Abstract
Type 2 diabetes (T2D) is a serious disease. The gut microbiota (GM) has recently been identified as a new potential risk factor in addition to well-known diabetes risk factors. To investigate the GM composition in association with the dietary patterns in patients with different glucose tolerance, we analyzed 92 patients: with normal glucose tolerance (n=48), prediabetes (preD, n=24), and T2D (n=20). Metagenomic analysis was performed using 16S rRNA sequencing. The diet has been studied by a frequency method with a quantitative evaluation of food intake using a computer program. Microbiota in the samples was predominantly represented by Firmicutes, in a less degree by Bacteroidetes. Blautia was a dominant genus in all samples. The representation of Blautia, Serratia was lower in preD than in T2D patients, and even lower in those with normal glucose tolerance. After the clustering of the samples into groups according to the percentage of protein, fat, carbohydrates in the diet, the representation of the Bacteroides turned to be lower and Prevotella abundance turned to be higher in carbohydrate cluster. There were more patients with insulin resistance, T2D in the fat–protein cluster. Using the Calinski–Harabasz index identified the samples with more similar diets. It was discovered that half of the patients with a high-fat diet had normal tolerance, the others had T2D. The regression analysis showed that these T2D patients also had a higher representation of Blautia. Our study provides the further evidence concerning the structural modulation of the GM in the T2DM pathogenesis depending on the dietary patterns.
Keywords: gut microbiota, 16S rRNA, impaired glucose metabolism, insulin resistance, type 2 diabetes, dietary patterns
Introduction
The epidemic of type 2 diabetes (T2D) gave rise to the numerous studies concerning the mechanisms of T2D. Different factors, associated with lifestyle, environment, and genetics and contributing to the development and progression of T2D, have been identified in the experimental, clinical, and epidemiological studies. It was found that the sociodemographic and environmental factors had the greatest effect (1, 2). However, despite the intensive study and regularly updated guidelines for the T2D management, an increase in the number of T2D patients is considered one of the main problems of the modern world (3, 4). The International Diabetes Federation (5) experts predict a 55% increase in the number of T2D patients reaching up to 592 million by 2035 (5). Recently, it has been established that the human gut microbiota (GM) plays key roles in health maintenance. The GM is altered in the development of chronic inflammation, T2DM, atherosclerosis, hypertension, and many other conditions.
The GM composition has gained more attention with the development of culture-independent techniques. Most of the microorganisms reside in the gastrointestinal tract. Up to 90% of the GM belongs to Bacteroidetes, Firmicutes, and Actinobacteria phyla (6, 7). The GM reacts to physiological, dietary, and climatogeographic factors by changes in its qualitative and quantitative composition (8). For example, it was found that switching from a diet rich in fat and carbohydrates to a diet low in fat and rich in plant polysaccharides (9), as well as switching from a high-fat diet to a diet with a low glycemic index, caused some substantial changes in the GM on the following day (10). Despite most of the studies were descriptive, some specific taxonomic and functional markers, associated with the diet and the development of T2D, have been revealed. Therefore, microbiota has been considered as a suitable target for studying the T2D mechanisms.
The aim of this study was to investigate the GM composition in association with the dietary patterns in patients with different glucose tolerance.
Materials and methods
Patients from Moscow and Moscow Region (the Caucasian race) aged from 25 to 75 years old with different glucose tolerance who had passed the preventive outpatient examination in the FGBI National Research Center for Preventive Medicine (NRCPM) in 2012–2013 were included in the cross-sectional study.
Patients were divided into three groups depending on the glucose metabolism deviation degree
Patients without glucose intolerance, who had passed the preventive outpatient examination.
Patients with prediabetes (preD) (impaired fasting glucose or impaired glucose tolerance or the level of HbA1c from 5.7 to 6.5%.
Patients with newly diagnosed T2D with disease duration <12 months after the diagnosis and the HbA1c level from 6.5 to 9.0%.
Patients were also divided into diet clusters depending on the diet nutrients (by using the k-means clustering).
Criteria for exclusion from the study
The criteria used for exclusion from the study were as follows: type 1 and other specific types of diabetes; regular intake of any drug (including antibiotics during the last 3 months, hypoglycemic drugs); severe diabetic microangiopathy (preproliferative and proliferative diabetic retinopathy, CKD 3b–5 stages); cardiovascular diseases: chronic heart failure class II–IV (New York Heart Association), valvular heart disease; chronic liver and kidney failure; cancer; pregnancy; lactation; moderate and severe anemia; infectious diseases; acute gastrointestinal tract diseases; the operations on the abdominal organs; diagnosed lactase intolerance; diagnosed allergic reaction to any food; a history of organ transplantation; diseases of the oral cavity and dentofacial system; refusal to participate in the study.
Patients with newly diagnosed T2D were included after the outpatient examination. They did not use any hypoglycemic drugs by that time. Also patients (n=5) who did not take hypoglycemic drugs due to different reasons (not high glucose levels, lack of awareness of the disease seriousness, or refusal of medical intervention, etc.) were included. All patients were administered a certain therapy after comprehensive examination.
Ethical aspects
All the patients signed a legal informed consent form to participate in the study. The local ethics committee FGBI NRCPM Ministry of Healthcare, Russian Federation, minutes of the LEC, meeting number #8, 29 November 2011, approved this study protocol. Patient data privacy had been provided using the code identification numbers to correlate with patient records in the computer files.
Patients screening
All the patients underwent a careful clinical assessment during screening. The assessment included medical history, physical examination, height and weight measurements to calculate BMI, and systolic and diastolic blood pressure measurement. The following examinations were performed: blood sampling and urine sampling for laboratory tests (clinical and biochemical) and electrocardiogram. Persons with abnormalities in the blood tests, electrocardiogram, etc. were considered not eligible for inclusion in the study. The first and second groups were performed an oral glucose tolerance test (OGTT).
It should be emphasized that all the participants had underwent a very strong selection process. Selecting was quite labor intensive. We tried to find healthy people without any clinical chronic illnesses. We excluded patients who had used any drugs as well. Only 97 of 858 screened patients had met the inclusion criteria. Glucose metabolism, low-grade inflammation, dietary patterns, and the GM taxonomic composition were estimated in all study participants. Five participants were excluded during the metagenome sequencing because of low quality reads.
Glucose metabolism assessment
The glucose concentration was measured by using the glucose oxidase method on a Sapphire 400 analyzer (Niigata Mechatronics, Tokyo, Japan) by means of DiaSys Diagnostic Kits (DiaSys Diagnostic and Systems, Holzheim, Germany). The HbA1c level was measured by liquid chromatography on a Sapphire 400 analyzer according to the manufacturer's standard procedure.
Insulin level was measured using the chemiluminescence method. HOMA-IR calculation was performed according to the formula: (concentration of fasting blood glucose (mmol/l))×(concentration of fasting blood insulin (mU/l))/22.5. Insulin resistance (IR) was diagnosed if HOMA-IR >2.5 (11).
A 75 g OGTT was performed with blood glucose measurement before glucose intake and 2 h later. Impaired glucose tolerance is considered the state in which the fasting glucose level <7.0 mmol/l, and 2 h later the OGTT ≥7.8 and <11.1 mmol/l. Impaired fasting glucose is considered the state in which the fasting glucose level ≥6.1 and <7.0 mmol/l, and 2 h later OGTT <7.8 mmol/l.
Dietary patterns evaluation
The diet has been studied by a frequency method with a quantitative evaluation of food intake using a standardized computer program ‘Analysis of Human Nutrition’ (version 1.2.4 FGBI Research Institute of Nutrition 2003–2006). Chemical composition, quantity and quality of consumed food, total caloric intake, and the risks of the insufficient or excessive intake of the key vitamins and minerals were estimated. Analysis was made taking into account the ‘Normal physiological needs for energy and nutrients in different population groups in the Russian Federation’ (Guidelines 2.3.1.2432–08).
Assessment of the GM
The collected stool samples (2 ml) were frozen and stored at −20 °C and then thawed; the DNA was extracted from each sample; sequencing of the variable V3–V4 16S rRNA gene regions was performed (after the total DNA isolation and library preparation) by using an MiSeq Reagent Kit v2 (300 cycles) and MiSDefault (Illumina, San Diego, CA, USA) device according to the manufacturer's recommendations.
DNA extraction
Silica beads of diameter 0.1 mm (300 mg) and 0.5 mm (100 mg) were added to a stool sample (125 mg); then 1200 μl of lysis buffer were added (500 mM NaCl, 50 mM Tris–HCl pH 8, 50 mM EDTA, and 4% SDS). The mixture was vortexed for 20 s and homogenized using MiniBeadBeater (BioSpec Products, Bartlesville, OK, USA) for 3 min. The lysate was incubated at 70 °C for 15 min, then centrifuged at 21,910 g for 20 min. Supernatant was transferred to a new 2 ml tube and put on ice; the pellet was added to a lysis buffer and the homogenization process was repeated once. The obtained supernatants were combined in equal volume (700 μl in three tubes for each sample). Two volumes of 96% ethanol (1400 μl) and 1/10 volume of 3 M AcNa (70 μl) were added. The mixture was incubated at −20 °C for 60 min, then centrifuged at 21,910 g for 20 min in 4 °C. The edge supernatant was poured over; 500 μl of 80% ethanol were added to pellet. The mixture was vortexed and centrifuged at 21,910 g for 5 min in 4 °C. The pellet was dried for 30 min and resuspended in 200–400 μl of TE-buffer. The mixture was incubated at 37 °C for 30 min, then centrifuged at 21,910 g for 15 min. The supernatants were transferred and combined in new 1.5 ml tube. One microliter of RNAse A (5 mg/ml) was added to each sample and the mixture was incubated at 37 °C for 60 min. The obtained DNA solution was stored at −20 °C.
Sequencing library preparation
The sequencing libraries were prepared using ‘16S Metagenomic Sequencing Library Preparation: Preparing 16S Ribosomal RNA Gene Amplicons for the Illumina MiSeq System’ protocol (Part Number 15044223 Rev. B, http://ngs.biodiv.tw/NGSCore/wp-content/uploads/application%20forms/16s-metagenomic-library-prep-guide-15044223-b.pdf) using the Nextera XT Index Kit (Illumina) with a dual indexing strategy.
Bioinformatic processing
Quality reads filtering and taxonomic classification were performed using QIIME Software (12). Taxonomic composition of the samples was evaluated using reference-based approach according to the database of 16S rRNA gene sequences Greengenes v. 13.5 (http://greengenes.secondgenome.com/downloads/database/13_50 using RDP Classifier. As a result of the classification, read counts of operational taxonomic units (OTU, taxonomic unit classified to the genus, species or strain, determined by the 16S rRNA gene homology) were; the classifier output was transformed to the form of the OTU and genus relative abundance matrices. All statistical analyses were performed in R programming language (version 3.1.0). Statistical comparison of the groups of samples was performed using Mann–Whitney test (corrected for multiple comparisons using Benjamini–Hochberg method) and generalized linear models (13).
UniFrac dissimilarity metric (14) was used for the construction of multidimensional scaling (MDS) plots; ggplot2 package was used for the illustrations.
The horizontal line in the box-and-whisker plots marks the median; the rectangle lower and upper bounds represent the first and third quartiles respectively; ‘whiskers’ correspond to the distance between quartiles multiplied by 1.5. The values beyond the ‘whiskers’ are considered dropouts and are marked as points.
Results
A total of 92 patients (26 men and 66 women) were included in this study. All the patients were divided into three groups according to the criteria mentioned above. The average duration of preD was 0.22±0.031 years and T2D was 0.78±0.069 years.
Table 1 shows that the average values of age, BMI, waist-to-hip ratio, fasting glucose, and HbA1c were significantly higher in patients with preD and T2D than in healthy individuals. Patients with preD and T2D did not differ in the energy value of the daily diet and in the amount of consumed proteins, fats, and carbohydrates.
Table 1.
Characteristics of patients and nutrients.
| Parameters | Normal glucose tolerance (n=49) | preD (n=25) | T2D (n=23) | P norm-violation/preD – T2D |
|---|---|---|---|---|
| Characteristics | ||||
| Sex (male/female) | 11/38 | 7/18 | 10/13 | 0.24/0.46 |
| Age (years) | 48.31±14.137 | 55±9.92 | 58.35±8.77 | 0.01/0.28 |
| BMI (kg/m2) | 25.13±3.93 | 29±5.88 | 30.5±5.72 | 0.0002/0.48 |
| Waist-to-hip ratio (cm) | 0.81±0.07 | 0.78±0.38 | 0.94±0.09 | 4.7×10−05/0.003 |
| Fasting glucose (mmol/l) | 4.92±0.41 | 5.69±1.18 | 7.76±1.78 | 1.3×10−08/6.4×10−05 |
| HbA1c (%) | 4.92±0.809 | 5.44±0.81 | 6.5±1.58 | 6.4×10−06/0.004 |
| Nutrients | ||||
| Energy (kcal) | 2172.6±463.7 | 2325±809.5 | 1984.4±834.9 | 0.25/0.02 |
| Carbohydrates (g) | 205.1±59.3 | 248±135.5 | 168.1±75 | 0.64/0.02 |
| Proteins (g) | 75.1±20.7 | 81.8±21.5 | 73.8±28.7 | 0.72/0.08 |
| Fats (g) | 101.9±27.9 | 100.4±27.1 | 102.1±51.1 | 0.45/0.3 |
Patients with T2D had higher levels of fasting glucose, HbA1c and waist-to-hip ratio (higher in T2D), as well as in the energy value of the daily diet and the amount of carbohydrates consumed than those with preD.
Sequencing
After the stool samples sequencing, we obtained an average of 1 02 582±46 284 metagenomic 16S rRNA reads per sample. After removal of short- and low-quality reads (QV <20), 1 02 581±39 210 high quality reads (87±2% of the initial amount) were included in the analysis. Out of those, 87.40±7.4% was classified and 97.41±0.9% was classified to the genus level.
The table of the OTU became the result of the reads classification (14).
Dominant phyla and genera
The dominant bacteria in all the samples were Bacteroidetes (12.7±9.86%) and Firmicutes (57.09±13.6%). About 50% of the total microbial abundance was represented by five genera: Blautia, Bacteroides, Prevotella, Faecalibacterium, and Clostridium. Blautia was the most dominant genus. Regression analysis revealed that the representation of Blautia and Serratia in preD patients was higher than in patients with normal glucose tolerance, and even higher in T2D.
Association between microbiota composition and dietary patterns
Using the regression analysis, we examined the correlation between the GM composition and the dietary pattern characteristics. The median of the average daily carbohydrate consumption in all patients was 207±89.7 g, fat consumption was 101.78±29.4 g, and calorie consumption was 2009±648.72 kcal. All numbers are presented in Table 1. In our study calories were mainly coming from the foods rich in fat and protein.
We discovered that an increase in the proportion of carbohydrates in the diet was associated with significantly increased representation of Prevotella (P=0.001), and high sugar consumption was associated with decreased representation of Catenibacterium genus (P=0.003).
Bifidobacterium abundance was significantly reduced in the participants with the high-calorie food (P=0.0001), high cholesterol (P=0.0006), and ethanol (P=0.01) consumption. A positive correlation between high intake of starches and an increased Bifidobacterium species (P=0.0007) was observed in our cohort. Conversely, high intake of starches was associated with the decreased representation of Blautia genus (P=0.0003).
Bacteria associated with diet and preD and T2D
After stratification by diet, it was revealed that the glucose intolerance was associated with some of microbiota members.
Blautia genus: high abundance of this genus was increased in the participants with glucose intolerance, even while consuming fewer carbohydrates (P=0.0004) and fat (P=0.0008) in comparing with healthy donors.
Serratia genus: among the participants, consuming an equal amount of carbohydrates (P=0.003) and calories (P=0.002), the abundance of this genus was higher in donors with glucose intolerance compared with healthy donors.
Verrucomicrobia phylum: decreased representation of Verrucomicrobia phylum was associated with glucose intolerance in donors consuming an equal (>2000 kcal, P=0.0005) and different (healthy >2000, patients with glucose intolerance <2000 kcal, P=0.008) amount of calories per day as well.
Diet clusters
All the samples, for which we had data both on nutrition and on the taxonomic composition of the GM, were divided into clusters according to the percentage of protein, fat, and carbohydrate in the diet. However, taking into account a fairly wide nutrient composition variation, we also used the Calinski–Harabasz index to find the optimal number of dietary clusters.
Clustering made according to the percentage composition of protein, fat, and carbohydrates in the diet (k-medoids clustering basing on Bray–Curtis dissimilarity)
To analyze nutrition types, all the samples were divided into two clusters by using the k-means clustering. The first cluster contained 64 and the second one contained 22 samples. These clusters differed significantly in the protein, fat, and carbohydrate percentage (Table 2).
Table 2.
Characteristics of dietary clusters.
| Nutrients | 1st cluster (n=64) | 2nd cluster (n=22) | P |
|---|---|---|---|
| Protein (%) | 18.81±2.82 | 24.5±4.62 | 8.92×10−07 |
| Fat (%) | 24.49±3.98 | 33.47±3.68 | 2.03×10−10 |
| Carbohydrates (%) | 56.71±5.48 | 42.03±5.61 | 3.33×10−12 |
The first cluster exhibited higher content of carbohydrates in the diet (56.7±5.5% vs 42±5.6%); the second one was characterized by a higher content of fat and protein (correspondingly 33.5±3.7% (fat) and 24.5±4.6% (protein) vs 24.5±4% (fat) and 18.8±2.8% (protein)). Analysis showed that the clusters did not differ in age, sex, and BMI (P=0.5, 0.25, and 0.4 respectively).
The relationship between the dietary clusters and glucose intolerance
Normal glucose metabolism prevalence was similar in both dietary clusters (55 and 50%, in the 1st and 2nd respectively). Although, the second cluster comprised more subjects with T2D (P=0.002) (Table 3, Fig. 1) and IR (P=0.03) than the first one.
Table 3.
Characteristics of patients in clusters.
| Glucose tolerance status | 1st cluster (n=64) | 2nd cluster (n=22) |
|---|---|---|
| Normal carbohydrate metabolism | 35 (54.7%) | 11 (50%) |
| Insulin resistance | 19 (29.7%) | 4 (18.2%) |
| Type 2 diabetes | 10 (15.6%) | 7 (31.8%) |
Figure 1.
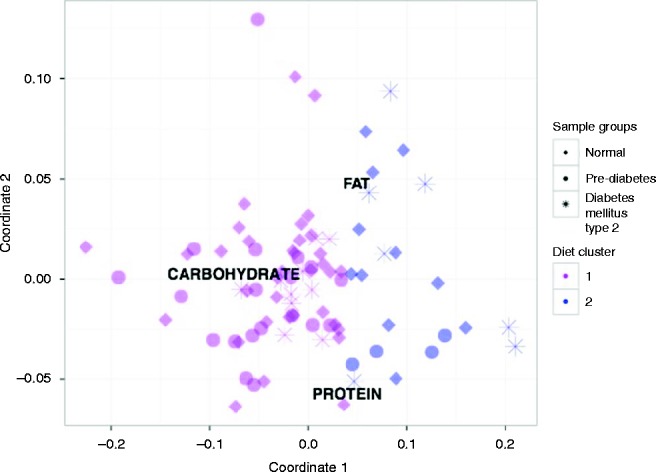
Multidimensional scaling chart of the samples according the diet composition. Note: the pink color indicates the first cluster samples, blue color indicates the second cluster samples. Icons indicate the donor clinical status (healthy, PreD, and T2D).
Bacteria associated with the dietary clusters
These dietary clusters had different relative abundance of two bacterial genera – Bacteroides and Prevotella. Bacteroides level was higher, and Prevotella level was lower in the ‘fat–protein’ cluster (Table 4).
Table 4.
Bacterial genera in which the dietary clusters differed.
| Genera | 1st cluster (n=64) | 2nd cluster (n=22) | P |
|---|---|---|---|
| Bacteroides | 8.36±7.53 | 11.91±9.85 | 0.00011 |
| Prevotella | 4.16±7.3 | 1.91±3.71 | 0.0004 |
Bacteria associated with the dietary clusters and glucose intolerance
Neither generalized linear modeling nor Mann–Whitney test revealed any differences in the taxonomic GM composition between the healthy donors and donors with IR and T2D in the ‘carbohydrate’ dietary cluster. There were also no significant differences in the taxonomic GM composition among the patients with T2D in each of the dietary clusters.
In the ‘fat–protein’ dietary cluster we found the difference in Prevotella genus representation among patients with normal glucose tolerance (2.77±4.06), IR (5.79±10.22), and T2D (3.68±6.13), P=0.0007.
Dietary clusters made by means of the Calinski–Harabasz index
Taking into account a fairly wide nutrient composition variation, we also used Calinski–Harabasz index in order to identify samples with similar diets. As a result, ten clusters had been found (Fig. 2).
Figure 2.
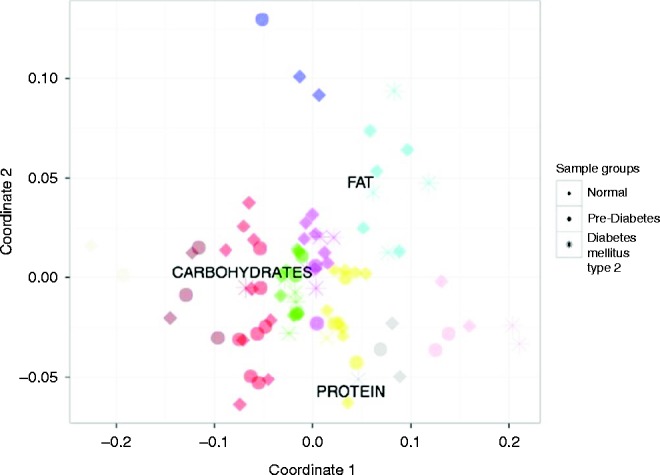
Multidimensional scaling chart of the samples according the diet composition. Note: colors indicate the clusters; icons indicate the donor clinical status (healthy, PreD, and T2D).
Among all clusters, particularly conspicuous was the cluster denoted with cyan color: donors in this cluster consumed a lot of fat, but a half of them had a normal glucose metabolism and the others had T2D. There were no age, sex, and calorie differences between these two groups, as well as any differences in nutrients. They differed only in BMI that was greater in patients with T2D (P=0.03).
Regression analysis showed that the samples from T2D patients had higher Blautia genus representation (P=0.0001). This is consistent with the findings of the previous regression analysis in our study.
Discussion
In order to establish possible associations between the GM changes and glucose intolerance, we had conducted a comprehensive patient examination, which included the GM 16S rRNA sequencing. Participants from Moscow and Moscow region were thoroughly examined. This is the first research on this theme conducted in this area. In our study, the GM composition differed from the usual data, described in the literature, where the most represented bacterial genera were Bacteroides, Prevotella, or some members of the Firmicutes such as Faecalibacterium and Rumminococcus (15). In our study, the microbiota of the samples was predominantly represented by Firmicutes and to a less degree by Bacteroidetes phylum. About 50% of all bacteria were represented by five genera: Blautia, Bacteroides, Prevotella, Faecalibacterium, and Clostridium.
The first data on the GM impact on the glucose metabolism were published in 2004 (16). Published data on the age and gender differences in the GM are contradictory. Several studies have shown that there were no significant gender differences in the GM composition (17), while some others have found the relationship between the GM and sex (18). Perhaps, the male and the female ratio limited our study; however, there were no differences in the GM composition between men and women.
It has been shown that the age-related changes in the intestinal microbiota composition included a lower diversity, an increase in the total number of facultative anaerobes, shifts in the dominant species within some genera (19). Patients older than 60 years accounted for 37% of all cases in our study. The only difference was in the Prevotella genus representation (0.06) between the age groups: under 30 years, 31–45 years, 46–59 years, 60–75 years.
The biggest studies of the association between the GM and T2D were conducted in Sweden (20) and China (21). In published works, there is little information available concerning the drugs prescribed to patients, and it is difficult to analyze and assess such findings. Realizing the fact that any infectious and chronic diseases as well as almost any therapy may influence the GM composition, we analyzed the group of patients who did not use any drugs and who were apparently healthy. For example, in the Swedish study it was shown that patients treated with metformin had an increased level of Enterobacteriaceae and low levels of Clostridium and Eubacterium (not significantly) (17). Despite some differences related to ethnicity, diet, and hypoglycemic drugs intake, these two large studies found that the butyrate-producing bacteria (Roseburia and Faecalibacterium prausnitzii) representation was lower in T2D patients (17, 21). It is known that butyrate-producing bacteria compete with Gram-negative bacteria, maintain microflora balance, and inhibit the growth and reproduction of pathogenic strains, most of which have proteolytic metabolism. Most of the colonic butyrate producers are Gram-positive Firmicutes. However, earlier clinical and experimental studies showed that obesity and glucose intolerance were associated with an increase in Firmicutes. (22). Blautia genus also belongs to Firmicutes phylum but does not produce butyrate. In our study, it was the most represented genus (23). According to Tuovinen (24), Blautia, especially Blautia coccoides, may activate secretion of TNFa, cytokines to an even greater extent than the lipopolysaccharide. In our study, Blautia was the most represented genus, and its representation was higher in patients with T2D (P=0.006), also Blautia representation (P=0.0006) was higher in preD patients than in patients with normal glucose tolerance, and even higher in T2D. However, a number of studies discuss the role of ‘Eubacterium rectale–Blautia coccoides group’ in fermenting oligosaccharides and producing butyrate (25, 26). Apparently, Eubacterium rectale bacteria mostly perform these functions, since Blautia coccoides bacteria do not produce butyrate and do not ferment starches.
According to the recent data, T2D is associated with the high abundance of Bacteroides and low abundance of Prevotella genus (both of them belong to Bacteroidetes phylum) (22). Although, neither the Swedish nor the Chinese study confirmed these data (17, 21). In another study, there was an increase in the Bacteroides and Prevotella representation and a reduction of Firmicutes and Clostridia abundance in patients with T2D comparing with the healthy control group. Moreover, the Bacteroidetes/Firmicutes ratio, and Bacteroides–Prevotella/Blautia coccoides–Eubacterium rectale ratio was positively correlated with plasma glucose concentration (27). In our study, Prevotella genus representation in the second ‘fat–protein’ cluster was increased in patients with glucose intolerance (P=0.0007). Different authors showed that the number of Bifidobacteria was reduced in patients with T2D and obesity (28). Glucose metabolism improvement and chronic inflammation reduction were found in patients who had been administered Bifidobacterium probiotics (29). Published in 2015 study suggested the role of Bifidobacterium in the glucagon-like peptide-1 synthesis (30) Akkermansia muciniphila (Verrucomicrobia) bacteria are also associated with the activity of the intestinal l-cells secreting the glucagon-like peptide-1. They form a close symbiotic community of cells, lining the inner surface of the intestine, and strengthen the intestinal barrier by reducing its permeability (31). In our study, low Verrucomicrobia representation was associated with glucose intolerance, regardless of calorie intake. Verrucomicrobia was also lower in middle age patients with glucose intolerance compared with healthy elderly of the same age (P=0.0005).
It is known that diet affects the GM. For example, high-fat diet decreases the number of Bifidobacteria (32). In our study, Bifidobacterium representation was significantly lower in patients consuming high-calorie food, as well as much cholesterol, or ethanol. On the contrary, high starch consumption increased Bifidobacterium, but reduced Blautia representation. Our data are consistent with the literature, since it is known that Bifidobacteria ferment resistant starch. Blautia hydrolyze some oligosaccharides and short-chain carbohydrates (which virtually do not reach the colon) with the formation of ethanol, hydrogen and succinate, but did not hydrolyze starch (28). Bifidobacterium reducing had not been found in patients with T2D; although high Blautia genus abundance was associated with glucose intolerance, even in patients who consumed fewer carbohydrates and fats. There is evidence that reducing the amount of carbohydrates in the diet leads to a decrease in the representation of some butyrate-producing bacteria – Roseburia and Eubacterium (33). The diet rich in fiber was associated with high bacteria diversity and high Prevotella abundance (34). Reduction of Prevotella during the exclusion of carbohydrates from the diet was observed within 24 h (35). In our research, the representation of Prevotella genus was positively correlated with the proportion of carbohydrates in the diet. After separation of the samples into two dietary clusters, we found the decreased Bacteroides and the increased Prevotella representation in the ‘carbohydrate’ cluster compared with the ‘fat–protein’ cluster. It is assumed that Prevotella genus may be useful for the survival in conditions of insufficient nutrition, but is associated with obesity in individuals consuming so-called ‘Western’ diet (35). We found that the ‘fat–protein’ cluster contained more samples from subjects with IR (P=0.03) and T2D (P=0.002). To identify samples with more similar diets, we had divided all the samples into ten clusters. Donors from one of the clusters eat a lot of fat, a half of them had a normal glucose metabolism, and the others had T2D. They differed only in BMI, which was greater in patients with T2D (P=0.03). Regression analysis showed that T2D patients in this cluster had higher Blautia genus abundance (P=0.0001). Perhaps, a combination of certain diet and microbiota composition may cause some metabolic disturbances. In our cohort, high Blautia abundance together with a high-fat diet was associated with T2D. This finding surely needs further investigation, but the sample here is too small.
Conclusion
Relevant knowledge concerning the structure and biological properties of the microbiota, as well as its relationship with the macroorganism, is constantly expanding. Although the GM role in the energy metabolism regulation is not completely clear and the literature data are contradictory, there are grounds to assume that nutrition is the main way to influence the microbiota composition. We examined participants from Moscow and Moscow region and revealed that the representation of Blautia and Serratia was higher in patients with T2DM. Also we analyzed dietary patterns and found some associations between the diet, composition of the GM, and glucose metabolism as well. Thus, our study provides further evidence about the structural modulation of the GM in the T2DM pathogenesis depending on the dietary patterns.
Acknowledgements
The authors would like to thank the National Research Centre for Preventive Medicine for financial support of this research.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
This research did not receive any specific grant from any funding agency in the public, commercial or not-for-profit sector.
References
- 1.Grarup N, Sandholt C, Hansen T, Pedersen O. Genetic susceptibility to type 2 diabetes and obesity: from genome-wide association studies to rare variants and beyond. Diabetologia. 2014;57:1528–1541. doi: 10.1007/s00125-014-3270-4. [DOI] [PubMed] [Google Scholar]
- 2.Lyssenko V, Jonsson A, Almgren P, Pulizzi N, Isomaa B, Tuomi T, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. New England Journal of Medicine. 2008;359:2220–2232. doi: 10.1056/NEJMoa0801869. [DOI] [PubMed] [Google Scholar]
- 3.Buse JB. Progressive use medical therapies in type 2 diabetes. Diabetes Spectrum. 2000;13:211–228. [Google Scholar]
- 4.American Diabetes Association. Economic Costs of Diabetes in the US in 2012. Diabetes Care. 2013;36:1033–1046. doi: 10.2337/dc12-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.International Diabetes Federation. Diabetes atlas. 6th edn 2013. (available from: http://www.diabetesatlas.org/)
- 6.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zoetendal E, Rajilic-Stojanovic M, de Vos WM. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut. 2008;57:1605–1615. doi: 10.1136/gut.2007.133603. [DOI] [PubMed] [Google Scholar]
- 9.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon1 JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Science Translational Medicine. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madeira IR, Carvalho CN, Gazolla FM, de Matos HJ, Borges MA, Bordallo MA. Cut-off point for homeostatic model assessment for insulin resistance index established from receiver operating characteristic curve in the detection of metabolic syndrome in overweight pre-pubertal children. Arquivos Brasileiros de Endocrinologia and Metabologia. 2008;52:9. doi: 10.1590/S0004-27302008000900010. [DOI] [PubMed] [Google Scholar]
- 12.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marschner IC. glm2: fitting generalized linear models with convergence problems. R Journal 2011 3(2) 12–15. (available at: https://journal.r-project.org/archive/2011-2/RJournal_2011-2_Marschner.pdf).
- 14.Lozupone C, Hamady M, Knight R. UniFrac – an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. PNAS. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dominianni C, Sinha R, Goedert JJ, Pei Z, Yang L, Hayes RB, Ahn J. Sex, body mass index, and dietary fiber intake influence the human gut microbiome. PLoS ONE. 2015;10:e0124599. doi: 10.1371/journal.pone.0124599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gavini F, Cayuela C, Antoine J-M, Lecoq C, Lefebvre B, Membréa J-M, Neuta C. Differences in the distribution of bifidobacterial and enterobacterial species in human faecal microflora of three different (children, adults, elderly) age groups. Microbial Ecology in Health and Disease. 2001;13:40–45. doi: 10.1080/089106001750071690. [DOI] [Google Scholar]
- 20.Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, Nielsen J, Bäckhed F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 21.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 22.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Almeida M, Arumugam M, Batto J-M, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 23.Liu C, Finegold SM, Song Y, Lawson PA. Reclassification of Clostridium coccoides, Ruminococcus hansenii, Ruminococcus hydrogenotrophicus, Ruminococcus luti, Ruminococcus productus and Ruminococcus schinkii as Blautia coccoides gen. nov., comb. nov., Blautia hansenii comb. nov., Blautia hydrogenotrophica comb. nov., Blautia luti comb. nov., Blautia producta comb. nov., Blautia schinkii comb. nov. and description of Blautia wexlerae sp. nov., isolated from human faeces. International Journal of Systematic and Evolutionary Microbiology. 2008;58:1896–1902. doi: 10.1099/ijs.0.65208-0. [DOI] [PubMed] [Google Scholar]
- 24.Tuovinen E, Keto J, Nikkilä J, Mättö J, Lähteenmäki K. Cytokine response of human mononuclear cells induced by intestinal Clostridium species. Anaerobe. 2013;19:70–76. doi: 10.1016/j.anaerobe.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Barcenilla A, Pryde SE, Martin JC, Duncan SH, Stewart CS, Henderson C, Flint HJ. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Applied and Environmental Microbiology. 2000;66:1654–1661. doi: 10.1128/AEM.66.4.1654-1661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simunek J, Brandysova V, Koppova I, Simunek J., Jr The antimicrobial action of chitosan, low molar mass chitosan, and chitooligosaccharides on human colonic bacteria. Folia Microbiologica. 2012;57:341–345. doi: 10.1007/s12223-012-0138-1. [DOI] [PubMed] [Google Scholar]
- 27.Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, Al-Soud WA, Sørensen SJ, Hansen LH, Jakobsen M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE. 2010;5:9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu X, Hui H, Cai D. Differences in fecal Bifidobacterium species between patients with type 2 diabetes and healthy individuals. Nan fang yi ke da xue xue bao. 2012;32:531–533, 564. [PubMed] [Google Scholar]
- 29.Stenman LK, Waget A, Garret C, Klopp P, Burcelin R, Lahtinen S. Potential probiotic Bifidobacterium animalis ssp. lactis 420 prevents weight gain and glucose intolerance in diet-induced obese mice. Beneficial Microbes. 2014;5:437–445. doi: 10.3920/BM2014.0014. [DOI] [PubMed] [Google Scholar]
- 30.Wei P, Yang Y, Li T, Ding Q, Sunet H. An engineered Bifidobacterium longum secreting a bioative penetratin-Glucagon-like peptide 1 fusion protein enhances Glucagon-like peptide 1 absorption in the intestine. Journal of Microbiology and Biotechnology. 2015;8:603–605. [PubMed] [Google Scholar]
- 31.Belzer C, de Vos WM. Microbes inside – from diversity to function: the case of Akkermansia. ISME Journal. 2012;6:1449–1458. doi: 10.1038/ismej.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turnbaugh P, Bäckhed F, Fulton L, Gordon J. Diet induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host and Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rampelli S, Candela M, Turroni S, Biagi E, Collino S, Franceschi C, O'Toole PW, Brigidi P. Functional metagenomic profiling of intestinal microbiome in extreme ageing. Aging. 2013;5:902–912. doi: 10.18632/aging.100623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeffery IB, O'Toole PW. Diet-microbiota interactions and their implications for healthy living. Nutrients. 2013;17:234–252. doi: 10.3390/nu5010234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Durban A, Abellan JJ, Latorre A, Moya A. Effect of dietary carbohydrate restriction on an obesity-related prevotella-dominated human fecal microbiota. Ashdin Publishing Metagenomics. 2013;2:4. [Google Scholar]



 This work is licensed under a
This work is licensed under a