Abstract
AIM: To determine the expression of microRNA-210 (miR-210) in hepatocellular carcinoma (HCC) and to examine its role using HCC cells.
METHODS: The expression of miR-210 was determined in 21 pairs of HCC samples and the corresponding surrounding non-tumor tissues. The effects of miR-210 on proliferation and cell cycle progression were examined using HepG2 and HuH7 cells. Over-expression and inhibition of miR-210 was achieved by transfection of the cells with miR-210 mimic or inhibitor. Luciferase reporter constructs were used to identify the miR-210 interacting site on Yes1. Yes1 expression was examined after miR-210 transfection, as well as in the HCC samples.
RESULTS: miR-210 was significantly up-regulated by 3.4 fold (P < 0.01) in the tumor samples. The over-expression of miR-210 significantly reduced cell proliferation compared to the mock-treated cells (68.9% ± 7.4% and 53.6% ± 5.0%, P < 0.05 for the HepG2 and HuH7 cells respectively). Analysis of the HuH7 cells transfected with miR-210 mimic by flow cytometry showed that the cells took a longer time to reach the G2/M phase. The interaction between miR-210 and the 3’UTR of the Yes1 transcript was confirmed using a luciferase reporter assay. Over-expression of miR-210 reduced the expression of Yes1 protein in both HuH7 and HepG2 cells. Tumors with a greater than four-fold increase in the expression of miR-210 showed consistently lower expressions of Yes1 in the tumors. In nocodazole-treated cells with a significant G2/M cell population, Yes1 protein was significantly reduced and pre-inhibition of miR-210 in HuH7 cells was able to prevent the reduction of Yes1 protein expression. Knock-down of Yes1 by siRNA also led to reduced cell proliferation (70.8% ± 7.5%, P < 0.05 in the HuH7 cells).
CONCLUSION: Up-regulation of miR-210 inhibits cell proliferation. Yes1 is a target of miR-210 and affects cell proliferation in HCC.
Keywords: MicroRNA-210, Hepatocellular carcinoma, Proliferation, Yes1
Core tip: In this study, miR-210 is significantly up-regulated in hepatocellular carcinoma (HCC). Over-expression of miR-210 decreased cell proliferation and delayed cell cycle progression of HCC cells. The tyrosine kinase Yes1 is shown to be a target of miR-210 and is down-regulated in HCC. Knock-down of Yes1 by siRNA also significantly reduced cell proliferation. These results increase the understanding of the multiple roles of miR-210 in liver cancer growth and metastasis.
INTRODUCTION
MicroRNAs (miRNAs) are small, RNA molecules of approximately 20-24 nucleotides in length. Most miRNA genes are transcribed by RNA Polymerase II[1]. The primary transcripts generated are processed in the nucleus to smaller approximately 70-nucleotide stem-looped precursor molecules (pre-miRNAs)[2,3]. The pre-miRNAs are then transported to the cytoplasm[4] where the pre-miRNAs are further processed by Dicer (an RNase III family member)[5,6], releasing approximately 22-nucleotide miRNA-miRNA* duplexes. The duplex is incorporated into the RNA-induced silencing complex (RISC) where it unwinds. Subsequently, one strand of the duplex will serve as the mature miRNA while the other strand (miRNA*) is degraded[7,8]. The mature miRNA that is incorporated into RISC then directs the RISC to its targets, resulting in two possible outcomes. When there is extensive base-pairing between the miRNA and its target RNA, cleavage of the target occurs at a single phosphodiester bond between nucleotides 10 and 11 of the miRNA[9,10]. In vertebrates including humans, most known mRNA targets base-pair only partially to their corresponding miRNAs, such that translation of the target RNA is repressed without cleavage of the target [9,11].
To date, there have been many studies documenting the differential expression of miRNAs in cancers including that of miR-210. It is now established that the expression of miR-210 is induced by hypoxia-inducible factor (HIF-1α)[12-15]. Work has also been carried out to determine the targets of miR-210 and these have been shown to affect many biological processes including angiogenesis, mitochondrial metabolism, DNA repair, apoptosis and cell cycle.
Targets of miR-210 include ephrin A3 and neuronal pentraxin 1[15,16]. Down-regulation of neuronal pentraxin 1 by miR-210 confers protection against hypoxic injury in cortical neurons. However, in vivo, the role of miR-210 in regulating ephrin-A3 expression is unclear as both are up-regulated in ischemic brain[15]. In vitro, the down-regulation of ephrin-A3 by miR-210 is essential for the response of endothelial cells to vascular endothelial growth factor (VEGF)-induced capillary-like formation and chemotaxis[16]. Hypoxia is also a strong inducer of VEGF, and miR-210 was predicted to target VEGF[17]. In addition, RAD52, a key factor in homology-dependent repair, is also a target of miR-210 and this probably accounts for one of the mechanisms leading to the reduced DNA repair activity in hypoxic cells[18]. miR-210 has also been shown to affect mitochondrial oxidative phosphorylation and cause a shift to glycolysis by targeting transcripts encoding various electron transport chain proteins[13,19-23].
The over-expression of miR-210 promotes survival by preventing apoptosis through targeting caspase-8-associated protein 2 and apoptosis-inducing factor, mitochondrion-associated 3[24,25]. In addition, miR-210 also disrupts mitosis by targeting mitosis-related genes[26]. Hence, when expression of miR-210 is low, activation of cell cycle occurs through the up-regulation of other miR-210 targets, E2F3 and fibroblast growth factor receptor-like 1[13,27,28]. However, the over-expression of miR-210 can down-regulate the MYC antagonist MNT, thereby bypassing hypoxia-induced cell cycle arrest[29,30]. Thus like other miRNAs, miR-210 can interact with multiple targets and the possibility of tissue-specific and cell type dependent effects of miR-210 must always be considered.
Both over-expression and down-regulation of miR-210 have been reported in human tumors. Enhanced expression of miR-210 has been observed in breast cancer and is inversely correlated with overall patient survival[12,31,32] but is significantly correlated to cancer aggression and metastatic capacity[33]. In contrast, gene copy loss of miR-210 was observed in epithelial ovarian cancer and melanoma cancer[13,34]. This has been attributed to the location of the miR-210 gene within the minimal region of loss of heterozygosity, LOH11B on 11p15.5. The over-expression of miR-210 has also been reported for hepatocellular carcinoma (HCC)[35-37]. In addition, the up-regulation of miR-210 has been observed in cirrhotic livers indicating the possibility of miR-210 playing important role(s) in the diseased liver[37]. This study thus further examine the role of miR-210 using the hepatocarcinoma cell lines HepG2 and HuH7 and consequently, Yes1, a member of the Src family of non-receptor tyrosine kinases, was identified as a target for miR-210.
MATERIALS AND METHODS
Human liver tissues
Surgical liver tumor tissues and the surrounding non-tumor tissues were obtained from National University Hospital, Singapore. The necessary ethics approval was obtained from the National Health Group Domain Specific Review Board (Reference code: 2012/00394) before the collection and analysis of tissues.
Cell culture
The HepG2 and HuH7 human liver cell lines were obtained from American Type Culture Collection (Manassas, United States), and from RIKEN Bioresource Center (Japan) respectively. Cells were cultured in DMEM, supplemented with 10% fetal bovine serum, 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mmol/L glutamine, 1 mmol/L sodium pyruvate and 0.1 mmol/L MEM nonessential amino acids at 37 °C under 5% CO2. Primary human hepatocytes were purchased from BD Biosciences (San Jose, CA, United States) and cultured according to the manufacturer’s protocol.
Quantitative RT-PCR
Total RNA was extracted from cultured cells or from liver tissues using the TRIzol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s protocols. TaqMan® MicroRNA Individual Assay for miR-210, the TaqMan® MicroRNA Reverse Transcription Kit and the TaqMan® Universal PCR Master Mix without AmpErase® UNG were purchased from Applied Biosystems (Foster City, CA, United States) and used to detect and quantify mature miR-210. This reverse transcription-real-time PCR was carried out as previously described[38] starting with 10 ng of total RNA in each reverse transcription reaction. The 5S rRNA was also quantified via reverse transcription- real time PCR and served as a control for normalization. The 5S rRNA primers and probe were obtained from Sigma-Proligo (The Woodlands, TX, United States). The sequences of these are listed in Table 1.
Table 1.
Primers used for RT-PCR
| Primer sequences | |
| 5S rRNA | For: CGCCCGATCTCGTCTGAT |
| Rev: GGTCTCCCATCCAAGTACTAACCA | |
| Probe: TCGGAAGCTAAGCAGGGTCGGGC | |
| Yes1 mRNA | For: GGACAAGGATGTTTCGGCGA |
| Rev: GATCTCGGTGAATATAGTTC | |
| GAPDH mRNA | For: GAAGGTGAAGGTCGGAGTC |
| Rev: GAAGATGGTGATGGGATTTC |
The real-time PCR reactions were carried out using an ABI PRISM 7300 sequence detection system with the ABI PRISM 7300 SDS software (Applied Biosystems). The relative amount of miR-210 to 5S rRNA was calculated using the equation: 2-ΔCT, where CT is the cycle threshold value and ΔCT = (CTmiR-210 - CT5S rRNA)[39]. To facilitate data presentation, relative gene expression was multiplied by 106. The relative gene expression for miR-210 was calculated for the liver samples (mean ± SE) and the cultured cells (mean ± SD).
For the detection and quantification of Yes1 mRNA, 0.2 μg of total RNA was reverse-transcribed using the Reverse Transcription System (Promega, Madison, WI, United States) with gene-specific reverse primer from Sigma-Proligo (The Woodlands, TX, United States). Real-time PCR amplification was done using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, United States). The GAPDH transcript was used as a normalization control. The sequences of the Yes1 and GAPDH primers are listed in Table 1.
Effects of miRNAs on cell proliferation
HepG2 or HuH7 cells (8 × 103 cells) were seeded into each well of a 48-well plate. The cells were allowed to attach and recover for 24 h. miRIDIAN miRNA mimic or inhibitor (Dharmacon, Lafayette, CO, United States) were diluted in Opti-MEM I reduced serum medium. Lipofectamine 2000 was also diluted 100 times with Opti-MEM I reduced serum medium. Equal volumes of the two solutions were mixed and incubated at 25 °C for 20 min. The cells were rinsed with Opti-MEM I reduced serum medium before the introduction of 200 μL of the miRNA-Lipofectamine 2000 solution (50 nmol/L) to each well. The plates were then incubated at 37 °C for 4 h. Control transfections were carried out with either miRIDIAN microRNA Mimic Negative Control CN-001000-01 or miRIDIAN microRNA Inhibitor Negative Control IN-002005-01 (Dharmacon, Lafayette, CO, United States) while mock transfections were carried out as described but without any mimics or inhibitors. After 4 h, the transfection reagent was removed and replaced with DMEM medium. 40 μL of MTS/PES reagent [3,(4,5-dimethylthiazol-2-yl)-5-(3-carboxy methoxy phenyl)-2-(4-sulfophenyl)-2H tetrazolium/phenazine ethosulfate] (Promega, Madison, WI, United States) was added to each well 72 h after transfection. Following an one-hour incubation at 37 °C, the absorbance at 490 nm was measured. The A490nm of mock transfected cells was set as 100%.
Similarly, for the small interfering RNA (siRNA)-mediated Yes1 knockdown, Silencer Select Validated siRNA for Yes1 (s14955) and the Silencer Select Negative Control No. 1 (4390843) (Ambion, Austin, TX, United States) were transfected at 5 nmol/L for 4 h, after which the transfection reagent was removed and replaced with DMEM medium. At 72 h post-transfection, 40 μL of MTS/PES reagent was added to each well. Following an one-hour incubation at 37 °C, the absorbance at 490 nm was measured. The A490nm of mock transfected cells was set as 100%.
Synchronization of cells
HuH7 cells were synchronized in G1 phase by incubating the cells in 4 mmol/L thymidine for 24 h followed by another 24 h in complete DMEM. To synchronize HuH7 cells at S phase, thymidine double-block was performed by incubating the cells in thymidine for 24 h followed by a 16-h recovery in normal complete medium and another 24-h incubation with thymidine; or alternatively cells were treated with 2.5 mmol/L hydroxyurea in complete medium for 48 h. HepG2 and HuH7 cells were synchronized at pro-metaphase by incubating the cells in 1 μg/mL nocodazole in complete medium for 24 h. This was followed by a mitotic shake-off, and the suspended cells were collected. HepG2 cells were synchronized in G1 phase by incubating mitotic shake-off cells for 4 h in complete medium. To obtain synchronized HepG2 cells at S phase, the cells in G1 phase were further treated with 4 mmol/L thymidine for 24 h.
To elucidate the mechanism involved in the decrease in cell proliferation by miR-210, HuH7 cells were transfected with 50 nmol/L mimic negative control or 50 nmol/L miR-210 mimic followed by sequential incubation with thymidine as described to synchronize the cells. The cells were released from the block by washing away the thymidine and replacing with fresh medium to allow cells to resume cell cycle progression. Samples were collected at different time points for flow cytometry analysis.
Cell cycle analysis
Cells were harvested by trypsinization, centrifuged and fixed with ice-cold 70% ethanol for 2 h, washed with phosphate-buffered saline (PBS), and re-suspended in 0.4 mL of PBS containing 0.1% Triton-X, 20 μg/mL propidium iodide and 0.2 mg/mL RNase A. After a final incubation at 37 °C for at least 30 min, cells were analyzed using a FACSCanto II flow cytometer (Becton Dickinson, Franklin Lakes, NJ, United States). A total of 10000 events were counted for each sample. Data were analyzed using the WinMDI 2.8 software.
Yes1 and MCM8 3’UTR constructs
A 775-bp SpeI-HindIII fragment containing the 3’UTR of the Yes1 cDNA (corresponding to nt-1854 to nt-2612 of the GenBank sequence NM_005433) was generated by reverse transcription-PCR using total RNA extracted from HuH7 cells. Primers used were: Forward: 5’-CGAACTAGTTCAAGTAGCCTATTTTATATG-3’ and Reverse: 5’-GGAAAGCTTCAATGCAACCTCATACAAG-3’. This was then cloned into the pMIR- REPORT Luciferase miRNA Expression Reporter Vector (Ambion, Austin, TX, United States) to generate the pMIR-luciferase Yes1/3’UTR (Luc-YES1) construct and the cloned fragment was verified by sequencing. This construct was used to generate the mutant fragment of 3’UTR of the Yes1 lacking the seed sequence of the miRNA binding site at position 1865-1869 of the sequence NM_005433, and the mutant Luc-Yes1mt construct was verified by sequencing. Similarly, a 561-bp SpeI-HindIII fragment containing the 3’UTR of the MCM8 cDNA (corresponding to nt-2907 to nt-3451 of the GenBank sequence NM_032485) was cloned into the pMIR- REPORT Luciferase miRNA Expression Reporter Vector (Ambion, Austin, TX, United States) to generate the pMIR-luciferase MCM8/3’UTR (Luc-MCM8) construct and the cloned fragment was verified by sequencing. Primers used were: Forward: 5’-TAAACTAGTTCACCAAGTTAGGGCCTCC-3’ and Reverse: 5’-GGAAAGCTTGGCTACCACTACAATTTTTT-3’.
Luciferase reporter assay
HuH7 cells (7 × 104 cells) were seeded into each well of a 24-well plate and allowed to recover for 24 h. 300 μL of miRNA mimic-Lipofectamine 2000 (50 nmol/L) was then added to each well and the plates were incubated at 37 °C. After 3 h the miRNA mimic was removed and the cells were transfected with 25 ng of reporter construct pMIR-luciferase Yes1/3’UTR, or the mutant Luc-Yes1mt, or pMIR-luciferase MCM8/3’UTR and 2.5 ng of pRL-CMV Renilla luciferase control plasmid (Promega, Madison, WI, United States) with Lipofectamine 2000 as the transfection reagent. The transfection solution was removed 3 h later and replaced with DMEM. The cells were lysed with the Passive Lysis Buffer 24 h after transfection and assayed for the firefly luciferase and the Renilla luciferase activities using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, United States). The firefly luciferase activity was normalized to that of the Renilla luciferase activity for each well, and the data was expressed as relative luciferase activity.
Western blot analysis
The cultured cells and liver tissues were lysed in ice-cold 2% Triton X-100 in PBS containing the Halt Protease Inhibitor Single-use Cocktail (Pierce, Rockford, IL, United States). 20 μg of protein from each lysate was separated on a SDS-PAGE gel and transferred onto nitrocellulose membrane. The membranes were incubated with monoclonal antibodies for Yes1 (610376, BD Transduction Laboratories, Lexington, KY, United States), β-actin (Calbiochem, San Diego, CA, United States) or GAPDH (Cell Signaling, Danvers, MA, United States) diluted at 1:5000 each at 4 °C overnight. The membranes were then washed and incubated with the respective goat anti-mouse or anti-rabbit secondary antibodies (Pierce, Rockford, IL, United States) for 1 h at 25 °C. The membranes were washed again and the bound antibodies were detected with the SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL, United States). The chemiluminescent signals were captured on X-ray films and quantified using the Syngene Gbox-HR gel documentation system (Syngene, Cambridge, United Kingdom).
RESULTS
Up-regulation of miR-210 in HCC and in hepatocarcinoma cell lines
The over-expression of miR-210 has been previously described for HCC[35-37]. To confirm this, 21 pairs of HCC and the corresponding non-tumor liver samples were used (Table 2). The expression of miR-210 in the tumor liver samples was 3.4 fold increased over that of the corresponding non-tumor samples (P < 0.01; Figure 1A). The expression of miR-210 was also determined for primary hepatocytes and HCC-derived HepG2 and HuH7 cells. In the hepatocytes, the relative miR-210 expression level was 0.13 ± 0.01 while that for HepG2 and HuH7 cells were 4.37 ± 1.48 and 2.39 ± 0.54 respectively (Figure 1B).
Table 2.
Clinical information of the patients
| Patient | Intrahepatic metastasis | Cirrhosis | Age (yr) | Sex | Race |
| 117 | - | - | 54 | M | Indian |
| 187 | - | + | 61 | M | Chinese |
| 195 | - | + | 65 | M | Indonesian |
| 211 | - | - | 57 | F | Malaysian |
| 233 | - | - | 53 | M | Chinese |
| 247 | - | - | 44 | F | Chinese |
| 198 | + | - | 57 | F | Chinese |
| 201 | + | - | 56 | M | Chinese |
| 206 | + | - | 65 | M | Chinese |
| 216 | + | - | 52 | F | Indian |
| 223 | + | - | 47 | F | - |
| 227 | + | - | 44 | M | Chinese |
| 228 | + | - | 47 | F | Russian |
| 229 | + | - | 75 | F | Chinese |
| 235 | + | - | 64 | F | Chinese |
| 239 | + | - | 53 | M | - |
| 241 | + | + | 65 | M | Chinese |
| 244 | + | - | 45 | F | Chinese |
| 255 | + | - | 64 | M | Chinese |
| 259 | + | - | 45 | M | Indian |
| 261 | + | - | 69 | M | Chinese |
Figure 1.
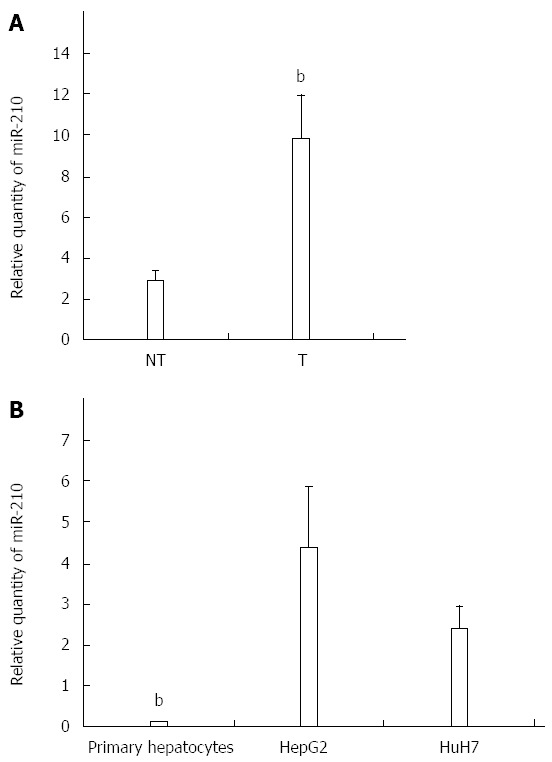
miR-210 expression is up-regulated in hepatocellular carcinoma. Reverse transcription-real time PCR analysis of miR-210 in (A) hepatocellular carcinoma (HCC) tumor (T) and paired non-tumor (NT) samples and (B) primary hepatocytes, HepG2 cells and HuH7 cells. Data shown are expressed as mean ± SE for the HCC paired samples with bP < 0.01, Student’s paired t-test analysis for comparison between tumor and paired non-tumor samples; and mean ± SD for the primary hepatocytes and cell lines with bP < 0.01, Student’s t-test analysis for comparison between primary hepatocytes to either HepG2 or HuH7 cells.
Effects of miR-210 on cell proliferation
miR-210 was over-expressed or blocked by the introduction of 50 nmol/L of the miRIDIAN miR-210 mimic or inhibitor to the HepG2 or HuH7 cells. The over-expression of miR-210 in HepG2 significantly reduced cell proliferation to 68.9% ± 7.4% compared to mock-treated cells (P < 0.05; Figure 2A). However, the inhibition of miR-210 in HepG2 cells did not affect cell proliferation. In HuH7 cells, over-expression of miR-210 significantly reduced cell proliferation to 53.6% ± 5.0% compared to mock-treated cells, while inhibition of miR-210 significantly increased cell proliferation to 145.0% ± 10.8% compared to mock-treated cells (P < 0.05; Figure 2B).
Figure 2.
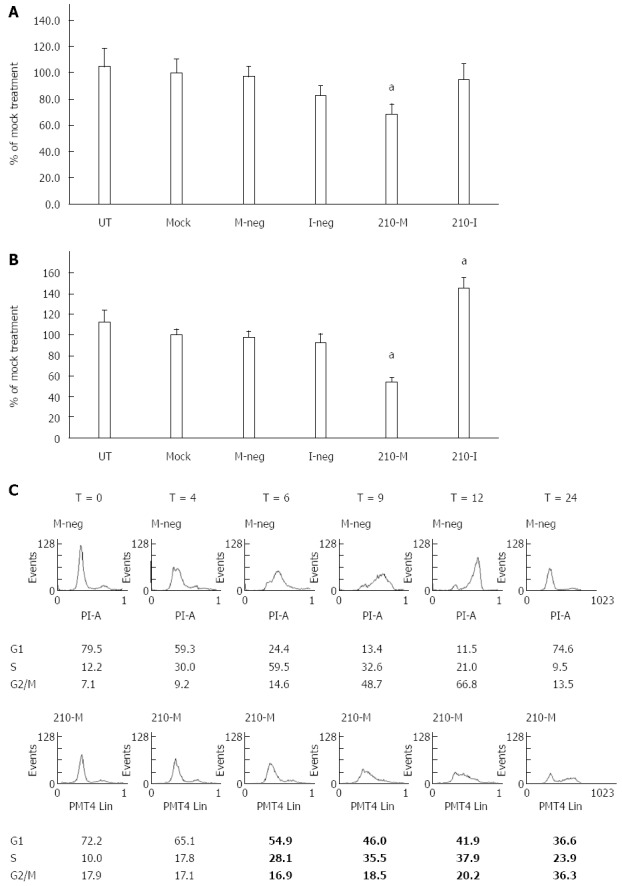
Effects of miR-210 on proliferation of hepatocellular carcinoma cells. A and B: HepG2 cells (A) or HuH7 cells (B) were left untreated (UT), or mock transfected with Lipofectamine 2000 (Mock), or transfected with Mimic Negative Control (M-Neg), or Inhibitor Negative Control (I-Neg), or microRNA-210 Mimic (210-M), or microRNA-210 Inhibitor (210-I). Cell proliferation was determined using the MTS assay. Data shown are expressed as mean ± SD (n = 4). aP < 0.05, Student’s t-test analysis for comparison to the mock treatment; C: Flow cytometry analysis of HuH7 cells at various time points (T = 0 h - T = 24 h) following transfection with either M-neg or 210-M and synchronization of the cells. Percentage of total cell population in each phase is shown below each graph.
To elucidate the mechanism involved in the decrease of cell proliferation, HuH7 cells transfected with miR-210 mimic or the negative control were synchronized and allowed to resume cell cycle progression. Analysis of the cells transfected with miR-210 mimic by flow cytometry did not show any significant accumulation of cells with sub-G1 DNA content which is suggestive of apoptotic cell death (Figure 2C). Cells treated with the miRNA mimic negative control entered the G2/M phase by 12 h. In contrast, cells transfected with miR-210 took a longer time to reach the G2/M phase with < 40% of the cells in this phase at 24 h. Taken together, these data indicate that over-expression of miR-210 inhibits cell proliferation by delaying cell cycle progression.
Yes1 is a target of miR-210
To identify putative targets of miR-210 a search of the computational target prediction programs miRBase and miRanda[40,41] was carried out. The list of relevant targets was narrowed down to those that would affect cell proliferation or cell cycle progress. Based on these criteria, MCM8 and Yes1 were selected for further study. The putative binding sites for miR-210 for these two transcripts are shown in Figure 3.
Figure 3.

Genes targeted by miR-210 as predicted by miRBase/miRanda and selected for further analysis in this study.
MCM8 is an MCM2-7-related protein. It functions as a DNA helicase during replication elongation[42]. Yes1 is a member of the Src family of non-receptor tyrosine kinases and has been shown to be involved in the regulation of many cellular processes, including cell proliferation, survival and differentiation[43,44]. To demonstrate the direct interaction between miR-210 and the predicted target transcripts, luciferase reporter constructs containing portions of the MCM8 3’UTR or Yes1 3’UTR with the predicted miR-210 interacting sites were generated. In the presence of miR-210 mimic, the relative luciferase activity was significantly reduced for the Luc-YES1 construct but not the Luc-MCM8 construct (P < 0.05; Figure 4A). As a control, no reduction was observed with the Luc-YES1mt mutant construct with deletions in the seed sequence of the miRNA binding site (Figure 4A).
Figure 4.
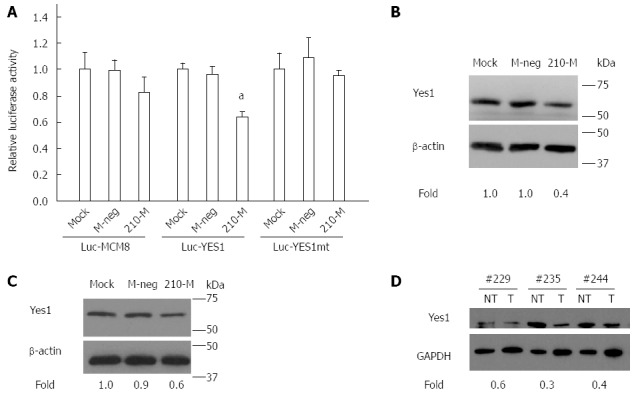
Regulation of Yes1 by miR-210. A: Relative luciferase activity of HuH7 cells which were mock-transfected with Lipofectamine 2000 (Mock), or transfected with Mimic Negative Control (M-Neg), or microRNA-210 Mimic (210-M) followed by transfection with the Luc-MCM8 or Luc-Yes1 or Luc-Yes1mt reporter constructs and the pRL-CMV Renilla luciferase control plasmid. Data shown are expressed as mean ± SD. Assays were carried out in triplicate and as two independent experiments. aP < 0.05, Student’s t-test analysis for comparison to the corresponding control; B and C: Western blot analysis of Yes1 protein expression (top panel) in HuH7 cells (B) or HepG2 cells (C) treated with Mock, M-Neg, or 210-M. β-actin was used as the control for normalization (bottom panel). Yes1 expression was normalized to that of β-actin and the fold change was determined by comparison with the levels in Mock transfected cells; D: Western blot analysis of Yes1 expression (top panel) in HCC tumor (T) and paired non-tumor (NT) samples. GAPDH was used as the control for normalization (bottom panel). Yes1 expression was normalized to that of GAPDH and the fold change was determined by comparison with the paired non-tumor samples.
Subsequently, experiments were carried out to determine whether miR-210 can regulate the expression of Yes1 protein in HCC. Over-expression of miR-210 in HuH7 and HepG2 cells led to reduced expression of Yes1 protein (Figure 4B and C). In the HCC samples, tumors with higher expression of miR-210 (> 4-fold increase) showed consistently lower expressions of Yes1 compared to the corresponding paired non-tumor samples (Figure 4D).
Expressions of miR-210 and Yes1 protein during cell cycle
The expressions of miR-210 and Yes1 protein during the different phases of the cell cycle were also examined. Treatment of HepG2 or HuH7 cells with thymidine, nocodazole or hydroxyurea as described led to enrichment of cells (> 75%) in the different phases of the cell cycle (data not shown). miR-210 was specifically up-regulated only in the nocodazole-arrested HuH7 cells which were enriched with cells in the G2/M phase (Figure 5A). Yes1 transcript and protein were expressed in all phases but its protein expression was greatly reduced in the nocodazole-arrested cells (Figure 5B-D). This reduction in Yes1 expression and the presence of different forms is similar to that described in earlier studies[45,46].
Figure 5.
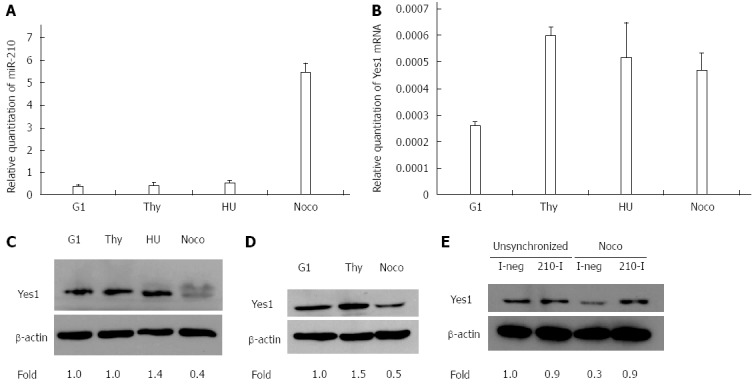
Expression of miR-210 and Yes1 in the different phases of the cell cycle. A: Expression of miR-210 in HuH7 cells; B: Expression of Yes1 transcript in HuH7 cells; C and D: Western blot analysis of Yes1 protein expression (top panel) and in HuH7 cells (C) and HepG2 cells (D). Cells were enriched in the different cell cycle phases corresponding to G1 phase, S phase with thymidine double block treatment (Thy) or hydroxyurea treatment (HU), and G2/M phase with nocodazole treatment and mitotic shake-off (Noco); E: Western blot analysis of Yes1 protein expression (top panel) was performed with unsynchronized or nocodazole-treated (Noco) HuH7 cells following transfection with Inhibitor Negative Control (I-neg) or microRNA-210 Inhibitor (210-I). β-actin was used as the control for normalization (bottom panel). Yes1 expression was normalized to that of β-actin and the fold change was determined by comparison with the levels in G1-enriched cells (for C, D) or the unsynchronized cells transfected with I-neg (for E).
To examine the influence of miR-210 on Yes1 protein levels during cell cycle, HuH7 cells were transfected with miR-210 inhibitor or with the inhibitor negative control for 4 h before nocodazole treatment. After incubating the cells with nocodazole for 24 h to synchronize the cells, the cells were harvested for the analysis of Yes1 protein expression. The decrease of Yes1 protein expression was reversed by pre-treatment with miR-210 inhibitor (Figure 5E) indicating that Yes1 is indeed a target of miR-210.
Silencing of Yes1 reduces proliferation of HCC cells
The effect of silencing Yes1 on cell proliferation was also examined. Yes1 in HuH7 cells was knocked down by siRNA targeting the open reading frame of Yes1, and the cell proliferation was significantly reduced to 70.8% ± 7.5% compared to mock-treated cells (P < 0.05; Figure 6), suggesting that the silencing of Yes1 can contribute to the decreased cell proliferation effect, similar to that observed when miR-210 was over-expressed.
Figure 6.
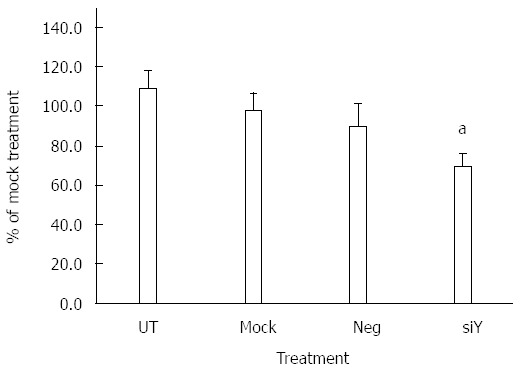
Silencing of Yes1 reduces proliferation of hepatocellular carcinoma cells. HuH7 cells were untreated (UT), or mock transfected with Lipofectamine 2000 (Mock), or transfected with either siRNA negative control (Neg) or siRNA targeting Yes1 (siY). Cell proliferation was determined using the MTS assay. Data shown represent mean ± SD (n = 4). aP < 0.05, Student’s t-test analysis for comparison to mock treatment.
DISCUSSION
In this study, we confirm the up-regulation of miR-210 in HCC tumors as well as in HCC-derived cells. This observation is consistent with earlier studies which had reported the over-expression of miR-210[35-37]. miR-210 has also been identified as one of twelve miRNAs which was consistently deregulated during the development of liver cirrhosis and subsequently during the progress from cirrhosis to HCC[37]. Thus miR-210 is likely to play an important role in disease progression in the liver.
To further examine the role of miR-210, its effect on HCC-derived HepG2 and HuH7 cells was determined. In both, over-expression of miR-210 led to significant reduction in cell proliferation. Yes1, a member of the Src family of non-receptor tyrosine kinases, was identified as a target for miR-210. The ability of miR-210 to interact with the 3’UTR of the Yes1 transcript was observed using a luciferase reporter construct and the reduced expression of Yes1 protein was evident following the over-expression of miR-210.
The Yes1 tyrosine kinase was identified as the cellular homologue of the oncogenic viral yes gene product[47,48]. This 62 kDa non-receptor tyrosine kinase is expressed in most tissues[49]. Yes1 has been implicated in a variety of signaling pathways, which include mediating cytokine and growth factor responses, alterations in the cytoskeleton, cell cycle progression, apoptosis and differentiation[43,44]. Our data shows that the expression of Yes1 was consistently high in G1 phase and S phase but was reduced in the G2/M cell population. Significant reduction in Yes1 expression during mitosis has also been previously described by Park et al[45]. However, it was unclear what regulates this reduced expression. In this study, we provide evidence that miR-210 contributes to the decreased Yes1 protein expression during cell cycle. In nocodazole-treated cells with a significant G2/M cell population, Yes1 protein is significantly reduced while that of miR-210 is significantly raised, and inhibition of miR-210 is able to restore Yes1 protein expression. In addition, the decreased Yes1 protein expression was not due to a significant reduction in the Yes1 transcript. Hence, it is likely that miR-210 represses the translation of Yes1. Interestingly, miR-210 has also been shown to regulate several other mitosis-related genes including Plk1, Cdc25B, Cyclin F, Bub1B and Fam83D[26].
Inhibition of Yes1 through knockdown using siRNA or the over-expression of miR-210 led to reduced cell proliferation. Similar knockdown studies of Yes1 by siRNA in human retinal microvascular endothelial cells showed decreased VEGF-induced cell proliferation, with no changes in DNA fragmentation[50]. The over-expression of miR-210 also led to delayed cell cycle progression with a significant delay at the G1/S transition. This effect is not unexpected as there is robust expression of Yes1 in cells in the G1 and S phases and hence, it is likely that Yes1 may be important in either one or both of these phases. Indeed it has been observed that members of the Src family are required to simulate entry of cells into S phase[51] and silencing of Yes1 expression has been shown to reduce cell growth through G1 cell cycle arrest via the inactivation of β-catenin signaling[52]. It is also likely that the over-expression of miR-210 may affect other targets such as E2F3 leading to the observed effect of significant delay in G1/S progression[13].
Each miRNA can potentially interact with multiple targets. This is likely to be the case for miR-210 in the context of the diseased liver and HCC. miR-210 has been shown to also target the hepatitis B virus S protein and pre-S protein coding regions and this may serve to regulate virion production in the chronic infection state or in the latency state[53]. The up-regulation of miR-210 expression over that of healthy liver has been observed in cirrhotic liver[37]. This study and that of others have reported increased expression of miR-210 in HCC[35-37]. Yes1 has also been detected in cirrhotic livers and in HCC[54,55]. In this study, Yes1 was expressed in both tumor and non-tumor liver samples. However, decreased Yes1 protein was observed in HCC tumors with high miR-210 expression. Similar to an earlier study by Ying et al[56], this study too provide evidence that over-expression of miR-210 led to reduced cell proliferation. In addition, this present study showed that this effect on cell growth was mediated though Yes1. It is thus likely that in cirrhotic livers and in HCC, the increase in miR-210 expression may help keep in check cell cycle progression and cell proliferation by repressing various miR-210 targets including Yes1.
Over-expression of miR-210 also promotes hypoxia-induced migration and invasion. In an earlier study by Ying et al[56], miR-210 was shown to mediate HCC metastasis through the down-regulation of vacuole membrane protein 1. miR-210 has been shown to also target apoptosis-inducing factor, mitochondrion-associated 3 in human hepatoma cells thereby preventing apoptosis[25]. Thus it is evident that miR-210 does have multiple roles in the liver. Its roles in hypoxia, apoptosis, cell proliferation and metastasis are likely to be important for cancer development and progression and more work has to be done to fully understand the roles of miR-210 in the context of liver cirrhosis and HCC.
ACKNOWLEDGMENTS
The authors thank Dr Aung Myat O for his assistance in the collection of the liver tissues for this study.
COMMENTS
Background
MicroRNAs (miRNAs) are small, RNA molecules involved in the process of silencing gene expression. The mature miRNAs of 20-24 nucleotides direct the RNA-induced silencing complex (RISC) to silence the expression of their complement target mRNAs. To date, there have been many studies documenting the differential expression of miRNAs in cancers including that of miR-210. The over-expression of miR-210 has been reported for hepatocellular carcinoma (HCC) and cirrhotic livers, indicating the possibility of miR-210 playing important role(s) in the diseased liver.
Research frontiers
miR-210 is hypoxia-inducible, and the over-expression of miR-210 in HCC was previously reported to promote metastasis through the down-regulation of vacuole membrane protein 1. miR-210 has also been shown to also target apoptosis-inducing factor, mitochondrion-associated 3 in human hepatoma cells thereby preventing apoptosis.
Innovations and breakthroughs
This present study showed that over-expression of miR-210 in HCC inhibited cell proliferation and cell cycle progression, and this effect on cell growth was mediated though Yes1, a member of the Src family of non-receptor tyrosine kinases.
Applications
It is evident that miR-210 is frequently over-expressed in cirrhotic livers and HCC. The increase in miR-210 expression may help keep in check cell cycle progression and cell proliferation by repressing various miR-210 targets including Yes1.
Terminology
Yes1 is a non-receptor tyrosine kinase that is expressed in most tissues. Yes1 has been implicated in a variety of signalling pathways, which include mediating cytokine and growth factor responses, alterations in the cytoskeleton, cell cycle progression, apoptosis and differentiation.
Peer-reviewer
Authors said that miR-210 is significantly up-regulated in HCC and over-expression of miR-210 decreased cell proliferation and delayed cell cycle progression of HCC cells via down-regulation of Yes1. This paper has been well described to suggest that Yes1 is a target of miR-210 and affects cell proliferation in HCC.
Footnotes
Supported by Biomedical Research Council and Ministry of Education (Tier 1) awarded to Tan TM.
Institutional review board statement: The study was reviewed and approved by the National Health Group Domain Specific Review Board.
Institutional animal care and use committee statement: This is not applicable. No animal was used for this study.
Conflict-of-interest statement: To the best of our knowledge, no conflict of interest exists.
Data sharing statement: No additional data are available.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: May 23, 2015
First decision: July 19, 2015
Article in press: October 20, 2015
P- Reviewer: Yang H, Yu DY S- Editor: Ma YJ L- Editor: A E- Editor: Liu XM
References
- 1.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 3.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 5.Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 6.Billy E, Brondani V, Zhang H, Müller U, Filipowicz W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc Natl Acad Sci USA. 2001;98:14428–14433. doi: 10.1073/pnas.261562698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 8.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hutvágner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 10.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 11.Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, Harris AL, Gleadle JM, Ragoussis J. hsa-miR-210 is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res. 2008;14:1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 13.Giannakakis A, Sandaltzopoulos R, Greshock J, Liang S, Huang J, Hasegawa K, Li C, O’Brien-Jenkins A, Katsaros D, Weber BL, et al. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol Ther. 2008;7:255–264. doi: 10.4161/cbt.7.2.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M, et al. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pulkkinen K, Malm T, Turunen M, Koistinaho J, Ylä-Herttuala S. Hypoxia induces microRNA miR-210 in vitro and in vivo ephrin-A3 and neuronal pentraxin 1 are potentially regulated by miR-210. FEBS Lett. 2008;582:2397–2401. doi: 10.1016/j.febslet.2008.05.048. [DOI] [PubMed] [Google Scholar]
- 16.Fasanaro P, D’Alessandra Y, Di Stefano V, Melchionna R, Romani S, Pompilio G, Capogrossi MC, Martelli F. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283:15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hua Z, Lv Q, Ye W, Wong CK, Cai G, Gu D, Ji Y, Zhao C, Wang J, Yang BB, et al. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crosby ME, Kulshreshtha R, Ivan M, Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009;69:1221–1229. doi: 10.1158/0008-5472.CAN-08-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan SY, Zhang YY, Hemann C, Mahoney CE, Zweier JL, Loscalzo J. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab. 2009;10:273–284. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fasanaro P, Greco S, Lorenzi M, Pescatori M, Brioschi M, Kulshreshtha R, Banfi C, Stubbs A, Calin GA, Ivan M, et al. An integrated approach for experimental target identification of hypoxia-induced miR-210. J Biol Chem. 2009;284:35134–35143. doi: 10.1074/jbc.M109.052779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Favaro E, Ramachandran A, McCormick R, Gee H, Blancher C, Crosby M, Devlin C, Blick C, Buffa F, Li JL, et al. MicroRNA-210 regulates mitochondrial free radical response to hypoxia and krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PLoS One. 2010;5:e10345. doi: 10.1371/journal.pone.0010345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Z, Li Y, Zhang H, Huang P, Luthra R. Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene. 2010;29:4362–4368. doi: 10.1038/onc.2010.193. [DOI] [PubMed] [Google Scholar]
- 23.Puisségur MP, Mazure NM, Bertero T, Pradelli L, Grosso S, Robbe-Sermesant K, Maurin T, Lebrigand K, Cardinaud B, Hofman V, et al. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 2011;18:465–478. doi: 10.1038/cdd.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim HW, Haider HK, Jiang S, Ashraf M. Ischemic preconditioning augments survival of stem cells via miR-210 expression by targeting caspase-8-associated protein 2. J Biol Chem. 2009;284:33161–33168. doi: 10.1074/jbc.M109.020925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang W, Sun T, Cao J, Liu F, Tian Y, Zhu W. Downregulation of miR-210 expression inhibits proliferation, induces apoptosis and enhances radiosensitivity in hypoxic human hepatoma cells in vitro. Exp Cell Res. 2012;318:944–954. doi: 10.1016/j.yexcr.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 26.He J, Wu J, Xu N, Xie W, Li M, Li J, Jiang Y, Yang BB, Zhang Y. MiR-210 disturbs mitotic progression through regulating a group of mitosis-related genes. Nucleic Acids Res. 2013;41:498–508. doi: 10.1093/nar/gks995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biswas S, Roy S, Banerjee J, Hussain SR, Khanna S, Meenakshisundaram G, Kuppusamy P, Friedman A, Sen CK. Hypoxia inducible microRNA 210 attenuates keratinocyte proliferation and impairs closure in a murine model of ischemic wounds. Proc Natl Acad Sci USA. 2010;107:6976–6981. doi: 10.1073/pnas.1001653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsuchiya S, Fujiwara T, Sato F, Shimada Y, Tanaka E, Sakai Y, Shimizu K, Tsujimoto G. MicroRNA-210 regulates cancer cell proliferation through targeting fibroblast growth factor receptor-like 1 (FGFRL1) J Biol Chem. 2011;286:420–428. doi: 10.1074/jbc.M110.170852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Z, Sun H, Dai H, Walsh RM, Imakura M, Schelter J, Burchard J, Dai X, Chang AN, Diaz RL, et al. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle. 2009;8:2756–2768. doi: 10.4161/cc.8.17.9387. [DOI] [PubMed] [Google Scholar]
- 30.Devlin C, Greco S, Martelli F, Ivan M. miR-210: More than a silent player in hypoxia. IUBMB Life. 2011;63:94–100. doi: 10.1002/iub.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 32.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foekens JA, Sieuwerts AM, Smid M, Look MP, de Weerd V, Boersma AW, Klijn JG, Wiemer EA, Martens JW. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci USA. 2008;105:13021–13026. doi: 10.1073/pnas.0803304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, Zhuang SM. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135–1142. doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- 37.Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM, Dejean A. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci USA. 2010;107:264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Tan W, Neo TW, Aung MO, Wasser S, Lim SG, Tan TM. Role of the miR-106b-25 microRNA cluster in hepatocellular carcinoma. Cancer Sci. 2009;100:1234–1242. doi: 10.1111/j.1349-7006.2009.01164.x. [DOI] [PubMed] [Google Scholar]
- 39.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 40.miRBase. Available from: http://microrna.sanger.ac.uk/targets/v4/
- 41.microRNA.org. Available from: http://cbio.mskcc.org/microrna-previous/
- 42.Maiorano D, Cuvier O, Danis E, Méchali M. MCM8 is an MCM2-7-related protein that functions as a DNA helicase during replication elongation and not initiation. Cell. 2005;120:315–328. doi: 10.1016/j.cell.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 43.Clump DA, Qazi IH, Sudol M, Flynn DC. c-Yes response to growth factor activation. Growth Factors. 2005;23:263–272. doi: 10.1080/08977190500199360. [DOI] [PubMed] [Google Scholar]
- 44.Summy JM, Sudol M, Eck MJ, Monteiro AN, Gatesman A, Flynn DC. Specificity in signaling by c-Yes. Front Biosci. 2003;8:s185–s205. doi: 10.2741/1011. [DOI] [PubMed] [Google Scholar]
- 45.Park J, Cartwright CA. Src activity increases and Yes activity decreases during mitosis of human colon carcinoma cells. Mol Cell Biol. 1995;15:2374–2382. doi: 10.1128/mcb.15.5.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuga T, Nakayama Y, Hoshino M, Higashiyama Y, Obata Y, Matsuda D, Kasahara K, Fukumoto Y, Yamaguchi N. Differential mitotic activation of endogenous c-Src, c-Yes, and Lyn in HeLa cells. Arch Biochem Biophys. 2007;466:116–124. doi: 10.1016/j.abb.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 47.Yoshida M, Kawai S, Toyoshima K. Unifected avian cells contain structurally unrelated progenitors of viral sarcoma genes. Nature. 1980;287:653–654. doi: 10.1038/287653a0. [DOI] [PubMed] [Google Scholar]
- 48.Sukegawa J, Semba K, Yamanashi Y, Nishizawa M, Miyajima N, Yamamoto T, Toyoshima K. Characterization of cDNA clones for the human c-yes gene. Mol Cell Biol. 1987;7:41–47. doi: 10.1128/mcb.7.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sudol M, Hanafusa H. Cellular proteins homologous to the viral yes gene product. Mol Cell Biol. 1986;6:2839–2846. doi: 10.1128/mcb.6.8.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Werdich XQ, Penn JS. Src, Fyn and Yes play differential roles in VEGF-mediated endothelial cell events. Angiogenesis. 2005;8:315–326. doi: 10.1007/s10456-005-9021-x. [DOI] [PubMed] [Google Scholar]
- 51.Twamley-Stein GM, Pepperkok R, Ansorge W, Courtneidge SA. The Src family tyrosine kinases are required for platelet-derived growth factor-mediated signal transduction in NIH 3T3 cells. Proc Natl Acad Sci USA. 1993;90:7696–7700. doi: 10.1073/pnas.90.16.7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sato A, Sekine M, Virgona N, Ota M, Yano T. Yes is a central mediator of cell growth in malignant mesothelioma cells. Oncol Rep. 2012;28:1889–1893. doi: 10.3892/or.2012.2010. [DOI] [PubMed] [Google Scholar]
- 53.Zhang GL, Li YX, Zheng SQ, Liu M, Li X, Tang H. Suppression of hepatitis B virus replication by microRNA-199a-3p and microRNA-210. Antiviral Res. 2010;88:169–175. doi: 10.1016/j.antiviral.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 54.Nonomura T, Masaki T, Morishita A, Jian G, Uchida N, Himoto T, Izuishi K, Iwama H, Yoshiji H, Watanabe S, et al. Identification of c-Yes expression in the nuclei of hepatocellular carcinoma cells: involvement in the early stages of hepatocarcinogenesis. Int J Oncol. 2007;30:105–111. [PubMed] [Google Scholar]
- 55.Feng H, Masaki T, Nonomura T, Morishita A, Jian G, Nakai S, Deguchi A, Uchida N, Himoto T, Iwama H, et al. Activation of c-Yes in hepatocellular carcinoma: a preliminary study. World J Gastroenterol. 2006;12:5743–5745. doi: 10.3748/wjg.v12.i35.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ying Q, Liang L, Guo W, Zha R, Tian Q, Huang S, Yao J, Ding J, Bao M, Ge C, et al. Hypoxia-inducible microRNA-210 augments the metastatic potential of tumor cells by targeting vacuole membrane protein 1 in hepatocellular carcinoma. Hepatology. 2011;54:2064–2075. doi: 10.1002/hep.24614. [DOI] [PubMed] [Google Scholar]


