Abstract
Plant resistance proteins mediate pathogen recognition and activate innate immune responses to restrict pathogen proliferation. One common feature of these proteins is an NB-ARC domain. In this study, we characterized a gene encoding a protein with an NB-ARC domain from wild Chinese grapevine Vitis pseudoreticulata accession “Baihe-35-1,” which was identified in a transcriptome analysis of the leaves following inoculation with Erysiphe necator (Schw.), a causal agent of powdery mildew. Transcript levels of this gene, designated VpCN (GenBank accession number KT265084), increased strongly after challenge of grapevine leaves with E. necator. The deduced amino acid sequence was predicted to contain an NB-ARC domain in the C-terminus and an RxCC-like domain similar to CC domain of Rx protein in the N-terminus. Ectopic expression of VpCN in Arabidopsis thaliana resulted in either a wild-type phenotype or a dwarf phenotype. The phenotypically normal transgenic A. thaliana showed enhance resistance to A. thaliana powdery mildew Golovinomyces cichoracearum, as well as to a virulent bacterial pathogen Pseudomonas syringae pv. tomato DC3000. Moreover, promoter::GUS (β-glucuronidase) analysis revealed that powdery mildew infection induced the promoter activity of VpCN in grapevine leaves. Finally, a promoter deletion analysis showed that TC rich repeat elements likely play an important role in the response to E. necator infection. Taken together, our results suggest that VpCN contribute to powdery mildew disease resistant in grapevine.
Keywords: wild Chinese Vitis, VpCN, disease resistance, powdery mildew, promoter analysis
Introduction
Plants have evolved multiple mechanisms to protect themselves against pathogens (Jones and Dangl, 2006). The first line of defense is microbe-associated molecular pattern (MAMP)-triggered immunity (MTI) following MAMP perception by membrane-resident pattern recognition receptors (Maekawa et al., 2011). MTI is thought to limit the growth of invasive pathogens. The second line of defense is plant innate immunity, which is activated by the specific recognition of pathogen-derived effectors by intracellular host resistance (R) proteins, and is termed effector-triggered immunity (ETI) (Chisholm et al., 2006). ETI typically leads to a hypersensitive response (HR) and gives rise to a faster and stronger defensive response than MTI-triggered immunity (Cesari et al., 2013). Understanding the function of R proteins, and the mechanisms by which they recognize pathogen effectors, can potentially lead to the development of a long-term strategy for the control and prevention of pathogen invasion.
Over the past few decades, numerous R genes have been cloned from model plants and important crops (Pan et al., 2000b; Collier and Moffett, 2009; Sekine et al., 2012). Most R proteins contain a nucleotide binding (NB) domain and a C-terminal leucine-rich repeat (LRR) domain, and belong to the so-called NB-LRR protein family (Ooijen et al., 2008). The most conserved domain in NB-LRR proteins is an NB domain that is found in proteins such as human Apaf-1, plant R proteins and Caenorhabditis elegans Ced-4 (ARC), and as such is referred to as the NB-ARC domain (Ooijen et al., 2008; van der Biezen and Jones, 1998). As a consequence of determining its three-dimensional structure, Albrecht and Takken (2006) proposed that the NB-ARC domain can be further divided into three sub-domains (NB, ARC1, and ARC2). Several conserved motifs have been identified thoughtout the NB-ARC domain in R proteins, such as Walker B, GxP, hhGRExE, Walker A or P-loop, MHD, and RNBS-A–D (Meyers et al., 1999; Pan et al., 2000a; Ooijen et al., 2008). Crystal structure analysis of the NB-ARC domain has led to the suggestion that it may function as a molecular switch to regulate signaling pathways through conformational changes (Riedl et al., 2005; Takken et al., 2006). It has also been shown that the nucleotide binding of the NB-ARC domain in the R proteins, I-2, and Mi-1, requires a P loop, since a P-loop mutant abolished the binding capacity (Tameling et al., 2010). Likewise, the oligomerization of an NB-ARC-LRR protein in the presence of its elicitor requires an intact P-loop in the NB-ARC domain (Mestre and Baulcombe, 2006).
Plant NB-LRR proteins can be divided into two distinct classes: the TNL and the CNL type, based on the domains present at their N terminus. Those that possess a Toll and human interleukin-1 receptor (TIR) domain are referred to as TIR-NB-ARC-LRR or TNL proteins, while those carrying a predicted coiled-coil (CC) domain are classified as CC-NB-ARC-LRR, or CNL proteins (Pan et al., 2000a; Lukasik-Shreepaathy et al., 2012). The potato (Solanum tuberosum) Rx protein is a typical CC-NB-ARC-LRR protein mediates resistance to potato virus X (PVX)(Kohm et al., 1993; Bendahmane et al., 1999), the CC domain of RX protein has a four bundle structure and forms a heterodimer with RanGAP2 WPP domain (Hao et al., 2013). The N-termini of the CC and TIR domains are thought to mediate downstream immune responses. It has been reported that in CNL proteins, the CC domain of NRG1 is capable of independently inducing defense responses (Collier et al., 2011), and in TIR proteins the TIR domain plays a crucial role in the cell death signaling pathway (Zhang et al., 2004; Weaver et al., 2006).
The identification and functional characterization of NB-ARC domain R proteins is of considerable interest in developing novel sources of disease resistance in crop plants that are threatened by phytopathogens. For example, Erysiphe necator is a fungus that causes powdery mildew (PM) disease in grapevine worldwide, resulting in serious losses in both grape yield and quality. The most economically important cultivated grapevine is V. vinifera, which is highly susceptible to PM (Gadoury et al., 2012). To combat the pathogen, fungicides are widely used, which causes environmental and financial pressure on grape growers and reduces wine quality. Thus, developing new grape cultivars with enhanced disease resistance mechanisms is of considerable interest. The wild Chinese Vitis, “Baihe-35-1,” is an accession of wild Chinese V. pseudoreticulata W. T. Wang that possesses high resistance to multiple fungi, and particularly to E. necator (Wang et al., 1995; Lin et al., 2006; Yu et al., 2011). To elucidate the resistance mechanisms involved in the defense response to fungal infection in this species, we previously performed an RNA-seq based transcriptome analysis V. pseudoreticulata “Baihe-35-1” that had been inoculated with E. necator (Weng et al., 2014). Among the pathogen induced genes, one was predicted to encode an NB-ARC domain protein.
In this current study, we report the isolation of the full length cDNA of this gene, which we designated VpCN, and its functional characterization following ectopic expression in Arabidopsis thaliana. Conclusions regarding its role in conferring Chinese Wild V. pseudoreticulata “Baihe-35-1” with disease resistance to powdery mildew are presented.
Materials and methods
Plant materials and growth conditions
Grapevines (Chinese wild V. pseudoreticulata accession Baihe-35-1 and V. vinifera cv. “Red globe”) were maintained in the grape germplasm resources orchard, Northwest A&F University, Yangling Shaanxi, China. A. thaliana (ecotype type, Columbia-0) was grown in a growth chamber under the following conditions: 22°C, 50% humidity, a 16/8 h day/night intensity of 125 μmolm−2 s−1 provided by cool white fluorescent bulbs.
Cloning and sequence analysis
Total RNA was extracted from grapevine as previously described (Zhang et al., 2003). First strand cDNA was synthesized from 1 μg of total RNA with the PrimerScript™ II 1st Strand cDNA Synthesis kit (TaKaRa Bio Inc., Dalian, China), according to the manufacturer's instructions. LA Taq (Takara Bio. Inc.) was used to amplify the ORF sequence of VpCN. The PCR products were cloned into the T-easy vector (Promega, USA), sequenced (Beijing Genomics Institute, Beijing, China) and submitted to GenBank (accession number KT265084). The VpCN cDNA sequence was analyzed using BLAST (http://Ncbi.nlm.Nih.gov/blast) in the NCBI database. Grapevine DNA extraction was conducted as previously described (Yu et al., 2013), primers for amplify promoter sequence were designed according to acquired sequence from Grape Genome Database (12 ×; http://www.genoscope.cns.fr), after cloning into the T-easy vector and sequencing, the promoter sequence was analyzed using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002). The deduced amino acid sequence of VpCN was aligned with closely related proteins and a phylogenetic tree was generated using neighbor joining algorithm with 1000 bootstrapping with the ClustalW tool in the MegAlign program (Version 5.07, DNASTAR Inc.) (Figure 1D). A structural model of the NB-ARC domain of VpCN was constructed using the structure of PDB 4m9x.1.C (Huang et al., 2013) in SWWISS-MODEL (Figure 1C). Real time PCR was conducted using SYBR@Premix EX Taq™II (Tli RNaseH Plus) (Takara Bio. Inc.) in a 20 μl volume reaction following the manufacturer's instructions using the CFX96TM real-time system (Bio-Rad, Hercules, CA, USA). The amplification cycles were as follows: initial denaturation at 94°C for 30 s, 40 cycles at 95°C 5 s, 60°C for 30 s. For melting curve analysis: 40 cycles at 95°C for 15 s followed by a constant increase from 60–95°C. The grapevine Actin 1 (GenBank Accession number AY680701) was used as reference gene.
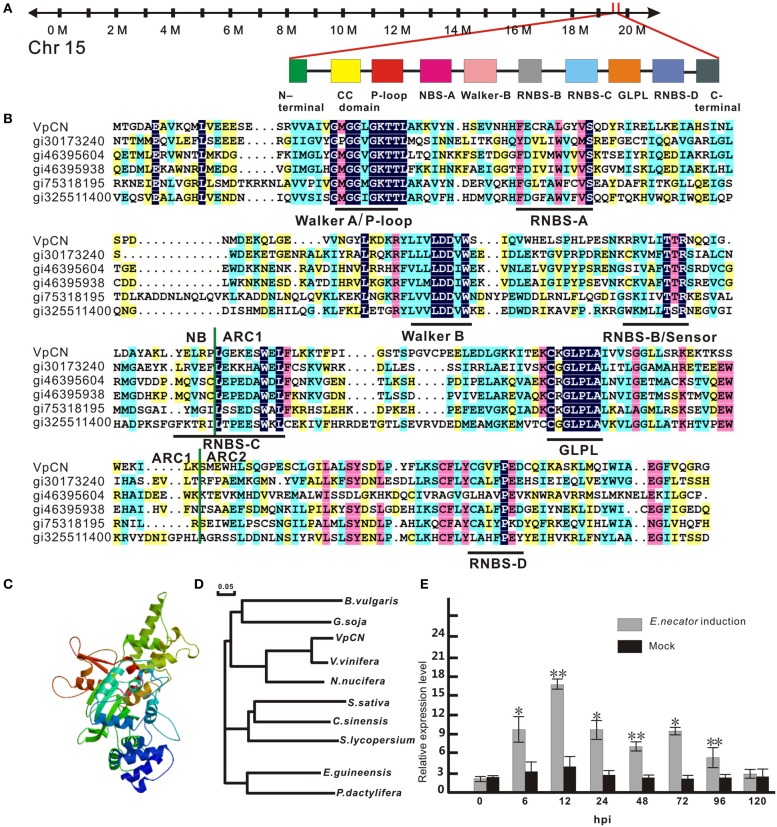
Figure 1.
Sequence analysis of VpCN and transcript level detection. (A) Schematic map of VpCN location and major motifs. (B) Multiple sequence alignment of the NB, ARC1 and ARC2 subdomains of NB-ARC in VpCN with closely related proteins. Domain borders are indicate as vertical green lines. Motifs are labeled by horizontal dark lines below the aligned sequences. gi30173240 (Bent et al., 1994), gi46395604 (Bevan et al., 1998), gi46395938 (Theologis et al., 2000), gi75318159 (Ori et al., 1997), gi325511400 (Theologis et al., 2000) (C) Structural model of the NB-ARC domain of VpCN. (D) Phylogenetic tree of VpCN and related proteins from other plant species. The tree was generated using the ClustalW function in the MegAlign program: Vitis vinifera (GenBank accession no. XP010661747), Nelumbo nucifera (GenBank accession no. XP0102588251), Glycine soja (GenBank accession no. KHN19144), Elaeis guineensis (GenBank accession no. XP010913221), Solanum lycopersicum (GenBank accession no. XP010319316), Beta vulgaris subsp. Vulgaris (GenBank accession no. XP010669409), Phoenix dactylifera (GenBank accession no. XP008791188), Camelina sativa (GenBank accession no. XP010426119), Citrus sinensis (GenBank accession no. XP006470644). The scale bar represents 0.05 substitutions per site. (D) Structure model of NB-ARC in VpCN. (E) Analysis of VpCN expression in response to E. necator inoculation. The third to fifth fully expanded young grapevine leaves beneath the apex were selected for samples. The experiment encompass three independent biological replicates, for each biological replicate three leaves haversted from three plant and three technical replicates were performed. Data represent means of three biological replicates ±SE, asterisksin indicate statistical significance in comparison with control (Student'st-test, significance levels of *P < 0.05, **P < 0.01 are indicated).
Construction of vectors for ectopic expression and A. thaliana transformation
To generate 35S:VpCN, the open reading frame (ORF) region of VpCN was cloned into the binary vector, pCAMBIA 2300 (CAMBIA company), downstream of the CaMV 35S promoter. The construct was introduced into Agrobacterium tumefaciens, strain GV3101, via electroporation, and the transformed A. tumefaciens was used to transform A. thaliana using the floral dip method (Clough and Bent, 1998). Transgenic plants were screened on MS (Murshige and Skoog, 1962) medium containing 60 mg/mL kanamycin, PCR amplification was performed to identify transgenic plants with gene specific primers.
Construction of VpCN promoter:: GUS gene fusion vectors and A. tumefaciens mediated transient expression assays
To generate the VpCN promoter:GUS vector, the VpCN promoter was cloned into the T-easy vector, digested with BamHI and PstI, and finally cloned into the binary vector pC0380GUS. 35S:GUS was used as a positive control (Xu et al., 2010). Four pVpCN promoter fragments with different 5′ deletions were amplified (Supplement Table 1). All the constructs were introduced into A. tumefaciens strain GV3101 via electroporation. The A. tumefaciens mediated transient expression assays were performed as previously described (Guan et al., 2011). A. tumefaciens GV3101 lines harboring the different constructs were grown in liquid Yeast Extract Phosphate (YEP) (Smith and Goodman, 1975) medium (supplemented with 100 μgml−1 kanamycin, 60 μgml−1 gentamycin, and 30 μgml−1 rifampicin) to an OD600 of 0.6, and harvested by centrifugation at 5000 × g for 10 min, before being resuspended in filtration solution (10 mM 2-(N-morpholino) ethanesulfonic acid (MES), pH 5.7, 10 mM MgCl2 and 15 μM acetosyringone) and adjusted to an OD600 of 0.6 for infiltration of young grapevine leaves using a vacuum infiltration method (Santos-Rosa et al., 2008). After infiltration, the leaves were kept in a chamber at 16/8 h day/night cycle at 23°C with 70% humidity for 48 h, before inoculation with E. necator (Guan et al., 2011; Yu et al., 2013).
Pathogen inoculation procedures
E. necator infected leaves were collected from a highly PM-susceptible wild Chinese wild V. Adstricta, Hance clone “Taishan-2.” Leaves of the Chinese wild V. pseudoreticulata “Baihe-35-1” were inoculated by touching the adaxial epidermis of leaves with sporulating colonies on the surface of pathogen leaves, the inoculation were repeated three times (Guan et al., 2011). The samples were harvested 0, 6, 12, 24, 48, 72, 96, and 120 h after inoculation.
A. thaliana powdery mildew G. cichoracearum was maintained on highly susceptible pad4 A. thaliana mutant plants. The infection was conducted as previously described (Tang and Innes, 2002). The susceptibility or resistance phenotypes were scored 8 days after infection (Nie et al., 2011). Analyses of pathogenesis-related 1 (PR1) gene expression were performed using qRT-PCR using the same PCR program as for the VpCN analysis. The A. thaliana tubulin gene (GenBank Accession number NM_179953) was used as a reference. Rosett leaves from 4 week old Arabidopsis were harvested at 0, 12, 24, 36, and 48 h after inoculation.
P. st DC3000 cells grown in King's B medium (supplemented with 100 μgml−1 kanamycin and 30 μgml−1 rifampicin) to an OD600 of 0.6, harvested by centrifugation for 5000 × g for 10 min and re-suspended in 10 mM MgSO4,adjusted to optical density at OD600 of 0.02. The bacterial suspension containing 0.025% Silwet-77, and the mixture were hand infiltrated into the abaxial side of the A. thaliana leaves using a needless 1 ml syringe (Fan et al., 2008). P. st DC3000 bacterial growth were assessed 3 and 5 days after infection as described (Ahn et al., 2007).
Trypan blue staining
For trypan blue staining, A. thaliana leaves were collected 12 hpi (hours post-inoculation) and boiled in alcoholic lactophenol trypan blue solution (20 mL of ethanol, 10 mL of phenol, 10 mL of water, 10 mL of lactic acid [83%], and 30 mg of trypan blue). Stained leaves were cleared in chloral hydrate (2.5 g dissolved in 1 mL of water) for 3 h, before placing under a coverslip in 50% glycerol (Koch and Slusarenko, 1990; Frye and Innes, 1998).
Peroxide assay
Peroxide (H2O2) was assayed using a hydrogen peroxide kit, according to the manufacturer's instructions (Nanjing Bio Ins., Nanjing, China). Quantification of dead cells was performed 12 hpi by staining leaf discs (0.5 mm in diameter) with 0.2% Evans blue (Sigma) for 30 min, followed by several washes with water to remove excess stain (Mino et al., 2002; Ahn et al., 2007). One milliliter of 50% methanol supplemented with 1% SDS was added and the samples were incubated at 50°C for 1 h. Absorbance at OD600 was determined by ultraviolet spectrophotometry after a 10-fold dilution of the extracts (Ahn et al., 2007). The nitro blue terazolium (NBT) staining was performed as described (Kim et al., 2011).
Callose accumulation
To observe callose accumulation, leaves (3 dpi) were immersed in destaining solution (10 ml phenol, 10 ml glycerin, 10 ml lactic acid, 10 ml H2O, and 80 ml ethanol) and kept in an oven at 60°C for 1 h to remove chlorophyll. The samples were washed to remove the destaining solution, and stained with 0.1% aniline. The fluorescence of callose was detected using an epifluorescence microscope (E800, Nikon) with a V-2A filter (Reuber et al., 1998; Ahn et al., 2007). For quantitative determination of callose, A. thaliana, leaves (3 dpi) were immersed in ethanol for 2–3 days to remove the chlorophyll, before centrifugation at 5000 × g for 10 min. The supernatant was discarded and the pellet resuspended in 0.4 ml DMSO. One hundred microliter of the supernatant was supplemented with loading mixture [400 μl 0.1% (w/v) aniline blue, 590 mL 1 M glycine/NaOH (pH 9.5), 210 mL 1 M HCl] and 200 μl 1 M NaOH. The control samples were not supplemented with aniline. The samples were incubated in a water bath 50°C for 20 min and cooled to room temperature before detection with a fluorescence spectrophotometer (F-4600, Hitachi, Tokyo, Japan) under 393 nm excitation, 479 nm emission and a voltage of 400 v. The fluorescence of the samples was determined by subtracting the fluorescence value of the control from those of the samples (Kohler et al., 2000).
GUS staining, histochemical and fluorometric assays for determining GUS activity
A histochemical β-glucuronidase (GUS) assay of leaves was carried out as previously described (Jefferson, 1987). Briefly, leaves were immersed in GUS staining solution at 37°C for 24 h, before washing in 70% ethanol at 37°C and viewing macroscopically (Guan et al., 2011; Yu et al., 2013). GUS fluorescence was determined quantitatively according to Jefferson (1987). Protein concentrations in grapevine extracts was normalized by dilution with extraction buffer according to Bradford (1976). GUS activity was expressed as pmol 4MU (Sigma-Aldrich China, Shanghai, China) per minute per mg of protein. Sample fluorescence was detected with an infinite 200® PRO (Tecan Trading AG, Switzerland). Three independent experiments were performed.
Results
VpCN expression during powdery mildew infection
To identify potential resistance mechanisms and resistance related genes in the response of wild Chinese V. pseudoreticulata to powdery mildew, we previously performed a transcriptome analysis of the “Baihe-35-1” using RNA-seq (Weng et al., 2014). We observed that the expression of VpCN (GenBank accession number KT265084) was strongly induced by inoculation with E. necator. To verify this, we performed quantitative real-time PCR (qPCR) analysis of VpCN expression in V. pseudoreticulata leaves that had been inoculated with E. necator, and observed 4.2-fold greater VpCN transcript levels than in leaves prior to inoculation. Subsequently, VpCN expression decreased but remained at a higher level than in mock inoculated plants (Figure 1E).
Cloning and sequence analysis of VpCN
To investigate the putative role of VpCN in providing resistance to pathogens, we first designed primers based on a cDNA sequence obtained from the Grape Genome Database (12 ×; http://www.genoscope.cns.fr), and isolated and designated the gene VpCN (GenBank accession number KT265084). The VpCN gene is located on chromosome 15 (Figure 1A), has an ORF of 1773 bp (Supplement Figure 1) and is predicted to encode a protein of 590 amino acids with a molecular mass of 67,390 Da and a theoretical pI value of 5.45. The amino acid sequence was further predicted to contain a RxCC-like domain in the N-terminus from residue 6–119, a Ran GTPase-acting protein 2 (RanGAP2) interaction site in the RxCC-like domain and an NB-ARC domain spanning residues 129–414. The NB-ARC sub-domains, NB, ARC1, and ARC2 were all present. Furthermore, several conserved motifs, such as a P-loop, RNBS A–D, and a GLPL (Figure 1B) were detected. In addition to a RxCC-like domain and an NB-ARC domain, we also found an AAA domain and a PLN03210 domain in the predicted amino acid sequence (picture not shown). A structure-based multiple amino acids sequence alignment was performed to compare the NB-ARC domain of VpCN with those of other closely related plant R proteins, including RPS2 (gi30173240) (Bent et al., 1994) and I-2 (gi75318159) (Ori et al., 1997). The amino acids sequence identity between the VpCN and the A. thaliana RPS2 NB-ARC domain was shown to be 33%, while the VpCN and I-2 NB-ARC domains had a 29%, sequence identity, concentrated on the conserved motifs of the NB-ARC subdomains (Figure 1B).
Ectopic expression VpCN in A. thaliana enhance resistance to powdery mildew
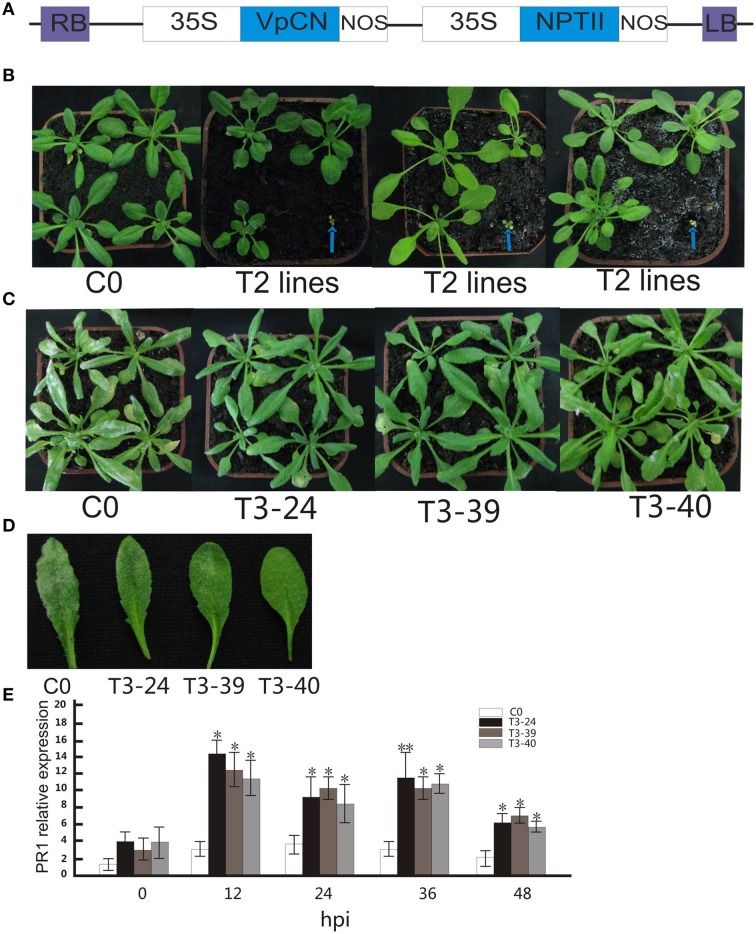
We next transformed the VpCN in A. thaliana under the control of the constitutive 35S promoter (Figure 2A). A total of 42 independent transgenic T1 lines were obtained and the presence of the transgene confirmed by PCR using VpCN specific primers. The T2 progeny segregated so that 39 lines displayed wild type morphology while three lines exhibited a dwarfed phenotype and morphological abnormalities, such as small yellow leaves, stunted growth, and chlorotic tissue (Figure 2B). These dwarf lines eventually died. The lines with a wild type phenotype were challenged with G. cichoracearum, and three transgenic lines with higher resistance were chosen for the generation of homozygous T3 generation lines. The transgenic lines displayed few visible white powdery areas on their leaves at 8 dpi, whereas the wild-type (Col-0) exhibited abundant powdery mildew development (Figures 2C,D). To determine whether the enhanced resistance to G. cichoracearum in the transgenic lines was related to an increase in the expression of a known defense gene, we evaluated PR1 (Pathogenesis Related 1) (Friedrich et al., 1996) transcript levels at 0, 12, 24, 36, and 48 hpi. Three transgenic plants displayed higher PR1 transcript abundance after pathogen inoculation than wild type plants, reaching a maximum level at 12 hpi. The PR1 transcript levels of transgenic plants were ~4–5-fold higher after inoculation than in wild type at all time points (Figure 2E).
Figure 2.
Generation of CaMV 35S promoter-VpCN constructs used for transformation of Arabidopsis thaliana, morphology of wild type and transgenic Arabidopsis thaliana plants, with transgenic plants showing enhanced disease resistance to G. cichoracearum after ectopic expression of VpCN. (A) Structure of the CaMV 35S promoter-VpCN ectopic expression construct. LB, left border; RB, right border; 35S, CaMV 35S promoter; NOS, terminator; NPT II, aminoglycoside-3′- phosphotransferase. (B) Indicate T2 transgenic plants displayed either normal phenotypes or dwarfism. Blue arrows indicate the dwarf phenotype in 4 week old plants. (C) Transgenic A. thaliana leaves developed fungal spores 8 dpi with G. cichoracearum. (D) Disease symptoms developed on the leaves of transgenic lines and wild type plants 8 dpi with G. cichoracearum. (E) A. thaliana PR1 transcript levels in T3 lines and wild-type after inoculation with G. cichoracearum. Total RNA was extracted from A. thaliana leaves 0, 12, 24, 36, and 48 h post-inoculation (hpi) with G. Cichoracearum. The experiment encompass three independent biological replicates, for each biological replicate six rosette leaves were harvested from three plant and three technical replicates were performed. Data represent means of three biological replicates ±SE, asterisksin indicate statistical significance in comparison with WT (Student's t-test, significance levels of *P < 0.05, **P < 0.01 are indicated).
Ectopic expression of VpCN results in enhanced protection against the bacterial pathogen, P. st DC3000
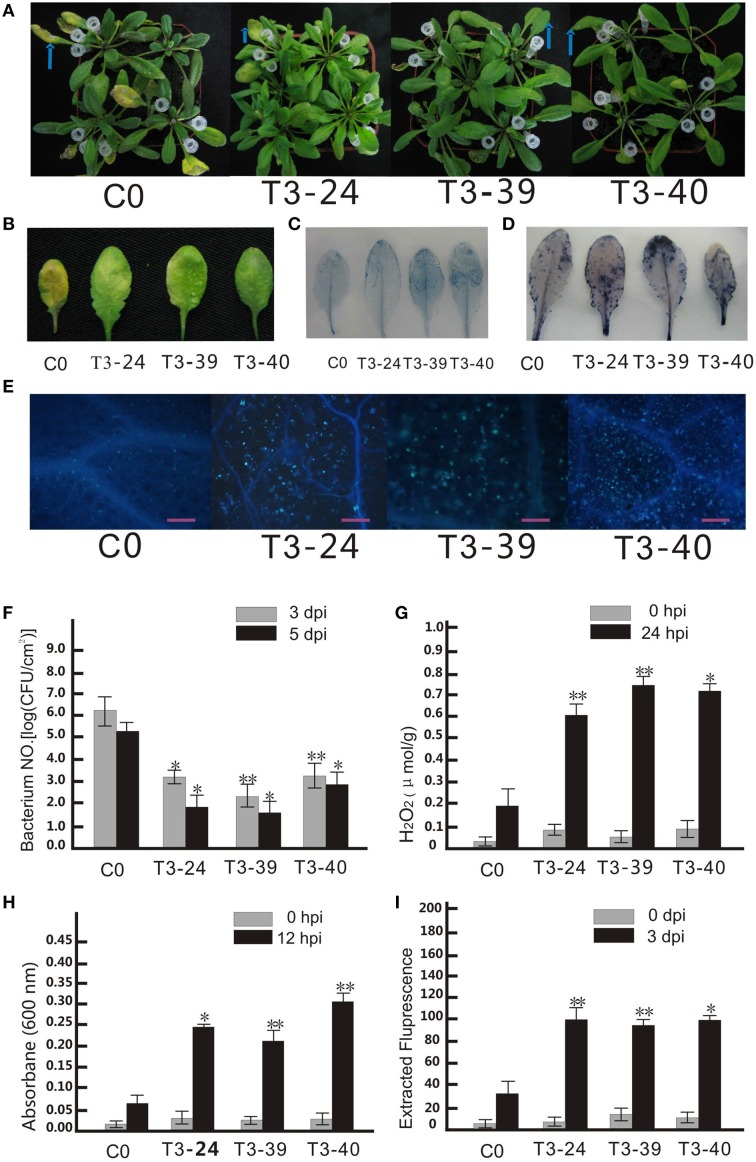
Since amino acid sequence of VpCN was predicted to contain a PLN03210 domain, which has been shown to be correlated with resistance to Pseudomonas syringae pv. glycinea race 6 (Kim et al., 2009), we hypothesized that it might function in providing resistance to bacterial infection. To test this, transgenic and control plants were challenged with the bacterial P. st DC3000 pathogen by leaf infiltration (Figure 3A). Most infiltrated wild type leaves exhibited water-soaking at 1 dpi, turned yellow and finally wilted at 5 dpi. In contrast, the transgenic plants infected with the pathogen showed fewer symptoms (Figure 3B), and when the growth of P. st DC3000 in the inoculated plants was quantified, it was found that the bacterial number in the transgenic plants was significantly lower than in the wild type plants (Figure 3F). To observe the effect of VpCN expression on cell death, trypan blue staining was performed of leaves and we observed that cell death was more widespread in the transgenic lines than the wild type plants (Figure 3C). Additionally, cell death quantification by Evans blue staining followed by spectrophotometric analysis, showed a 5-6 fold higher level cell death in the transgenic plants (Figure 3H). Nitroblue tetrazolium (NBT) staining for the superoxide anion also showed higher accumulation in the transgenic plants (Figure 3D), as did quantitative measurements of H2O2 (Figure 3G). Finally, the accumulation of the (1,3)-β-glucan polymer callose, which is known to be involved in plant defense responses (Brown et al., 1998), was visualized by aniline blue staining of wild type and transgenic plants after treated with P. st DC3000 (Figure 3E). Greater accumulation of callose was observed in the transgenic plants than in wild, and when callose levels were quantified, it was confirmed that the transgenic lines contained significantly (P < 0.05) more callose (Figure 3I).
Figure 3.
Ectopic expression of VpCNin Arabidopsis thaliana enhanced disease resistance to Pseudomonas syringae pv. tomato DC3000. (A) P. st DC3000 was diluted to OD600 0.02 and injected into the middle of a leaf with needleless syringes. The injected leaves were marked with white pipette tips, and pictures taken 3 dpi. (B) Disease symptoms developed on the leaves of transgenic lines and wild type plants 3 dpi with P.st DC3000. (C) Transgenic plants and wild type leaves were stained with trypan blue 12 hpi with P.st DC3000. (D) Transgenic plants and wild type leaves were stained with nitro blue terazolium (NBT). (E) Microscopic observation of callose deposition after 3 dpi. Bars = 50 μm. (F) The numbers of bacterial cells in the leaves were determined at 3 and 5 dpi. (G) Detection of H2O2 concentration in Arabidopsis leaf samples harvested at 24 hpi. (H) Quantification of dead cells at 12 hpi. (I) Quantification of callose from A. thaliana leaves at 3 dpi. The experiment encompass three independent biological replicates, for each biological replicate six rosette leaves were harvested from three plant and three technical replicates were performed. Data represent means of three biological replicates ±SE, asterisksin indicate statistical significance in comparison with WT (Student's t-test, significance levels of *P < 0.05, **P < 0.01 are indicated).
Isolation and analysis of the VpCN promoter sequence
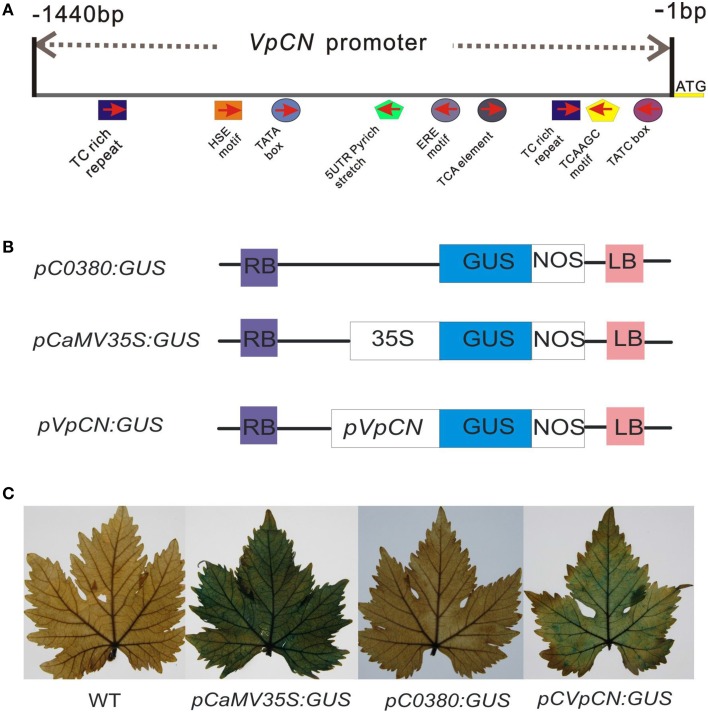
A 1440 bp upstream sequence was cloned using wild Chinese V. pseudoreticulata “Baihe-35-1” genomic DNA by PCR, regulatory cis-acting elements predicted showed that several putative regulatory elements involved in the activation of defense-related genes, including 72 predicted TATA boxes, 32 CAAT boxes, and two TC-repeat elements, which are known to be involved in defense and stress responses, a TCA element, which is involved in salicylic acid (SA) responses, a TGACG motif, which is associate with methyl jasmonate-response, an HSE element, which is involved in heat stress responses and two TATC elements, which are related to gibberellin responses (Figure 4A). Additional predicted cis-regulatory elements included light response elements (TCCC-motif, MRE, I-box, GT1-motif, GAG-motif, GA-motif, G-box, CATT motif, Box-I, AT1-motif, and Box-4), as well as others cis-elements (5UTR Py-rich stretch, circadian element and, TATC box). Several of the predicted cis-elements are known to be involved in responses to environmental stresses, further suggesting that the VpCN promoter may play a role in defense responses.
Figure 4.
The main predicted cis-acting elements in the pVpCN promoter sequence, structure of the VpCN promoter fused to the GUS reporter gene and GUS staining of the transient constructs in transformed grapevine leaves. (A) Schematic diagram of the main predicted cis-acting elements in the VpCN promoter sequence of Chinese wild V. pseudoreticulata. (B) The pVpCN promoter was fused to the GUS gene. The plasmid pCaMV35S:GUS was used as a positive control and pC0380:GUS was used as a negative control. (C) The fully expanded grapevine leaves of V. vinifera “Red globe” were collect from a grape germplasm resources orchard and used for agroinfiltration.
Promoter::GUS (glucuronidase) assays
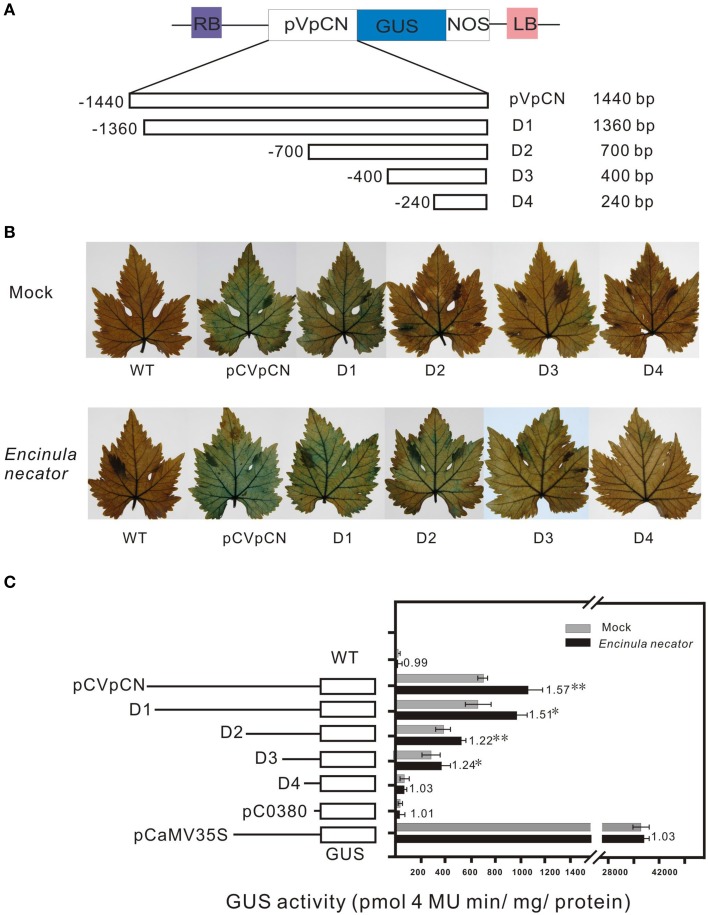
To test the activity of the VpCN promoter, the 1440-bp promoter fragment was fused to a reporter gene encoding β-glucuronidase (GUS), generating the construct pCVpCNGUS. As a positive control, a CaMV35S::GUS (PC35SGUS) construct was used and a construct with no promoter was used as a negative control (pC0380GUS) (Xu et al., 2010; Figure 4B). All the constructs were expressed transiently in grapevine leaves, which were subsequently subjected to GUS staining. Leaves transformed with the PC35SGUS construct showed strong GUS activity, while no activity was detected in wild type (WT) and very little in PC0380GUS. pCVpCNGUS transformed leaves showed GUS activity but at a lower level than leaves transformed with PC35SGUS (Figure 4C), and when leaves were infected with E. necator 2 dpi prior to GUS staining, the infected leaves exhibited stronger GUS activity than mock-inoculated control leaves. To further determine the location of the pathogen-responsive cis-regulatory region, we generated four promoter deletion fragments and fused them to GUS (−1360, −700, −400, and −240 bp) (Figure 5A). When the GUS activity was quantified fluorescently, the highest levels were measured in grapevines containing the −1440 bp fragment, where it was induced 1.57-fold after treatment with E. necator compared to mock controls. Leaves transformed with −1360, −700, and, −400 promoter fragments exhibited a relative low level of GUS activity; however, they showed increased GUS activity after being challenged with E. necator (Figure 5B). Since the leaves transformed with the −240 bp fragment showed no significant difference in GUS activity before and after treatment with E. necator (Figure 5C), the −400 bp promoter fragment was deduced to be the minimal promoter region required for the response to E. necator infection.
Figure 5.
Schematic map of the pVpCN promoter-GUS gene fusion deletion constructs, histochemical analysis of GUS expression in transiently transformed V. vinifera “Red globe” leaves after inoculation with E.necator, and fluorometric analysis of GUS activity in the transiently transformed grapevine leaves. (A) The GUS gene was driven by the VpCN promoter deletions, the exact locations of the promoter fragments are shown in Supplement Figure 2. The deletion size is indicated at the far right. (B) GUS staining was carried out 2 days after treatment with sterile water (upper panel) or E. necator (lower panel). (C) The various deletion fragments of the VpCN promoter fused to GUS and relative GUS activity driven in the transiently transformed grapevine leaves. The dark bars indicate the average GUS activity for deletion constructs in transiently transformed grapevine leaves treated with E. necator, the gray bars indicate the mock treatment (sterile water). Numbers adjacent to the bars indicate the fold difference in GUS activity leaves harboring the various constructs challenged with E. necator relative to the mock samples. The mean GUS activity (±SD) is averaged from three independent experiments (n = 3), the errors bars indicate the stand deviation. Significant difference between treatment and mock conditions was analyzed using one sided paired t-test (** and * meaning P < 0.0.1 or P < 0.05, respectively).
Discussion
We previously reported the leaf transcriptome of wild Chinese grape (V. Pseudoreticulata, “Baihe-35-1”) that had been inoculated with E. necator, and showed that expression of a unigene corresponding to VpCN was strongly induced by the infection (Weng et al., 2014). Here, we isolated the ORF sequence of VpCN and ectopically expressed it in A. thaliana. This resulted in enhanced disease resistance to the pathogens G. cichoracearum and P. st DC3000. The deduced amino acid sequence of the corresponding protein is predicted to contain an RxCC-like and an NB-ARC domain. Most currently known R proteins have a NB-ARC domain and the CC domain is thought to initiate signaling (Radirdan et al., 2008). Given the rapid and strong up-regulation of VpCN transcript accumulation in wild Chinese Vitis after treatment with E. necator, we suggest that VpCN may play a role in the early defense signaling pathways in pathogen recognition. In addition to these two domains, the deduced amino acid sequence also contained a PLN03210 domain, which is thought to contribute to the identification of resistance signaling components and to convey resistance to P. syringae (Kim et al., 2009), suggesting that VpCN may also be associated with bacterial disease resistance.
Several studies have already demonstrated that over-expression of an R-gene can cause growth retardation, spontaneous cell death, and constitutive defense activation (Tao et al., 2000; Bendahmane et al., 2002; Stokes et al., 2002; Mohr et al., 2010; Nandety et al., 2013) due an over activation of the ETI system. In this study, three independent transgenic lines exhibited dwarfism and stunted growth, as well as other morphological defects, although since these plants eventually died, we were unable to investigate whether they also exhibited enhanced resistance to G. Cichoracearum. In agree with these results we suggest that VpCN ectopic expression may active ETI system and cause constitutive defense in three transgenic plants and cause growth retardation, spontaneous cell death. Further studies will investigate whether the three dwarf and lethal phenotypes is caused by toxic effects of high level of VpCN expression or the co-suppression between VpCN and Arabidopsis endogenous genes with VpCN-homologous sequences.
There have been several reports suggesting that over-expression of R genes enhances disease resistance due to constitutive SA accumulation, PR gene expression and active defense responses (Keller et al., 1999; Tang et al., 1999; Kim et al., 2001; Shirano et al., 2002; Stokes et al., 2002). In this study, ectopic expression of VpCN in A. thaliana enhanced disease resistance to G. cichoracearum, and when the PR1 transcript levels was assessed, a 4-5 fold increase in expression was observed in 12 hpi in transgenic plants compared to WT, and these levels remained higher over the time course. These results suggest that ectopic expression of VpCN in A. thaliana activate defense responses after pathogen inoculation.
The production of reactive oxygen species (ROS), mainly in the form of a superoxide burst and H2O2 accumulation, is thought to enhance plant defense responses and to be essential for the establishment of plant immunity (Alvarez et al., 1998; Grant and Loake, 2000; Punja, 2004; Choi and Hwang, 2011; Kim and Hwang, 2014). In agreement with these results, we found that higher levels of anions and H2O2 in the transgenic plants than in WT after challenging with P. st DC3000. This suggests that ectopic expression of VpCN triggers an oxidative burst to induce plant immunity to P. st DC3000; however, further studies are needed to investigate how oxidative burst and H2O2 accumulation is mediated by VpCN. High concentrations of ROS can result in HR-like cell death (Kovtun et al., 2000; Wang et al., 2007; Zhang et al., 2012), and over-expression of a TIR-NB-LRR gene from wild north American grapevine in V. vinifera wine grape cultivars was reported to lead to HR-like cell death after inoculation with E. necator (Feechan et al., 2013). Moreover, over-expression of a RPP1A truncation in A. thaliana induced elicitor-independent HR-like cell death (Weaver et al., 2006). In this study, an increase in ROS ( and H2O2) accumulation followed by H2O2 induced HR-like cell death was observed after ectopic expression of VpCN in A. thaliana, when the transgenic plants were inoculated with P. st DC3000.
Callose-containing cell-wall appositions, called papillae, provide a physical barrier that slows pathogen invasion at the site of pathogen attack (Luna et al., 2011). Callose deposition is typically triggered by conserved pathogen-associated molecular patterns (PAMPs) and contributes to the innate immunity (Brown et al., 1998; Luna et al., 2011). Ellinger et al. (2013) demonstrated that over-expression of PMR4 in transgenic plants promoted early callose accumulation at attempted fungal penetration sites, which provided complete resistance to G. cichoracearum, and the non-adapted PM agent, B. graminis. In this study, transgenic plants displayed more callose deposition than WT plants in response to treatment with P. st DC3000, suggesting that callose deposition may contribute to the enhanced disease resistance to the pathogen displayed by the transgenic plants.
To elucidate the molecular basis of VpCN transcript induction after inoculation with E. necator, the VpCN promoter was isolated and its activation investigated using A. tumefaciens-mediated transient expression of VpCN in V. vinifera leaves. Bioinformatic analysis of the promoter sequence revealed two TC-rich repeats (′5-ATTCTCTAAC-3′), which are thought to be involved in defense and stress responses (Diaz-De-Leon et al., 1993). We hypothesized that these might be involved in the response to E. necator, and generated four promoter deletion constructs to test this idea. Plants harboring a −1360, −700, or −400 bp region of the promoter sequence, all of which contain two or one TC rich repeat elements (Supplement Figure 2), showed increased GUS activity after challenge with E. necator. However, plants containing only a −240 bp region sequence, which has no TC-rich repeat elements (Supplement Figure 2), showed no significant change in GUS activity after inoculation with E. necator. Thus, we propose that the TC-rich repeat elements may play a role in the VpCN promoter activity in response to E. necator infection. This study suggests that VpCN is a disease resistance gene, and we will investigate that whether the VpCN is interact with AVR protein (effector) from Erysiphe necator. Further functional studies to the VpCN with other proteins and downstream defense signaling involved in the powdery mildew disease resistance will be helpful in understanding the molecular mechanisms of powdery mildew disease resistance in Chinese wild V. pseudoreticulata.
Author contributions
XW and ZW designed the experiments. ZW, LY, RW, and ZL performed the experiments. XW, ZW, and CL analyzed the results and wrote the manuscript. All authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31272136) and the Program for Innovative Research Team of Grape Germplasm Resources and Breeding (2013KCT-25). We thank PlantScribe (http://www.plantscribe.com/) for editing the manuscript.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.01087
List of primer sequence used in this study. F, Forward primer; R, Reverse primer.
Sequence analysis VpCN from Chinese wild V. pseudoreticulata W. T. Wang “Baihe-35-1.” The ORF sequence of VpCN is 1773 bp and encodes a polypeptide of 590 amino acids. The Rx-CC-like domain is labeled by single underline and the NB-ARC domain by a double underline.
Sequence analysis of the VpCN promoter. Motifs with significant similarity to previously identified cis-acting elements are shaded and the names are given under each element. Sequences labeled in yellow correspond to primer design positions. Arrow heads represent the start point of the 5-deleted promoter derivatives.
References
- Ahn L. P., Kim S., Lee Y. H., Suh S. C. (2007). Vitamin B1-induced priming is dependent on hydrogen peroxide and the NPRl gene in Arabidopsis. Plant Physiol. 143, 838–848. 10.1104/pp.106.092627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht M., Takken F. L. W. (2006). Update on the domain architectures of NLRs and R proteins. Biochem. Biophys. Res. Commun. 339, 459–462. 10.1016/j.bbrc.2005.10.074 [DOI] [PubMed] [Google Scholar]
- Alvarez M. E., Pennell R. I., Meijer P. J., Ishikawa A., Dixon R. A., Lamb C. (1998). Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92, 773–784. 10.1016/S0092-8674(00)81405-1 [DOI] [PubMed] [Google Scholar]
- Bendahmane A., Farnham G., Moffett P., Baulcombe D. C. (2002). Constitutive gain-of-function mutants in a nucleotide binding site–leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 32, 195–204. 10.1046/j.1365-313X.2002.01413.x [DOI] [PubMed] [Google Scholar]
- Bendahmane A., Kanyuka K., Baulcombe D. C. (1999). The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11, 781–792. 10.1105/tpc.11.5.781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent A. F., Kunkel B. N., Dahlbeck D., Brown K. L., Schmidt R., Giraudat J., et al. (1994). RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 265, 1856–1860. 10.1126/science.8091210 [DOI] [PubMed] [Google Scholar]
- Bevan M., Bancroft I., Bent E., Love K., Goodman H., Dean C., et al. (1998). Analysis of 1.9Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 391, 485–493. 10.1038/35140 [DOI] [PubMed] [Google Scholar]
- Bradford M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dyebinding. Anal. Biochem. 72, 248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Brown I., Trethowan J., Kerry M., Mansfiled J., Bolwell G. P. (1998). Localization of components of the oxidative cross-linking of glycoproteins and of callose synthesis in papillae formed during the interaction between non-pathogenic strains of Xanthomonas campestris and French bean mesophyll cells. Plant J. 15, 333–343. 10.1046/j.1365-313X.1998.00215.x [DOI] [Google Scholar]
- Cesari S., Thilliez G., Ribot C., Chalvon V., Michwl C., Jauneau A., et al. (2013). The rice resistance protein RGA4/RGA5 recognizes th Magnaporthe oryzae effectors AVR1-CO39 by direct binding. Plant Cell 25, 1463–1481. 10.1105/tpc.112.107201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm S. T., Coaker G., Day B., Staskawicz B. J. (2006). Host-microbe interactions: shaping the evolution of plant immune response. Cell 124, 803–814. 10.1016/j.cell.2006.02.008 [DOI] [PubMed] [Google Scholar]
- Choi D. S., Hwang B. K. (2011). Proteomics and functional analyses of pepper abscisic acid-responsive 1 (ABR1), which is involved in cell death and defense signaling. Plant Cell 23, 823–842. 10.1105/tpc.110.082081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Collier S. M., Hamel L. P., Moffett P. (2011). Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR Protein. Mol. Plant Microbe Interact. 24, 918–931. 10.1094/MPMI-03-11-0050 [DOI] [PubMed] [Google Scholar]
- Collier S. M., Moffett P. (2009). NB-LRRs works a “bait and switch” on pathogens. Trends Plant Sci. 14, 521–529. 10.1016/j.tplants.2009.08.001 [DOI] [PubMed] [Google Scholar]
- Diaz-De-Leon F., Klotz K. L., Lagrimini M. (1993). Nucleotide sequence of the tobacco (Nicotiana tabacum) anionic peroxides gene. Plant Physiol. 101, 1117–1118. 10.1104/pp.101.3.1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinger D., Naumann M., Falter C., Zwikowics C., Jamrow T., Manisseri C., et al. (2013). Elevated early callose deposition results in complete penetration to powdery mildew in Arabidopsis. Plant Physiol. 161, 1433–1444. 10.1104/pp.112.211011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Crooks C., Lamb C. (2008). High-throughput quantitative luminescence assay of the grow thin planta of Pseudomonas syringae chromosomally tagged with Photorhabdus luminescens lux CDABE. Plant J. 53, 393–399. 10.1111/j.1365-313X.2007.03303.x [DOI] [PubMed] [Google Scholar]
- Feechan A., Anderson C., Torregrosa L., Jermakow A., Mestre P., Wiedemann M. S., et al. (2013). Genetic dissection of a TIR-NB-LRR locus from the wild North American grapevine species Muscadinia rotundifoliaidentifies paralogous genes conferring resistance to major fungal and oomycete pathogens in cultivated grapevine. Plant J. 76, 661–674. 10.1111/tpj.12327 [DOI] [PubMed] [Google Scholar]
- Friedrich L., Lawton K., Ruess W., Masnet P., Specker N., Rella M. G., et al. (1996). A benzothiadiazole derivative induces systemic acquired resistance in tobacco. Plant J. 10, 61–70. 10.1046/j.1365-313X.1996.10010061.x [DOI] [Google Scholar]
- Frye C. A., Innes W. (1998). An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10, 947–956. 10.1105/tpc.10.6.947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadoury D. M., Cadle-Davidson L., Wilcox W. F., Dry I. B., Seem R. C., Milgroom M. G. (2012). Grapevine powdery mildew (Erysiphe necator): a fascinating system for the study of the biology, ecology and epidemiology of an obligate biotroph. Mol. Plant Pathol. 13, 1–16. 10.1111/j.1364-3703.2011.00728.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant J. J., Loake G. J. (2000). The Role of reactive oxygen intermediates and cognate redox signaling in disease resistance. Plant Physiol. 124, 21–29. 10.1104/pp.124.1.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X., Zhao H. Q., Xu Y., Wang Y. J. (2011). Transient expression of glyoxal oxidase from the Chinese wild grape Vitis pseudoreticulata can suppress powdery mildew in a susceptible genotype. Protoplasma 248, 415–423. 10.1007/s00709-010-0162-4 [DOI] [PubMed] [Google Scholar]
- Hao W., Collier S. M., Moffett P., Chai J. J. (2013). Structural basis for the interaction between the potato virus X resistance protein (Rx) and its cofactor Ran GTPase-activating protein 2 (RanGAP2). J. Biol. Chem. 288, 35868–35876. 10.1074/jbc.M113.517417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W. J., Jiang T. Y., Choi W., Pang Y. X., Hu Q., Xu Y. H., et al. (2013). Mechanistic insights into CED-4-mediated activation of CED-3. Genes Dev. 27, 2039–2048. 10.1101/gad.224428.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R. (1987). Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405. 10.1007/BF02667740 [DOI] [Google Scholar]
- Jones J. D., Dangl K. L. (2006). The plant immune systems. Nature 444, 323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Keller H., Pamboukdjian N., Ponchet M., Poupet A., Delon R., Verrier J. L., et al. (1999). Pathogen-induced elicitin production in transgenic tobacco generates a hypersensitive response and nonspecific disease resistance. Plant Cell 11, 223–235. 10.1105/tpc.11.2.223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. S., Hwang B. K. (2014). An important role of the pepper phenylalanine ammonia-lyase gene (PAL1) in salicylic acid-dependent signaling of the defense response to microbial pathogens. J. Exp. Bot. 65, 2295–2306. 10.1093/jxb/eru109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Ahn I. P., Park C., Park S. G., Park S. Y., Jwa N. S., et al. (2001). Molecular characterization of the cDNA encoding an acidic isoform of PR-1 gene protein in rice. Mol. Cells 11, 115–121. Available online at: http://europepmc.org/abstract/MED/11266113 [PubMed] [Google Scholar]
- Kim S. H., Kwon S. I., Saha D., Anyanwu N. C., Gassmann W. (2009). Resistance to the Pseudomonas syringae effector HopA1 is governed by the TIR-NBS-LRR protein RPS6 and is enhanced by mutations in SRFR1. Plant Physiol. 150, 1723–1732. 10.1104/pp.109.139238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Woo D. H., Kim J. M., Lee S. Y., Chung W. S., Moon Y. H. (2011). Arabidopsis MKK4 mediates osmotic-stress response via its regulation of MPK3 activity. Biochem. Biophys. Res. Commun. 412, 150–154. 10.1016/j.bbrc.2011.07.064 [DOI] [PubMed] [Google Scholar]
- Koch E., Slusarenko A. (1990). Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2, 437–445. 10.1105/tpc.2.5.437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A., Schwindling S., Conrath U. (2000). Extraction and quantitative determination of callose from Arabidopsis leaves. Biotechniques 28, 1084–1086. [DOI] [PubMed] [Google Scholar]
- Kohm B. A., Goulden M. G., Gilbert J. E., Kavanagh T. A., Baulcombe D. C. (1993). A potato virus X resistance gene mediates an induced, nonspecific resistance in protoplasts. Plant Cell 5, 913–920. 10.1105/tpc.5.8.913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun Y., Chiu W. L., Tena G., Sheen J. (2000). Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. U.S.A. 97, 2940–2945. 10.1073/pnas.97.6.2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M., Déhais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30:325. 10.1093/nar/30.1.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Wang X. P., Wang Y. J. (2006). cDNA clone, fusion expression and purification of the novel gene related to ascorbate peroxidase from Chinese wild Vitis pseudoreticulata in E.coli. Mol. Biol. Rep. 33, 197–206. 10.1007/s11033-006-0008-5 [DOI] [PubMed] [Google Scholar]
- Lukasik-Shreepaathy W., Slootweg E., Richter H., Goverse A., Cornelissen B. J. C., Takken F. L. W. (2012). Dual regulatory roles of the extended N terminus for activation of the tomato Mi-1.2 resistance protein. Mol. Plant Microbe Interact. 25, 1045–1057. 10.1094/MPMI-11-11-0302 [DOI] [PubMed] [Google Scholar]
- Luna E., Pastor V., Robert J., Flors V., Mauch-Mani B., Ton J. (2011). Callose deposition: a multifaceted plant defense response. Mol. Plant Microbe Interact. 24, 183–193. 10.1094/MPMI-07-10-0149 [DOI] [PubMed] [Google Scholar]
- Maekawa T., Cheng W., Spiridon L. N., Töller A., Lukasik E., Saijo Y. (2011). Coiled-coil domain-dependent homo-dimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe 9, 187–199. 10.1016/j.chom.2011.02.008 [DOI] [PubMed] [Google Scholar]
- Mestre P., Baulcombe D. C. (2006). Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell 18, 491–501. 10.1105/tpc.105.037234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B. C., Dickerman A. W., Michelmore R. W., Sivaramakrishnan S., Sobral B. W., Young N. D. (1999). Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 20, 317–333. 10.1046/j.1365-313X.1999.t01-1-00606.x [DOI] [PubMed] [Google Scholar]
- Mino M., Maekawa K., Ogawa K., Yamagishi H., Inoue M. (2002). Cell death processes during expression of hybrid lethality in interspecific F1 hybrid between Nicotiana gossei domin and Nicotiana tabacum. Plant Physiol. 130, 1776–1787. 10.1104/pp.006023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr T. J., Mammarella N. D., Hoff T., Woffenden B. J., Jelesko J. G., McDowell J. M. (2010). The Arabidopsis downy mildew resistance gene RPP8 is induced by pathogens and salicylic scid and is regulated by W Box cis elements. Mol. Plant Microbe Interact. 23, 1303–1315. 10.1094/MPMI-01-10-0022 [DOI] [PubMed] [Google Scholar]
- Murshige T., Skoog F. (1962). A revised medium for rapid growth bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. 10.1111/j.1399-3054.1962.tb08052.x [DOI] [Google Scholar]
- Nandety R. S., Caplan J. L., Cavanaugh K., Perroud B., Wroblewski T., Michelmore R. W., et al. (2013). The role of TIR-NBS and TIR-X proteins in plant basal defense response. Plant Physiol. 162, 1459–1472. 10.1104/pp.113.219162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie H. Z., Wu X. Y., Yao C. P., Tang D. Z. (2011). Suppression of edr2-mediated powdery mildew resistance, cell death and ethylene-induced senescenceby mutations in ALD1 in Arabidopsis. J. Genet. Genomics 38, 137–148. 10.1016/j.jgg.2011.03.001 [DOI] [PubMed] [Google Scholar]
- Ooijen G. V., Mayr G., Albrecht M., Cornelissen B. J. C., Takken F. L. W. (2008). Transcomplementation, but not physical association of the CC-NB-ARC and LRR domains of tomato R protein Mi-1.2 is altered by mutations in the ARC2 sub-domain. Mol. Plant 1, 401–410. 10.1093/mp/ssn009 [DOI] [PubMed] [Google Scholar]
- Ori N., Eshed Y., Paran I., Presting G., Aviv D., Tanksley S., et al. (1997). The I2C family from the wilt disease resistance locus I2 belongs to the nucleotide binding, leucine-rich repeat superfamily of plant resistance genes. Plant Cell 9, 521–532. 10.1105/tpc.9.4.521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q., Liu Y. S., Budai-Hadrian O., Sela M., Carmel-Goren L., Zamir D., et al. (2000b). Comparative genetics of nucleotide binding site-leucine rich repeat resistance gene homologues in the genomes of two dicotyledons: tomato and Arabidopsis. Genetics 155, 309–322. Available online at: http://www.genetics.org/content/155/1/309.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q., Wendel J., Fluhr R. (2000a). Divergent evolution of plant NBS-LRR resistance gene homologues in dicot and cereal genomes. J. Mol. Evol. 50, 203–213. 10.1007/s002399910023 [DOI] [PubMed] [Google Scholar]
- Punja Z. K. (2004). Fungal Disease Resistance in Plants. New York, NY; London; Oxford: Food Products Press® An important of the Haworth Press, Inc. [Google Scholar]
- Radirdan G. J., Collier S. M., Sacco M. A., Baldwin T. T., Boettrich T., Moffett P. (2008). The coiled-coil and nucleotide binding domains of the potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell 20, 739–751. 10.1105/tpc.107.056036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuber T. L., Plotnikova J. M., Dewdney J., Rogers E. E., Wood W., Ausubel F. M. (1998). Correlation of defense gene induction defects with powder mildew susceptibility in Arabidopsis enhanced disease susceptibility mutants. Plant J. 16, 473–485. 10.1046/j.1365-313x.1998.00319.x [DOI] [PubMed] [Google Scholar]
- Riedl S. J., Li W., Chao Y., Schwarzenbacher R., Shi Y. (2005). Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature 434, 926–933. 10.1038/nature03465 [DOI] [PubMed] [Google Scholar]
- Santos-Rosa M., Poutaraud A., Merdinoglu D., Mestre P. (2008). Development of a transient expression system in grapevine viaagro-infiltration. Plant Cell Rep. 27, 1053–1063. 10.1007/s00299-008-0531-z [DOI] [PubMed] [Google Scholar]
- Sekine K. T., Tomita R., Takeuchi S., Atsumi G., Saitoh H., Miroyuki H., et al. (2012). Functional differentiation in the leucine-rich repeat domains of closely related plant virus-resistance proteins that recognize common avr proteins. Mol. Plant Microbe Interact. 25, 1219–1229. 10.1094/MPMI-11-11-0289 [DOI] [PubMed] [Google Scholar]
- Shirano Y., Kachroo P., Shah J., Klessig D. F. (2002). A gain-of-function mutation in an Arabidopsis Toll Interleukin1 receptor–nucleotide binding site–leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 14, 3149–3162. 10.1105/tpc.005348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L. D., Goodman N. L. (1975). Improved culture method for the isolation of Histoplasma capsulatum and Blastomyces dermatitidis from contamincated specimens. Am. J. Clin. Pathol. 63, 276–280. Available online at: http://europepmc.org/abstract/med/1115035 [DOI] [PubMed] [Google Scholar]
- Stokes T. L., Kunkel B. L., Richards E. J. (2002). Epigenetic variation in Arabidopsis disease resistance. Genes Dev. 16, 171–182. 10.1101/gad.952102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken F. L. W., Albrecht M., Tameling W. I. L. (2006). Resistance proteins: molecular switches of plant defence. Curr. Opin. Plant Biol. 9, 383–390. 10.1016/j.pbi.2006.05.009 [DOI] [PubMed] [Google Scholar]
- Tameling W. I. L., Nooijen C., Ludwig N., Boter E., Goverse A., Shirasu K., et al. (2010). RanGAP2 mediates nucleocytoplasmic partitioning of the NB-LRR immune receptor Rx in the solanaceae, There by dictating Rx function. Plant Cell 22, 4176–4194. 10.1105/tpc.110.077461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D. Z., Innes R. W. (2002). Over-expression of a kinase-deficient form of the EDR1gene enhances powdery mildew resistance and ethylene-induced senescence in Arabidopsis. Plant J. 32, 975–983. 10.1046/j.1365-313X.2002.01482.x [DOI] [PubMed] [Google Scholar]
- Tang X. Y., Xie M. T., Kim Y. J., Zhou J. M., Klessig D. F., Martin G. B. (1999). Over expression of Pto activates defense responses and confers broad resistance. Plant Cell 11, 15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Yuan F. H., Leister R. T., Ausubel F. M., Katagiri F. (2000). Mutational analysis of the Arabidopsis nucleotide binding site-leucine-rich repeat resistance gene RPS2. Plant Cell 12, 2541–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theologis A., Ecker J. R., Palm C. J., Federspiel N. A., Kaul S., White O., et al. (2000). Sequence and analysis of chromosome 1 of the plant Arabidopsis thaliana. Nature 408, 816–820. 10.1038/35048500 [DOI] [PubMed] [Google Scholar]
- van der Biezen E. A., Jones J. D. G. (1998). The NB-ARC domain: a novel signaling motif shared by plant resistance gene products and regulators of cell death in animals. Curr. Biol. 8, 226–227. 10.1016/S0960-9822(98)70145-9 [DOI] [PubMed] [Google Scholar]
- Wang W. M., Devoto A., Turner J. G., Xiao S. Y. (2007). Expression of the membrane-associated resistance protein RPW8 enhances basal defense against biotrophic pathogens. Mol. Plant Microbe Interact. 20, 966–976. 10.1094/MPMI-20-8-0966 [DOI] [PubMed] [Google Scholar]
- Wang Y., Liu Y., He P., Chen J., Lamikanra O., Lu J. (1995). Evaluation of foliar resistance to Uncinula necatorin Chinese wild Vitis species. Vitis 3, 159–164. [Google Scholar]
- Weaver L. M., Swidersk M. R., Li Y., Jone J. D. G. (2006). The Arabidopsis thaliana TIR-NB-LRR R proteins, RPP1A; Protein localization and constitutive activation of defense by truncated alleles in tobacco and Arabidopsis. Plant J. 47, 829–840. 10.1111/j.1365-313X.2006.02834.x [DOI] [PubMed] [Google Scholar]
- Weng K., Li Z. Q., Liu Q. R., Wang L., Wang Y. J., Xu Y. (2014). Transcriptome of Erysiphe necator-infected Vitis pseudoreticulata leaves provides insight into grapevine resistance to powdery mildew. Hortic. Res. 1:14049. 10.1038/hortres.2014.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W. R., Yu Y. H., Ding J. H., Hua Z. H., Wang Y. J. (2010). Characterization of a novel stilbene synthase promoter involved in pathogen-and stress-inducible expression from Chinese wild Vitis pseudoreticulata. Planta 231, 475–487. 10.1007/s00425-009-1062-8 [DOI] [PubMed] [Google Scholar]
- Yu Y. H., Xu R. W., Wang J., Wang L., Yao W. K., Xu Y., et al. (2013). A core functional region of the RFP1 promoter from Chinese wild grapevine is activated by powdery mildew pathogen and heat stress. Planta 237, 293–303. 10.1007/s00425-012-1769-9 [DOI] [PubMed] [Google Scholar]
- Yu Y. H., Xu W. R., Wang S. Y., Xu Y., Li H. E., Wang Y. J., et al. (2011). VpRFP1, a novel C4C4-type RING finger protein gene from Chinese wild Vitis pseudoreticulata, functions as a transcriptional activator in defence response of grapevine. J. Exp. Bot. 62, 5671–5682. 10.1093/jxb/err253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. J., Wang Y. J., Wang X. P., Yang K. Q., Yang J. X. (2003). An improved method for rapidly extracting total RNA from Vitis. J. Fruit Sci. 53, 771–787. Available online at: http://www.gskk.cbpt.cnki.net/WKA/WebPublication/paperDigest.aspx? [Google Scholar]
- Zhang L., Li Y. Z., Lu W., Meng F., Wu C. A., Guo X. Q. (2012). Cotton GhMKK5 affects disease resistance, induces HR-like cell death, and reduces the tolerance to salt and drought stress in transgenic Nicotiana benthamiana. J. Exp. Bot. 63, 3935–3951. 10.1093/jxb/ers086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Dorey S., Swiderski M., Jones J. D. (2004). Expression of RPS4 in tobacco induced an AvrRPS4-independent HR that requires EDS1, SGT1 and HSP90. Plant J. 40, 213–224. 10.1111/j.1365-313X.2004.02201.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of primer sequence used in this study. F, Forward primer; R, Reverse primer.
Sequence analysis VpCN from Chinese wild V. pseudoreticulata W. T. Wang “Baihe-35-1.” The ORF sequence of VpCN is 1773 bp and encodes a polypeptide of 590 amino acids. The Rx-CC-like domain is labeled by single underline and the NB-ARC domain by a double underline.
Sequence analysis of the VpCN promoter. Motifs with significant similarity to previously identified cis-acting elements are shaded and the names are given under each element. Sequences labeled in yellow correspond to primer design positions. Arrow heads represent the start point of the 5-deleted promoter derivatives.