Abstract
Genetic information is traditionally thought to be transferred from parents to offspring. However, there is evidence indicating that gene transfer can also occur from microbes to higher species, such as plants, invertebrates, and vertebrates. This horizontal transfer can be carried out by small RNAs (sRNAs). sRNAs have been recently reported to move across kingdoms as mobile signals, spreading silencing information toward targeted genes. sRNAs, especially microRNAs (miRNAs) and small interfering RNAs (siRNAs), are non-coding molecules that control gene expression at the transcriptional or post-transcriptional level. Some sRNAs act in a cross-kingdom manner between animals and their parasites, but little is known about such sRNAs associated with plants. In this report, we provide a brief introduction to miRNAs that are transferred from plants to mammals/viruses and siRNAs that are transferred from microbes to plants. Both miRNAs and siRNAs can exert corresponding functions in the target organisms. Additionally, we provide information concerning a host-induced gene silencing system as a potential application that utilizes the transgenic trafficking of RNA molecules to silence the genes of interacting organisms. Moreover, we lay out the controversial views regarding cross-kingdom miRNAs and call for better methodology and experimental design to confirm this unique function of miRNAs.
Keywords: miRNA, siRNA, horizontal gene transfer, plant-microbe interactions, controversy of cross-kingdom miRNA
Introduction
The fundamental concept of gene transfer is that it occurs from parents to offspring. In addition to this vertical transfer, horizontal gene transfer has also been shown to exist in bacteria and simple eukaryotes (Gogarten et al., 2002; Anderson, 2009). Recently, studies have indicated that genes from viruses, prokaryotes and fungi can be transferred to higher species, a phenomenon that has received a great attention (Yue et al., 2012; Crisp et al., 2015). As the products of gene transcription, small RNAs (sRNAs) have been reported to move horizontally between different species. sRNAs of approximately 19–25 nucleotides in length belong mainly to two classes: microRNAs (miRNAs) and siRNAs. Acting as regulatory molecules, sRNAs are involved in a wide range of biological processes that are essential for organ morphogenesis, genome modification, and adaptive responses to biotic and abiotic stresses (Reinhart et al., 2002; Carrington and Ambros, 2003; Lai, 2003; Bartel, 2004). In plants and animals, sRNAs direct the cleavage of endogenous mRNAs or repress their translation (Hamilton et al., 2002; Llave et al., 2002). In addition, sRNAs protect plants and animals from viral infections through the RNA interference (RNAi) system (Wang et al., 2004). It is believed that RNAi also functions in communication among different kingdoms. Recently, both animals and plants have been reported to exchange sRNAs with parasites, pathogens, or symbiotic organisms. Many studies have reported the introduction of cross-kingdom sRNAs between animals and parasites. For example, miRNAs traffic from human sickle cells to malarial parasites (LaMonte et al., 2012) and from helminth nematodes to mouse cells (Buck et al., 2014). However, compared with animals, trafficking of sRNAs has not been widely observed between plants and other organisms. Additionally, plant sRNAs are mobilized through the phloem and are carried to distinct target cells, where the sRNAs induce a reduction of gene expression (Molnar et al., 2010). Therefore, it is of great value to investigate the mobility of sRNAs between different species.
According to computational analysis and experimental validation, certain types of plant-associated sRNAs have been shown to cross kingdoms and play a role in improving immunity and the defense against viruses or the aggravation of viral symptoms. In this report, we describe the processes and effects of plant-derived miRNAs that move to animals/viruses and microbe-derived siRNAs that move to plants, including host-induced gene silencing (HIGS) system that utilize sRNAs to silence parasite genes in plants. Moreover, we discuss the competing view of cross-kingdom miRNAs and introduce the main techniques employed to measure exogenous miRNAs, and call for better methodology and experimental design to confirm this unique function of miRNAs.
Roles of Horizontally Transferred miRNAs
miRNAs Transferred from Plants to Animals
Dietetically absorbed plant miRNAs have been confirmed to exist stably in human plasma (Liang et al., 2014a). Thus, it is an intriguing question whether these evolutionarily conserved plant miRNAs can enter into mammalian cells and exert physiological functions. Indeed, it has been demonstrated by high-throughput sequencing that certain miRNAs from plants such as Zea mays (Z. mays), Arabidopsis thaliana (A. thaliana), Oryza sativa (O. sativa), and Citrus trifoliate (C. trifoliate) can exist stably in human plasma and breast milk exosomes, including 35 miRNAs from 25 MIR families (Lukasik and Zielenkiewicz, 2014). Targets of the aforementioned miRNAs included the mRNAs of proteins associated with transcription factors (e.g., low-density lipoprotein receptor, LDLR), immune system functions (e.g., zinc finger e-box-binding homeobox 1, ZEB1), saccharometabolism (e.g., glycogen debranching enzyme, GDE) and hormone responses (e.g., melanocortin receptor 4, MC4R; Table 1, Figure 1). Due to the vital role of breast milk in infant growth and nutrition, it is of great value to explore the effects of foreign miRNAs on infants. It has been proven that plant miRNAs are consistently present in the umbilical cord blood and amniotic fluid of humans (Li et al., 2015). This suggests that those plant miRNAs may transfer through the placenta to the fetus. Moreover, a greater number of immune-related miRNAs have been detected in the colostrum than in mature milk (Gu et al., 2012). Therefore, certain immune-related exogenous miRNAs that may exist in the colostrum are expected to influence an infant’s immune system.
Table 1.
Trans-kingdom small RNAs.
| Number/Name of sRNAs | Derivation | Target species | Target genes/ORFs | Reference |
|---|---|---|---|---|
| 35 miRNAs | Z. mays, A. thaliana, | H. sapiens | LDLR, ZEB1, GDE, | Lukasik and Zielenkiewicz, 2014 |
| (e.g., zma-miR156a, | O. sativa, C. trifoliate, | MC4R, etc. | ||
| ath-miR319b, osa-miR444.2, | etc. | |||
| ctr-miR167) | ||||
| 38 miRNA/miRNA∗ | S. lycopersicum | CMV-Fny | 2a, 3a ORFs | Feng and Chen, 2013 |
| (e.g., miR171b∗, miR156) | ||||
| 37 miRNA/miRNA∗ | S. lycopersicum | CMV-Q | 2a, 3a ORFs | Feng and Chen, 2013 |
| (e.g., miR171b∗, miR395) | ||||
| 8 miRNA/miRNA∗ | S. lycopersicum | ToLCNDV | AC1, AC2, AC3, | Naqvi et al., 2010 |
| (e.g., miR1918, miR156b∗, miR164a, miR172b∗, miR166a∗) | AV1, AV2, BV1 ORFs | |||
| miR168a | O. sativa | H. sapiens/M. musculus | LDLRAP1 | Zhang et al., 2012a |
| miR2911 | L. ponica | IAVs | PB2 and NS1 | Zhou et al., 2014 |
| vsiR1378 | GFkV | V. vinifers | S2P metalloprotease | Miozzi et al., 2013 |
| vsiR6978 | GRSPaV | V. vinifers | VPS55 | Miozzi et al., 2013 |
| siR221 | TMV-Cg | A. thaliana | CPSE30 | Qi et al., 2009 |
| siR118 | TMV-Cg | A. thaliana | TRAPα | Qi et al., 2009 |
| vsRNA (termed as sRCC1) | CaMV | A. thaliana | At1g76950 (awaits functional characterization) | Moissiard and Voinnet, 2006 |
| Bc-siR3.2 | B. cinerea | A. thaliana | MPK2 and MPK1 | Weiberg et al., 2013 |
| Bc-siR3.1 | B. cinerea | A. thaliana | PRXIF | Weiberg et al., 2013 |
| Bc-siR5 | B. cinerea | A. thaliana | WAK | Weiberg et al., 2013 |
| Bc-siR3.2 | B. cinerea | S. lycopersicum. | MAPKKK4 | Weiberg et al., 2013 |
| Y-sat derived siRNA | CMV | N. tabacum | ChlI | Shimura et al., 2011; Smith et al., 2011 |
Predicted sRNAs of shadow area and validated sRNAs are listed in the first column. The direction of trans-kingdom sRNAs above is from ‘Derivation’ to ‘Target Species.’ Some predicted sRNAs are of large quantities and we do not spread them in the table, just give the number and representative ones, each corresponding to their target genes/ORFs, except of the underlined miR156b∗ targeting two ORFs, AC2, and AC3.
FIGURE 1.
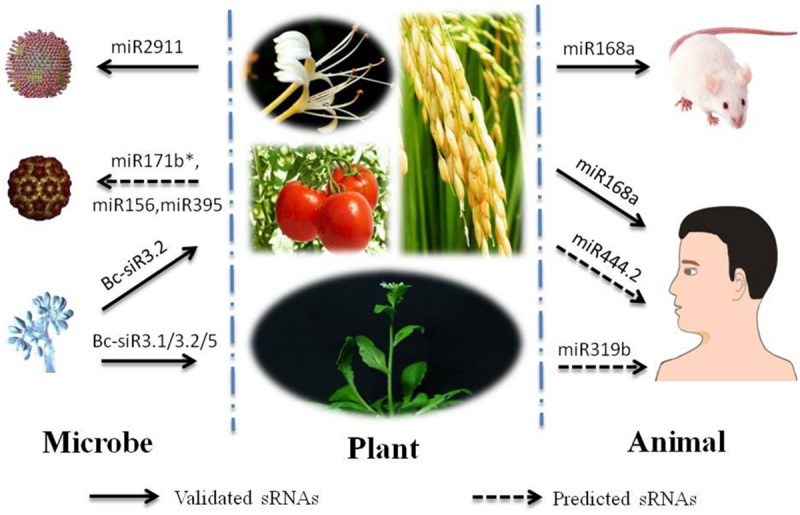
Trafficking of typical cross-kingdom small RNAs (sRNAs) involved with plants. Nine organisms are selected for depiction of cross-kingdom sRNAs involved with plants. Among them, three organisms of ‘Microbe’ column top-down are Influenza A viruses (IAV), CMV and Botrytis cinerea. Four organisms of ‘Plant’ column are Loniceraja ponica (L. ponica), Solanum lycopersicum, and Oryza sativa on the first line and Arabidopsis thaliana on the second line. Two organisms of ‘Animal’ column are Mus musculus (M. musculus) and Homo sapiens(H. sapiens). Cross-kingdom sRNAs are listed in Table 1. The arrows under the cross-kingdom sRNAs point to target species.
In another study, Zhang et al. (2012a) demonstrated that single-stranded mature miRNAs present in rice can exist stably in the sera and tissues of various animals and humans. One of these miRNAs, osa-miR168a, has been shown to be selectively packed into microvesicles (MVs; Liang et al., 2010) that were shed from intestinal epithelial cells and then released into the circulatory system. These MVs efficiently delivered the miRNAs into recipient cells, in which osa-miR168a suppressed the expression of a target gene, low-density lipoprotein receptor adapter protein 1 (LDLRAP1), in the livers of humans and mice, thereby decreasing the removal of low-density lipoprotein from the plasma (Table 1, Figure 1). In addition to miR168a, another type of plant miRNA, miR172, has been observed in the stomach, intestine, serum, and feces of mice after being fed total RNA extracted from Brassica oleracea (Liang et al., 2014b), which suggests that plant miRNAs can survive in the circulatory system and gastrointestinal (GI) tract in mice. Interestingly, synthetic tumor suppressor miRNAs that mimic plant miRNAs can be absorbed by GI tract and functions in reducing tumor burden of mice (Mlotshwa et al., 2015). This method could be utilized to produce edible plants that contain therapeutic tumor miRNAs, which may be applied as clinical small molecules for patient treatment.
miRNAs Transferred from Plants to Viruses
In addition to plant miRNAs that can transfer into the bodies of humans and mice, there are other types of miRNAs that have been speculated to move in a trans-kingdom manner from plants to viruses. Feng and Chen (2013) predicted a total of 38 and 37 tomato miRNA/miRNA∗ sequences that mostly shared high complementarity with the open reading frames (ORFs) of the genomic RNAs of CMV-Fny (severe subgroup 1A strain) and CMV-Q (mild CMV subgroup strain), respectively, which may result in repression of translation or the cleavage of target genes. Importantly, some of these genes act in CMV replication (e.g., CMV protein 2a) and movement (e.g., CMV protein 3a; Table 1, Figure 1). It has been assumed that plants utilize the mechanism of post-transcriptional gene silencing (PTGS) to prevent the replication and spread of CMV virions. Similar results have been obtained in Tomato leaf curl New Delhi virus (ToLCNDV), in which six encoded ORFs of DNA-A and DNA-B were shown to be targeted by eight miRNA/miRNA∗ sequences (Naqvi et al., 2010; Table 1).
Computational prediction alone cannot demonstrate that certain plant miRNAs crossover to viruses. Plant miR2911, the only miRNA that exists stably in honeysuckle decoction (HS decoction) due to its special high G-C content, has been shown to target the genes of Influenza A viruses (IAVs) with the help of MVs in humans and mice (Zhou et al., 2014). Drinking HS decoction results in a significant increase in miR2911 levels in the plasma and lungs of mice. Plant miR2911 can directly bind to the target genes PB2 and NS1, which are essential for influenza replication, thereby inhibiting their amplification (Table 1, Figure 1). Notably, the transport of miR2911 has been shown to be similar to that of miR168a, as both are packed into MVs and go through the GI tract. Afterwards, these miRNAs are transported to target cells through the circulatory system.
The Probable Process of the Horizontal Transfer of miRNAs from Plants to Mammals
When we ingest plant materials, they are preliminarily crushed into debris by the mechanical action of the oral cavity and stomach and simultaneously catabolized into glucose by various digestive enzymes. It has been assumed that, in this process, mature miRNAs are released from the destroyed plant cells and transferred to the small intestine in the gut (Zhang et al., 2012a). Along with AGO2, these miRNAs have been observed to selectively pack into shedding vesicles or exosomes (both called MVs) and are secreted to the outer space by epithelial cells (Cocucci et al., 2009; Elhassan et al., 2012; Zhang et al., 2012a; Zhou et al., 2014). It is worth noting that not all of the miRNA-AGO2 complexes are packed into MVs when entering into the intestinal epithelial cells, and only a minority of the complexes in the MVs can gain access to the recipient cells (Collino et al., 2010). Indeed, exosomal miRNAs are derived from a particular subset of genes (Valadi et al., 2007). In some cases, the packaging of miRNAs into MVs is driven by particular substances, such as an antigen (Mittelbrunn et al., 2011). Via endocytosis and exocytosis, MVs translocate through the vascular wall and are transported to the target cells via the circulatory system, with the ensuing step of releasing the miRNA-AGO2 complexes. Nevertheless, MVs do not interact with all types of cells and interact only with those cells that the MVs specifically recognize. Because foreign miRNAs have high G-C content and specific 2′-O-methylated 3′ ends (Yu et al., 2005), and their transportation is with the help of binding to AGO2 and shielding in MVs (Mitchell et al., 2008; Arroyo et al., 2011; Zhang et al., 2012a; Zhou et al., 2014), they can exist stably under different temperatures, acidification and RNase activity during the process of digestion and transportation.
Roles of Horizontally Transferred siRNAs
siRNAs Transferred from Viruses/Fungi to Plants
Virus-derived siRNAs bind to plant transcripts. vsiRNAs from Grapevine fleck virus (GFkV) and Grapevine rupestris stem pitting-associated virus (GRSPaV) have been predicted to target plant transcripts according to genome-wide identification (Miozzi et al., 2013). It has been reported that 24/26 different grapevine transcripts could be targeted by 27/30 vsiRNAs from the GFkV genome and GRSPaV genome, respectively. To test whether the decrease in grapevine transcripts described above was related to the infection of viruses, Miozzi et al. (2013) carried out quantitative real-time polymerase chain reaction (qRT-PCR) and 5′-RACE analyses. The results supported the idea that a lower accumulation of transcripts, such as S2P metalloprotease and vacuolar protein-sorting 55 (VPS55), was associated with the cleavage of vsiRNA from GFkV (vsiR1378) and GRSPaV (vsiR6978) (Table 1). The former transcript is implicated in the control of regulated intramembrane proteolysis, and the latter is involved in the regulation of the endosomal trafficking of proteins. In the same manner, Tobacco Mosaic Virus (TWV-Cg) siR221 and siR118 target cleavage and polyadenylation specificity factor (CPSE30) and translocon-associated protein alpha (TRAPα) in A. thaliana, respectively (Qi et al., 2009) (Table 1). Two targets of the siRNAs were validated through modified RNA ligase-mediated 5′-RACE experiments. In addition to the RNA viruses described above, sRNAs from the DNA virus Cauliflower Mosaic Virus (CaMV) can also silence plant transcripts (Moissiard and Voinnet, 2006). The CaMV-derived sRNAs mostly come from the polycistronic 35S RNA sequence, which exhibits an extensive secondary structure known as a translational leader. By employing the entire leader sequence in a BLAST search against cDNAs and ESTs from Arabidopsis, three transcripts (At4g05190, At4g17710, and Atlg76950) were retrieved. One of these transcripts, Atlg76950, was confirmed to bind the corresponding vsiRNA (termed as sRCC1; Table 1).
Fungus-derived siRNAs bind to plant transcripts as well. Botrytis cinerea (B. cinera) sRNAs (Bc-sRNAs) that silence plant genes involved in immunity are an example. Normally, pathogens deliver protein effectors into plant cells to suppress plant immunity. However, sRNAs derived from B. cinera may also act as effectors (Weiberg et al., 2013). In A. thaliana, three Bc-sRNAs (Bc-siR3.1, Bc-siR3.2, and Bc-siR5) that structurally mimic plant sRNAs can be loaded into the plant AGO1 protein, after which the Bc-sRNAs target genes with complementary sequences, such as mitogen-activated protein kinase 2 (MPK2) and MPK1 (by Bc-siR3.2); an oxidative stress-related gene, peroxiredoxin (PRXIIF; by Bc-siR3.1); and cell wall-associated kinase (WAK; by Bc-siR5), which are involved in the plant’s immunity against B. cinera. Similar results have been obtained in Solanum lycopersicum, where MAPKKK4 was targeted by Bc-siR3.2 (Table 1, Figure 1).
siRNAs from CMV Satellite RNAs Transferred to Plants
Satellite RNAs (satRNAs), a type of subviral RNA, are encapsulated by their helper viruses, such as CMV. SatRNAs are dispensable for the replication of the genome/subgenome of viruses but have the ability to aggravate or attenuate disease symptoms (Simon et al., 2004). CMV exhibits a tripartite genome whose components are termed RNA1, RNA2, and RNA3, which are mainly required for its replication, virulence, and movement (Kouakou et al., 2013). CMV also contains subgenomic RNAs (RNA4 and RNA4A), which are each responsible for the translation of the coat protein and protein 2b. In addition to genomic and subgenomic RNAs, CMV strains encompass satRNAs, and one type of satRNA, Y satRNAs (Y-sat), can produce siRNAs that may be associated with yellowing symptoms caused by an RNAi mechanism in Nicotiana tabacum (N. tabacum). Because subviral RNA has no ability to encode proteins, it is reasonable to postulate that RNA silencing mediates satellite pathogenicity (Wang et al., 2004). Shimura et al. (2011) and Smith et al. (2011) demonstrated that CMV Y-sat-derived siRNAs could interfere with the mRNA of the host magnesium protoporphyrin chelatase subunit I (ChlI) gene, leading to inhibition of chlorophyll biosynthesis, thus causing the yellow phenotype (Table 1).
Host-Induced Gene Silencing (Higs) Acts as a Tool in the Defense Against Biotic Stress
It has been noted that plant inverted-repeat transgenic constructs, usually with a sense-intron-antisense palindromic structure, can be employed to produce dsRNAs and siRNAs and further silence the transcripts of parasitic organisms. Hence, such RNAi constructs could be designed to test whether HIGS can affect the interaction between plants and parasitic organisms. Based on experimental data, dsRNAs and siRNAs derived from transgenic constructs in host cells would be transferred to fungi/nematodes to achieve silencing of their genes (Nowara et al., 2010; Ibrahim et al., 2011; Yin et al., 2011; Koch et al., 2013; Ghag et al., 2014; Vega-Arreguín et al., 2014). HIGS, which is as an effective transgenic tool, has been used extensively to protect plants from infection by parasitic organisms (Nowara et al., 2010; Nunes and Dean, 2012; Ghag et al., 2014). It is still unknown how these RNA molecules are transferred to the interacting organisms. Nowara et al. (2010) considered it likely that these molecules may travel to the fungi via an exosomal pathway. On the one hand, exosomes are accumulating at plant-fungus contact sites and vesicle fusion/budding have been observed at the haustoria complex, which is responsible for the transfer of nutrients to the fungi. On the other hand, plant multivesicular bodies have been shown to contain small RNAs as well as components of the silencing machinery (Valadi et al., 2007). However, how RNA molecules gain access to the bodies of nematodes and target specific genes remains unknown and requires further elucidation.
Controversies Related to Horizontally Transferred miRNAs that Remain to be Verified
The crossover of miRNAs is not universal between plants and animals. For example, insignificant plant miRNA levels have been detected in the plasma of healthy athletes and mice fed with fruits/vegetables (Snow et al., 2013). In addition, some plant miR168 family members were nearly undetectable when organisms were fed monocot plants in which miR168 levels were relatively higher (Zhang et al., 2012b). Besides, conflicting studies identified low levels of the aforementioned osa-miR168a that did not result in an RNAi-mediated decrease of LDLRAP1 in mouse livers (Dickinson et al., 2013; Witwer et al., 2013). Due to the controversies described above, we mainly focus on the research of Zhang et al. (2012a), who support the cross-kingdom function of miRNAs, and the research of Dickinson et al. (2013), who dispute this function, to examine the causes of the controversy. The experimental methods both show that deep sequencing and qRT-PCR are the main tools for measuring exogenous miRNAs. However, Zhang’s group conducted a more detailed experiment. The sequencing results obtained by Dickson may present a bias in plants, as only a thousand reads per million raw reads of rice sRNAs were detected in rice-containing chow, which is inconsistent with previous high-throughput sequencing studies showing that the miRNA reads obtained from rice generally represent 10% of total reads (Zhu et al., 2008; Jeong et al., 2011; Yi et al., 2013). Thus, it is not surprising that plant miRNAs cannot be detected in mouse sera and livers. As plant miRNAs bear 2′-O-methylated 3′ ends, which can result in a decreased ligation efficiency (Munafo and Robb, 2010), the deep sequencing results reported by Dickson need to be further verified, such as oxidized deep sequencing used by Zhang’s group to test genuine plant miRNAs, with the exception of those based on the qRT-PCR technique (Chen et al., 2013; Dickinson et al., 2013).
Apart from the two major controversial studies, many other studies involving cross-kingdom miRNAs have been conducted using similar methods. Some authors have been unable to detect or have only detected very small amounts of plant-derived miRNAs in mammals (Snow et al., 2013; Witwer et al., 2013), while others, who hold different views, claim that plant miRNAs exist in silkworms but do not play roles in physiological progress (Ling et al., 2015). It is worth noting that new techniques, known as next generation sequencing (NGS) and digital droplet PCR (dPCR), has been applied to test exogenous miRNAs successfully (Wang et al., 2012; Ling et al., 2015). Thus, the existing techniques and experimental design need to be modified for searching more exogenous miRNAs. Regarding the functions of exogenous miRNAs, studies have shown that the concentration of miRNAs affects their ability to target corresponding genes (Mullokandov et al., 2012). Therefore, the concentration of miRNAs in the species of origin has been a major limitation in the detection of miRNAs in other kingdoms thus far. Moreover, Dietary MicroRNA Database (DMD) presents for researchers the types and functions of dietary derived miRNAs, which will be a great tool to explore more of dietary miRNAs in the future (Chiang et al., 2015).
Conclusion
The mobility of sRNA molecules is the key to understanding how sRNA molecules function in regulatory roles between one kingdom and another. In this process, the transportation of sRNAs through the MV pathway is shared both from plants to mammalians or from plants to fungi. Trafficking sRNAs derive from various species, including plants, viruses, and fungi. In most cases, crossover by these RNAs occurs in host-parasites interactions. Unlike miRNAs transferred from plants to mammals, these interactions are not one-sided, but bidirectional. It should be noted that the movement of RNA molecules has been used as a transgenic tool to control plant disease, such as through HIGS. Therefore, the elucidation of cross-kingdom sRNA mechanisms between two interacting organisms is of great interest. The horizontal transfer of sRNAs extends our understanding of sRNAs. In humans, this unique feature may be utilized to control viruses, such as Influenza A viruses, and improve infant immunity when consuming colostrum. For plants, the horizontal transfer of sRNAs can be used to control diseases caused by parasitic organisms. Although the number of plant-associated trafficking sRNAs is fewer compared with those of animals, and controversial views associated with dietary-derived miRNAs await validation, we still remain confident that an increasing number of foreign sRNAs will be studied and utilized in the future.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (31272167, 31471880, and 61472061).
References
- Anderson J. O. (2009). Gene transfer and diversification of microbial eukaryotes. Annu. Rev. Microbiol. 63 177–191. 10.1146/annurev.micro.091208.073203 [DOI] [PubMed] [Google Scholar]
- Arroyo J. D., Chevillet J. R., Kroh E. M., Ruf I. K., Pritchard C. C., Gibson D. F., et al. (2011). Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U.S.A. 108 5003–5008. 10.1073/pnas.1019055108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 281–297. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Buck A. H., Coakley G., Simbari F., McSorley H. J., Quintana J. F., LeBihan T., et al. (2014). Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat Commun 5:5488 10.1038/ncomms6488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington J. C., Ambros V. (2003). Role of microRNAs in plant and animal development. Science 301 336–338. 10.1126/science.1085242 [DOI] [PubMed] [Google Scholar]
- Chen X., Zen K., Zhang C. Y. (2013). Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat. Biotechnol. 31 967–969. 10.1038/nbt.2741 [DOI] [PubMed] [Google Scholar]
- Chiang K., Shu J., Zempleni J., Cui J. (2015). Dietary MicroRNA database (DMD): an archive database and analytic tool for food-borne microRNAs. PLoS ONE 6:e0128089 10.1371/journal.pone.0128089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocucci E., Racchetti G., Meldolesi J. (2009). Shedding microvesicles: artefacts no more. Trends Cell Biol. 19 43–51. 10.1016/j.tcb.2008.11.003 [DOI] [PubMed] [Google Scholar]
- Collino F., Deregibus M. C., Bruno S., Sterpone L., Aghemo G., Viltono L., et al. (2010). Microvesicles Derived from adult human bone marrow and tissue specific mesenchymal stem cells shuttle selected pattern of miRNAs. PLoS ONE 5:e11803 10.1371/journal.pone.0011803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp A., Boschetti C., Perry M., Tunnacliffe A., Micklem G. (2015). Expression of multiple horizontally acquired genes is a hallmark of both vertebrate and invertebrate genomes. Genome Biol. 19:50 10.1186/s13059-015-0607-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson B., Zhang Y. J., Petrick J. S., Heck G., Ivashuta S., Marshall W. S. (2013). Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat. Biotechnol. 31 965–967. 10.1038/nbt.2737 [DOI] [PubMed] [Google Scholar]
- Elhassan M. O., Christie J., Duxbury M. S. (2012). Homo sapiens systemic RNA interference-defective-1 transmembrane family member 1(SIDT1) protein mediates contact-dependent small RNA transfer and MicroRNA-21-driven chemoresistance. J. Bio Chem. 287 5267–5277. 10.1074/jbc.M111.318865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J. L., Chen J. S. (2013). In silico analysis the complementarity of tomato microrna/microrna sequences with cucumber mosaic virus (CMV) genomic RNAs. J. Nanosci. Nanotechnol. 13 4421–4426. 10.1166/jnn.2013.7161 [DOI] [PubMed] [Google Scholar]
- Ghag S. B., Shekhawat U. K., Ganapathi T. R. (2014). Host-induced post-transcriptional hairpin RNA-mediated gene silencing of vital fungal genes confers efficient resistance against Fusarium wilt in banana. Plant Biotechnol. J. 12 541–553. 10.1111/pbi.12158 [DOI] [PubMed] [Google Scholar]
- Gogarten J. P., Doolittle W. F., Lawrence J. G. (2002). Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 19 2226–2238. 10.1093/oxfordjournals.molbev.a004046 [DOI] [PubMed] [Google Scholar]
- Gu Y. R., Li M. Z., Wang T., Liang Y., Zhong Z. J., Wang X. Y., et al. (2012). Lactation-related microRNA expression profiles of porcine breast milk exosomes. PLoS ONE 7:e43691 10.1371/journal.pone.0043691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton A., Voinnet O., Chappell L., Baulcombe D. (2002). Two classes of short interfering RNA in RNA silencing. EMBO J. 21 4671–4679. 10.1093/emboj/cdf464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim H. M. M., Alkharouf N. W., Meyer S. L. F., Aly M. M., Gamal El-Din A. E. K. Y., Hussein E. H. A., et al. (2011). Post-transcriptional gene silencing of root-knot nematode in transformed soybean roots. Exp. Parasitol. 127 90–99. 10.1016/j.exppara.2010.06.037 [DOI] [PubMed] [Google Scholar]
- Jeong D. H., Park S., Zhai J. X., Gurazada S. G. R., De P. E., Meyers B. C., et al. (2011). Massive analysis of rice small RNAs: mechanistic implications of regulated MicroRNAs and variants for differential target RNA cleavage. Plant Cell 12 4185–4207. 10.1105/tpc.111.089045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch A., Kumar N., Weber L., Keller H., Imani J., Kogel K. H. (2013). Host-induced gene silencing of cytochrome P450 lanosterol C14α-demethylase-encoding genes confers strong resistance to Fusarium species. Proc. Natl. Acad. Sci. U.S.A. 110 19324–19329. 10.1073/pnas.1306373110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouakou T. K., Caroline D. C., Thérèse A. A., Olivier P., Philippe L., Haissam J. (2013). Role of satellite RNAs in Cucumber mosaic virus-host plant interactions. A review. Biotechnol. Agron. Soc. Environ. 17 644–650. [Google Scholar]
- Lai E. C. (2003). microRNAs: runts of the genome assert themselves. Curr. Biol. 13 925–936. 10.1016/j.cub.2003.11.017 [DOI] [PubMed] [Google Scholar]
- LaMonte G., Philip N., Reardon J., Lacsina J. R., Majoros W., Chapman L., et al. (2012). Translocation of sickle cell erythrocyte microRNAs into Plasmodium falciparum inhibits parasite translocation and contributes to malaria resistance. Cell Host Microbe 12 187–199. 10.1016/j.chom.2012.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhang Y. J., Li D. M., Liu Y. C., Chu D. P., Jian X. H., et al. (2015). Small non-coding RNAs transfer through mammalian placenta and directly regulate fetal gene expression. Protein Cell 6 391–396. 10.1007/s13238-015-0156-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G. F., Zhu Y. L., Sun B., Shao Y. H., Jing A. H., Wang J. H., et al. (2014a). Assessing the survival of exogenous plant microRNA in mice. Food Sci. Nutr 2 380–388. 10.1002/fsn3.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Zhang S., Fu Z., Wang Y., Wang N., Liu Y., et al. (2014b). Effective detection and quantification of dietetically absorbed plant microRNAs in human plasma. J. Nutr. Biochem. 26 505–512. 10.1016/j.jnutbio.2014.12.002 [DOI] [PubMed] [Google Scholar]
- Liang H. W., Chen X., Zeng K., Zhang C. Y. (2010). A signaling molecule mediating intercellular communication——Microvesicle-transported microRNA. J. Mol. Diagn. Ther. 2 417–423. 10.1016/j.ajpath.2013.06.032 [DOI] [Google Scholar]
- Ling J., Zhang D. Y., Xiang Z. H., He N. J. (2015). Nonfunctional ingestion of plant miRNAs in silkworm revealed by digital droplet PCR and transcriptome analysis. Sci. Rep. 5:12290 10.1038/srep12290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C., Kasschau K. D., Rector M. A., Carrington J. C. (2002). Endogenous and silencing-associated small RNAs in plants. Plant Cell 14 1605–1619. 10.1105/tpc.003210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukasik A., Zielenkiewicz P. (2014). In silico identification of plant miRNAs in mammalian breast milk exosomes – A small step forward? PLoS ONE 9:e99963 10.1371/journal.pone.0099963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miozzi L., Gambino G., Burgyan J., Pantaleo V. (2013). Genome-wide identification of viral and host transcripts targeted by viral siRNAs in Vitis vinifera. Mol. Plant Pathol. 14 30–43. 10.1111/j.1364-3703.2012.00828.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. S., Parkin R. K., Kroh E. M., Fritz B. R., Wyman S. K., Pogosova-Agadjanyan E. L., et al. (2008). Circulating microRNAs as stable blood-based markers for cancerdetection. Proc. Natl. Acad. Sci. U.S.A 108 10513–10518. 10.1073/pnas.0804549105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelbrunn M., Gutierrez-Vazquez C., Villarroya-Beltri C., Gonzalez S., Sanchez-Cabo F., Gonzalez M. A., et al. (2011). Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2:282 10.1038/ncomms1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlotshwa S., Pruss G. J., MacArthur J. L., Endres M. W., Davis C., Hofseth L. J., et al. (2015). A novel chemopreventive strategy based on therapeutic microRNAs produced in plants. Cell Res. 25 521–524. 10.1038/cr.2015.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moissiard G., Voinnet O. (2006). RNA silencing of host transcripts by cauliflower mosaic virus requires coordinated action of the four Arabidopsis Dicer-like proteins. Proc. Natl. Acad. Sci. U.S.A. 103 19593–19598. 10.1073/pnas.0604627103 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Molnar A., Melnyk C. W., Bassett A., Hardcastle T. J., Dunn R., Baulcombe D. C. (2010). Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328 872–887. 10.1126/science.1187959 [DOI] [PubMed] [Google Scholar]
- Mullokandov G., Baccarini A., Ruzo A., Jayaprakash A. D., Tung N., Israelow B., et al. (2012). High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat. Methods 9 840–846. 10.1038/NMETH.2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munafo G. B., Robb G. B. (2010). Optimization of enzymatic reaction conditions for generating representative pools of cDNA from small RNA. RNA 16 2537–2552. 10.1261/rna.2242610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi A. R., Choudhury N. R., Mukherjee S. K., Haq Q. M. (2010). In silico analysis reveals that several tomato microRNA/microRNA∗ sequences exhibit propensity to bind to tomato leaf curl virus (ToLCV) associated genomes and most of their encoded open reading frames (ORFs). Plant Physiol. Biochem. 49 13–17. 10.1016/j.plaphy.2010.09.013 [DOI] [PubMed] [Google Scholar]
- Nowara D., Gay A., Lacomme C., Shaw J., Ridout C., Douchkov D., et al. (2010). HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 22 3130–3141. 10.1105/tpc.110.077040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes C. C., Dean R. A. (2012). Host-induced gene silencing: a tool for understanding fungal host interaction and for developing novel disease control strategies. Mol. Plant Pathol. 13 519–529. 10.1111/j.1364-3703.2011.00766.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Bao F. S., Xie Z. (2009). Small RNA deep sequencing reveals role for Arabidopsis thaliana RNA-dependent RNA polymerases in viral siRNA biogenesis. PLoS ONE 4:e4971 10.1371/journal.pone.0004971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B. J., Weinstein E. G., Rhoades M. W., Bartel B., Bartel D. P. (2002). MicroRNAs in plants. Gene Dev. 16 1616–1626. 10.1101/gad.1004402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura H., Pantaleo V., Ishihara T., Myojo N., Inaba J. I., Sueda K., et al. (2011). A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLoS Pathog. 7:e1002021 10.1371/journal.ppat.1002021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon A. E., Roossinck M. J., Havelda Z. (2004). Plant virus satellite and defective interfering RNAs, New paradigms for a new century. Annu. Rev. Phytopathol. 42 415–437. 10.1146/annurev.phyto.42.040803.140402 [DOI] [PubMed] [Google Scholar]
- Smith N. A., Eamens A. L., Wang M. B. (2011). Viral small interfering RNAs target host genes to mediate disease symptoms in plants. PLoS Pathog. 7:e1002022 10.1371/journal.ppat.1002022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow J. W., Hale A. E., Isaacs S. K., Baggish A. L., Chan S. Y. (2013). Ineffective delivery of diet-derived microRNAs to recipient animal organisms. RNA Biol. 10 1107–1116. 10.4161/rna.24909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J. J., Lötvall J. O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9 654–659. 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- Vega-Arreguín J. C., Jalloh A., Bos J. I., Moffett P. (2014). Recognition of an Avr3a homologue plays a major role in mediating non-host resistance to Phytophthora capsiciin Nicotiana species. Mol. Plant Microbe Interact. 27 770–780. 10.1094/MPMI-01-14-0014-R [DOI] [PubMed] [Google Scholar]
- Wang K., Li H., Yuan Y., Etheridge A., Zhou Y., Huang D., et al. (2012). The complex exogenous RNA spectra in human plasma: an interface with human gut biota? PLos ONE 7:e51009 10.1371/journal.pone.0051009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. B., Bian X. Y., Wu L. M., Liu L. X., Smith N. A., Isenegger D., et al. (2004). On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc. Natl. Acad. Sci. U.S.A. 101 3275–3280. 10.1073/pnas.0400104101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiberg A., Wang M., Lin F. M., Zhao H. W., Zhang Z. H., Kaloshian I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342 118–123. 10.1126/science.1239705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer K. W., McAlexander M. A., Queen S. E., Adams R. J. (2013). Real-time quantitative PCR and droplet digital PCR for plant miRNAs in mammalian blood provide little evidence for general uptake of dietary miRNAs. RNA Biol. 10 1080–1086. 10.4161/rna.25246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R., Zhu Z. X., Hu J. H., Qian Q., Dai J. C., Ding Y. (2013). Identification and expression analysis of micrornas at the grain filling stage in rice (Oryza sativa L.) via deep sequencing. PLoS ONE 3:e57863 10.1371/journal.pone.0057863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C., Jurgenson J. E., Hulbert S. H. (2011). Development of a host-induced RNAi system in the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. Mol. Plant Microbe Interact. 24 554–561. 10.1094/MPMI-10-10-0229 [DOI] [PubMed] [Google Scholar]
- Yu B., Yang Z. Y., Li J. J., Minakhina S., Yang M. C., Padgett R. W., et al. (2005). Methylation as a crucial step in plant microRNA biogenesis. Science 307 932–935. 10.1126/science.1107130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J. P., Hu X. Y., Sun H., Yang Y. P., Huang J. L. (2012). Widespread impact of horizontal gene transfer on plant colonization of land. Nat. Commun. 3:1152 10.1038/ncoms2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Hou D., Chen X., Li D., Zhu L., Zhang Y., et al. (2012a). Exogenous plant MiR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 22 107–126. 10.1038/cr.2011.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wiggins B. E., Lawrence C., Petrick J., Ivashuta S., Heck G. (2012b). Analysis of plant-derived miRNAs in animal small RNA datasets. BMC Genomics 13:381 10.1186/1471-2164-13-381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Li X., Liu J., Dong L., Chen Q., Liu J., et al. (2014). Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res. 25 39–49. 10.1038/cr.2014.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q. H., Spriggs A., Matthew L. (2008). A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Res. 9 1456–1465. 10.1101/gr.075572.107 [DOI] [PMC free article] [PubMed] [Google Scholar]


