Abstract
Cancer cells adapt their metabolism during tumorigenesis. We studied two isogenic breast cancer cells lines (highly metastatic 4T1; nonmetastatic 67NR) to identify differences in their glucose and glutamine metabolism in response to metabolic and environmental stress. Dynamic magnetic resonance spectroscopy of 13C-isotopomers showed that 4T1 cells have higher glycolytic and tricarboxylic acid (TCA) cycle flux than 67NR cells and readily switch between glycolysis and oxidative phosphorylation (OXPHOS) in response to different extracellular environments. OXPHOS activity increased with metastatic potential in isogenic cell lines derived from the same primary breast cancer: 4T1 > 4T07 and 168FARN (local micrometastasis only) > 67NR. We observed a restricted TCA cycle flux at the succinate dehydrogenase step in 67NR cells (but not in 4T1 cells), leading to succinate accumulation and hindering OXPHOS. In the four isogenic cell lines, environmental stresses modulated succinate dehydrogenase subunit A expression according to metastatic potential. Moreover, glucose-derived lactate production was more glutamine dependent in cell lines with higher metastatic potential. These studies show clear differences in TCA cycle metabolism between 4T1 and 67NR breast cancer cells. They indicate that metastases-forming 4T1 cells are more adept at adjusting their metabolism in response to environmental stress than isogenic, nonmetastatic 67NR cells. We suggest that the metabolic plasticity and adaptability are more important to the metastatic breast cancer phenotype than rapid cell proliferation alone, which could 1) provide a new biomarker for early detection of this phenotype, possibly at the time of diagnosis, and 2) lead to new treatment strategies of metastatic breast cancer by targeting mitochondrial metabolism.
Introduction
Breast cancer is the most prevalent type of cancer among women in the United States [1], and mortality is primarily caused by metastatic disease. The complex mechanisms of breast cancer invasion and metastasis [2] are intrinsically related to the malignant cell type [3], their interaction with stromal cells [4], [5], and changes in the tumor microenvironment, related to poor perfusion, intermittent hypoxia, transient nutrient deprivation, and acidity [6], [7]. Cancer cells adapt to dynamic stresses and proliferate by reprogramming their metabolism to support synthesis of an expanding biomass [8], [9]. Due to oncogene-driven upregulation of key glycolytic enzymes [10], most cancer cells exhibit aerobic glycolysis known as the Warburg effect [11]. This metabolic phenotype has been studied by noninvasive techniques, such as 18F-fluorodeoxyglucose positron emission tomography and magnetic resonance spectroscopy (MRS of 13C-labeled substrates) [12], [13]. Due to enhanced glycolysis, tumor cells synthesize high levels of lactate and export H+, resulting in acidification of the microenvironment, which in turn promotes invasion and dissemination [14], [15]. Recent studies with two isogenic murine breast cancer cell lines derived from the same spontaneous breast tumor, 4T1 and 67NR [16], have shown differences in lactate dehydrogenase (LDH) A expression during normoxia and hypoxia [17]. However, other studies have highlighted the importance of oxidative phosphorylation (OXPHOS) in tumorigenesis and progression [18], [19], [20].
Because it is well recognized that tumor cells are often hypoxic and nutritionally deprived in vivo[7], we have monitored in real time the metabolic changes in live 4T1 and 67NR cells under conditions that reflect these common, often transient, physiologic stresses. Our hypothesis is that the metabolism of metastases-forming breast cancer cells (4T1) is more adaptable to changes in the microenvironment than the metabolism of isogenic, nonmetastatic cells (67NR), thus providing 4T1 cells with a distinct advantage to grow, invade, and proliferate under different conditions. Previous metabolic studies using cell extracts have demonstrated marked differences in basal glucose consumption, lactate production, and oxygen consumption between these two cell lines grown in standard bidimensional tissue culture conditions [17], [21]. However, these studies have not investigated the — potentially reversible — metabolic adaptations of these cancer cells to in vivo tumor conditions, which include changing microenvironmental stresses during tridimensional growth. We used a magnetic resonance (MR)–compatible cell perfusion system and time-course MRS of 13C isotopomers to investigate how living cancer cells adapt their metabolism and growth to selective supply/deprivation of glucose and glutamine under both aerobic and hypoxic conditions. In contrast to standard 2D tissue culture studies, the MR-compatible cell perfusion system allows high-density 3D cancer cell growth and exposing cells dynamically and reversibly to various tissue growth environments in a single sample, more similar to the cellular microenvironment of small (< 100 mm3), well-perfused tumors. Moreover, compared with indirect metabolic measurements based on dynamic extracellular pH and O2 changes (e.g. Seahorse XF analyzer studies), the cell perfusion system allows measuring real-time changes in intra- and extracellular metabolite levels and cellular bioenergetic profiles by sequential multinuclear (13C, 31P) MRS. Our assessment of the dynamic interplay between various environmental stresses and tumor cell metabolic response clearly demonstrates that 4T1 cells are more capable of adapting their metabolic responses to changes in the microenvironment than 67NR cells. This is largely accomplished in 4T1 cells by their greater plasticity and ability to more effectively metabolize glucose through either glycolysis or OXPHOS than 67NR cells, providing greater adaptability to a changing tumor and metastatic microenvironment.
Materials and Methods
Cell Lines
The 67NR, 168FARN, 4T07, and 4T1 cell lines were initially derived from a spontaneous breast tumor growing in a BALB/c mouse [16]. These cell lines were kindly provided by Dr. Fred Miller (Karmanos Cancer Institute, Detroit, MI) and grown in Dulbecco’s modified Eagle’s media containing 25 mM glucose (Glc), 6 mM glutamine (Gln), 100 U/ml of penicillin, 100 μg/ml of streptomycin, and 10% fetal calf serum, referred to as DMEcompl. Cells were cultured in 5% CO2 / 95% air at 37 °C in a humidified chamber, split every 2 to 3 days, and used up to passage 10.
Cell Perfusion Studies
For MR cell perfusion studies, 4T1 and 67NR cells were grown to > 70% confluence on microcarrier beads. The cell perfusion studies were carried out on a 500-MHz AVANCE III Bruker MR system equipped with a 10-mm BBO probe for 1H and 31P/13C acquisition (Bruker BioSpin, Billerica, MA). Three different experimental studies were performed for 4T1 and 67NR cell lines, as detailed in Table 1, each consisting of three independent cell perfusion experiments per cell line. During the experiments, the cells were continuously perfused with media containing either 98% to 99% 1-13C-glucose (CLM-420-0; Cambridge Isotope Labs, Billerica, MA) or 99% 3-13C-glutamine (604941-SPEC; Sigma-Aldrich, St. Louis, MO), whereas 13C MR spectra and 31P MR spectra were acquired repeatedly to assess metabolism of the 13C-labeled nutrients and cellular energy status, respectively. Tumor cell–conditioned medium (TCM) samples were also collected at the end of the cell perfusion studies A (Table 1) and stored at −20°C until high-resolution 1H-decoupled 13C MRS studies were performed. Additional details about the cell perfusion system, MR data acquisition, processing, and isotopomer analysis are provided in the Supplementary Experimental Procedures.
Table 1.
MR Cell Perfusion Experiments
| Study | Label (13C) | Stage | Experiment Time (h) | Medium (mM) |
Oxygenation | |
|---|---|---|---|---|---|---|
| Glucose | Glutamine | |||||
| A | – | A-1 | 0-1 | 25 | 6 | Ox |
| Glc C1 | A-2 | 1-6 | 25 | 2 | Ox | |
| Glc C1 | A-3 | 6-32 | 25 | 6 | Ox | |
| B | – | B-1 | 0-1 | 25 | 6 | Ox |
| Gln C3 | B-2 | 1-6 | 25 | 6 | Ox | |
| Gln C3 | B-3 | 6-11 | 25 | 6 | H | |
| Gln C3 | B-4 | 11-16 | 0 | 6 | Ox | |
| Gln C3 | B-5 | 16-21 | 0 | 6 | H | |
| – | B-6 | 21-22 | 25 | 6 | Ox | |
| C | – | C-1 | 0-1 | 25 | 6 | Ox |
| Glc C1 | C-2 | 1-6 | 25 | 0 | Ox | |
| Glc C1 | C-3 | 6-11 | 25 | 0 | H | |
| Glc C1 | C-4 | 11-16 | 25 | 2 | H | |
| Glc C1 | C-5 | 16-21 | 25 | 2 | Ox | |
| – | C-6 | 21-22 | 25 | 6 | Ox | |
Three different studies were carried out: A, B, and C. The first experimental hour of each experiment was reserved for loading the sample into the MR spectrometer, MR probe tuning, and matching, followed by sample shimming. Study A consisted of well-oxygenated cells perfused continuously for 31 hours with 99% 1-13C-glucose–containing culture medium, initially with 2 mM glutamine (1-6 hours) and then with 6 mM glutamine (6-32 hours). In separate experiments, cells were studied during four sequential 5-hour stress conditions in the presence of either 99% 3-13C-glutamine (study B) or 99% 1-13C-glucose (study C) in the culture/perfusion medium. In study B, the experiment with 3-13C-Gln–labeled medium consisted of the four conditions/stages, B2 to B5, evaluating the effects of the presence or absence of 25 mM glucose under well-oxygenated or hypoxic conditions on glutamine metabolism. In study C, the experiments with 99% 1-13C-glucose–labeled culture medium were carried out in four stages/environmental conditions: C-2 to C-5. The perfusion media for stages C-4 and C-5 contained in addition to 2 mM Gln also a basal level of pyruvate (1 mM). Abbreviations: Glc C1: 1-13C-glucose, Gln C3: 3-13C-glutamine, Ox: well-oxygenated conditions, H: hypoxia.
Cell Culture Studies
The four isogenic cell lines (67NR, 168FARN, 4T07, and 4T1) were seeded in 24-well cell culture microplates (5 × 104 cells/well). When cells reached 70% confluence, DMEcompl was replaced with different media containing 1-13C-glucose, as described for cell perfusion studies A and C (Table 1: DMEcompl with 6 mM glutamine and DMEcompl where 6 mM glutamine was replaced with 2 mM and 0 mM glutamine, respectively). Cells were grown for 5 hours either in normoxia (20% oxygen) or in hypoxia (1% oxygen), at 37°C and 5% CO2, followed by collection of TCM2D, cell harvest, and cell count for each sample. TCM2D samples were stored at −20°C. High-resolution 13C MR spectra of TCM2D samples were acquired to determine glucose consumption and lactate excretion in 2D tissue culture and compare them with the results obtained from TCM samples. Additional details regarding the MR data acquisition, processing, and analysis are provided in the Supplementary Experimental Procedures.
Seahorse Bioscience XF96 Extracellular Flux Analyzer Experiments
Both 4T1 and 67NR cells were seeded separately in the same 96-well plate at 1.0 × 104 and 2.0 × 104 cells per well, respectively. After attachment and growth for 18 hours, the DMEcompl medium in each well was changed to XF assay medium (102365-100; Seahorse Bioscience, Billerica, MA) with the same glutamine and glucose concentration as in DMEcompl and incubated for 1 hour in a CO2-free atmosphere. The cells were then studied in an XF96 Analyzer (Seahorse Bioscience, Billerica, MA), measuring the extracellular acidification rate (ECAR; studied during the first 27 minutes) and the oxygen consumption rate (OCR; 2-hour studies) in each well. During the OCR studies, the specific respiratory chain inhibitors oligomycin (inhibition of ATP synthase), Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP; H+ ionophore, uncoupler), and antimycin A plus rotenone (inhibition of complexes I and III, respectively, disabling the electron transport completely in the respiratory chain) (Sigma-Aldrich, St. Louis, MO) were sequentially injected into each well at 30, 60, and 90 minutes, respectively [22]. The time course of OCR values collected during data acquisition were normalized to the total protein content in each well. The latter were determined at the end of the experiment from the cell lysates (RIPA buffer; Thermo Scientific, Rockford, IL) using the bicinchoninic acid protein assay (BCA kit; Thermo Scientific Pierce, Rockford, IL). Measurements were summarized by taking the geometric mean across wells for each cell line, at each time point, and in each replicate of the experiment. The geometric mean was used because of the skewed distribution of data across wells within each experimental condition. In the specific case of ECAR, the logarithm of ratios between paired 4T1 and 67NR results was calculated based on data skewness and because a ratio of 1 translates into a log-ratio of 0 when applying a one-sample t test across experimental replicates. The same analysis was applied to the log-ratio of the geometric means across the four time points and to the linear contrast, estimating the slope of log-ratios versus time points. Finally, an additional experiment was carried out to compare OCR measurements between all four isogenic cell lines according to the protocol previously described. Results were analyzed as above using the geometric means of the dynamic readings in each microplate well (4T1, n = 21; 4T07, n = 22; 168FARN, n = 23; 67NR, n = 22).
Expression and Activity of Succinate Dehydrogenase (SDH)
In both 4T1 and 67NR cells, SDH protein expression was assessed by Western blot of the SDH subunit A (SDH-A) using a monoclonal antibody and a methodology analogous to our previous studies [17]. We also used a polyclonal antibody to assess SDH-A expression by Western blot in the four isogenic cell lines: Cells were exposed for 5 hours to media with different concentrations of glucose (25 and 0 mM) and glutamine (6, 2, and 0 mM) under normoxia and hypoxia, paralleling the media conditions in MR cell perfusion studies as described in Table 1. The Western blot protocols are provided in the Supplementary Experimental Procedures. All results are averaged over three independent experiments. The total SDH activity of 4T1 cells and 67NR was assessed using the Complex II Enzyme Activity Microplate Assay Kit (ab109908; Abcam, Cambridge, MA). The activity of SDH was measured in parallel for fresh lysates of 4T1 and 67NR cells, as described by the manufacturer.
Statistical Analyses
Statistical analyses were carried out with the t test (unless indicated otherwise): unpaired when comparing 4T1 and 67NR data acquired under the same experimental conditions (⁎ indicates statistical significance: P < .05) and paired when comparing the same cell line in consecutive stages during the same study, as specified for each experiment (# indicates statistical significance: P < .05). Results are displayed as mean ± standard error (SEM). In graphs, error bars that are smaller than the symbol size are not visible.
Results
Glycolytic Activity of 4T1 and 67NR Cells
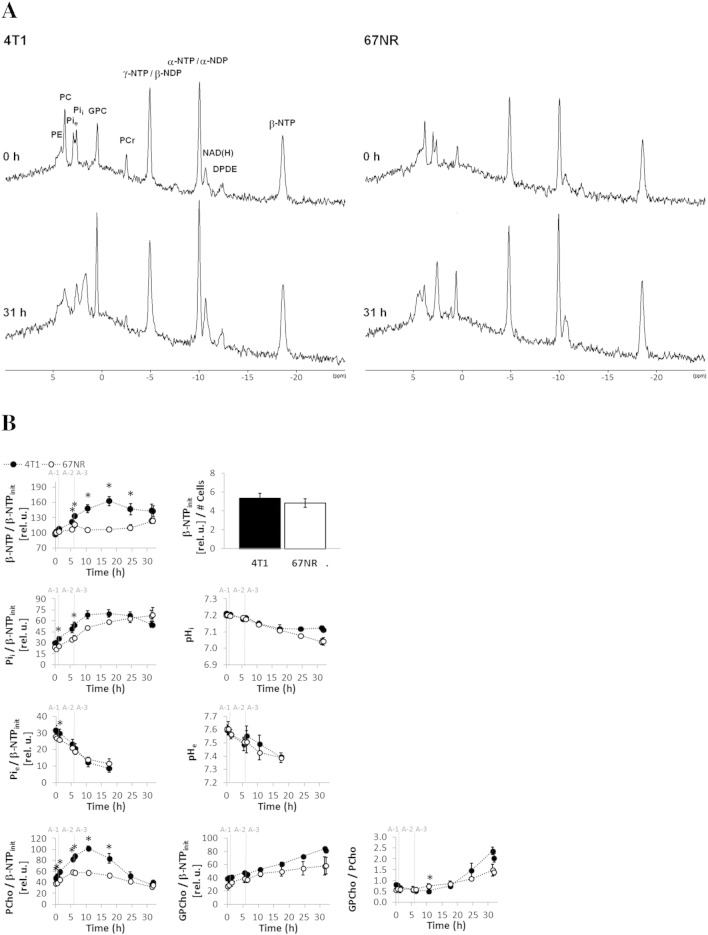
We used an MR-compatible cell perfusion system to investigate the differences in glucose metabolism between 4T1 and 67NR cells in the presence of 6 mM and 2 mM glutamine (DMEcompl versus more physiologic glutamine concentrations, respectively) (Study A, Table 1). The bioenergetic profile and phospholipid metabolism were measured dynamically during all cell perfusion studies by 31P MRS and were characteristic for each cell line (Figure S1). The metabolism of 1-13C-glucose (Figure 1) was measured by 13C MRS. Multiple 13C-labeled metabolites synthesized de novo by the tumor cells from 1-13C-glucose were detected (Figure 1A). Their peak areas were normalized to the initial number of cells (Figure 1, B and C) by referencing to the corresponding β-NTPinit levels (Figure S1). Because glucose was labeled only in the C1 position, 50% of the metabolic pool downstream of fructose 1,6-diphosphate incorporated the 13C-label and was detected.
Figure S1.
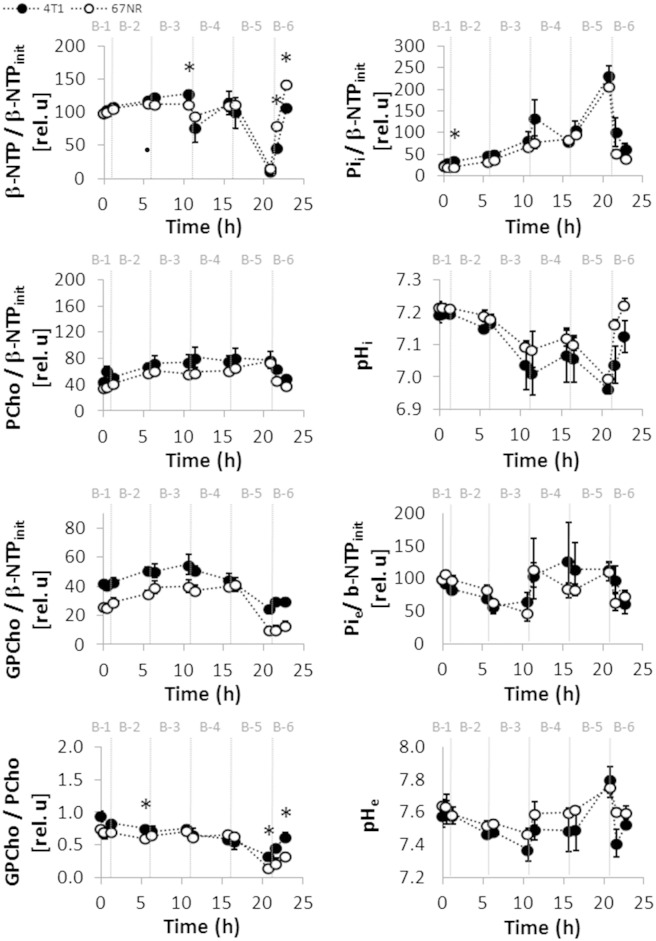
Dynamic bioenergetic profile, pH, and phospholipid metabolism of live 4T1 and 67NR cells. The cells were studied dynamically for 31 hours in an MR-compatible cell perfusion system as described in Table 1, Study A (corresponding to 13C MR studies in Figure 1). (A) Representative 1D 31P MR spectra acquired from 4T1 (left side) and from 67NR (right side) cells, each before (top) at baseline and after (bottom) perfusion with 1-13C-glucose at the end of the experiment. Peak assignments are shown in the top-left spectrum: Phosphoethanolamine (PE), phosphocholine (PCho), extracellular inorganic phosphate (Pie), intracellular inorganic phosphate (Pii), glycerophosphocholine (GPCho), phosphocreating (PCr), nucleoside diphosphate (NDP), nucleoside triphosphate (NTP), overlapping signals from the nicotinamide adenine dinucleotides NAD +/NADH and NADP +/NADPH [NAD(H)], diphosphodiester (DPDE). All spectra are displayed with 30-Hz exponential line broadening. (B) Time-course changes for β-NTP (mostly ATP [12]supp), Pii, Pie, PCho, and GPCho normalized to β-NTPinit are shown on the left, from top to bottom. Right column from top to bottom: Initial β-NTP (β-NTPinit) from 31P MRS normalized to the number of cells—determined by Crystal Violet nuclei counting [3], [5]supp—at time of loading of the cells; pHi and pHe, determined from the chemical shift of the corresponding Pii and Pie signals [3], [9]supp; GPCho/PCho ratio. β-NTPinit (β-NTP at time 0 hour), as a fraction of the initial number of cells when loading the cell perfusion system, was similar between the two cell lines. Whereas 67NR cells demonstrate slightly increasing β-NTP levels over the entire time, in 4T1 cells, β-NTP increases up to 1.7-fold (of β-NTPinit) within the first 18 hours and plateaus from 18 to 32 hours. At that point, 4T1 cells (initially > 70% confluent on the microcarriers) had time to double in the 3D cell perfusion system and therefore are close to or have likely reached confluence, resulting in the plateau of their β-NTP. These observations indicate that the changes observed in the energy status between these two cell lines during the first 15 hours of the cell perfusion experiments may be attributable to their differences in cell proliferation, as has been observed previously with a glioma cell line [1], [4], [13]supp. During the cell perfusion experiments, both 4T1 and 67NR cells produce and accumulate PCho in the first 11 hours, 4T1 cells (two-fold increase) significantly more so than 67NR cells (1.5-fold increase), consistent with the metastatic phenotype and faster proliferation of 4T1 cells as observed previously for other tumor cell lines [14], [15], [16]supp. Cellular GPCho levels increased significantly in both cell lines; the slight differences between 4T1 and 67NR cells were not significant. The GPCho/PCho ratio, which has been associated with slower proliferation in breast cancer cell lines and human patient samples [14]supp, decreased in 4T1 cells during the first 11 hours of perfusion (Table S1), and an upward trend after that time was noted, both agreeing with the shorter cell doubling time of 4T1 than 67NR cells and the respective changes detected in β-NTP. Beyond 18 hours, pHi plateaus in 4T1 cells while still continuing to decrease in 67NR cells. Because at these later time points the Pie signal decreased to levels at which it could not be resolved from the Pii signal, pHe could not be resolved from pHi. Average values (mean ± SEM, n = 3 each) are shown for 4T1 (closed circles) and 67NR cells (open circles). Significant differences (P < .05, unpaired t test) between 4T1 and 67NR cells are denoted by *. The statistical significance for the same cell line between different time points (P < .05, paired t test) are shown in Table S1, Study A.
Figure 1.
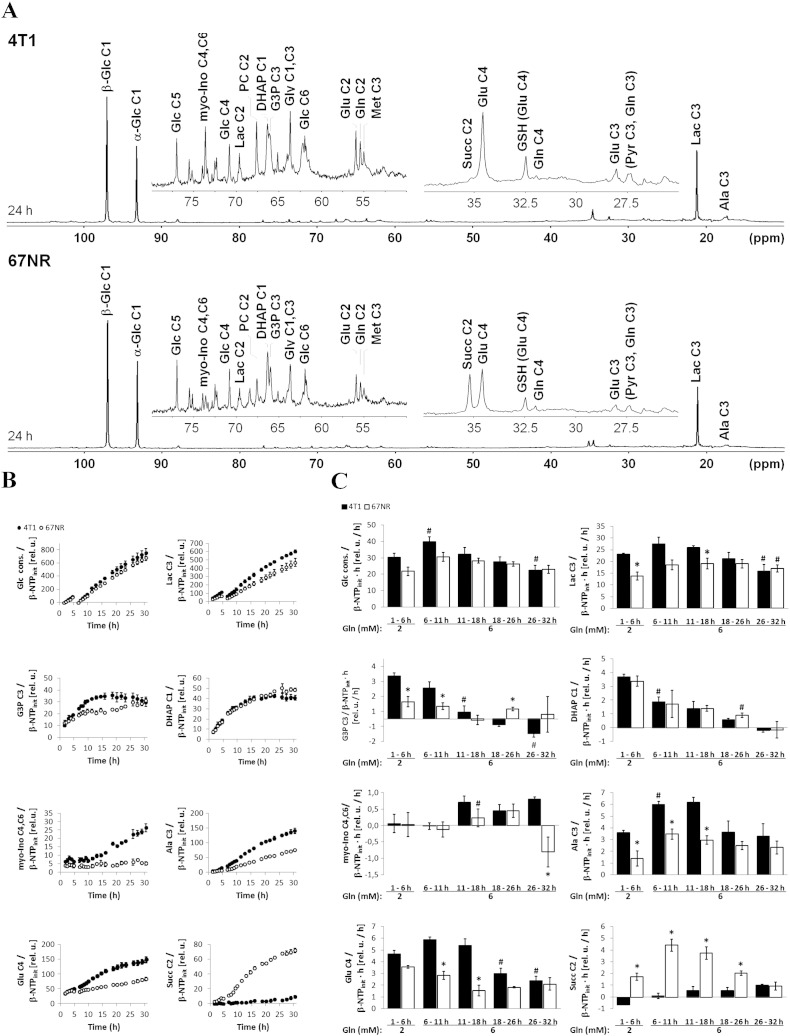
Basal glucose metabolism of live 4T1 and 67NR cells. MR cell perfusion studies (n = 3 for each cell line). (A) Representative nuclear Overhauser effect–enhanced, 1D 1H-decoupled 13C MR spectra acquired from 4T1 (top) and 67NR (bottom) cells after 24-hour perfusion with culture medium containing 1-13C-glucose (Glc), as detailed in Study A, Table 1. The regions from 80 to 50 ppm and 40 to 25 ppm are shown enlarged above the full spectra. All spectra are displayed with 10-Hz exponential line broadening. Signal assignments of metabolites, with C1-6 depicting the position of the incorporated 13C label: alanine (Ala), dihydroxyacetone phosphate (DHAP), α- and β-glucose (α-Glc and β-Glc, respectively), glutamate (Glu), glutamine (Gln), glutathione (GSH-Glu), glycerol (Gly), glycerol-3-phosphate (G3P), lactate (Lac), methionine (Met), myo-inositol (myo-Ino), pyruvate (Pyr), succinate (Succ). (B) Average time-course changes for the major peak areas, normalized to the initial β-NTP values (β-NTPinit, determined from 31P MRS; Figure S1). (C) Average 13C-labeling rates of metabolites at different time intervals for the data shown in B.
As shown in Figure 1, 4T1 cells demonstrated significantly higher glycolytic activity than 67NR cells, independent of glutamine concentrations in the perfusion medium (2 mM, 1-6 hours, P < .05; and 6 mM, 6-32 hours, P < .05). During the initial 10 hours of perfusion with 13C-labeled medium, the glucose consumption rate was slightly higher in 4T1 cells than in 67NR cells and increased significantly in 4T1 cells when changing the medium from 2 mM to 6 mM glutamine (after 6 hours) (Figure 1C). Under both experimental conditions, the glycolytic-lactate synthesis rates were also approximately 1.5-fold higher in 4T1 than in 67NR cells (1-6 hours, P < .05; 6-11 hours, P = .06), closely matching the difference in doubling times between the two cell lines. In standard tissue culture medium (25 mM glucose and 6 mM glutamine), 4T1 cells have a doubling time of 13.6 ± 1.5 hours (n = 13, mean ± SE) and proliferate significantly more rapidly than 67NR cells [doubling time of 22.4 ± 1.8 hours (n = 16, mean ± SE)] (P < .05, Mann-Whitney U test). Similar doubling times were measured when the cells were grown under more physiological concentrations of glucose (5 mM) [23]. The differences in proliferation were also reflected in the bioenergetic and phosphomonoester profile of the two cell lines, with β-NTP and PCho both increasing faster in the 4T1 cells than in 67NR cells during cell growth in the perfusion system (Figure S1). The initial rates (1-11 hours) of glucose consumption and total de novo extracellular lactate (Lac) derived from 13C-labeled glucose (Figure S2; see Supplementary Experimental Procedures) were similar to those obtained in previous studies of glioma and EMT-6 cells [24], [25].
Figure S2.
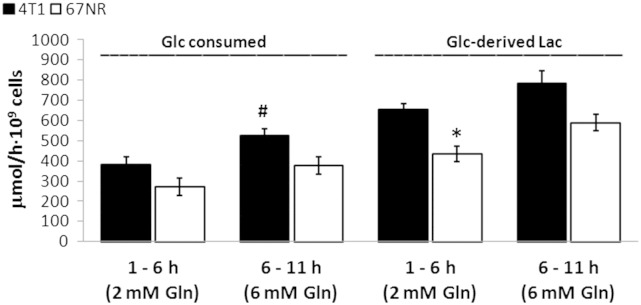
Absolute rates of 1-13C-glucose consumption and de novo 1-13C-glucose–derived lactate synthesis in 4T1 and 67NR cells. Average values (n = 3) determined from the cell perfusion data shown in Figure 1 during the initial 10-hour perfusion with 2 mM (1-6 hours) and 6 mM (6-11 hours) glutamine-containing media. Metabolite concentrations at the two time points were calculated based on concentrations determined from medium samples collected at the end of each experiment (32 hours), as detailed in supplemental experimental procedures. *Statistically significant differences between cell lines during the same time interval (P < .05, unpaired t test); #statistically significant change versus previous interval for the same cell line (P < .05, paired t test). Error bars indicate the standard error of the means.
The 13C-labeled pools of glycerol-3-phosphate [3-13C-G3P, obtained by reduction of dihydroxyacetone phosphate (1-13C-DHAP) and essential for membrane phospholipid synthesis] and myo-inositol [4(6)-13C-myo-Ino, precursor of phosphatidyl inositol phosphates] were also observed in 4T1 and 67NR cells (Figure 1, B and C). Compared with 67NR cells, the higher levels of 3-13C-G3P and 4(6)-13C-myo-Ino in 4T1 cells also indicate their higher glycolytic turnover and membrane biosynthesis requirements, in both 2-mM and 6-mM glutamine medium, and relate to their invasive/metastatic phenotypes, in agreement with Smirnova et al. [26].
The synthesis rate of glucose-derived alanine (3-13C-Ala, an alternative end-product of glycolysis obtained by transamination of 3-13C-pyruvate with glutamate) was higher in 4T1 than in 67NR cells. In response to increasing glutamine from 2 mM to 6 mM, the synthesis rate of 3-13C-Ala increased 1.7-fold in 4T1 cells and 2.5-fold in 67NR cells (Figure 1C). In subsequent cell perfusion experiments, under metabolic stress (Table 1, Study C: 0 and 2 mM glutamine, under hypoxia and normoxia), both cell lines were essentially unable to use glucose for alanine synthesis when fully deprived of glutamine (Figure S3, 1-11 hours). These observations are consistent with the central role of glutamine as a nitrogen donor for biosynthesis of amino acids and nucleotides [27], [28] and the greater metabolic demands of the faster-growing, metastatic 4T1 cells.
Figure S3.
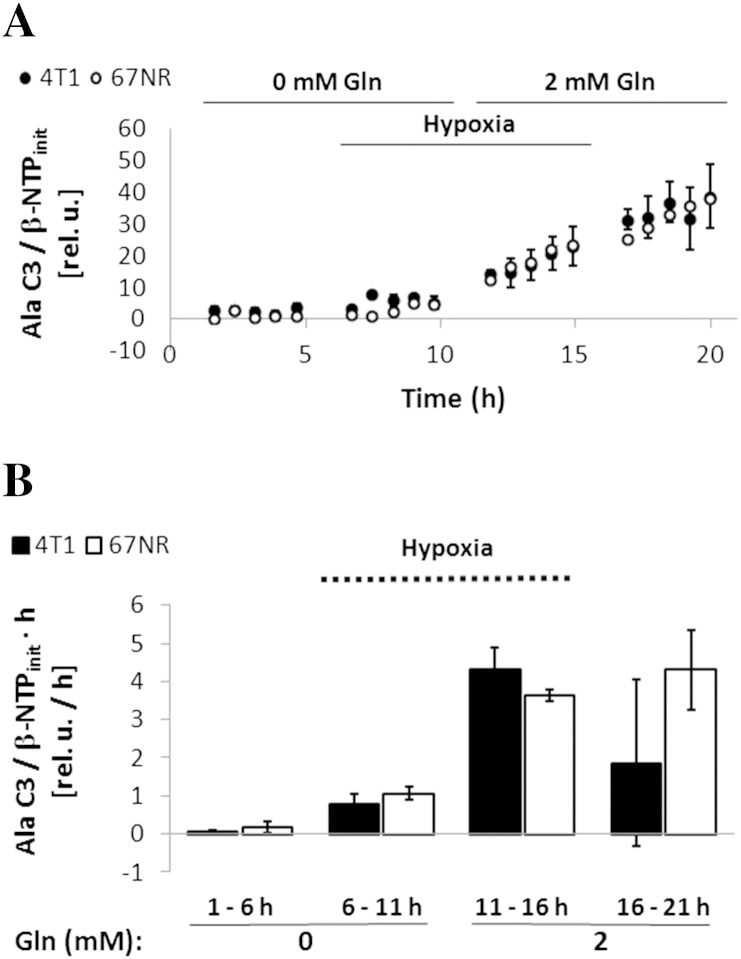
Labeling of alanine-C3 with 13C in cell perfusion stress studies with 1-13C-glucose. Normalized average time-course changes of alanine-C3 (A) and respective 13C-labeling rates at different time intervals (B), determined from 13C MR spectra, in response to environmental conditions as detailed in Table 1 (Study C). Error bars indicate the standard error of the means. Open circles: 67NR, closed circles: 4T1.
Consistent with ongoing glycolytic activity and synthesis of glucose-derived lactate in proliferating live cells, we observed in our cell perfusion studies intra- and extracellular acidification during the first 18 hours, with pHi decreasing from 7.2 to 7.1 in both cell lines (P < .05, paired t test) and pHe from 7.6 to 7.4 (P < .05 only for 67NR cells, paired t test) (Supplementary Experimental Procedures and Figure S1B). The basal glycolytic activity of 4T1 and 67NR cells was also measured for cells in 2D tissue culture by the ECAR (Figure 2A). The ECAR measurements were significantly higher for 4T1 than for 67NR at each successive time point (P = .021, .023, .041, and .05, respectively), and also for the geometric mean across time points (P = .034). The mean of geometric ratios, between paired 4T1 and 67NR results, indicated 1.4-fold higher ECAR in 4T1 (Figure 2A), consistent with the ratios of the glucose-derived lactate synthesis rates for 4T1 and 67NR obtained with the cell perfusion system within the first 18 hours (1.7, 1.5, and 1.4), respectively (Figure 1C).
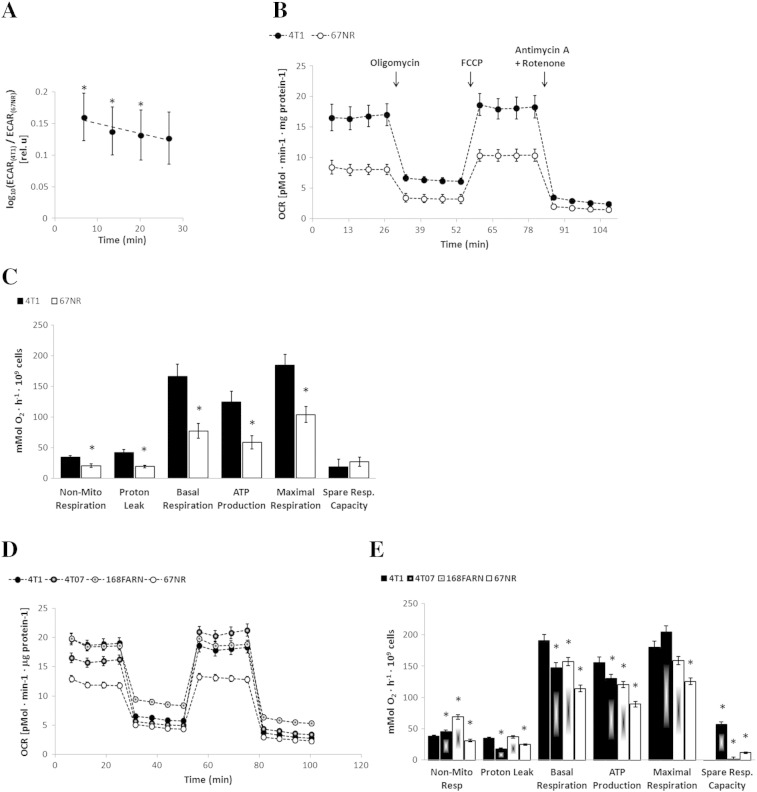
Figure 2.
Mitochondrial respiration of breast cancer cells with different metastatic potential. Extracellular flux analyzer experiments (Seahorse Bioscience XF96) for different cell lines using XF assay medium containing 25 mM glucose and 6 mM glutamine: 4T1 and 67NR: n = 4 (A–C); 4T1, 4T07, 168FARN, and 67NR: n = 1 (D–E). (A) The logarithm of ratios of ECARs (mpH · min− 1 · mg protein− 1) of 4T1 and 67NR cells, ECAR(4T1)/ECAR(67NR), plotted repeatedly over 27 minutes. (B) OCR (pMol · min− 1 · mg protein− 1) measurements for 4T1 and 67NR cells, respectively. (C) For 4T1 and 67NR cells, the following mitochondrial respiration parameters were calculated from the OCR curves shown in B: nonmitochondrial respiration, proton leak, basal respiration, ATP production, maximal respiration, and spare respiratory capacity. An additional experiment, performed as described for B and C, included four isogenic cell lines and is shown in D and E, respectively. In A, * indicates significant differences between 4T1 and 67NR cells at successive time points (P = .021, .023, .041, and .05, respectively) and for the geometric mean across time points (P = .034) based on one-sample t tests of the log-ratios across experimental replicates.
OXPHOS and Metastatic Potential
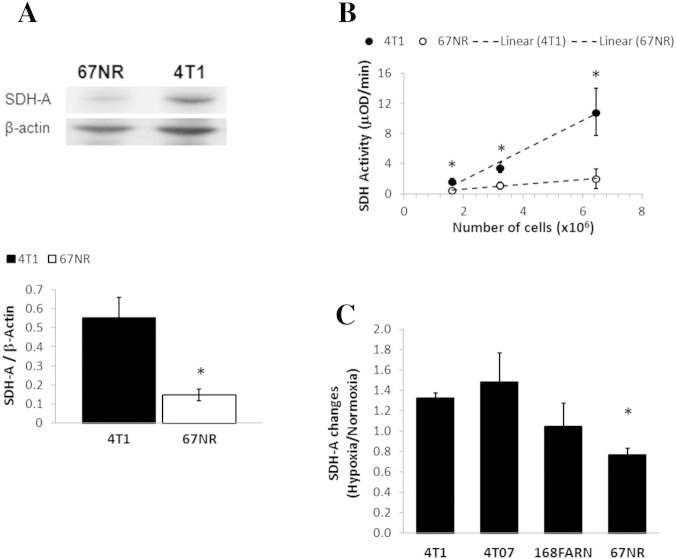
To investigate the role of OXPHOS in each cell line, we carried out mitochondrial functional analyses under 2D growth conditions (25 mM Glc, 6 mM Gln) by selective inhibition of different respiratory complexes while measuring the effects on the cellular OCR (Figure 2B). Consistent with the 31P MRS results on live, perfused cells (Figure S1B), 4T1 cells showed a significantly higher basal respiration rate (2.3-fold) and ATP production (2.2-fold) than 67NR cells (Figure 2C). When extending these measurements to two additional cell lines from the same isogenic breast cancer family (168FARN and 4T07), we noticed higher basal respiration and ATP production with increasing metastatic ability: 4T1 (overt metastasis) > 4T07 and 168FARN (micrometastasis) > 67NR (nonmetastatic) (Figure 2, D and E).
Tricarboxylic Acid (TCA) cycle activity in 4T1 and 67NR cells
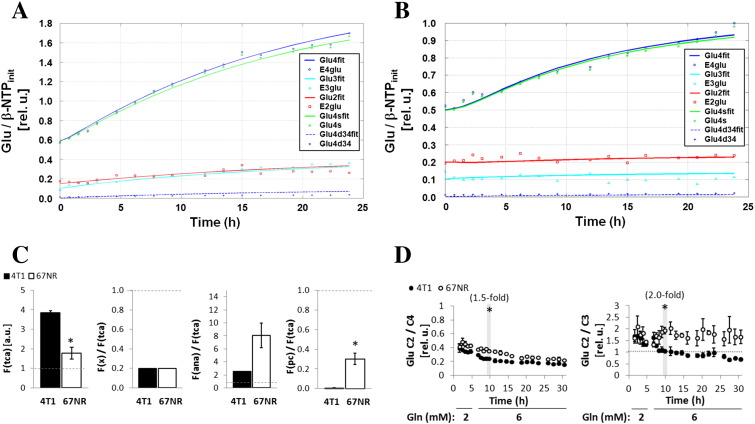
Both 4T1 and 67NR cells incorporated 1-13C-glucose into 4-13C-glutamate (Figure 1, A and B) via pyruvate dehydrogenase, indicative of mitochondrial TCA cycle activity in both cell lines. However, the synthesis rate of 4-13C-glutamate was two-fold higher in 4T1 cells than in 67NR cells at 6 to 11 hours of perfusion with 1-13C-glucose in the perfusate (Figure 1C). To further investigate the TCA cycle activity and metabolite contributions to anaplerosis [29], we considered a dynamic flux analysis based on the time-course changes of the different 13C-glutamate isotopomers (Figure 3, A and B). This model estimated a 2.2 ± 0.6-fold higher TCA cycle flux [F(tca)] in 4T1 cells than in 67NR cells (Figure 3C), which is consistent with the differences in OCR measurements (Figure 2, B and C). The TCA flux analysis model projected identical relative exchange fluxes between α-ketoglutarate and glutamate in both cell lines [F(x)/F(tca), Figure 3C]. However, the total exchange estimated at the levels of succinyl-CoA, fumarate, and oxaloacetate (OAA), i.e., anaplerotic/cataplerotic fluxes [F(ana)], was 3.2 ± 0.8-fold higher in 67NR cells than in 4T1 cells, although not significantly. Moreover, the estimated pyruvate carboxylase flux F(pc) (OAA–pyruvate exchange) was significantly higher in 67NR cells than in 4T1 cells (Figure 3C). Accordingly, the time-course changes in the 2-13C/4-13C glutamate ratio (<<1, Figure 3D) suggest that pyruvate is channeled to the TCA cycle mostly via pyruvate dehydrogenase [30] in both cell lines, whereas the changes in 2-13C-glutamate to 3-13C-glutamate ratio (Figure 3D) reflect differences in pyruvate carboxylase activity according to glutamine availability. In the presence of 2 mM glutamine (Figure 3D, 1-6 hours), both cell lines displayed similar 2-13C-glutamate to 3-13C-glutamate ratios (~ 1.5-2), suggesting similar pyruvate carboxylase activity. However, after increasing glutamine to 6 mM (Figure 3D, 6-11 hours), the 2-13C-glutamate to 3-13C-glutamate ratio remained essentially unchanged in 67NR cells but significantly decreased in 4T1 cells (~ 1), suggesting a reduction in pyruvate carboxylase activity, likely due to more glutamine availability to replenish the TCA cycle at the level of OAA. Altogether, our results indicate a rate-limiting step of the TCA cycle downstream of α-ketoglutarate in 67NR cells. This is summarized in our proposed model (Figure 4, A and B).
Figure 3.
TCA cycle flux analysis of 4T1 and 67NR cells. Representative examples of flux fitting for 4T1 (A) and 67NR (B) cells during the 6- to 32-hour perfusion period with 99% 1-13C-Glc and 6 mM glutamine based on the time courses of labeled glutamate isotopomers C4 (E4glu), C3 (E3glu), and C2 (E2glu), and the estimated glutamate C4 singlet (Glu4s, 98%) and doublet (after one turn of the TCA cycle: Glu4d34, 2%). The solid lines are the fits of the kinetic model to the data. Dashed lines are the predicted Glu4d34 from the model. (C) From left to right: estimations of TCA cycle flux [F(tca)], α-ketoglutarate to glutamate flux [F(x)] relative to F(tca), anaplerotic flux [F(ana), total exchange estimated at the levels of succinyl-CoA, fumarate, and OAA; center panel] relative to F(tca), and pyruvate carboxylase flux [F(pc) relative to F(tca)]. (D) Experimental time-course data for 4T1 and 67NR cells illustrate the relative fluxes estimated by the model (shown in C) in presence of 2 mM glutamine (1-6 hours) and 6 mM glutamine (6-32 hours): left panel, Glu C2/C4 ratios; right panel, Glu C2/C3 ratios. Significantly higher Glu C2/C4 and Glu C2/C3 ratios in 67NR than 4T1 cells, indicative of a higher F(pc)/F(tca) and F(ana)/F(tca), respectively.
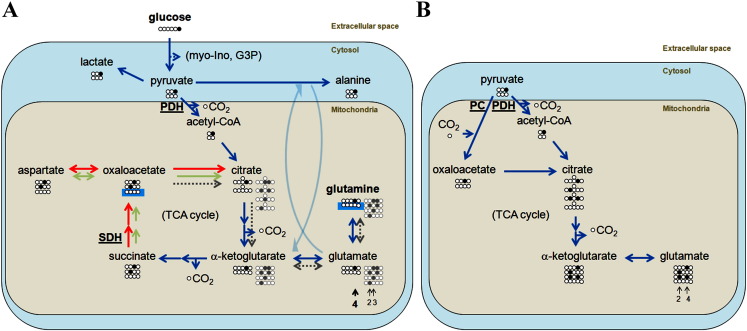
Figure 4.
Metabolic model for 4T1 and 67NR cells. (A) Extracellular 1-13C-Glc (carbons C1-6 shown as spheres; 13C-labeled carbon in black) in the medium is taken up by the cells via GLUT transporters. Glucose is then metabolized through the glycolytic pathway, labeling 50% of the de novo pools of myo-inositol (C2, C4), DHAP (C1), and glycerol-3-phosphate (C1), which could be detected in the cell perfusion system experiments (Figure 1). Pyruvate (C3 labeling) can be further metabolized in the cytosol to lactate (C3 labeling), regenerating the NAD+ consumed during glycolysis, or to alanine (C3 labeling) by transamination of glutamate with pyruvate. Alternatively, pyruvate can be channeled to the mitochondria and converted to acetyl-CoA (C2 labeling) via pyruvate dehydrogenase (PDH). Acetyl-CoA enters the TCA cycle and condenses with OAA (initial unlabeled pool) to produce citrate, which then transfers its label to α-ketoglutarate (C4). The exchange between α-ketoglutarate (fast turnover) and glutamate (C4 labeling) makes the latter a marker of TCA cycle activity. The C4 label of α-ketoglutarate is then transferred to succinate C2 (or C3; not distinguishable) and then to OAA C2 (C3). Because succinate C2 (C3) accumulates in 67NR cells, it is likely that their TCA cycle is restricted at the level of SDH, leading to lower flux to OAA (green arrows; higher flux expected in 4T1 cells, displayed by red arrows). After this first turn of the TCA cycle, a significant part of the intermediates has been removed for biosynthetic purposes; the remaining OAA can be used for the second turn — reactions indicated by the dotted gray arrows and the resulting molecules by the gray spheres (13C label, gray; unlabeled, white). In this case, the resulting labeling pattern of glutamate is more complex and generates an additional 1:1 pool of C2 and C3 labeling (highlighted by short gray arrows). (B) Besides PDH, an alternative way for pyruvate to enter the TCA cycle is through pyruvate carboxylase (PC). This anaplerotic pathway becomes more important when glutamine is not sufficiently available to cells or they cannot use it to regenerate OAA. In this case, the first turn of the TCA cycle will generate a 1:1 labeling of glutamate in positions C4 and C2 (indicated by short gray arrows).
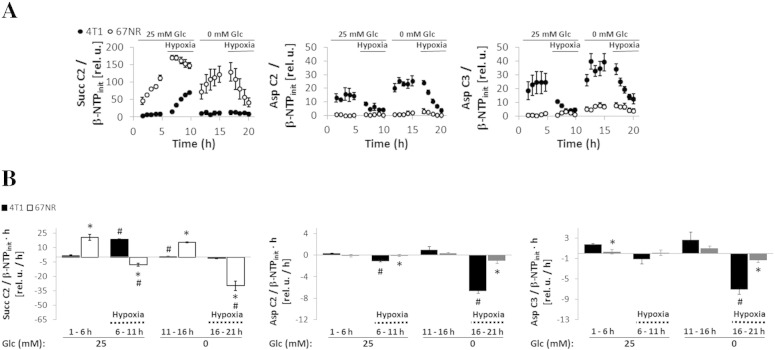
TCA Cycle Restriction in 67NR Cells
The nonmetastatic 67NR cells showed a strong accumulation of 1-13C-glucose–derived succinate (2-13C-Succ) under full nutrient conditions, whereas glucose-derived succinate was essentially not detected in 4T1 cells (Figure 1, A–C). High-resolution MR analysis of TCM samples collected at the end of each experimental stage (Supplementary Experimental Procedures) also showed that 67NR cells extruded 5.8-times more succinate than 4T1 cells (data not shown). The accumulation of glucose-derived succinate observed in 67NR cells was further investigated by perfusing each cell line with 3-13C-glutamine during regular growth conditions (1-6 hours), under reversible hypoxia (6-11 and 16-21 hours), and under glucose starvation (11-21 hours) (Table 1, Study B). The time-course changes in phospholipid metabolism, bioenergetic profiles, pHi, or pHe essentially showed the same trends in both cell lines under glucose deprivation or hypoxia (Figure S4).
Figure S4.
Bioenergetic profile, pH, and phospholipid metabolism of 4T1 and 67NR cells during the cell perfusion stress studies B, detailed in Table 1. Time-course changes of β-NTP, intracellular (i) and extracellular (e) Pi and pH, PCho, GPCho, and GPCho/PCho for the studies with 3-13C-glutamine (corresponding to 13C MR studies in Figure 5). The different stages detailed in Table 1 are indicated in each plot and separated by dashed lines. Error bars indicate the standard error of the means. *Statistically significant differences between 4T1 and 67NR cells during the same time interval (P < .05, unpaired t test). Statistically significant changes (P < .05, paired t test) between different time points for the same cell line are shown in Table S1 (Study B, with 3-13C-glutamine).
3-13C-glutamine–derived succinate levels (2-13C-Succ) were always significantly higher in 67NR cells than in 4T1 cells in the cell perfusion system for all the microenvironment conditions (Study B, Table 1) tested (Figure 5A). 67NR cells synthesized 2-13C-succinate under aerobic conditions, either from 1-13C-glucose (Figure 1, A–C) or from 3-13C-glutamine (Figure 5, A and B; 1-6 and 11-16 hours). The accumulation of 2-13C- succinate from 3-13C-glutamine in 4T1 cells could only be detected during hypoxia with glucose-containing perfusate (Figure 5, A and B; 6-11 hours). This is consistent with a reductive turnover of the TCA cycle, as recently shown in other cancer cell lines growing in hypoxia [31] or with defective mitochondria [32]. In 4T1 cells (and not in 67NR cells), 13C-labeled aspartate (2-13C and 3-13C, in equilibrium with OAA labeled in the same positions) was detected only when oxygen was available (Figure 5A, 1-6 and 11-16 hours) and not under hypoxia (Figure 5A, 6-11 and 16-21 hours).
Figure 5.
Cell perfusion stress studies with 3-13C-glutamine. Normalized average time-course changes of metabolite levels (A) and 13C-labeling rates at different time intervals (B) for succinate C2, aspartate C2, and aspartate C3 determined from 13C MR spectra under environmental conditions as detailed in Table 1 (Study B).
Full glucose deprivation under well-oxygenated conditions caused a slight (not statistically significant) increase in the levels of 3-13C-glutamine–derived aspartate in 4T1 cells and of succinate in 67NR cells (Figure 5A, 1-6 vs. 11-16 hours), with no corresponding differences between the rates of synthesis (Figure 5B). These findings indicate that the glucose concentration does not have a significant effect on the oxidation of glutamine through the TCA cycle in both cell lines. This backs up a prior observation in a human lymphoma cell line that supports the existence of an alternative carbon source pathway into the TCA cycle driven by glutamine and independent of glucose [33]. Our results, together with the flux model predictions based on glutamate isotopomer analysis (Figure 3), suggest that the lower TCA cycle activity observed in 67NR cells is due to a restriction in TCA cycle flux at the level of SDH (respiratory complex II) (Figure 4).
SDH-A Protein Expression Pursuant to Metastatic Ability and Cell Line-dependent Response to Environmental Stresses
We investigated if the lower TCA cycle activity in 67NR cells and the accumulation of succinate could be explained by a deficiency in SDH expression and/or activity. Western blots showed a significant four-fold lower expression of SDH-A (FAD-containing subunit that binds succinate) in 67NR cells than 4T1 cells (Figure 6A). The enzyme activity of SDH was also significantly lower (six-fold) in 67NR cells than in 4T1 cells (Figure 6B). Because an impairment of SDH should directly affect mitochondrial respiration, these results are consistent with the approximately two-fold higher basal respiration rate and ATP production observed in 4T1 cells compared with 67NR cells (Figure 2C). The expression of SDH-A in regular medium conditions (25 mM Glc, 6 mM Gln) was noted to increase with metastatic ability across the isogenic cell line panel for oxygenated and hypoxic cells (Table 2). Hypoxia itself affected SDH-A expression depending on the metastatic potential (Table 2, Figure 6C): downregulation in 67NR (nonmetastatic), upregulation in 4T1 (metastatic) (P = .003), and intermediate effects in the other two cell lines characterized by local micrometastases. Relative to 4T1 cells, glutamine deprivation (0 mM and 2 mM Gln) lowered SDH-A protein expression in the three less metastatic cell lines more than glucose deprivation (Table 2). These results suggest a better ability of 4T1 cells to use the TCA cycle, even under stress, than the other three cell lines.
Figure 6.
SDH expression and activity in breast cancer cells with different metastatic potential. (A) Protein expression of SDH-A was higher in 4T1 than 67NR cells, as shown in a representative Western blot of 4T1 and 67NR cells (left panel) and quantified by averaging the SDH-A/β-actin ratios over three independent experiments (right, bar graph). (B) SDH activity assessed kinetically by absorbance spectroscopy at 600 nm in 4T1 and 67NR cells (dots are average values of at least three independent experiments) and linear fittings for each cell line (slope = SDH activity): 4T1, 1.97 ± 0.28 (R2 = 0.98); 67NR, 0.32 ± 0.03 (R2 = 0.99). (C) Ratio of cellular SDH-A/β-actin expression in hypoxia to normoxia for 4T1, 4T07, 168FARN, and 67NR cell lines exposed to culture medium containing 25 mM Glc and 6 mM Gln.
Table 2.
SDH-A Expression in Four Isogenic Breast Cancer Cell Lines under Environmental Stress
| Medium |
Cell Lines |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Glc |
Gln |
67NR |
168FARN |
4T07 |
4T1 |
||||
| (mM) | (mM) | (Ox) | (H) | (Ox) | (H) | (Ox) | (H) | (Ox) | (H) |
| 0 | 6 | 0 | + 1 | + 3 | + 2 | − 1 | − 2 | (ref) | + 2 |
| 25 | 6 | − 1 | − 2 | 0 | 0 | − 1 | + 1 | (ref) | + 2 |
| 25 | 2 | − 3 | − 3 | − 2 | − 3 | − 3 | − 3 | (ref) | 0 |
| 25 | 0 | − 2 | − 3 | − 2 | 0 | − 2 | − 2 | (ref) | − 1 |
For each independent experiment (n = 3), SDH-A expression levels assessed by Western blot were normalized to β-actin and ranked separately for each condition [glucose and glutamine concentrations and effect of hypoxia (H)] with respect to expression levels of well-oxygenated (Ox) 4T1 cells. Legend: + 3 > 150%; + 2 > 125%; + 1 > 110%; 0 ± 10%; − 1 < 90%; − 2 < 75%; − 3 < 50%.
Metabolic Adaption to Nutrient (Glucose, Glutamine) Supplyin 4T1 than in 67NR Cells
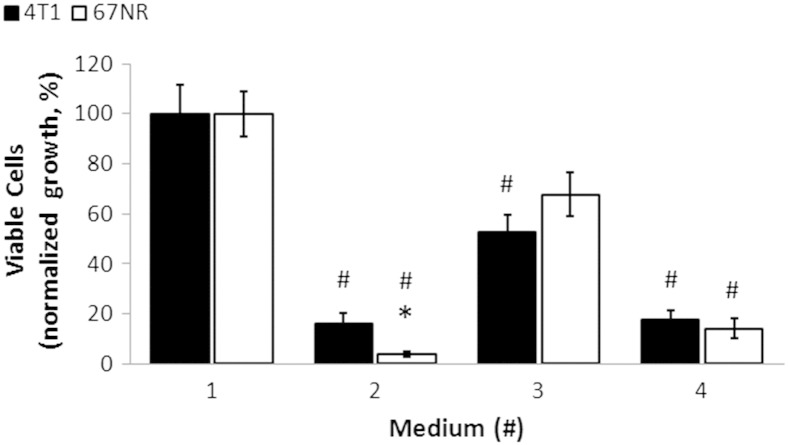
To further understand the metabolic differences between 4T1 and 67NR cells, we investigated the effects of glucose and glutamine deprivation on glucose metabolism. Using standard 2D tissue culture conditions (Figure S5), we found that the cell growth significantly decreased by approximately five-fold in both cell lines when they were fully deprived of either glutamine or glucose. In response to partial glutamine deprivation (2 mM), an approximately two-fold decrease was observed in both cell lines. Therefore, both 4T1 and 67NR cells depend on glucose and glutamine to grow.
Figure S5.
Effects of environmental stresses on cell growth. In vitro cell growth response of 4T1 and 67NR cells to 48-hour exposure to different metabolite stresses (media: #2, 25 mM Glc and 0 mM Gln; #3, 25 mM Glc and 2 mM Gln; #4, 0 mM Glc and 6 mM Gln) as percentage of standard tissue culture conditions (medium #1, 25 mM Glc and 6 mM Gln). Cells were all grown in 20% O2/5% CO2 at 37°C. Error bars indicate the standard error of the means (n = 5 for each condition). #Significant differences of cell growth for stress versus standard tissue culture conditions (100%) for each cell line (P < .05, paired t Test). *Significant differences of cell growth between cell lines under the same culture conditions (P < .05, unpaired t test).
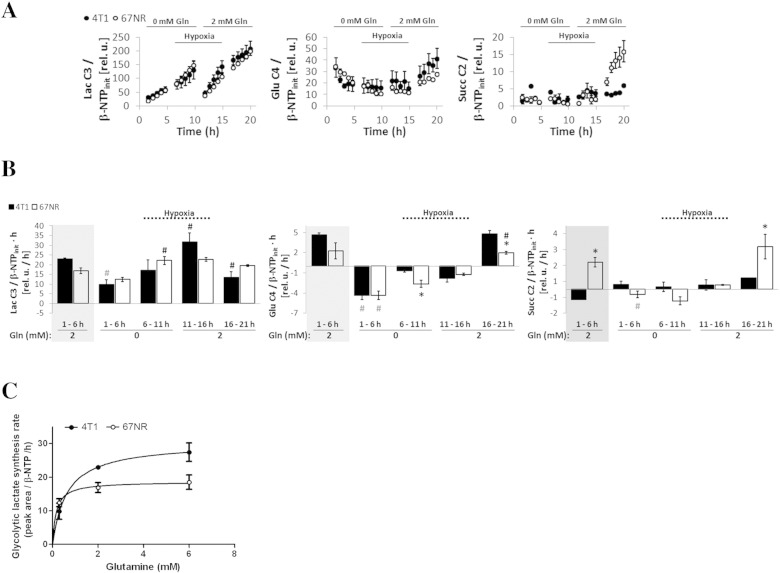
For the cell perfusion experiments performed under cellular stress conditions (Table 1, Study C), we selected 2 mM glutamine over 6 mM to more closely simulate the glutamine levels observed in vivo (~0.5 mM Gln, [34], [35]). Under aerobic conditions, metastatic 4T1 cells perfused with medium containing 2 mM glutamine (Figure 1, B and C; 1-6 hours) responded to full glutamine deprivation by significantly reducing their glycolytic lactate synthesis (2.3-fold) to levels similar to those observed in 67NR cells under the same conditions (Figure 7, A and B; 1-6 hours). In comparison, the 67NR cells did not significantly alter their lactate synthesis rate with glutamine deprivation under comparable conditions. This differential response to glutamine deprivation is likely related to its importance as a major anaplerotic precursor of the TCA cycle together with higher TCA cycle function in 4T1 cells compared with 67NR cells.
Figure 7.
Cell perfusion stress studies with 1-13C-glucose as detailed in Table 1 (Study C). Normalized average time-course changes (A) and labeling rates at different time intervals (B) of lactate C3, glutamate C4, and succinate C2 as determined from 13C MR spectra. In B, the rates in the gray boxes are from the experiments shown in Figure 1C and displayed again for comparison. (C) The values obtained from experiments C-2, A-2, and A-3 (detailed in Table 1) and displayed in A (1-6 hours, 0 mM Gln) and Figure 1C (1-6 hours, 2 mM Gln; and 6-11 hours, 6 mM Gln) were fitted according to the Michaelis-Menten asymptotic model. Because the cellular washout of glutamine is slow (data not shown), when changing from 6 mM to 0 mM glutamine-containing perfusion media, a 5% residual level of glutamine (0.3 mM) was estimated in the cell microenvironment at the beginning of the study. Error bars indicate the standard error of the means.
Based on our experimental data and our proposed metabolic model (Figure 4), glutamine deprivation would have a higher impact in replenishing the OAA pool in 4T1 cells than in 67NR cells. As a result, glutamine-deprived 4T1 cells would become more dependent on glucose-derived carbon (pyruvate) entering the TCA cycle and would establish a metabolic phenotype that is closer to the basal metabolic phenotype of 67NR cells. A corresponding reduction of the synthesis rate of lactate would be expected, as we observed (Figure 7, A and B). However, full glutamine deprivation under aerobic conditions reduced TCA cycle flux ~ 2.3-fold in both cell lines, as evidenced by the significant decrease of glucose-derived glutamate (Figure 7, A and B; 1-6 hours, 0 mM Gln). Our results show that the synthesis rate of glucose-derived lactate is higher in 4T1 cells (Vmax = 30.26 ± 0.12) than in 67NR cells (Vmax = 18.64 ± 0.47) and more dependent on glutamine in the former (Km = 0.63 ± 0.01) than in the latter (Km = 0.16 ± 0.03) cells. Thus, the aerobic synthesis rate of lactate from glucose [Vm(4T1/67NR) ~ 1.6] requires approximately a four-times higher glutamine concentration in the medium for metastatic 4T1 cells than for nonmetastatic 67NR cells [Km(4T1/67NR) ~ 4] (Figure 7C). This effect is also apparent for the entire panel of isogenic cell lines in 2D cell culture (Figure 8A), as increased glucose-derived lactate synthesis rates were associated with higher metastatic ability and stronger dependency on glutamine levels.
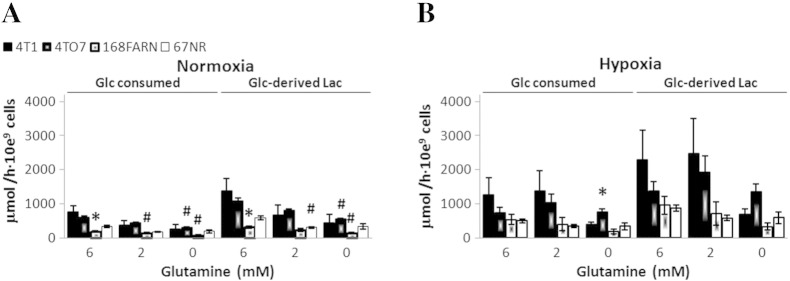
Figure 8.
Absolute rates of 1-13C-glucose consumption and de novo 1-13C-glucose–derived lactate synthesis in breast cancer cells of different metastatic potential and under different stresses. Each cell line was incubated with 1-13C-glucose for 5 hours, either in 20% O2 (A) or in 1% O2 (B), and TCM2D samples were collected for high-resolution 13C-MR studies. Extracellular 1-13C-glucose consumption and 1-13C-glucose–derived lactate synthesis rates were averaged over three independent experiments for each condition. # Statistically significant change with respect to same cell line in medium containing 6 mM Gln.
Interchangeability of Glycolysis and OXPHOS in 4T1 cells
When 4T1 and 67NR cells are fully deprived of glutamine and subsequently exposed to hypoxia, the glycolytic lactate synthesis rate increased by ~ 1.8-fold in both cell lines (Figure 7, A and B; 6-11 hours), although this increase was significant only in 67NR cells. This observation is expected because 1) hypoxia is a driver of increased glucose uptake and glycolysis [36] and 2) less glucose is being consumed by the TCA cycle under hypoxic conditions, as shown by the low levels of de novo glutamate synthesis (Figure 7, A and B; 6-11 hours). After replenishing glutamine-starved, hypoxic 4T1 cells with glutamine (2 mM), glycolytic synthesis of lactate significantly increased 1.9-fold (P = .011, paired t test), whereas it remained unchanged in 67NR cells exposed to the same nutrient conditions (Figure 7, A and B; 11-16 hours). Although the glutamine-containing medium used in these studies contained pyruvate (1 mM), similar changes in the synthesis rates of lactate and glutamate were obtained for cells in pyruvate-free medium under the same environment stress conditions studied above (results not shown). Furthermore, MR analysis of TCM2D samples from the four isogenic cell lines grown in hypoxia also indicates a higher lactate synthesis rate in more metastatic cell lines with higher glutamine availability (Figure 8B).
In the last stage of the cell perfusion experiments, shown in Figure 7, A and B, reoxygenation (16-21 hours) of the nutrient-replenished 4T1 cells significantly decreases (2.5-fold) the synthesis rate of glucose-derived lactate, whereas no change was observed in 67NR cells. During this period, the synthesis rate of glucose-derived glutamate was 2.4-fold higher in 4T1 cells than in 67 NR cells, consistent with a stronger reactivation of TCA cycle flux in 4T1 cells following the reintroduction of oxygen and glutamine. Thus, less glucose transitioned to lactate in 4T1 cells, and de novo succinate from glucose accumulated in 67NR cells (but not in 4T1 cells) (Figure 7, A and B). These observations agree with the results of the initial cell perfusion experiments performed under standard growth conditions (Figure 1).
No significant differences in Pii, Pie, or pHi were detected between 4T1 and 67NR cells during the periods of full glutamine deprivation and hypoxia (Figure S6, 1-11 hours). Whereas significant differences in extracellular pH were detected during initial glutamine deprivation (Figure S6, 1-6 hours), the time-course changes in both cell lines followed the same trend. During these periods, the nonmetastatic 67NR cells consistently accumulated more PCho, decreased GPCho/PCho ratios, and had significantly higher levels of β-NTP than 4T1 cells (Figure S6, Table S1). These results suggest that, compared with 4T1 cells, the energy metabolism of 67NR cells is less sensitive to short-term glutamine deprivation and hypoxic stress because they rely less on OXPHOS than 4T1 cells. The slight decrease of β-NTP levels detected in 4T1 cells (compared with initial values; Table S1, Study C) could also reflect an attempt of the metastatic phenotype to maintain a high glycolytic flux by removing ATP-induced inhibition on phosphofructokinase [37].
Figure S6.
Bioenergetic profile, pH, and phospholipid metabolism of 4T1 and 67NR cells during the cell perfusion stress studies C, detailed in Table 1. Time-course changes of β-NTP, intracellular (i) and extracellular (e) Pi and pH, PCho, GPCho, and GPCho/PCho for the studies with 1-13C-glucose (corresponding to 13C MR studies in Figure 7). The different stages detailed in Table 1 are indicated in each plot and separated by dashed lines. Error bars indicate the standard error of the means. *Statistically significant differences between 4T1 and 67NR cells during the same time interval (P < .05, unpaired t test). Statistically significant changes (P < .05, paired t test) between different time points for the same cell line are shown in Table S1 (study C, with 1-13C-glucose).
Discussion
We investigated the dynamic flux of glucose through glycolysis and the TCA cycle, and the flux of glutamine through the TCA cycle. Experiments were performed on a panel of isogenic murine cell lines derived from the same spontaneous breast tumor, focusing mostly on the highly metastatic (4T1) and nonmetastatic (67NR) variants [16]. We found significant differences in metabolism between these two cell lines. The more aggressive cell line (4T1) had higher glycolytic, OXPHOS, and TCA cycle activity than 67NR cells to meet their energy and anabolic requirements for rapid proliferation and the formation of metastases. 4T1 cells adapted more readily than 67NR cells to a changing microenvironment. This is accomplished by a greater ability to regulate glucose metabolism through glycolysis and/or oxidative metabolism. This “plasticity” of 4T1 cells is also supported by our previous experiments [17], [23]. For instance, 4T1 cells, which have higher basal levels of LDH-A expression than 67NR cells [17], significantly increased their OCR in response to LDH-A knockdown and reduced aerobic glycolysis [23].
Further, we identified a restriction of TCA cycle flux in 67NR cells but not in 4T1 cells. This was found to be due to lower expression of SDH in 67NR in comparison to 4T1 cells. Thus, 67NR cells accumulated significantly more succinate and displayed significantly lower OXPHOS activity than 4T1 cells under standard growth conditions. These data agree well with the study by Lu et al. [21], which used targeted liquid chromatography–mass spectrometry to assess the metabolomic profile of a panel of nonmetastatic and metastatic murine breast cancer cell lines, including 67NR and 4T1. Under standard tissue culture conditions, they also reported higher succinate levels in 67NR than in 4T1 cells (Table S2, [21]). The accumulation of succinate has also been associated with a “pseudohypoxic” cellular microenvironment that stabilizes HIF-1α [38]. Stabilization of HIF-1α is associated with upregulation of angiogenesis-related proteins, e.g., VEGF [39]. This would agree with our previous observations [17] and others [40], demonstrating that 1) 67NR tumors have better vascular perfusion than 4T1 tumors at comparable tumor sizes and 2) 67NR cells are better adapted to growing in hypoxia [17]. Although SDH has been proposed as a tumor suppressor gene [38], no evidence of its epigenetic silencing has been found in clinical breast cancers [41]. Thus, the presence of a fully functional TCA cycle (including SDH and OXPHOS) may be a hallmark of metastatic breast cancer.
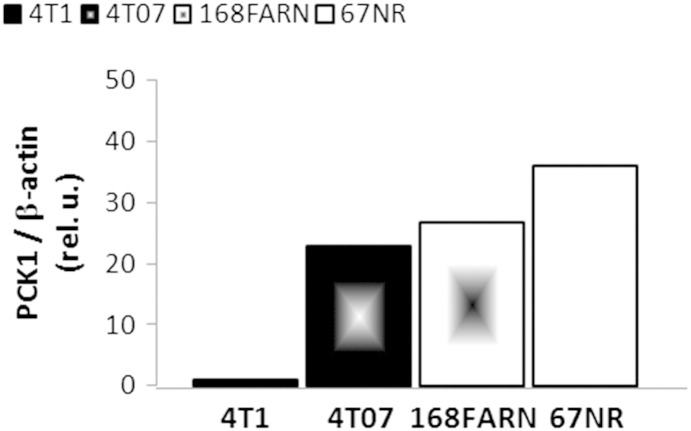
Our results suggest lower SDH-A expression in less aggressive cell lines either in replete environmental conditions or under stress (glutamine deprivation and hypoxia). This agrees with the study by Kim et al. showing an association between low SDH expression and low-grade histology in a panel of 721 breast cancers [42]. With relatively less efficient mitochondrial OXPHOS activity, less aggressive cell lines could potentially reduce their TCA cycle by channeling glucose into other, more efficient pathways. This would also agree with data from our group indicating upregulation of phosphoenolpyruvate carboxykinase (PCK1) aligning with decreased metastatic ability (67NR > 168FAR > 4T07 >> 4T1, Figure S7). Together with our estimates of relatively lower pyruvate carboxylase flux in 4T1 cells, this would suggest lower gluconeogenic potential in more aggressive breast cancer phenotypes, in agreement with the loss of fructose-1,6-bisphosphatase in basal-like breast cancer [43]. Moreover, higher relative levels of SDH-A in hypoxic 4T1 cells could be a response to the increased levels of glutamine-derived succinate (reductive carboxylation pathway [44]), whereas SDH-A decreases in hypoxic 67NR cells. Therefore, although more adaptable to changes in their microenvironment, more aggressive cell lines may preserve a relatively higher basal potential for aerobic metabolism under stress, enabling faster adaptation to new microenvironments.
Figure S7.
Protein expression of PCK1 in four isogenic cell lines: 4T1, 4 T07, 168FARN, and 67NR. 105 cells were seeded on six-well plates and grown for 3 days, at which point they were harvested and counted. Cells were then used to assess the expression of PCK1 (monoclonal antibody #12940; Cell Signaling Technology, Danvers, MA) following the same Western blot protocol described in the “Methods” section for SDH-A. The preliminary results are from a single experiment.
As summarized in Table S2, recent data support an association between mitochondrial function and metastatic potential in breast cancer, and an association with a short metastasis-free survival [45]. The ability to effectively use OXPHOS in addition to glycolysis has been demonstrated in aggressive breast cancer cell lines [46], [47], [48] and proposed as a key adaptation for developing brain metastasis [19]. A recent model of tumor metabolism also suggested that stromal cells fuel breast cancer cell growth and the development of metastasis through mitochondrial metabolism [20]. Lu et al. showed that TCA cycle intermediates increase with metastatic potency in isogenic 67NR, 168FARN, 4T07, and 4T1 cells [21]. In our studies with the same panel of cell lines, we found this to be related to higher OXPHOS, indicating that higher metastatic potential is associated with less reliance on aerobic glycolysis alone. The higher utilization of OXPHOS in primary 4T1 tumors may contribute to the development of necrosis due to outgrow of their vascular oxygen supply [17]. Our previous attempts to inhibit 4T1 cancer growth and metastasis focused on LDH-A knockdown alone [23]. These new observations suggest that mitochondrial metabolism (OXPHOS) is also a potential target for treatment in this breast cancer model. Thus, combined metabolic inhibition of both the glycolytic and OXPHOS pathways may prove to be more effective in the treatment of metastatic breast cancer.
Glutamine is an essential nitrogen source for the synthesis of nucleotides, amino acids, and ATP, and a major anaplerotic precursor for the TCA cycle [27]. Glutamine addiction is frequent in cancer cells [49] and has been linked to Myc overexpression [33]. Previous work has shown that Myc expression is 2.3-fold higher in 4T1 than in 67NR cells [50]. Singh et al. observed in a panel of aggressive, glutamine-addicted breast cancer cell lines (including 4T1) a correlation between glutamine addiction and metastatic ability [51]. More recently, glutamine dependency was associated with higher invasion and metabolic “rewiring” in ovarian cancer cell lines; specifically, glutamine downregulated glycolysis (glucose uptake and lactate secretion) in highly invasive cancer cells but not in cancer cells with low invasive potential [52]. Similarly, we observed that lactate production by aerobic glycolysis (Warburg effect) is four times more glutamine dependent in 4T1 cells than in 67NR cells. However, in our study, glutamine stimulated the synthesis of glucose-derived lactate according to metastatic potential. Compared with the studies by Yang et al. [52], this could indicate 1) basal metabolic differences between the two cancer cell types (ovarian and breast) and/or 2) different effects of glutamine during the metabolic reprogramming toward more invasive or metastatic phenotypes. Because lactate is a major driver of invasion and dissemination [14], [15], our data suggest that glutamine deprivation might prevent production of lactate and inhibit/delay breast cancer metastases. Glutamine was found to recycle into OAA only in 4T1 cells (as shown by the labeling of aspartate from 3-13C-glutamine studies), whereas nonmetastatic 67NR cells are more dependent on glucose as an anaplerotic precursor of the TCA cycle via pyruvate carboxylase. Accordingly, the flux analysis of our cell perfusion data (in high-glutamine–containing medium) predicted an exchange at the level of pyruvate carboxylase only in 67NR cells and not in 4T1 cells. This also explains the marked dependency of glycolytic lactate synthesis on glutamine observed in 4T1 cells but not in 67NR cells. To sustain their higher ATP demands by OXPHOS, glutamine-deprived 4T1 cells must rely more on glucose to replenish the OAA pool of the TCA cycle (via pyruvate carboxylase). This is accomplished by increasing the amount (fraction) of glucose used for de novo synthesis of citrate. Both cell lines ultimately respond to full glutamine deprivation by reducing the TCA cycle flux and decreasing their lactate synthesis rates to similar levels. This observation agrees with the results reported by Richardson et al. in a panel of transformed MCF-10 breast cancer cells grown in glutamine-free medium [53], which showed no significant differences in lactate excretion rates with increasing malignancy and metastatic potential. Thus, limiting OXPHOS by targeting glutamine metabolism [28] may provide an additional therapeutic approach for metastases-prone or metastatic breast cancer, although other modulators of cellular adaptation should be considered, such as asparagine [54].
Conclusions
Whereas most previous investigations have focused on identifying gene signatures associated with the pathophysiology of metastases [55], our results suggest that the noninvasive evaluation of a tumor’s metabolic phenotype can provide useful information related to the metastatic propensity of the tumor. We show that 4T1 metastatic breast cancer is associated with metabolic plasticity in response to changing tumor cell microenvironments. Specifically, these cells use OXPHOS efficiently during optimal growth conditions (whereas nonmetastatic 67NR cells do not) and reversibly shut down OXPHOS during transient hypoxia and/or glutamine deprivation. These transitions were dynamically quantified for the first time with tumor cell perfusion experiments that closely mimic the in vivo tumor microenvironment. The observed metabolic differences and the ability to assess adaptability in living cells provide the opportunity to identify new biomarkers for early detection of the metastatic phenotype, possibly at the time of diagnosis. Metabolic therapies directed at inhibiting both the glycolytic and OXPHOS pathways are also suggested by these studies; such combination therapies may be more effective in the treatment of aggressive metastatic breast cancer.
The following are the supplementary data related to this article.
Supplementary Material.
Acknowledgements
The authors thank Anthony Mancuso for his advice with the metabolite assignment of selected MR peaks. This work was supported by National Institutes of Health grants R01 CA172846, P01 CA115675, NCI P30 CA008748 (Cancer Center Support Grant), R01 CA193256, and P01 CA094060 and the Breast Cancer Molecular Imaging Fund.
Footnotes
This work was supported by National Institutes of Health grants R01 CA172846, P01 CA115675, NCI P30 CA008748 (Cancer Center Support Grant), P01 CA115675 and P01 CA094060 and the Breast Cancer Molecular Imaging Fund.
References
- 1.Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220–241. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 2.Chiang AC, Massague J. Molecular basis of metastasis. N Engl J Med. 2008;359:2814–2823. doi: 10.1056/NEJMra0805239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fidler IJ, Kripke ML. Metastasis results from preexisting variant cells within a malignant tumor. Science. 1977;197:893–895. doi: 10.1126/science.887927. [DOI] [PubMed] [Google Scholar]
- 4.Bianchini G, Qi Y, Alvarez RH, Iwamoto T, Coutant C, Ibrahim NK, Valero V, Cristofanilli M, Green MC, Radvanyi L. Molecular anatomy of breast cancer stroma and its prognostic value in estrogen receptor-positive and -negative cancers. J Clin Oncol. 2010;28:4316–4323. doi: 10.1200/JCO.2009.27.2419. [DOI] [PubMed] [Google Scholar]
- 5.Rattigan YI, Patel BB, Ackerstaff E, Sukenick G, Koutcher JA, Glod JW, Banerjee D. Lactate is a mediator of metabolic cooperation between stromal carcinoma associated fibroblasts and glycolytic tumor cells in the tumor microenvironment. Exp Cell Res. 2012;318:326–335. doi: 10.1016/j.yexcr.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gatenby RA, Smallbone K, Maini PK, Rose F, Averill J, Nagle RB, Worrall L, Gillies RJ. Cellular adaptations to hypoxia and acidosis during somatic evolution of breast cancer. Br J Cancer. 2007;97:646–653. doi: 10.1038/sj.bjc.6603922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaupel P, Kallinowski F, Okunieff P. Blood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: a review. Cancer Res. 1989;49:6449–6465. [PubMed] [Google Scholar]
- 8.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 11.Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyon RC, Cohen JS, Faustino PJ, Megnin F, Myers CE. Glucose metabolism in drug-sensitive and drug-resistant human breast cancer cells monitored by magnetic resonance spectroscopy. Cancer Res. 1988;48:870–877. [PubMed] [Google Scholar]
- 13.Bohndiek SE, Brindle KM. Imaging and 'omic' methods for the molecular diagnosis of cancer. Expert Rev Mol Diagn. 2010;10:417–434. doi: 10.1586/erm.10.20. [DOI] [PubMed] [Google Scholar]
- 14.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 15.Webb BA, Chimenti M, Jacobson MP, Barber DL. Dysregulated pH: a perfect storm for cancer progression. Nat Rev Cancer. 2011;11:671–677. doi: 10.1038/nrc3110. [DOI] [PubMed] [Google Scholar]
- 16.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52:1399–1405. [PubMed] [Google Scholar]
- 17.Serganova I, Rizwan A, Ni X, Thakur SB, Vider J, Russell J, Blasberg R, Koutcher JA. Metabolic imaging: a link between lactate dehydrogenase A, lactate, and tumor phenotype. Clin Cancer Res. 2011;17:6250–6261. doi: 10.1158/1078-0432.CCR-11-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jose C, Bellance N, Rossignol R. Choosing between glycolysis and oxidative phosphorylation: a tumor's dilemma? Biochim Biophys Acta. 2011;1807:552–561. doi: 10.1016/j.bbabio.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 19.Chen EI, Hewel J, Krueger JS, Tiraby C, Weber MR, Kralli A, Becker K, Yates JR, III, Felding-Habermann B. Adaptation of energy metabolism in breast cancer brain metastases. Cancer Res. 2007;67:1472–1486. doi: 10.1158/0008-5472.CAN-06-3137. [DOI] [PubMed] [Google Scholar]
- 20.Bonuccelli G, Tsirigos A, Whitaker-Menezes D, Pavlides S, Pestell RG, Chiavarina B, Frank PG, Flomenberg N, Howell A, Martinez-Outschoorn UE. Ketones and lactate "fuel" tumor growth and metastasis: evidence that epithelial cancer cells use oxidative mitochondrial metabolism. Cell Cycle. 2010;9:3506–3514. doi: 10.4161/cc.9.17.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu X, Bennet B, Mu E, Rabinowitz J, Kang Y. Metabolomic changes accompanying transformation and acquisition of metastatic potential in a syngeneic mouse mammary tumor model. J Biol Chem. 2010;285:9317–9321. doi: 10.1074/jbc.C110.104448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Invernizzi F, D'Amato I, Jensen PB, Ravaglia S, Zeviani M, Tiranti V. Microscale oxygraphy reveals OXPHOS impairment in MRC mutant cells. Mitochondrion. 2012;12:328–335. doi: 10.1016/j.mito.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rizwan A, Serganova I, Khanin R, Karabeber H, Ni X, Thakur SB, Zakian KL, Blasberg RG, Koutcher JA. Relationships between LDH-A, lactate and metastases in 4T1 breast tumors. Clin Cancer Res. 2013;19:5158–5169. doi: 10.1158/1078-0432.CCR-12-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mancuso A, Zhu A, Beardsley NJ, Glickson JD, Wehrli S, Pickup S. Artificial tumor model suitable for monitoring 31P and 13C NMR spectroscopic changes during chemotherapy-induced apoptosis in human glioma cells. Magn Reson Med. 2005;54:67–78. doi: 10.1002/mrm.20545. [DOI] [PubMed] [Google Scholar]
- 25.Wehrle JP, Ng CE, McGovern KA, Aiken NR, Shungu DC, Chance EM, Glickson JD. Metabolism of alternative substrates and the bioenergetic status of EMT6 tumor cell spheroids. NMR Biomed. 2000;13:349–360. doi: 10.1002/1099-1492(200010)13:6<349::aid-nbm652>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 26.Smirnova T, Zhou ZN, Flinn RJ, Wyckoff J, Boimel PJ, Pozzuto M, Coniglio SJ, Backer JM, Bresnick AR, Condeelis JS. Phosphoinositide 3-kinase signaling is critical for ErbB3-driven breast cancer cell motility and metastasis. Oncogene. 2012;31:706–715. doi: 10.1038/onc.2011.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Owen OE, Kalhan SC, Hanson RW. The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem. 2002;277:30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- 30.Henry PG, Adriany G, Deelchand D, Gruetter R, Marjanska M, Oz G, Seaquist ER, Shestov A, Ugurbil K. In vivo13C NMR spectroscopy and metabolic modeling in the brain: a practical perspective. Magn Reson Imaging. 2006;24:527–539. doi: 10.1016/j.mri.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 31.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, DeBerardinis RJ. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang H. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15:110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scaglia F, Carter S, O'Brien WE, Lee B. Effect of alternative pathway therapy on branched chain amino acid metabolism in urea cycle disorder patients. Mol Genet Metab. 2004;81(Suppl. 1):S79–S85. doi: 10.1016/j.ymgme.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 35.Braulio VB, Ten Have GA, Vissers YL, Deutz NE. Time course of nitric oxide production after endotoxin challenge in mice. Am J Physiol Endocrinol Metab. 2004;287:E912–E918. doi: 10.1152/ajpendo.00540.2003. [DOI] [PubMed] [Google Scholar]
- 36.Semenza GL. Regulation of cancer cell metabolism by hypoxia-inducible factor 1. Semin Cancer Biol. 2009;19:12–16. doi: 10.1016/j.semcancer.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 37.Golinska M, Troy H, Chung YL, McSheehy PM, Mayr M, Yin X, Ly L, Williams KJ, Airley RE, Harris AL. Adaptation to HIF-1 deficiency by upregulation of the AMP/ATP ratio and phosphofructokinase activation in hepatomas. BMC Cancer. 2011;11:198. doi: 10.1186/1471-2407-11-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 39.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 40.Lou Y, McDonald PC, Oloumi A, Chia S, Ostlund C, Ahmadi A, Kyle A, Auf dem Keller U, Leung S, Huntsman D. Targeting tumor hypoxia: suppression of breast tumor growth and metastasis by novel carbonic anhydrase IX inhibitors. Cancer Res. 2011;71:3364–3376. doi: 10.1158/0008-5472.CAN-10-4261. [DOI] [PubMed] [Google Scholar]
- 41.Huang KT, Dobrovic A, Fox SB. No evidence for promoter region methylation of the succinate dehydrogenase and fumarate hydratase tumour suppressor genes in breast cancer. BMC Res Notes. 2009;2:194. doi: 10.1186/1756-0500-2-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim S, Kim do H, Jung WH, Koo JS. Succinate dehydrogenase expression in breast cancer. Springerplus. 2013;2:299. doi: 10.1186/2193-1801-2-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong C, Yuan T, Wu Y, Wang Y, Fan TW, Miriyala S, Lin Y, Yao J, Shi J, Kang T. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell. 2013;23:316–331. doi: 10.1016/j.ccr.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mullen AR, Hu Z, Shi X, Jiang L, Boroughs LK, Kovacs Z, Boriack R, Rakheja D, Sullivan LB, Linehan WM. Oxidation of alpha-ketoglutarate is required for reductive carboxylation in cancer cells with mitochondrial defects. Cell Rep. 2014;7:1679–1690. doi: 10.1016/j.celrep.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitaker-Menezes D, Martinez-Outschoorn UE, Flomenberg N, Birbe RC, Witkiewicz AK, Howell A, Pavlides S, Tsirigos A, Ertel A, Pestell RG. Hyperactivation of oxidative mitochondrial metabolism in epithelial cancer cells in situ: visualizing the therapeutic effects of metformin in tumor tissue. Cell Cycle. 2011;10:4047–4064. doi: 10.4161/cc.10.23.18151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guppy M, Leedman P, Zu X, Russell V. Contribution by different fuels and metabolic pathways to the total ATP turnover of proliferating MCF-7 breast cancer cells. Biochem J. 2002;364:309–315. doi: 10.1042/bj3640309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meadows AL, Kong B, Berdichevsky M, Roy S, Rosiva R, Blanch HW, Clark DS. Metabolic and morphological differences between rapidly proliferating cancerous and normal breast epithelial cells. Biotechnol Prog. 2008;24:334–341. doi: 10.1021/bp070301d. [DOI] [PubMed] [Google Scholar]
- 48.Fogal V, Richardson AD, Karmali PP, Scheffler IE, Smith JW, Ruoslahti E. Mitochondrial p32 protein is a critical regulator of tumor metabolism via maintenance of oxidative phosphorylation. Mol Cell Biol. 2010;30:1303–1318. doi: 10.1128/MCB.01101-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tao K, Fang M, Alroy J, Sahagian GG. Imagable 4T1 model for the study of late stage breast cancer. BMC Cancer. 2008;8:228. doi: 10.1186/1471-2407-8-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh B, Tai K, Madan S, Raythatha MR, Cady AM, Braunlin M, Irving LR, Bajaj A, Lucci A. Selection of metastatic breast cancer cells based on adaptability of their metabolic state. PLoS One. 2012;7:e36510. doi: 10.1371/journal.pone.0036510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang L, Moss T, Mangala LS, Marini J, Zhao H, Wahlig S, Armaiz-Pena G, Jiang D, Achreja A, Win J. Metabolic shifts toward glutamine regulate tumor growth, invasion and bioenergetics in ovarian cancer. Mol Syst Biol. 2014;10:728. doi: 10.1002/msb.20134892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richardson AD, Yang C, Osterman A, Smith JW. Central carbon metabolism in the progression of mammary carcinoma. Breast Cancer Res Treat. 2008;110:297–307. doi: 10.1007/s10549-007-9732-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang J, Fan J, Venneti S, Cross JR, Takagi T, Bhinder B, Djaballah H, Kanai M, Cheng EH, Judkins AR. Asparagine plays a critical role in regulating cellular adaptation to glutamine depletion. Mol Cell. 2014;56:205–218. doi: 10.1016/j.molcel.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fokas E, Engenhart-Cabillic R, Daniilidis K, Rose F, An HX. Metastasis: the seed and soil theory gains identity. Cancer Metastasis Rev. 2007;26:705–715. doi: 10.1007/s10555-007-9088-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material.