoker patients, and genes that behave differently between smoker and nonsmoker patients. To make sure there were sufficient counts for each gene in the test, only tags that had at least 1 count per million (cpm) in at least half of the sample size were kept in the analysis. Genes with Benjamini-Hochberg adjusted FDR< 0.05 and absolute values of logFC greater than 1 were reported as significant genes.
Gene function annotation and pathway analysis
Pathway analysis was conducted to infer the functional roles and relationships of the genes with varied expression in the analysis. logFC value of significant genes were submitted to Ingenuity Pathway Analysis (IPA) for pathway analysis.
Results
Data filtering process
We applied a stringent filter on the data to remove the gene tags with sparse count data. Genes with at least 1 count per million (cpm) in at least half of the sample size were kept in the analysis. There are 62,069 gene tags in the raw data and 17,757 of them were kept in the analysis after the filter. The biological coefficient of variation (BCV) in the 136 RNA-seq samples was about 0.4, which indicates that a good quality of this dataset as a typical BCV value from a well-controlled experiment is 0.4 for human data [13].
Differential gene expression analysis
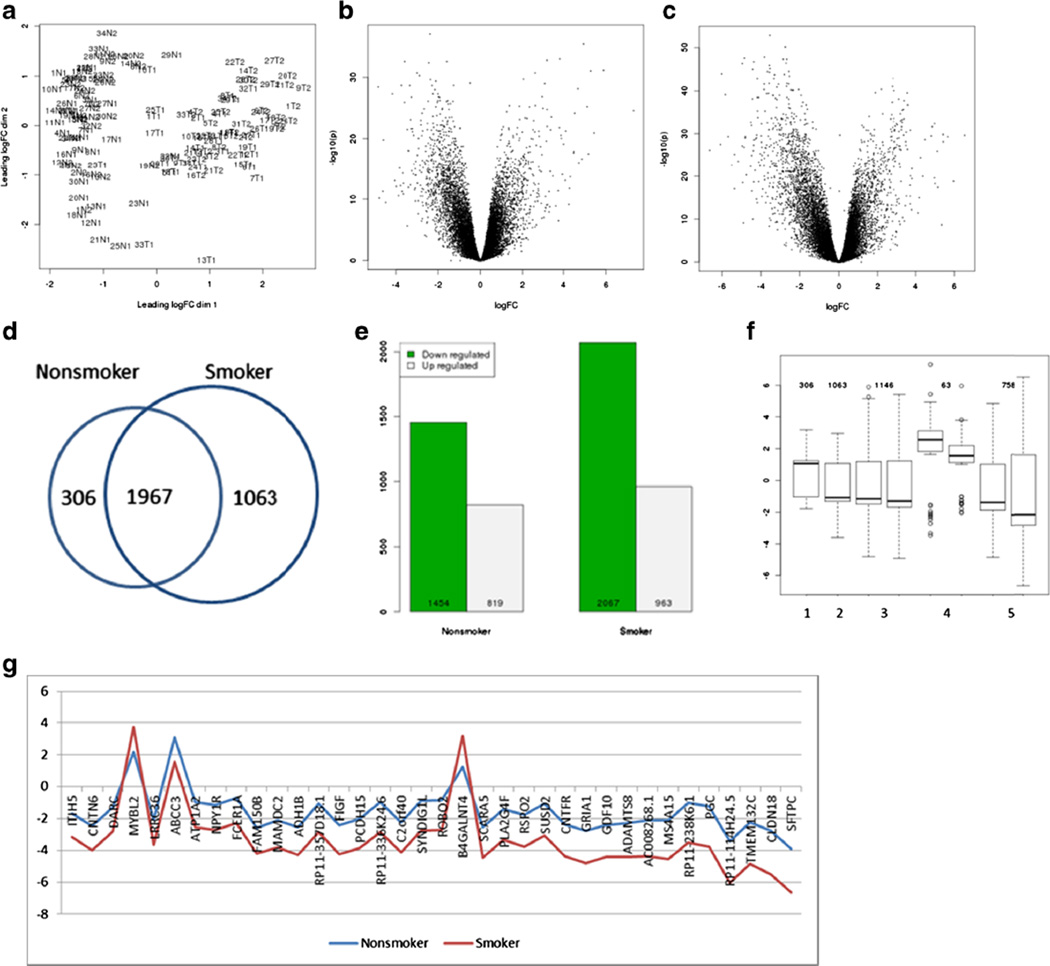
We used edgeR to identify the genes differently expressed between tumor and normal tissues as well as the genes that varied between nonsmoker and smoker patients. Figure 1a displays the cluster plots of 136 RNA-seq samples from 68 patients based on leading log-fold change. The multidimensional plots clearly divided the samples into normal and tumor groups. This plot together with SNV genotype calling from bam files confirmed the paired nature of the RNA-seq data. We filtered the gene tags with low count and conducted the differential analysis. Plot B-C in Fig. 1 displays the average log-counts per million (logCPM) against log2-fold change (logFC) in nonsmoker and smoker group, respectively. The significant genes identified in smoker group have a larger variation in terms of both −log10(p) and logFC change compared with nonsmokers. A total of 2273 and 3031 genes with FDR<0.05 and |logFC|>1 were identified in nonsmoker and smoker group, respectively, and 1967 of them are shared in both groups (Fig. 1d, Supplementary 2). About 95 % of the genes identified in nonsmoking group are also detected in smoking lung adenocarcinoma patients, but a larger number of genes were found to be significantly different between normal and tumor tissue in the smoker group. This finding suggests that a core set of genes are critical for both nonsmokers and smokers but additional genes are often affected in lung adenocarcinomas, especially in smoking patients. Sixty-eight percent of the significant genes were found downregulated in smoking group, and this number increased to 70 % in nonsmoking group (Fig. 1e).
Fig. 1.
Differential gene expression analysis. a Multi-dimensional scaling plot of the RNA sample-seq data. N1 and T1 denote normal and tumor samples from nonsmoker patients; N2 and T2 denote normal and tumor samples from smoker patients. The leading logFC is the average (root mean square) of the largest log-fold changes difference between each pair of samples. b, c Volcano plots of signals from differential analysis in nonsmoker and smoker patients, respectively. d Venn diagram shows the number of significant (FDR<0.05 and |logFC| >1) genes in nonsmoker and smoker patients. e Bar plot of down- and upregulated genes in non- and smoker group. f Box plot of logFC for significant genes with FDR<0.05 and |logFC|>1. 1, in only nonsmokers; 2, in only smokers; 3, in both group but with difference between logFC< 0.5; 4, in both group but with greater value in nonsmokers; 5, in both group but with greater value in smokers. 3–5, left and right box plot display nonsmoker and smoker patients, respectively. The numbers indicate number of genes in each group. g Genes significant in both groups but with big difference in logFC
One goal of this study was to identify the differences in expression profiles between smoking and nonsmoking groups. We divided the genes with FDR<0.05 and |logFC|> 1 into five groups: (1) genes identified in only nonsmoker patients, (2) genes identified in only smoker patients, (3) genes identified in both groups but with greater fold change in non-smoker patients, (4) genes identified in both group but with greater fold change in smoker patients, and (5) genes identified in both group with similar logFC value. Figure 1f displays the boxplot of logFC from these five groups of genes. Among the 306 genes with |logFC|>1 only identified in nonsmoker patients, 197 of them are upregulated in tumor tissues, whereas only 341 out of 1063 genes are upregulated in smoker patients. Among the genes common to both groups, 63 genes have a larger change of |logFC| in nonsmoker group and 51 of them are upregulated in tumor tissue; 758 genes have a larger value of |logFC| in smoker group and 564 of them are down-regulated in tumor tissues; 1146 genes are shared by both groups but with little difference in logFC change (difference less than 0.5). This finding suggests that although a common set of genes are affected in tumorigenesis of lung adenocarcinoma for both nonsmoker and smoker patients, they are more strikingly affected in ever smoking patients especially for those genes downregulated in tumor tissues.
Compared with p value, fold change by itself provides more meaningful biology-related information. Table 1 lists the top 20 genes with largest |logFC| value in both nonsmoking and smoking group, and 11 of them are shared in both lists. In the nonsmoking group, the SPINK1 gene has the biggest value of logFC of 7.31, which indicates a fold change of 159 in tumor tissue compared with normal tissue. The logFC value of this gene is 5.95 in smoker patients with lung adenocarcinoma. Gene expression and microarray profiling between paired tumor and normal tissue samples showed that SPINK1 was a candidate biomarker with increased expression in tumor tissues with non-small cell lung cancer, and it had the potential to characterize adenocarcinoma and squamous cell carcinoma [14]. The other top 20 genes, including TMPRSS4, SPP1, and MMP1, etc., have been shown to be related with poor prognosis, more aggressive phenotype, or early onset of lung cancer [15–17]. In the smoking group, the gene with largest logFC change is SFTPC, which has a logFC of −6.66. This gene is essential for lung function and associated with pulmonary fibrosis, a disease that can lead to lung cancer [18, 19]. CYP24A1 is another top gene with logFC of 5.66 in smoker patients, compared with 3.94 in nonsmoker patients. Kim and his colleagues analyzed the effect of smoking on CYP24A1 expression in 100 patients with lung adenocarcinoma and detected a significantly higher (p<0.004) expression of CYP24A1 in smoker patients compared with nonsmoker patients [20]. Our result provided evidence supporting their report.
Table 1.
Top 20 genes with largest |logFC| value in nonsmoker and smoker group with lung adenocarcinoma
| Top 20 in nonsmoker | Smoker2 | Top 20 in smoker | Nonsmoker4 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene1 | logFC | FDR | logFC | FDR | Gene3 | logFC | FDR | logFC | FDR |
| SPINK1 | 7.31 | 2.73E-25 | 5.95 | 1.48E-19 | SFTPC | −6.66 | 7.14E-27 | −3.89 | 2.57E-11 |
| TMPRSS4 | 5.89 | 7.77E-32 | 5.42 | 1.50E-30 | FAM83A | 6.53 | 2.28E-30 | 4.86 | 2.21E-17 |
| HABP2 | 5.44 | 1.25E-21 | 3.79 | 6.98E-13 | MYOC | −6.08 | 3.15E-28 | −4.06 | 8.38E-14 |
| SPP1 | 5.28 | 9.96E-32 | 4.97 | 4.20E-33 | RP11-114H24.5 | −6.02 | 1.40E-44 | −3.40 | 9.57E-19 |
| CXCL13 | 5.17 | 3.28E-18 | 5.15 | 4.09E-18 | SPINK1 | 5.95 | 1.48E-19 | 7.31 | 2.73E-25 |
| ATP10B | 5.11 | 5.65E-26 | 5.12 | 1.07E-26 | SERTM1 | −5.84 | 7.58E-40 | −4.84 | 3.72E-29 |
| GCNT3 | 4.94 | 3.79E-36 | 3.80 | 1.64E-22 | ITLN2 | −5.69 | 4.78E-30 | −4.60 | 1.77E-20 |
| FAM83A | 4.86 | 2.21E-17 | 6.53 | 2.28E-30 | CYP24A1 | 5.66 | 5.02E-30 | 3.94 | 8.15E-15 |
| SERTM1 | −4.84 | 3.72E-29 | −5.84 | 7.58E-40 | CLDN18 | −5.51 | 2.23E-23 | −2.78 | 3.38E-07 |
| ITLN1 | −4.80 | 6.43E-11 | −4.91 | 7.83E-10 | MMP12 | 5.43 | 7.33E-29 | 4.48 | 2.29E-18 |
| MMP1 | 4.73 | 9.45E-17 | 5.00 | 5.50E-19 | TMPRSS4 | 5.42 | 1.50E-30 | 5.89 | 7.77E-32 |
| B3GNT3 | 4.68 | 6.94E-29 | 4.47 | 2.45E-29 | SLC6A4 | −5.37 | 7.55E-18 | −4.63 | 4.81E-12 |
| SLC6A4 | −4.63 | 4.81E-12 | −5.37 | 7.55E-18 | LGI3 | −5.37 | 7.43E-26 | −3.79 | 5.84E-13 |
| ITLN2 | −4.60 | 1.77E-20 | −5.69 | 4.78E-30 | MUC5B | 5.35 | 2.13E-09 | 3.75 | 2.93E-07 |
| COL10A1 | 4.53 | 6.50E-25 | 4.25 | 3.43E-25 | CHIAP2 | −5.17 | 3.06E-24 | −3.68 | 4.80E-15 |
| AL121963.1 | 4.49 | 1.25E-18 | 4.37 | 1.03E-20 | CXCL13 | 5.15 | 4.09E-18 | 5.17 | 3.28E-18 |
| HAS1 | −4.49 | 3.42E-10 | −4.42 | 9.77E-11 | ATP10B | 5.12 | 1.07E-26 | 5.11 | 5.65E-26 |
| MMP12 | 4.48 | 2.29E-18 | 5.43 | 7.33E-29 | MMP1 | 5.00 | 5.50E-19 | 4.73 | 9.45E-17 |
| CR2 | 4.32 | 1.97E-14 | 3.20 | 1.63E-9 | SPP1 | 4.97 | 4.20E-33 | 5.28 | 9.96E-32 |
| CEACAM5 | 4.27 | 1.49E-12 | 3.01 | 9.56E-8 | GPM6A | −4.96 | 5.82E-47 | −3.75 | 1.97E-29 |
Genes in bold text are shared in both top 20 lists
genes are sorted by |logFC| values in nonsmoker group
for the top 20 genes in nonsmoker group, their logFC and FDR values in smoker patients
genes are sorted by |logFC| values in smoker group
for the top 20 genes in smoker group, their logFC and FDR values in nonsmoker patients
The majority of the detected genes are concordantly significant in both nonsmoker and smoker patients, but some show a substantial difference in logFC value between these two groups. Figure 1h demonstrates the 36 genes that are significant in both smoking and nonsmoking patients but with a large difference in differential expression level between smoking and nonsmoking patients. Most of the 36 genes have negative logFC, and smoking patients have a larger fold change (red line) for most of the genes. For example, the SFTPC gene has a logFC of −3.89 in the nonsmoking group and −6.66 in smoking group. Deletion of SFTPC gene has been detected in non-small cell lung cancer from cDNA Mi-croarrays comparative genomic hybridization analysis [21, 22]. ADAMTS8 has been reported as a tumor suppressor gene that is usually downregulated or silenced in various tumor cell lines [23]. In our study, it has a logFC of −2.28 in nonsmoking and −4.43 in smoking patients. The big difference in gene expression level suggests generally more dysregulation in tumor tissues from smokers.
Differential analysis between nonsmoker and smoker patients
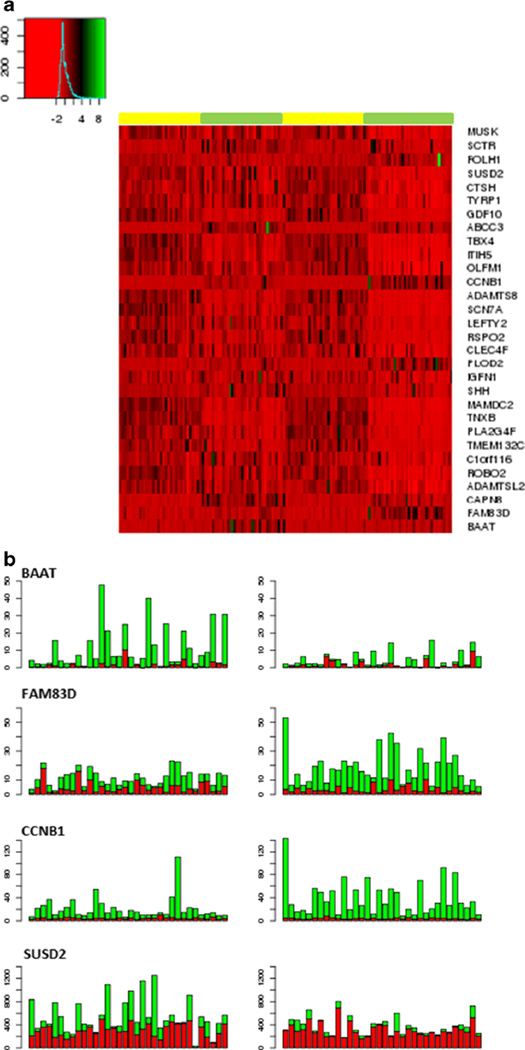
The GLM model in edgeR allows us to identify the genes that varied between nonsmoker and smoker patients with lung adenocarcinoma. We identified 175 genes whose expressions were statistically different between nonsmoker and smoker group (Supplementary 2). These genes can be further divided into three groups: genes exclusively significant in only non-smoker patients, genes exclusively significant in only smoker patients, and genes significant in both group but with big difference in logFC. Table 2 and heatmap in Fig. 2a displayed some of the genes identified in the between group comparison analysis. BAAT gene, which encodes a liver enzyme that involved in bile acid metabolism, was identified differentially expressed between tumor and normal tissues in only non-smoker patients. It has a logFC of 2.81 with FDR 1.41E-11, compared with logFC 0.76 with FDR 0.07 from smoker patients. Twenty-one out of 34 nonsmoker patients were detected with increased expression in tumor samples. We further validated the signal using independent RNA-seq from six non-smoker patients, and the logFC is 3.00 with FDR 3.89E-11. FAM83D gene, on the other hand, was over expressed in tumor tissues from only smoker patients with logFC 1.99 and FDR 6.46E-14.
Table 2.
Significant genes that behave differently between nonsmoker and smoker group
| Tests for differences between smokers and nonsmokers |
Difference in expression between tumor and normal for nonsmokers |
Difference in expression between tumor and normal for smokers |
||||
|---|---|---|---|---|---|---|
| GENE | logFC | FDR | logFC | FDR | logFC | FDR |
| Genes exclusively significant in nonsmokers | ||||||
| FGF12 | 1.22 | 3.93E-02 | −1.31 | 7.40E-07 | −0.09 | 7.49E-01 |
| CD207 | −1.94 | 2.47E-02 | 2.04 | 2.97E-07 | 0.10 | 8.28E-01 |
| HOXD1 | −1.76 | 4.23E-02 | 1.43 | 1.67E-04 | −0.34 | 4.36E-01 |
| GDF15 | −1.64 | 2.23E-02 | 2.09 | 1.16E-10 | 0.46 | 1.71E-01 |
| BAAT | −2.05 | 2.38E-02 | 2.81 | 1.41E-11 | 0.76 | 7.38E-02 |
| CLDN2 | −2.50 | 2.34E-02 | 3.18 | 8.96E-10 | 0.68 | 1.74E-01 |
| MUC1 | −1.16 | 4.23E-02 | 1.48 | 8.77E-09 | 0.32 | 2.33E-01 |
| CAPN8 | −1.84 | 6.22E-03 | 2.27 | 8.11E-14 | 0.43 | 2.16E-01 |
| Genes exclusively significant in smokers | ||||||
| FAM83D | 1.47 | 1.30E-02 | 0.53 | 6.33E-02 | 1.99 | 6.46E-14 |
| TYRP1 | −2.09 | 1.52E-05 | −0.09 | 7.85E-01 | −2.17 | 1.02E-15 |
| OLFM1 | −1.41 | 5.41E-03 | −0.35 | 1.55E-01 | −1.76 | 1.66E-13 |
| MYOCD | −1.54 | 2.16E-02 | −0.49 | 1.11E-01 | −2.04 | 9.23E-12 |
| CLEC4F | −1.59 | 7.45E-03 | −0.44 | 1.19E-01 | −2.03 | 8.62E-13 |
| ALPL | −1.85 | 2.23E-02 | −0.18 | 6.71E-01 | −2.02 | 2.53E-08 |
| IGFN1 | −1.91 | 8.40E-03 | 0.19 | 6.26E-01 | −1.72 | 4.20E-07 |
| SCGB3A2 | −2.36 | 3.08E-02 | 0.42 | 4.37E-01 | −1.94 | 8.55E-05 |
| SHH | −2.00 | 7.53E-03 | 0.21 | 5.80E-01 | −1.78 | 6.11E-07 |
| PTCHD1 | −2.11 | 3.12E-03 | −0.53 | 1.35E-01 | −2.64 | 1.82E-14 |
| DPCR1 | −2.31 | 2.26E-02 | 0.41 | 4.15E-01 | −1.90 | 4.01E-05 |
| RAC3 | 1.27 | 1.09E-02 | 0.47 | 5.20E-02 | 1.74 | 1.09E-14 |
| DMBT1 | −2.23 | 4.42E-02 | 0.17 | 7.58E-01 | −2.06 | 7.49E-05 |
| LRRK2 | −1.51 | 3.79E-02 | −0.50 | 1.47E-01 | −2.01 | 7.69E-10 |
| Genes significantly expressed in both groups but with big difference in logFC | ||||||
| ABCC3 | −1.55 | 8.08E-03 | 3.10 | 9.94E-30 | 1.55 | 2.13E-08 |
| KCNK5 | −1.14 | 2.70E-02 | 1.82 | 2.92E-15 | 0.68 | 4.03E-03 |
| CLDN18 | −2.73 | 2.84E-02 | −2.78 | 3.38E-07 | −5.51 | 2.23E-23 |
| PGC | −2.52 | 2.16E-02 | −1.23 | 1.29E-02 | −3.76 | 1.62E-14 |
| SUSD2 | −2.01 | 4.04E-03 | −1.05 | 2.39E-03 | −3.06 | 2.63E-21 |
| GDF10 | −2.04 | 2.57E-03 | −2.40 | 5.35E-14 | −4.44 | 7.96E-39 |
| CNTFR | −2.02 | 2.09E-02 | −2.36 | 1.45E-09 | −4.38 | 4.83E-28 |
| ADAMTS8 | −2.15 | 8.27E-05 | −2.28 | 2.55E-15 | −4.43 | 1.17E-45 |
| RSPO2 | −2.01 | 5.36E-03 | −1.78 | 7.31E-08 | −3.79 | 2.03E-27 |
| GRIA1 | −2.03 | 2.42E-02 | −2.78 | 5.77E-12 | −4.81 | 9.73E-30 |
| MS4A15 | −2.42 | 1.77E-02 | −2.12 | 4.35E-06 | −4.54 | 4.41E-23 |
| SFTPC | −2.77 | 2.69E-02 | −3.89 | 2.57E-11 | −6.66 | 7.14E-27 |
| TMEM132C | −2.64 | 4.87E-03 | −2.24 | 2.17E-07 | −4.88 | 1.50E-27 |
Fig. 2.
Differential analysis between nonsmoker and smoker patients with lung adenocarcinoma. a Heatmap of normalized count per million (CPM) for genes that behave differently between two groups. Yellow and green bar denote normal and tumor tissues, respectively. Nonsmoker group is on the left. b Bar plot of CPM for genes. Left, nonsmoker patients; right, smoker patients. Red and green colors denote CPM from normal and tumor tissues, respectively
CCNB1 gene plays an important role in cell cycle regulation, and deregulated expression of CCNB1 will result in uncontrolled cell growth and malignancy [24]. Overexpression of CCNB1 has been reported to be associated with various malignancies including NSCLC [25–30]. We detected over-expression of CCNB1 in both smoker and nonsmoker tumor samples but with significantly higher expression detected in tumor tissue from smoker patients (Supplementary 2, Fig. 2b). It has a logFC 1.41 from nonsmoker patients and 2.74 in smoker patients, and between group comparison test produced a FDR p value 0.004. This result suggested the involvement of CCNB1 gene in tumorigenesis of lung cancer, and smoking behavior plays an adverse effect in development of lung adenocarcinoma. Another example is SUSD2 gene; it has a logFC of −1.05 in nonsmoker group and −3.06 in smoker group, with FDR p value 0.004 in the between groups analysis (Supplementary 2, Fig. 2b). Thirty-three out of 34 tumor samples were detected with underexpression in tumor samples in smoker group compared with 23 out of 34 tumor samples in nonsmoker group. SUSD2 has been identified as a potential tumor suppressor gene and been shown to be involved in breast cancer and lung cancer [31, 32]. Our finding suggested that cigarette smoking was associated with SUSD2 expression in tumor tissues.
One hundred seventy-five genes were identified significantly different between nonsmoker and smoker patients. We further conducted a differential analysis between normal samples from nonsmoker and smoker patients to make sure those variations did not arise from the variation in the normal tissues. One hundred twenty-one genes were identified as significant in the analysis, and only three genes, SERPIND1, TNNT1, and PCSK2, were among the 175 genes that varied between smoker and nonsmoker patients. (Supplementary 2).
Pathway analysis
Differential expression analysis provided a large number of candidate genes. Gene function annotation analysis will help us better understand the relationship among those genes and extract biological meaning from a large list of genes. The three groups of genes, genes with FDR<0.05 and |logFC| >1 in smoking group or nonsmoking group and genes common to both groups as defined in Fig. 1d were submitted to IPA pathway analysis. The majority of the genes in the three groups are in various cancer-related pathways (Table 3). For example, 77 % genes that are common to both nonsmoker and smoker groups are related to cancer. The top five canonical pathways from each analysis were listed in Table 3. If most of the involved genes are upregulated, then this pathway is labeled U in the table and D if most genes are downregulated. Epidemiological studies showed that atherosclerosis is related with lung function and lung cancer [33–35]. The pathway analysis of genes common to both smokers and nonsmokers showed that the atherosclerosis signaling pathway was significant with p value 9.42E 10-8. An earlier study identified dysregulation of axon guidance signaling pathway is involved in NSCLC [36]. In our study, we identified this significant signaling pathway with p value 9.00E-05 in smoker patients (Table 3).
Table 3.
Pathway analysis for significant genes identified in gene differential expression analysis in both smoker and nonsmoker group
| Top canonical pathway | p value | Top upstream regulators | p value | Disease and disorders | p value | # molecul |
|---|---|---|---|---|---|---|
| Significant genes with FDR<0.05 and |logFC|>1 in nonsmoking group |
||||||
| Bladder cancer signaling (N) | 2.27E-03 | Estrogen receptor | 5.50E-07 | Cancer | 1.05E-02–1.66E-07 | 197 |
| Granulocyte adhesion and diapedesis (U) | 2.76E-03 | Tetrachlorodibenzodioxin | 7.55E-07 | Organismal injury and abnormalities |
1.05E-02–1.66E-07 | 197 |
| Sertoli cell-Sertoli cell junction signaling (U) | 2.85E-03 | NFkB (complex) | 1.53E-06 | Infectious disease | 3.85E-03–1.61E-06 | 27 |
| IL-12 signaling and production in macrophages (D) | 3.10E-03 | Growth hormone | 4.93E-06 | Inflammatory response | 1.05E-02–3.87E-06 | 58 |
| Role of macrophages, fibroblasts, and endothelial cells in rheumatoid arthritis (D) |
4.46E-03 | EGFR | 6.57E-06 | Dermatological diseases and conditions |
1.05E-02–7.98E-06 | 106 |
| Significant genes with FDR<0.05 and |logFC|>1 in smoking group |
||||||
| MSP-RON signaling pathway (D) | 8.10E-05 | lipopolysaccharide | 1.14E-15 | Cancer | 3.41E-04–4.63E-23 | 764 |
| Axonal guidance signaling (U) | 9.00E-05 | TGFB1 | 8.15E-15 | Organismal injury and abnormalities |
3.58E-04–4.63E-23 | 765 |
| Molecular mechanisms of cancer (N) | 2.10E-04 | Dexamethasone | 9.28E-15 | Gastrointestinal disease | 3.10E-04–2.09E-16 | 581 |
| EL-12 signaling and production in macrophages (D) | 1.18E-03 | dihydrotestosterone | 1.05E-12 | Dermatological diseases and conditions |
6.18E-05–5.52E-16 | 379 |
| FXR/RXR activation (D) | 1.97E-03 | Beta-estradiol | 3.46E-11 | Reproductive system disease | 3.58E-04–3.32E-11 | 395 |
| Significant genes common to both the nonsmoker and smoker groups |
||||||
| Granulocyte adhesion and diapedesis (D) | 3.80E-09 | TGFB1 | 2.92E-52 | Cancer | 6.08E-08–4.85E-69 | 1521 |
| Agranulocyte adhesion and diapedesis (D) | 8.19E-09 | Vegf | 8.12E-41 | Organismal injury and abnormalities |
6.08E-08–4.85E-69 | 1545 |
| Atherosclerosis signaling (N) | 9.42E-08 | Progesterone | 7.53E-40 | Reproductive system disease | 6.08E-08–4.85E-69 | 877 |
| Hepatic fibrosis/hepatic stellate cell activation (N) | 9.74E-08 | ERBB2 | 5.44E-39 | Neurological disease | 3.55E-08–8.67E-40 | 1160 |
| Eicosanoid signaling (D) | 1.48E-06 | Dexamethasone | 7.77E-34 | Cardiovascular disease | 6.70E-09–11.23E-28 | 757 |
The canonical pathway is labeled with U if most of the involved genes are upregulated and D if most genes are downregulated. Ef comparable of genes are upregulated and downregulated, then labeled with N
Discussion
Researchers have been interested in identification of gene expression variation associated with lung cancer, but most of them are limited to smokers or nonsmokers only. In 2012, Seo and his colleagues used a clustering method to identify differentially expressed genes in smoker and nonsmoker patients with lung adenocarcinoma, and they detected a list of genes with varied expression between tumor and normal tissue across the samples. In this study, we conducted differential gene expression analysis using pair-wise comparison between tumor and normal tissues in smoker and nonsmoker patients. Using GLM model, we combined all the RNA-seq data from 68 patients together and compared the gene expression profile between nonsmoker and smoker patients. We conducted a genome-wide transcriptome analysis in smoker and nonsmoker patients with lung adenocarcinoma; and we also identified some genes that were varied between nonsmoker and smoker group.
The results from our study show that although nonsmokers and smokers share some common genes and disease pathway in the development of lung adenocarcinoma, tobacco smoking plays an important role in deregulated gene expression in tumor tissues. Overall smoking exerts an adverse effect and deteriorates the dysregulation of gene expression in tumor tissues, especially for the genes downregulated in tumor tissues. Smoking, as a major environmental factor associated with lung cancer, has a broad and complicated impact on the patients. It is interesting to note that most of the genes identified are downregulated in tumor versus normal tissues, especially among the genes with larger logFC value (Fig. 1e). This trend has been found in many other studies [37–39]. One explanation is that most antioncogenes or tumor suppressor genes have higher mutation rate than oncogenes [40]. The mutation of those negative regulators will result in the inhibited expression of a series of downstream elements in the network. Our results also demonstrated a larger variation in gene expression levels in smoker patients which supports the finding from Seo’s study.
We identified overexpressed outlier genes in both smoking and nonsmoking group including SPINK1, SPP1, and FAM83A, etc. SPP1 gene has a logFC of 5.28 in nonsmoker and 4.97 in smoker patients. This finding validated the result from Han’s group in 2014. They conducted differential gene expression analysis using RNA-seq data from 88 patients with either adenocarcinoma or squamous cell carcinoma and identified SPP1 gene with biggest logFC of 4.11 [10]. Another recent meta-analysis on 1536 NSCLC tumor tissues and 340 normal tissues displayed that this gene was highly overexpressed in tumor tissue with OR of 6.427 [41]. The highly overexpression of SPP1 has also been shown to be related with chemotherapy response, NSCLC stages, and poor survival rate in advanced NSCLC patients [42–44]. Our results showed that SPP1 is a good candidate gene for early detection of lung cancer and potentially for therapy in both smoker and nonsmoker patients with NSCLC, given its consistent large difference between normal and tumor tissue.
In the comparison analysis between normal and tumor tissues, several genes were identified including BAAT, CAPN8, FAM83D, and RAC3, etc., were exclusively differentially expressed in either nonsmoker or smoker patients. FAM83D was detected differentially expressed in only smoker patients in our analysis. The family of sequence similarity 83 has many members, and FAM83A and FAM83B have been suggested to be related with lung cancer. There is evidence that cigarette smoking can induce the expression of FAM83A gene [45–48]. Our results suggested that FAM83D is another member in this family that is related with tobacco induced lung cancer. The genes identified in the study provide candidate genes for personalized disease treatment.
To validate the significant genes identified in nonsmoker group, we downloaded an independent RNA-seq data from six nonsmoker Korean patients with lung adenocarcinoma (GEO: GSE37765, [12]). SPINK1 gene has the largest logFC value of 7.31 with FDR 2.73E-25 in nonsmoker patients, and the validation test produced a logFC value of 5.63 with FDR 2.92E-26. BAAT gene was found exclusively differentially expressed in only nonsmokers, and it had a logFC of 2.81 with FDR 1.41E-11 in the current analysis; the validation analysis produced a logFC of 3.00 with FDR 3.89E-11. FAM83D gene was exclusively significant in only smoker patients in the current study; the validation analysis for nonsmokers still showed this gene non-significant between tumor and normal samples (Supplementary 2).
In summary, our study provided a transcriptome profile for both nonsmoker and smoker patients with lung adenocarcinoma. And, we also identified the genes that varied between nonsmoker and smoker patients and provided candidates for gene expression signature associated with nonsmoker or smoker patients. In the current study, the lack of quantitative smoking information limits out ability to test the association between smoking quantity and genes expression level. We also did not have information about disease stages or if the samples were taken from the primary or metastatic side and these factors may also affect gene expression level. All the samples in our study come from Asia, and additional analysis is needed to expand to other ethnicities. Further validation is needed for the signals from smoker group as well as the genes that behave differently between nonsmoker and smoker patients.
Supplementary Material
Acknowledgments
This research was partially supported by NIH research grant U19CA148127.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s13277-015-3576-y) contains supplementary material, which is available to authorized users.
Conflict of interest None
References
- 1.Navada S, Lai P, Schwartz AG, Kalemkerian GP. Temporal trends in small cell lung cancer: analysis of the national Surveillance Epidemiology and End-Results (SEER) database [abstract 7082] J Clin Oncol. 2006;24(18S):384S. [Google Scholar]
- 2.Sher T, Dy GK, Adjei AA. Small cell lung cancer. Mayo Clin Proc. 2008;83(3):355–367. doi: 10.4065/83.3.355. [DOI] [PubMed] [Google Scholar]
- 3. http://www.cap.org/apps/docs/reference/myBiopsy/LungAdenocarcinoma.pdf.
- 4.Powell HA, Iyen-Omofoman B, Hubbard RB, Baldwin DR, Tata LJ. The association between smoking quality and lung cancer in men and women. Chest. 2013;143(1):123–129. doi: 10.1378/chest.12-1068. [DOI] [PubMed] [Google Scholar]
- 5.Massion PP, Zou Y, Chen H, Jian A, Coulson P, et al. Smoking-related genomic signatures in non-small cell lung cancer. Am J Respir Crit Care Med. 2008;178:1164–1172. doi: 10.1164/rccm.200801-142OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers—a different disease. Nature. 2007;7:778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 7.Zhao S, Fung-Leung W, Bittner A, Ngo K, Liu X. Comparison of RNA-seq and microarray in transcriptome profiling of activated T cells. PLoS One. 2014 doi: 10.1371/journal.pone.0078644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng P, Chen Y, Li Y, Zhao Z, Gao H, Li D, et al. Comparison of the gene expression profiles between smokers with and without lung cancer using RNA-seq. Asian Pac J Cancer Prev. 2012;13(8):3605–3609. doi: 10.7314/apjcp.2012.13.8.3605. [DOI] [PubMed] [Google Scholar]
- 9.Beane J, Vick J, Schembri F, Anderlind C, Gower A, Campbell J, et al. Characterizing the impact of smoking and lung cancer on the air way transcriptome using RNA-seq. Cancer Prev Res (Phila) 2011;4(6):803–817. doi: 10.1158/1940-6207.CAPR-11-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han S, Kim W, Hong Y, Hong S, Lee S, Ryu D, et al. RNA sequencing identifies novel markers of non-small cell lung cancer. Lung Cancer. 2014;84(3):229–235. doi: 10.1016/j.lungcan.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 11.Seo J, Ju Y, Lee W, et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res. 2012;22:2109–2119. doi: 10.1101/gr.145144.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim SC, Jung Y, Park J, Cho S, et al. A high-dimensional, deep-sequencing study of lung adenocarcinoma in female never-smokers. PLoS One. 2013;8(2):e55596. doi: 10.1371/journal.pone.0055596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res. 2012:1–10. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lazar V, Suo C, Orear C, Oord VJ, et al. Integrated molecular portrait of non-small cell lung cancers. BMC Medical Genomics. 2013;6:53. doi: 10.1186/1755-8794-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larzabal L, Nguewa PA, Pio R, Blanco D, Scanchez B, et al. Overexpression of TMPRSS4 in on-small cell lung cancer is associated with poor prognosis in patients with squamous histology. Br J Cancer. 2011;105(10):1608–1614. doi: 10.1038/bjc.2011.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Z, Lin D, Yuan J, Xiao T, Zhang H, et al. Overexpression of osteopontin is associated with more aggressive phenotypes in human non-small cell lung cancer. Clin Cancer Res. 2005;11(13):4646–4652. doi: 10.1158/1078-0432.CCR-04-2013. [DOI] [PubMed] [Google Scholar]
- 17.Sauter W, Rosenberger A, Beckmann L, Kropp S, Mittelstrass K, et al. Matrix metalloproteinase 1 (MMP1) is associated with early-onset lung cancer. Cancer Epidemiol Biomarkers Prev. 2008;17(5):1127–1135. doi: 10.1158/1055-9965.EPI-07-2840. [DOI] [PubMed] [Google Scholar]
- 18.Knight SD, Presto J, Linse S, Johansson J. The BRICHOS domain. Amyloid fibril formation and their relationship. Biochemistry. 2013;52(43):7523–7531. doi: 10.1021/bi400908x. [DOI] [PubMed] [Google Scholar]
- 19.Ono S, Tanaka T, Ishida M, Kinoshita A, Fukuoka J, Takaki M. Surfactant protein C G100S mutation causes familial pulmonary fibrosis in Japanese kindred. Eur Respir J. 2011;38(4):861–869. doi: 10.1183/09031936.00143610. [DOI] [PubMed] [Google Scholar]
- 20.Kim SH, Chen G, Jeon CK, Zhao L, Colacino J, et al. Abstract 3124: Smoking effects on CYP24A1 in lung adenocarcinoma: epigenetic changes by smoking. Proceedings: AACR 103rd Annual meeting.2012. [Google Scholar]
- 21.Jiang F, Yin Z, Caraway NP, Li R, Katz RL. Genomic profiles in stage I primary non-small cell lung cancer using comparative genomic hybridization analysis of cDNA microarrays. Neoplasia. 2004;6:623–635. doi: 10.1593/neo.04142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li R, Wang H, Bekele BN, Jiang F. Identification of putative oncogenes in lung adenocarcinoma by a comprehensive functional genomic approach. Oncogene. 2005;18:2628–2635. doi: 10.1038/sj.onc.1209289. [DOI] [PubMed] [Google Scholar]
- 23.Choi CGC, Li J, Wang Y, Li L, Zhong L, Ma B, et al. The metalloprotease ADAMTS8 displays antirumor properties through antagonizing EGFR-MEK-ERK signaling and is silenced in cancinomas by CpG methylation. Mol Cancer Res. 2014;12:228–238. doi: 10.1158/1541-7786.MCR-13-0195. [DOI] [PubMed] [Google Scholar]
- 24.Doree M, Galas S. The cyclin-dependent protein kinases and the control of cell division. FASEB J. 1994;8:1114–1121. doi: 10.1096/fasebj.8.14.7958616. [DOI] [PubMed] [Google Scholar]
- 25.Yasuda M, Takesue F, Inutsuka S, et al. Overexpression of cyclin B1 in gastric cancer and its clinicopathological significance: an immunohistological study. J Cancer Res Clin Oncol. 2002;128:412–416. doi: 10.1007/s00432-002-0359-9. [DOI] [PubMed] [Google Scholar]
- 26.Korenaga D, Takesue F, Yasuda M, et al. The relationship between cyclin B1 overexpression and lymph node metastasis in human colorectal cancer. Surgery. 2001;131:114–120. doi: 10.1067/msy.2002.119362. [DOI] [PubMed] [Google Scholar]
- 27.Takeno S, Noguchi T, Kikuchi R, et al. Prognostic value of cyclin B1 in patients with esophageal squamous cell carcinoma. Cancer. 2002;94:2874–2881. doi: 10.1002/cncr.10542. [DOI] [PubMed] [Google Scholar]
- 28.Hassan KA, El-Naggar AK, Soria JC, et al. Clinical significance of cyclin B1 protein expression in squamous cell carcinoma of the tongue. Clin Cancer Res. 2001;7:2458–2462. [PubMed] [Google Scholar]
- 29.Soria JC, Jang SJ, Khuri FR, et al. Overexpression of cyclin B1 in early-stage non-small cell lung cancer and its clinical implication. Cancer Res. 2000;60:4000–4004. [PubMed] [Google Scholar]
- 30.Yoshida T, Tanaka S, Mogi A, Shitara Y, Kuwano H. The clinical significant of Cyclin B1 and Wee1 expression in non-small-cell lung cancer. Ann Oncol. 2004;15(2):252–256. doi: 10.1093/annonc/mdh073. [DOI] [PubMed] [Google Scholar]
- 31.Watson AP, Evans RL, Egland KA. Multiple functions of Sushi Domain Containing 2 (SUSD2) in breast tumorigenesis. Mol Cancer Res. 2012;11(1):74–85. doi: 10.1158/1541-7786.MCR-12-0501-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pio R, Blanco D, Pajares MJ, Aibar E, Olga D, et al. Development of a novel splice array platform and its application in the identification of alternative splice variants in lung cancer. BMC Genomics. 2010;11:352. doi: 10.1186/1471-2164-11-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schroeder EB, Welch VL, Couper D, Nieto FJ, Liao D, Rosamond WD, et al. Lung function and incident coronary heart disease: the atherosclerosis risk in communities study. Am J Epidemiol. 2003;158(12):1171–1181. doi: 10.1093/aje/kwg276. [DOI] [PubMed] [Google Scholar]
- 34.Taneda K, Namekata T, Hughes D, Suzuki K, Knopp R, Ozasa K. Association of lung function with atherosclerotic risk factors among Japanese Americans: Seattle Nikkei health study. Clin Exp Pharmacol Physiol. 2004;31(Suppl 2):S31–S34. doi: 10.1111/j.1440-1681.2004.04116.x. [DOI] [PubMed] [Google Scholar]
- 35.Dreyer L, Prescott E, Gyntelberg F. Association between atherosclerosis and female lung cancer—a Danish cohort study. Lung Cancer. 2003;42(3):247–254. doi: 10.1016/s0169-5002(03)00295-2. [DOI] [PubMed] [Google Scholar]
- 36.Lu TP, Tsai MH, Lee JM, Hsu CP, Chen PC, et al. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer Epidemiol Biomarkers Prev. 2010;19(10):2590–2597. doi: 10.1158/1055-9965.EPI-10-0332. [DOI] [PubMed] [Google Scholar]
- 37.Lu Y, Yi Y, Liu P, Wen W, James M, Wang D, et al. Common Human Cancer Genes Discovered by Integrated Gene-Expression Analysis. PLoS ONE. 2007;2(11):e1149. doi: 10.1371/journal.pone.0001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rohrbeck A, Borlak J. Cancer Genomics Identifies Regulatory Gene Networks. Associated with the Transition from Dysplasia to Advanced Lung Adenocarcinomas Induced by c-Raf-1. PLoS One. 2009 doi: 10.1371/journal.pone.0007315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campioni M, Ambrogi V, Pompeo E, Citro G, Castelli M, Spugnini EP, et al. Identification of genes down-regulated during lung cancer progression: A cDNA array study. J Exp Clin Cancer Res. 2008;27(1):38. doi: 10.1186/1756-9966-27-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knudson AG. Antioncogenes and human cancer. Proc Natl Acad Sci USA. 1993;90:10914–10921. doi: 10.1073/pnas.90.23.10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang T, Zhang DM, Zhao D, Hou XM, Liu XJ, Ling XL, et al. The prognostic value of osteopontin expression in non-small cell lung cancer: a meta-analysis. J Mol Histol. 2014;45(5):533–540. doi: 10.1007/s10735-014-9574-3. [DOI] [PubMed] [Google Scholar]
- 42.Jin Y, Tong D, Tang L, Chen J, Zhou J, Feng Z, et al. Expressions of osteopontin (OPN), αvβ3 andPim-1 associated with poor prognosis in non-small cell lung cancer (NSCLC) Chin J Cancer Res. 2012;24(2):103–108. doi: 10.1007/s11670-012-0103-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y, Liu H, Wu W, Li Y, Li J. Osteopontin genetic variants are associated with overall survival in advanced non-small-cell lung cancer patients and bone metastasis. J Exp Clin Cancer Res. 2013;32(45) doi: 10.1186/1756-9966-32-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hao Y, Liu J, Wang P, Wang F, Yu Z, Li M, et al. OPN polymorphism is related to the chemotherapy response and prognosis in advanced NSCLC. Int J Genomics. 2014 doi: 10.1155/2014/846142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okabe N, Ezaki J, Yamaura T, Muto S, Osugi J, et al. FAM83B is a novel biomarker for diagnosis and prognosis of lung squamous cell carcinoma. Int J Oncol. 2015;46(3):999–1006. doi: 10.3892/ijo.2015.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu L, Liao GQ, He P, Zhu H, Liu PH, et al. Detection of circulating cancer cells in lung cancer patients with a panel of marker genes. Biochem Biophys Res Commun. 2008;372(4):756–760. doi: 10.1016/j.bbrc.2008.05.101. [DOI] [PubMed] [Google Scholar]
- 47.Li Y, Dong X, Yin Y, Su Y, Xu Q, et al. BJ-TSA-9, a novel human tumor-specific gene, has potential as a biomarker as a biomarker of lung cancer. Beoplasia. 2005;7(12):1073–1080. doi: 10.1593/neo.05406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.GDS1348 / 002003020005 / FAM83A / Homo sapiens. Analysis of cultured normal bronchial epithelial cells 4 and 24 hours after exposure to 15 minutes of cigarette smoke in order to better understand molecular impact of tobacco exposure. http://www.ncbi.nlm.nih.gov/geo/gds/profileGraph.cgi?&dataset=A5KYKBJEB4MVOzwuoi&dataset=awhtm-la8vptmyyyxx$&gmin=0.511550&gmax=8.553317&absc=&uid=12856830&gds=1348&idref=002003020005&annot=FAM83A.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.