Abstract
AIM: To investigate associations between the tumor necrosis factor alpha (TNF-α) -1031 T>C, -863 C>A, -857 C>T, -308 G>A, and -238 G>A polymorphisms and HCC in Korea.
METHODS: Hepatocellular carcinoma (HCC) cases were diagnosed at CHA Bundang Medical Center from June 1996 to August 2008. The association between TNF-α polymorphisms and HCC was analyzed in 157 HCC patients and 201 controls using a polymerase chain reaction-restriction fragment length polymorphism assay. We investigated five TNF-α polymorphisms, which are TNF-α -1031 T>C, -863 C>A, -857 C>T, -308 G>A, and -238 G>A. The TNF-α genotype frequencies, genotype combinations and haplotypes were analyzed to disclose the association with HCC.
RESULTS: None of the TNF-α polymorphisms was significantly associated with HCC. However, nine genotype combinations had associations with increased likelihood of HCC. Among them, TNF-α -1031/-857/-238 TT/CC/GA (AOR = 18.849, 95%CI: 2.203-161.246, P = 0.007), TNF-α -1031/-308/-238 TT/GG/GA (AOR = 26.956, 95%CI: 3.071-236.584, P = 0.003), and TNF-α -1031/-238 TT/GA (AOR = 21.576, 95%CI: 2.581-180.394, P = 0.005) showed marked association with HCC. There were five haplotypes of TNF-α polymorphisms which were significantly associated with HCC. They are TNF-α -1031/-863/-857/-308/-238 T-C-C-G-A (OR = 25.824, 95%CI: 1.491-447.223, P = 0.0005), TNF-α -1031/-857/-308/-238 T-C-G-A (OR = 12.059, 95%CI: 2.747-52.950, P < 0.0001), TNF-α -1031/-857/-238 T-C-A (OR = 10.696, 95%CI: 2.428-47.110, P = 0.0001), TNF-α -1031/-308/-238 T-G-A (OR = 7.556, 95%CI: 2.173-26.280, P = 0.0002) and TNF-α -1031/-238 T-A (OR = 10.865, 95%CI: 2.473-47.740, P = 0.0001). Moreover, HCC Okuda stage III cases with the TNF-α -1031 CC genotype had better survival than those with the TT genotype (AOR = 5.795, 95%CI: 1.145-29.323).
CONCLUSION: Although no single TNF-α polymorphism is associated with HCC in this study, some TNF-α genotype combinations and haplotypes are associated with HCC. In addition, HCC Okuda stage III cases with the TNF-α -1031 TT genotype may have a better prognosis than those with the CC genotype.
Keywords: Tumor necrosis factor-alpha, Polymorphism, Single nucleotide, Carcinoma, Hepatocellular
Core tip: We genotyped five single nucleotide polymorphisms [Tumor necrosis factor alpha (TNF-α) -1031T>C, -863C>A, -857C>T, -308G>A, and -238G>A] in hepatocellular carcinoma (HCC) patients and control subjects. A number of genotype combinations and some haplotypes had association with HCC. In addition, HCC Okuda stage III cases with the TNF-α -1031 TT genotype had a better prognosis than those with the CC genotype.
INTRODUCTION
Primary liver cancer is the fifth most common cancer (fourth in men, sixth in women) in South Korea, and the second most common cause of cancer-related deaths[1]. Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer. HCC carcinogenesis is a multistep, multifactorial process. However, hepatitis B virus (HBV) and hepatitis C virus (HCV) are the best-known risk factors for HCC. Approximately 70%-80% of HCCs are related to these persistent viral infections[2-5]. Various inflammatory mediators related to chronic inflammation are known as cofactors in carcinogenesis[6]. Among these, tumor necrosis factor alpha (TNF-α) is the most important cytokine in inflammation-associated tumoriogenesis[7].
TNF-α is a potent, pleiotropic, proinflammatory cytokine. It is produced mainly by macrophages, but also by a broad variety of other cell types, including lymphoid cells, mast cells, endothelial cells, fibroblasts, and neurons[8]. The TNF-α gene lies in the class III region of the major histocompatibility complex (MHC) and is located on human chromosome 6p21.3[9]. Some single nucleotide polymorphisms (SNPs) in the TNF promoter have been identified, and they are thought to affect TNF-α production[10]. Past research has shown that the TNF-α -308 GG genotype[11] and TNF-α -857 T polymorphism[12] are associated with gastric cancer, the TNF-α -308 G>A polymorphism[13] is associated with vascular invasion of breast tumors, the TNF-α -308 G/A polymorphism[14] is associated with lung cancer, the TNF-α -308 G>A polymorphism[15] is associated with prostate cancer, the TNF-α -308 G>A polymorphism[16] is associated with oral squamous cell carcinoma, the TNF-α -238 G>A polymorphism[17] is associated with bone cancer, and the TNF-α -308 G>A polymorphism[18] is associated with invasive cervical cancer.
The results of past studies on the association between TNF-α polymorphisms and HCC have had conflicting results. A meta-analysis of 10 case-control studies reported that the TNF-α -308 GG genotype is associated with a modest decrease in HCC risk[19]. Similarly, a meta-analysis by Yang et al[20] reported that the TNF-α -308 G>A polymorphism is associated with increased HCC risk and that the TNF-α -238 G>A polymorphism is not associated with HCC. However, these meta-analyses did not investigate other TNF-α polymorphisms. Wei et al[21] performed a meta-analysis of 17 relevant studies and found that the TNF-α -308 G>A, -238 G>A, and -863 C>A polymorphisms are associated with HCC among Asians, while the TNF-α -857 C>T and -1031 T>C polymorphisms are not related to HCC risk. This meta-analysis did not investigate the association between TNF-α haplotypes and HCC risk, however.
We investigated the association between the TNF-α -1031 T>C, -863 C>A, -857 C>T, -308 G>A, and -238 G>A polymorphisms and HCC in Korea.
MATERIALS AND METHODS
Study population
A total of 157 HCC cases diagnosed at CHA Bundang Medical Center from June 1996 to August 2008 were enrolled. The control group consisted of 201 individuals randomly selected from a health screening program with age and gender matched to HCC patients. Exclusion criteria included a history of other cancer or other severe medical conditions. Individuals with only hypertension and diabetes were not excluded. The grade of hepatic impairment was classified by Child-Pugh scores. HCC clinical stage was evaluated on the basis of TNM classification and Okuda stage. The present study was approved by the Institutional Review Board of CHA Bundang Medical Center, and written informed consent was obtained from all case and control subjects in the study.
DNA extraction and genotyping
Genomic DNA was extracted from leukocytes using a G-DEX II Genomic DNA Extraction Kit (iNtRON Biotechnology, Seongnam, South Korea). The TNF-α -238 G>A (rs361525), TNF-α -308 G>A (rs1800629), TNF-α -857 C>T (rs1799724), TNF-α -863 C>A (rs1800630), and TNF-α -1031 T>C (rs1799964) polymorphisms were analyzed using a polymerase chain reaction (PCR)-restriction fragment length polymorphism (RFLP) assay. The -238 G>A polymorphism was analyzed using the forward primer 5’-AGA AGA CCC CCC TCG GAA CC and the reverse primer 5’-ATC TGG AGG AAG CGG TAG TG, and PCR products were cut using the restriction enzyme MspI. Genotyping of the -308 G>A polymorphism was performed using the forward primer 5’-AGG CAA TAG GTT TTG AGG GCC AT and reverse primer 5’-TCC TCC CTG CTC CGA TTC CG, and PCR products were cut using the restriction enzyme Nco. Genotyping of the -857 C>T polymorphism was performed using the forward primer 5′- AAG TCG AGT ATG GGG ACC CCC CGT TAA and the reverse primer 5′- CCC CAG TGT GTG GCC ATA TCT TCT T, and PCR products were cut using the restriction enzyme MspA1 I. To genotype the -863 C>A polymorphism, the forward primer 5’-GGC TCT GAG GAA TGG GTT and the reverse primer 5’- CTA CAT GGC CCT GTC TTC GTT ACG were used, and PCR products were cut using the restriction enzyme TaiI. To genotype the -1031 T>C polymorphism, the forward primer 5’-AGC AAG AGC TGT GGG GAG AA and the reverse primer 5’-CCT GTA ACC CAT TCC TCA GAG CC were used, and PCR products were cut using the restriction enzyme BbsI. These primers and restriction enzymes are shown at Table 1.
Table 1.
Used primer and restriction enzyme at each polymorphism
| Polymorphisms | Forward primer | Reverse primer | Restriction enzyme1 |
| -238 G>A | 5’-AGA AGA CCC CCC TCG GAA CC2-3’ | 5’-ATC TGG AGG AAG CGG TAG TG-3' | MspI |
| -308 G>A | 5’-AGG CAA TAG GTT TTG AGG GCC AT-3' | 5’-TCC TCC CTG CTC CGA TTC CG-3' | NcoI |
| -857 C>T | 5′-AAG TCG AGT ATG GGG ACC CCC CGT TAA-3' | 5′-CCC CAG TGT GTG GCC ATA TCT TCT T-3' | AseI |
| -863 C>A | 5'-GGC TCT GAG GAA TGG GTT | 5'-CTA CAT GGC CCT GTC TTC GTT ACG-3' | MnlI |
| -1031 T>C | 5’-AGC AAG AGC TGT GGG GAG AA-3' | 5’-CCT GTA ACC CAT TCC TCA GAG CC-3' | BbsI |
All of the restriction enzymes were available from New England Biolabs (MA, United States) and we used the reaction conditions recommended by the instructions;
The underlined bases in the primer were mismatched with the wild-type sequence in order to introduce the restriction enzyme site.
The five polymorphic regions were amplified using the following PCR conditions: 95 °C for 5 min, 38 cycles of denaturing at 95 °C for 30 s, annealing at 60 °C for 30 s, extension at 72 °C for 30 s, and a final extension at 72 °C for 5 min. Digestion with the appropriate restriction enzymes was performed at 37 °C for 16 h, as described by the manufacturer (New England BioLabs, Beverly, MA, United States).
Statistical analysis
We used χ2 tests to analyze baseline categorical variables and Mann-Whitney tests to analyze baseline continuous variables. Associations between TNF-α polymorphisms and HCC were estimated using adjusted odds ratios (AORs) and 95%CIs obtained from multiple logistic regression models adjusted for age, sex, hypertension, diabetes mellitus, body mass index, smoking status, and drinking status. The informations about hypertension, diabetes, smoking status and drinking status were obtained by questionnaires.
Survival time was calculated from the date of HCC diagnosis to the date of death or last follow-up. Survival analysis was performed using the Kaplan-Meier method, the log-rank test, and the Cox-proportional hazards regression model. The statistical significance level of all tests was set at P < 0.05. Analyses were performed using GraphPad Prism 4.0 (GraphPad Software Inc., San Diego, CA, United States) and Medcalc version 11.1.1.0 (Medcalc Software, Mariakerke, Belgium).
RESULTS
Study sample
Baseline characteristics of HCC cases and controls are shown in Table 2. The mean age of HCC cases was 56 years, and 120 were male (76.4%). HCC cases and controls were matched on age and sex (P = 0.069 and 0.570, respectively). Compared with controls, HCC cases were less likely to have hypertension (P = 0.002). However, no significant differences in obesity (BMI > 25 kg/m2) or in the prevalence of diabetes mellitus, smoking, or drinking were identified between cases and controls. HBV and HCV infections were identified in 125 (79.6%) and 16 (10.2%) HCC cases, respectively. Of the HCC cases, 12 (7.6%) underwent surgical resection. The Okuda stage distribution was as follows: stage I, 46 (29.3%); stage II, 68 (43.3%); and stage III, 43 (27.4%).
Table 2.
Baseline characteristics of study sample n (%)
| Variable | Cases | Controls | P value1 |
| n | 157 | 201 | |
| Age (yr, mean ± SD) | 56.00 ± 11.06 | 54.00 ± 11.22 | 0.061 |
| Sex (male, %) | 120 (76.4) | 148 (73.6) | 0.871 |
| Hypertension | 19 (12.1) | 50 (24.9) | 0.012 |
| Diabetes mellitus | 30 (19.1) | 26 (12.9) | 0.195 |
| BMI > 25 kg/m2 | 40 (25.5) | 51 (24.9) | 1.000 |
| Smoking | 84 (53.5) | 78 (38.8) | 0.106 |
| Drinking | 90 (57.3) | 93 (46.3) | 0.274 |
| Tumor size | |||
| < 5 cm | 68 (43.3) | - | - |
| ≥ 5 cm | 89 (56.7) | - | - |
| Portal vein thrombosis | |||
| No | 92 (58.6) | - | - |
| Yes | 65 (41.4) | - | - |
| Surgical resection | |||
| No | 145 (92.4) | - | - |
| Yes | 12 (7.6) | - | - |
| CTx/RTx | |||
| No | 27 (17.2) | - | - |
| Yes | 130 (82.8) | - | - |
| TNM stage | |||
| I | 34 (21.7) | - | - |
| II | 37 (23.5) | - | - |
| III | 51 (32.5) | - | - |
| IV | 35 (22.3) | - | - |
| Okuda stage | |||
| I | 46 (29.3) | - | - |
| II | 68 (43.3) | - | - |
| III | 43 (27.4) | - | - |
| CTP class | |||
| A | 83 (52.9) | - | - |
| B | 36 (22.9) | - | - |
| C | 38 (24.2) | - | - |
Derived from χ2 tests for categorical data and Mann-Whitney tests for continuous data. BMI: Body mass index; CTx: Chemotherapy; RTx: Radiotherpay; CTP: Chlid-Turcotte-Pugh.
Genotype frequencies of the TNF-α -1031 T>C, -863 C>A, -857 C>T, -308 G>A, and -238 G>A polymorphisms
The genotype distributions of the TNF-α -1031 T>C, -863 C>A, -857 C>T, -308 G>A, and -238 G>A polymorphisms in HCC cases, HCC cases with HBV, and controls are shown in Table 3. We found no significant differences in the genotype frequencies of the TNF-α -1031 T>C, -863 C>A, -857C>T, -308 G>A, or -238 G>A polymorphisms between cases and controls. We also found no significant difference in genotype frequencies of these polymorphisms between HCC cases with HBV and controls.
Table 3.
Genotype frequencies and hepatocellular carcinoma adjusted odd ratios for the TNF-α -1031 T>C, -863 C>A, -857 C>T, -308 G>A, and -238 G>A polymorphisms in hepatocellular carcinoma cases and controls n (%)
| Characteristics | Controls, n = 201 |
HCC cases |
HCC cases with HBV |
||||||
| Cases, n = 157 | AOR (95%CI)1 | P-value | FDR-P-value | Cases, n = 125 | AOR (95%CI)1 | P-value | FDR-P-value | ||
| TNF-α -1031 T>C, rs1799964 | |||||||||
| TT | 121 (60.2) | 99 (63.1) | 1.000 (reference) | 73 (58.4) | 1.000 (reference) | ||||
| TC | 70 (34.8) | 47 (29.9) | 0.796 (0.487-1.301) | 0.362 | 0.586 | 42 (33.6) | 0.922 (0.550-1.544) | 0.756 | 0.960 |
| CC | 10 (5.0) | 11 (7.0) | 1.308 (0.498-3.436) | 0.586 | 0.586 | 10 (8.0) | 1.724 (0.628-4.729) | 0.290 | 0.580 |
| Dominant (TT vs TC + CC) | 0.860 (0.540-1.369) | 0.524 | 0.586 | 1.013 (0.622-1.650) | 0.960 | 0.960 | |||
| Recessive (TT + TC vs CC) | 1.393 (0.540-3.595) | 0.493 | 0.586 | 1.745 (0.649-4.693) | 0.270 | 0.580 | |||
| HWE-P | 0.976 | 0.112 | 0.268 | ||||||
| TNF-α -863 C>A, rs1800630 | |||||||||
| CC | 129 (64.2) | 98 (62.4) | 1.000 (reference) | 74 (59.2) | 1.000 (reference) | ||||
| CA | 62 (30.8) | 53 (33.8) | 1.154 (0.709-1.878) | 0.565 | 0.672 | 46 (36.8) | 1.284 (0.767-2.150) | 0.341 | 0.750 |
| AA | 10 (5.0) | 6 (3.8) | 0.784 (0.255-2.414) | 0.672 | 0.672 | 5 (4.0) | 0.825 (0.253-2.690) | 0.750 | 0.750 |
| Dominant (CC vs CA + AA) | 1.109 (0.695-1.768) | 0.666 | 0.672 | 1.226 (0.749-2.007) | 0.418 | 0.750 | |||
| Recessive (CC + CA vs AA) | 0.734 (0.243-2.222) | 0.584 | 0.672 | 0.760 (0.238-2.428) | 0.643 | 0.750 | |||
| HWE-P | 0.477 | 0.723 | 0.513 | ||||||
| TNF-α -857 C>T, rs1799724 | |||||||||
| CC | 142 (70.6) | 116 (73.9) | 1.000 (reference) | 92 (73.6) | 1.000 (reference) | ||||
| CT | 53 (26.4) | 35 (22.3) | 0.841 (0.493-1.432) | 0.523 | 0.697 | 29 (23.2) | 0.840 (0.477- 1.478) | 0.545 | 0.893 |
| TT | 6 (3.0) | 6 (3.8) | 1.816 (0.428-7.714) | 0.419 | 0.697 | 4 (3.2) | 1.110 (0.226- 5.438) | 0.898 | 0.893 |
| Dominant (CC vs CT + TT) | 0.930 (0.558-1.548) | 0.780 | 0.78 | 0.888 (0.516-1.531) | 0.670 | 0.893 | |||
| Recessive (CC + CT vs TT) | 2.091 (0.494-8.860) | 0.317 | 0.697 | 1.500 (0.312-7.203) | 0.612 | 0.893 | |||
| HWE-P | 0.698 | 0.120 | 0.371 | ||||||
| TNF-α -308 G>A, rs1800629 | |||||||||
| GG | 181 (90.0) | 133 (84.7) | 1.000 (reference) | 108 (86.4) | 1.000 (reference) | ||||
| GA | 18 (9.0) | 23 (14.6) | 1.949 (0.940- 4.040) | 0.073 | 0.178 | 17 (13.6) | 1.779 (0.821-3.853) | 0.144 | 0.430 |
| AA | 2 (1.0) | 1 (0.6) | 0.936 (0.075-11.646) | 0.959 | 0.959 | 0 (0.0) | N/A | 0.995 | 0.995 |
| Dominant (GG vs GA + AA) | 1.841 (0.911-3.720) | 0.089 | 0.178 | 1.610 (0.758-3.419) | 0.215 | 0.430 | |||
| Recessive (GG + GA vs AA) | 0.861 (0.069-10.734) | 0.908 | 0.959 | N/A | 0.995 | 0.995 | |||
| HWE-P | 0.057 | 0.996 | 0.415 | ||||||
| TNF-α -238 G>A, rs 361525 | |||||||||
| GG | 180 (89.6) | 130 (82.8) | 1.000 (reference) | 104 (83.2) | 1.000 (reference) | ||||
| GA | 20 (10.0) | 26 (17.6) | 1.634 (0.821-3.251) | 0.162 | 0.324 | 20 (16.0) | 1.491 (0.713-3.117) | 0.288 | 0.576 |
| AA | 1 (0.5) | 1 (0.6) | N/A | 0.993 | 0.994 | 1 (0.8) | N/A | 0.993 | 0.993 |
| Dominant (GG vs GA + AA) | 1.732 (0.876-3.424) | 0.115 | 0.324 | 1.614 (0.780-3.340) | 0.197 | 0.576 | |||
| Recessive (GG + GA vs AA) | N/A | 0.994 | 0.994 | N/A | 0.993 | 0.993 | |||
| HWE-P | 0.587 | 0.807 | 0.972 | ||||||
Adjusted for age, sex, hypertension, diabetes mellitus, drinking status, and smoking status. HCC: Hepatocellular carcinoma; AOR: Adjusted odd ratios; HBV: Hepatitis B virus.
Associations between combinations of TNF-α polymorphisms and HCC
Next, we evaluated the combined effect of TNF-α polymorphisms on HCC risk. Using multiple logistic regression models, we found nine combined TNF-α genotypes associated with increased likelihood of HCC, as shown in Table 4. Some combinations showed marked association with HCC. The combinations were TNF-α -1031/-857/-238 TT/CC/GA (AOR = 18.849, 95%CI: 2.203-161.246, P = 0.007) TNF-α -1031/-308/-238 TT/GG/GA (AOR = 26.956, 95%CI: 3.071-236.584, P = 0.003) and TNF-α -1031/-238 TT/GA (AOR = 21.576, 95%CI: 2.581-180.394, P = 0.005).
Table 4.
TNF-α genotype combinations in hepatocellular carcinoma cases and controls1 n (%)
| Genotype | HCC cases, n = 157 | Controls, n = 201 | AOR (95%CI)2 | P-value |
| TNF-α -1031/-857/-238 | ||||
| TT/CC/GG | 56 (35.7) | 81 (40.3) | 1.000 (reference) | |
| TT/CC/GA | 11 (7.0) | 2 (1.0) | 18.849 (2.203-161.246) | 0.007 |
| TNF-α -1031/-308/-238 | ||||
| TT/GG/GG | 68 (43.3) | 103 (51.2) | 1.000 (reference) | |
| TT/GG/GA | 13 (8.3) | 2 (1.0) | 26.956 (3.071-236.584) | 0.003 |
| TT/GA/GG | 17 (10.8) | 14 (7.0) | 2.712 (1.085-6.778) | 0.033 |
| TNF-α -863/-308/-238 | ||||
| CC/GG/GG | 68 (43.3) | 99 (49.3) | 1.000 (reference) | |
| CC/GA/GG | 16 (10.2) | 13 (6.5) | 2.533 (1.007-6.371) | 0.048 |
| CA/GG/GA | 13 (8.3) | 5 (2.5) | 4.242 (1.243-14.473) | 0.021 |
| TNF-α -1031/-238 | ||||
| TT/GG | 86 (54.8) | 119 (59.2) | 1.000 (reference) | |
| TT/GA | 13 (8.3) | 2 (1.0) | 21.576 (2.581-180.394) | 0.005 |
| TNF-α -863/-238 | ||||
| CC/GG | 85 (54.1) | 114 (56.7) | 1.000 (reference) | |
| CA/GA | 13 (8.3) | 5 (2.5) | 3.669 (1.098-12.253) | 0.035 |
| TNF-α -308/-238 | ||||
| GG/GG | 107 (68.2) | 160 (79.6) | 1.000 (reference) | |
| GA/GG | 23 (14.6) | 18 (9.0) | 2.283 (1.078-4.836) | 0.031 |
| GA+AA/GG | 24 (15.3) | 20 (10.0) | 2.150 (1.041-4.441) | 0.039 |
1Only combinations significantly associated with HCC are presented in the table;
Adjusted for age, sex, hypertension, diabetes mellitus, drinking status, and smoking status. HCC: Hepatocellular carcinoma.
Haplotype frequencies of TNF-α polymorphisms
We then calculated haplotype frequencies to attain more comprehensive information about the association between TNF-α and HCC. TNF-α -1031 T>C, -863 C>A, -857 C>T, -308 G>A, and -238 G>A haplotype frequencies in HCC cases and controls are shown in Table 5. The TNF-α -1031/-863/-857/-308/-238 T-C-C-G-A haplotype was significantly associated with HCC (OR = 25.824, 95%CI: 1.491-447.223, P = 0.0005). Other haplotypes significantly associated with HCC included TNF-α -1031/-857/-308/-238 T-C-G-A (OR = 12.059, 95%CI: 2.747-52.950, P < 0.0001), TNF-α -1031/-857/-238 T-C-A (OR = 10.696, 95%CI: 2.428-47.110, P = 0.0001), TNF-α -1031/-308/-238 T-G-A (OR = 7.556, 95%CI: 2.173-26.280, P = 0.0002), and TNF-α -1031/-238 T-A (OR = 10.865, 95%CI: 2.473-47.740, P = 0.0001).
Table 5.
TNF-α -1031 T>C, -863 C>A, -857 C>T, -308 G>A, and -238 G>A haplotypes in hepatocellular carcinoma cases and controls1
| Haplotype | Overall | Control | Case | OR (95%CI) | P-value |
| TNF-α -1031/-863/-857/-308/-238 | |||||
| T-C-C-G-G | 0.5266 | 0.5399 | 0.5139 | 1.000 (reference) | |
| T-C-C-G-A | 0.0119 | 0.0000 | 0.0289 | 25.824 (1.491-447.223) | 0.0005 |
| T-C-C-A-G | 0.0582 | 0.0486 | 0.0647 | 1.428 (0.749-2.723) | 0.320 |
| T-C-T-G-G | 0.1334 | 0.1435 | 0.1232 | 0.891 (0.564-1.407) | 0.645 |
| T-C-T-A-G | 0.0056 | 0.0040 | 0.0072 | 1.360 (0.190-9.764) | 1.000 |
| T-A-C-G-G | 0.0171 | 0.0210 | 0.0000 | 0.080 (0.005-1.396) | 0.023 |
| T-A-C-G-A | 0.0109 | 0.0025 | 0.0226 | 4.761 (0.976-23.230) | 0.043 |
| T-A-C-A-G | 0.0010 | 0.0000 | 0.0066 | 6.796 (0.324-142.621) | 0.181 |
| T-A-T-G-G | 0.0123 | 0.0121 | 0.0133 | 1.088 (0.288-4.118) | 1.000 |
| T-A-T-A-G | 0.0009 | 0.0022 | 0.0000 | 0.453 (0.018-11.201) | 1.000 |
| C-C-C-G-G | 0.0133 | 0.0103 | 0.0162 | 2.176 (0.699-6.778) | 0.254 |
| C-C-C-G-A | 0.0414 | 0.0498 | 0.0318 | 0.605 (0.257-1.425) | 0.306 |
| C-C-T-G-A | 0.0042 | 0.0009 | 0.0066 | 6.796 (0.324-142.600) | 0.181 |
| C-A-C-G-G | 0.1631 | 0.1638 | 0.1646 | 1.088 (0.717-1.652) | 0.749 |
| TNF-α -1031/-857/-308/-238 | |||||
| T-C-G-G | 0.5430 | 0.5637 | 0.5105 | 1.000 (reference) | |
| T-C-G-A | 0.0235 | 0.0060 | 0.0531 | 12.059 (2.747-52.950) | < 0.0001 |
| TNF-α -1031/-857/-238 | |||||
| T-C-G | 0.6026 | 0.6128 | 0.5854 | 1.000 (reference) | |
| T-C-A | 0.0231 | 0.0060 | 0.0517 | 10.696 (2.428-47.110) | 0.0001 |
| TNF-α -1031/-308/-238 | |||||
| T-G-G | 0.6885 | 0.7187 | 0.6488 | 1.000 (reference) | |
| T-G-A | 0.0238 | 0.0063 | 0.0518 | 7.556 (2.173-26.280) | 0.0002 |
| TNF-α -1031/-238 | |||||
| T-G | 0.7546 | 0.7738 | 0.7297 | 1.000 (reference) | |
| T-A | 0.0233 | 0.0062 | 0.0505 | 10.865 (2.473- 47.740) | 0.0001 |
1Only data for haplotypes significantly associated with hepatocellular carcinoma are presented in the table. Haplotypes with frequencies of less than zero among cases or controls are not shown.
Cox regression analysis of HCC case survival
A stepwise Cox regression analysis of HCC-related survival is shown in Table 6. Survival in HCC cases was associated with sex, history of chemotherapy or radiotherapy, portal vein thrombosis, and TNF-α -1031 polymorphism status.
Table 6.
Results of stepwise Cox regression analysis of hepatocellular carcinoma survival
| Covariate | β | SEM | HR (95%CI) | P value |
| Sex | -1.318 | 0.480 | 0.268 (0.105-0.682) | 0.006 |
| Chemotherapy or radiotherapy | -1.914 | 0.500 | 0.148 (0.056-0.391) | 0.0001 |
| Portal vein thrombosis | 1.334 | 0.478 | 3.795 (1.495-9.631) | 0.005 |
| TNF-α -1031 TT vs CC | 1.772 | 0.733 | 5.881 (1.408-24.554) | 0.016 |
TNF-α polymorphisms and HCC case survival stratified by stage
TNF-α polymorphism-related survival was analyzed in HCC cases stratified by TNM and Okuda stage. Survival of cases with the TNF-α -1031 wild type (TT) genotype or a single T>C polymorphism was significantly better than those with the CC genotype (regression coefficient = 1.772, HR = 5.881, 95%CI: 1.408-24.554, P = 0.016), as shown in Figure 1 and Table 7. Other TNF-α polymorphisms did not affect HCC case survival at any TNM or Okuda stage.
Figure 1.
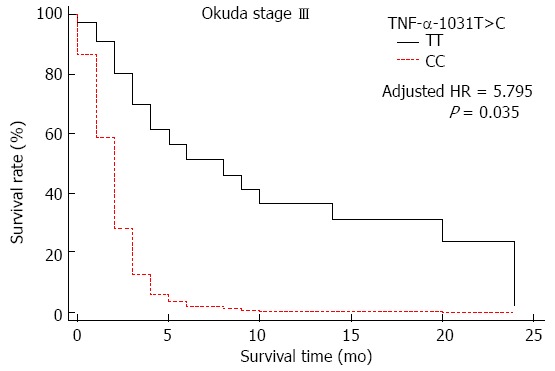
Survival curves for hepatocellular carcinoma Okuda stage III cases with the TNF-α-1031 (rs1799964) TT genotype (reference) and the CC genotype.
Table 7.
TNF-α genotypes and survival in Okuda stage III hepatocellular carcinoma cases
| Genotype |
Validation set |
Adjusted HR (95%CI)1 | P-value | |
| Cases (n = 43) | Deaths (n = 41) | |||
| TNF-α -1031 T>C, rs1799964 | ||||
| TT | 29 (67.4) | 27 (65.9) | 1.000 (reference) | |
| TC | 11 (25.6) | 11 (26.8) | 1.314 (0.448-3.856) | 0.621 |
| CC | 3 (7.0) | 3 (7.3) | 5.795 (1.145-29.323) | 0.035 |
| Dorminant model (TT vs TC + CC) | 1.805 (0.733-4.442) | 0.201 | ||
| Recessive model (TT + TC vs CC) | 3.761 (0.869-16.265) | 0.078 | ||
Adjusted for age, sex, hypertension, diabetes mellitus, drinking status, smoking status, portal vein thrombosis, tumor size, surgical resection, and chemotherapy or radiotherapy.
DISCUSSION
We performed this case-control study to investigate the associations between the TNF-α -1031 T>C, -863 C>A, -857 C>T, -308 G>A, and -238 G>A polymorphisms and HCC. Furthermore, we analyzed TNF-α polymorphism combinations and haplotypes. In this study, the TNF-α -1031 T>C, -863 C>A, -857 C>T, -308 G>A, and -238 G>A polymorphisms were not associated with HCC risk. This result is inconsistent with the findings of previous meta-analyses. An insufficient number of HCC cases and controls in our study could be the reason for this discordance; for example, we were able to analyze only 18 TNF-α -308 GA controls and 23 cases and 2 TNF-α -308 AA controls and 1 HCC case.
Past studies have analyzed combined genotypes or haplotypes to attain a more comprehensive understanding of the association between various polymorphisms and diseases[22,23]. In this study, we found significant associations between some TNF-α genotype combinations and HCC risk. All of the statistically significant genotype combinations included the GA genotype at TNF-α -238 or -308. The TNF-α -863 CA genotype accompanied by the TNF-α -238 GA genotype was also associated with increased HCC risk in some combinations. This result suggests that the TNF-α -238 GA and -308 GA genotypes increase HCC risk in specific genotype combinations. The TNF-α -863 CA genotype accompanied by the TNF-α -238 GA genotype was associated with increased HCC risk, but the TNF-α -863 CA genotype was not associated with increased HCC risk when accompanied by other genotype combinations. This suggests that the TNF-α -863 polymorphism increases the risk of HCC only in the presence of the TNF-α -238 GA genotype.
Associations between TNF-α haplotypes and various medical conditions, such as polycystic ovary syndrome (PCOS)[24], bladder cancer[25], gastric carcinoma[26,27], peptic ulcer[28], and anemia[29] have been reported. Chen et al[30] reported that the TNF-α -1031/-863/-857/-308/-238 T-C-C-G-A haplotype may account for decreased susceptibility to HCC. However, we found this haplotype was more common in HCC patients. All haplotypes significantly associated with HCC contained the TNF-α -238 A allele, suggesting it may play a role in HCC development. The HCC odds ratio for TNF-α haplotype -1031/-863/-857/-308/-238 T-A-C-G-G was 0.080, while the OR for haplotype T-A-C-G-A, in which the -238 allele changed from G to A, was 4.716. This result supports the hypothesis that the -238 A allele is associated with HCC risk, though the HCC odds ratio for haplotype T-A-C-G-A was not statistically significant.
To the best of our knowledge, no study has reported an association between HCC prognosis and TNF-α polymorphisms. TNF-α acts like a double-edged sword via its two distinct receptors: TNF receptor 1 (TNFR-1) and receptor 2 (TNFR-2)[31,32]. The major difference between the two receptors is the death domain in TNFR-1 absent in TNFR-2. On the one hand, TNFR-1 plays an important role in apoptotic cell death. On the other hand, TNF-α can stimulate the growth, proliferation, and angiogenesis of cancer cells by activating nuclear factor NF-κB[32]. Nakazaki et al[33] reported higher levels of TNF-α in patients with recurrent HCC than in patients without HCC recurrence or patients with metastatic liver carcinoma or gastrointestinal carcinoma. Liu et al[34] have reported that TNF-α overexpression is related to poor differentiation and microvascular invasion in HCC. Our study raises the possibility that specific TNF-α polymorphisms are related to HCC patient prognosis. The TNF-α polymorphism associated with treatment outcomes in our study (TNF-α -1031) may potentially be used in the clinic as a surrogate biomarker of prognosis.
In conclusion, although we found no direct association between TNF-α polymorphisms and HCC risk, some TNF-α genotype combinations and haplotypes were associated with HCC. In addition, HCC patients with the TNF-α -1031 TT genotype had a better prognosis than those with the CC genotype. An important limitation of our study was the small sample size, which prevents us from drawing definitive conclusions. Therefore, our results should be interpreted cautiously. However, to the best of our knowledge, ours is the first study to analyze the association between these five TNF-α polymorphisms, as well as their combinations and haplotypes, and HCC case status and prognosis. Our findings provide a more comprehensive understanding of TNF-α polymorphisms and HCC. Further studies with sufficient sample size are needed to clarify the role of TNF-α polymorphisms in HCC risk and prognosis.
COMMENTS
Background
Tumor necrosis factor alpha (TNF-α) is a pro-inflammatory cytokine that plays a role in inflammatory pathways leading to tumorigenesis. Some TNF-α polymorphisms have been found to be associated with hepatocellular carcinoma (HCC) susceptibility.
Research frontiers
Some TNF-α polymorphisms have been considered to have association with the susceptibility of HCC in many studies. But not all the five TNF-α polymorphisms are sufficiently studied and some of them are controversial. Thus to elucidate accurate genetic risk factors for HCC is important. Genetic factors affecting prognosis in patients with HCC is also important research area.
Innovations and breakthroughs
As far as I know, this study is first that analyzed the association between all the five TNF-α polymorphisms, as well as their combinations and haplotypes, and HCC. So it can provides more comprehensive understandings of TNF-α polymorphisms and HCC. Furthermore, our study raises the possibility that specific TNF-α polymorphisms are related to HCC patient prognosis.
Applications
Analyzing TNF-α polymorphisms in patients with hepatitis or cirrhosis may help predicting the risk of HCC in individual patient. And TNF-α -1031 may potentially be used in the clinic as a surrogate biomarker of prognosis.
Peer-review
It’s a case-control study investigating the associations between the TNF-α polymorphisms and HCC with analysis of TNF-α polymorphism combinations and haplotypes. In this study, although no direct association between TNF-α polymorphisms and HCC risk was found, some TNF-α genotype combinations and haplotypes were associated with HCC. A significant association was find between some TNF-α genotype combinations and HCC risk particularly the TNF-α -238GA and -308GA genotypes, and the TNF-α -863 polymorphism increases the risk of HCC only in the presence of the TNF-α -238 GA genotype.
Footnotes
Supported by Basic Science Research Program through National Research Foundation of Korea Grants funded by the Korean Government, No. NRF-2012R1A1A2007033 and No. 2009-0093821, South Korea.
Institutional review board statement: The present study was approved by the Institutional Review Board of CHA Bundang Medical Center, and written informed consent was obtained from all case and control subjects in the study.
Conflict-of-interest statement: The authors disclose no conflicts of interest.
Data sharing statement: All the dataset are available from the author at nkkim@cha.ac.kr
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: July 4, 2015
First decision: July 19, 2015
Article in press: October 20, 2015
P- Reviewer: Berkane S S- Editor: Ma YJ L- Editor: A E- Editor: Zhang DN
References
- 1.Jung KW, Won YJ, Kong HJ, Oh CM, Lee DH, Lee JS. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2011. Cancer Res Treat. 2014;46:109–123. doi: 10.4143/crt.2014.46.2.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsai JF, Jeng JE, Ho MS, Chang WY, Hsieh MY, Lin ZY, Tsai JH. Effect of hepatitis C and B virus infection on risk of hepatocellular carcinoma: a prospective study. Br J Cancer. 1997;76:968–974. doi: 10.1038/bjc.1997.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kremsdorf D, Soussan P, Paterlini-Brechot P, Brechot C. Hepatitis B virus-related hepatocellular carcinoma: paradigms for viral-related human carcinogenesis. Oncogene. 2006;25:3823–3833. doi: 10.1038/sj.onc.1209559. [DOI] [PubMed] [Google Scholar]
- 4.Suruki R, Hayashi K, Kusumoto K, Uto H, Ido A, Tsubouchi H, Stuver SO. Alanine aminotransferase level as a predictor of hepatitis C virus-associated hepatocellular carcinoma incidence in a community-based population in Japan. Int J Cancer. 2006;119:192–195. doi: 10.1002/ijc.21796. [DOI] [PubMed] [Google Scholar]
- 5.Levrero M. Viral hepatitis and liver cancer: the case of hepatitis C. Oncogene. 2006;25:3834–3847. doi: 10.1038/sj.onc.1209562. [DOI] [PubMed] [Google Scholar]
- 6.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshimura A. Signal transduction of inflammatory cytokines and tumor development. Cancer Sci. 2006;97:439–447. doi: 10.1111/j.1349-7006.2006.00197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwabe RF, Brenner DA. Mechanisms of Liver Injury. I. TNF-alpha-induced liver injury: role of IKK, JNK, and ROS pathways. Am J Physiol Gastrointest Liver Physiol. 2006;290:G583–G589. doi: 10.1152/ajpgi.00422.2005. [DOI] [PubMed] [Google Scholar]
- 9.Hajeer AH, Hutchinson IV. TNF-alpha gene polymorphism: clinical and biological implications. Microsc Res Tech. 2000;50:216–228. doi: 10.1002/1097-0029(20000801)50:3<216::AID-JEMT5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 10.Huizinga TW, Westendorp RG, Bollen EL, Keijsers V, Brinkman BM, Langermans JA, Breedveld FC, Verweij CL, van de Gaer L, Dams L, et al. TNF-alpha promoter polymorphisms, production and susceptibility to multiple sclerosis in different groups of patients. J Neuroimmunol. 1997;72:149–153. doi: 10.1016/s0165-5728(96)00182-8. [DOI] [PubMed] [Google Scholar]
- 11.Gorouhi F, Islami F, Bahrami H, Kamangar F. Tumour-necrosis factor-A polymorphisms and gastric cancer risk: a meta-analysis. Br J Cancer. 2008;98:1443–1451. doi: 10.1038/sj.bjc.6604277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohyama I, Ohmiya N, Niwa Y, Shirai K, Taguchi A, Itoh A, Hirooka Y, Wakai K, Hamajima N, Mori N, et al. The association between tumour necrosis factor-alpha gene polymorphism and the susceptibility to rugal hyperplastic gastritis and gastric carcinoma. Eur J Gastroenterol Hepatol. 2004;16:693–700. doi: 10.1097/01.meg.0000108315.52416.bf. [DOI] [PubMed] [Google Scholar]
- 13.Azmy IA, Balasubramanian SP, Wilson AG, Stephenson TJ, Cox A, Brown NJ, Reed MW. Role of tumour necrosis factor gene polymorphisms (-308 and -238) in breast cancer susceptibility and severity. Breast Cancer Res. 2004;6:R395–R400. doi: 10.1186/bcr802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stankovic MM, Nestorovic AR, Tomovic AM, Petrovic-Stanojevic ND, Andjelic-Jelic MS, Dopudja-Pantic VB, Nagorni-Obradovic LjM, Mitic-Milikic MM, Radojkovic DP. TNF-alpha-308 promotor polymorphism in patients with chronic obstructive pulmonary disease and lung cancer. Neoplasma. 2009;56:348–352. doi: 10.4149/neo_2009_04_348. [DOI] [PubMed] [Google Scholar]
- 15.Sáenz-López P, Carretero R, Cózar JM, Romero JM, Canton J, Vilchez JR, Tallada M, Garrido F, Ruiz-Cabello F. Genetic polymorphisms of RANTES, IL1-A, MCP-1 and TNF-A genes in patients with prostate cancer. BMC Cancer. 2008;8:382. doi: 10.1186/1471-2407-8-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta R, Sharma SC, Das SN. Association of TNF-alpha and TNFR1 promoters and 3’ UTR region of TNFR2 gene polymorphisms with genetic susceptibility to tobacco-related oral carcinoma in Asian Indians. Oral Oncol. 2008;44:455–463. doi: 10.1016/j.oraloncology.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Patio-Garcia A, Sotillo-Pieiro E, Modesto C, Sierrases-Maga L. Analysis of the human tumour necrosis factor-alpha (TNFalpha) gene promoter polymorphisms in children with bone cancer. J Med Genet. 2000;37:789–792. doi: 10.1136/jmg.37.10.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duarte I, Santos A, Sousa H, Catarino R, Pinto D, Matos A, Pereira D, Moutinho J, Canedo P, Machado JC, et al. G-308A TNF-alpha polymorphism is associated with an increased risk of invasive cervical cancer. Biochem Biophys Res Commun. 2005;334:588–592. doi: 10.1016/j.bbrc.2005.06.137. [DOI] [PubMed] [Google Scholar]
- 19.Qin H, Liu B, Shi T, Liu Y, Sun Y, Ma Y. Tumour necrosis factor-alpha polymorphisms and hepatocellular carcinoma: a meta-analysis. J Int Med Res. 2010;38:760–768. doi: 10.1177/147323001003800304. [DOI] [PubMed] [Google Scholar]
- 20.Yang Y, Luo C, Feng R, Bi S. The TNF-α, IL-1B and IL-10 polymorphisms and risk for hepatocellular carcinoma: a meta-analysis. J Cancer Res Clin Oncol. 2011;137:947–952. doi: 10.1007/s00432-010-0959-8. [DOI] [PubMed] [Google Scholar]
- 21.Wei Y, Liu F, Li B, Chen X, Ma Y, Yan L, Wen T, Xu M, Wang W, Yang J. Polymorphisms of tumor necrosis factor-alpha and hepatocellular carcinoma risk: a HuGE systematic review and meta-analysis. Dig Dis Sci. 2011;56:2227–2236. doi: 10.1007/s10620-011-1617-y. [DOI] [PubMed] [Google Scholar]
- 22.Chung YS, Jeon YJ, Shin DE, Min KT, Shin YS, Won KS, Koh YC, Hong SH, Kim NK. Methylenetetrahydrofolate reductase (MTHFR) and thymidylate synthase (TS) polymorphisms with osteoporotic vertebral compression fracture (OVCF) in postmenopausal Korean women. Genes Genom. 2012;34:257–263. [Google Scholar]
- 23.Kim SY, Jeon YJ, Oh SH, Kim HS, Kim OJ, Oh D, Jeung M, Shin BS, Park YS, Kim NK. Association of kinase insert domain receptor (KDR -604, 1192, and 1719) polymorphisms with cerebral white matter lesions. Genes Genom. 2012;34:485–492. [Google Scholar]
- 24.Deepika ML, Reddy KR, Yashwanth A, Rani VU, Latha KP, Jahan P. TNF-α haplotype association with polycystic ovary syndrome - a South Indian study. J Assist Reprod Genet. 2013;30:1493–1503. doi: 10.1007/s10815-013-0080-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahirwar DK, Mandhani A, Dharaskar A, Kesarwani P, Mittal RD. Association of tumour necrosis factor-alpha gene (T-1031C, C-863A, and C-857T) polymorphisms with bladder cancer susceptibility and outcome after bacille Calmette-Guérin immunotherapy. BJU Int. 2009;104:867–873. doi: 10.1111/j.1464-410X.2009.08549.x. [DOI] [PubMed] [Google Scholar]
- 26.Canedo P, Durães C, Pereira F, Regalo G, Lunet N, Barros H, Carneiro F, Seruca R, Rocha J, Machado JC. Tumor necrosis factor alpha extended haplotypes and risk of gastric carcinoma. Cancer Epidemiol Biomarkers Prev. 2008;17:2416–2420. doi: 10.1158/1055-9965.EPI-08-0413. [DOI] [PubMed] [Google Scholar]
- 27.Yang JJ, Ko KP, Cho LY, Shin A, Gwack J, Chang SH, Shin HR, Yoo KY, Kang D, Park SK. The role of TNF genetic variants and the interaction with cigarette smoking for gastric cancer risk: a nested case-control study. BMC Cancer. 2009;9:238. doi: 10.1186/1471-2407-9-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salagacka A, Zebrowska M, Jelen A, Mirowski M, Balcerczak E. Haplotype analysis of TNFA gene in peptic ulcer patients. Int J Hum Genet. 2014;14:9–15. [Google Scholar]
- 29.Atkinson SH, Rockett KA, Morgan G, Bejon PA, Sirugo G, O’Connell MA, Hanchard N, Kwiatkowski DP, Prentice AM. Tumor necrosis factor SNP haplotypes are associated with iron deficiency anemia in West African children. Blood. 2008;112:4276–4283. doi: 10.1182/blood-2008-06-162008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X, Zhang L, Chang Y, Shen T, Wang L, Zhuang H, Lu F. Association of TNF-alpha genetic polymorphisms with hepatocellular carcinoma susceptibility: a case-control study in a Han Chinese population. Int J Biol Markers. 2011;26:181–187. doi: 10.5301/JBM.2011.8580. [DOI] [PubMed] [Google Scholar]
- 31.Ryu SH, Chung YH. How to overcome multidrug resistance in chemotherapy for advanced hepatocellular carcinoma. Liver Int. 2010;30:496–498. doi: 10.1111/j.1478-3231.2010.02215.x. [DOI] [PubMed] [Google Scholar]
- 32.Aggarwal BB. Signalling pathways of the TNF superfamily: a double-edged sword. Nat Rev Immunol. 2003;3:745–756. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- 33.Nakazaki H. Preoperative and postoperative cytokines in patients with cancer. Cancer. 1992;70:709–713. doi: 10.1002/1097-0142(19920801)70:3<709::aid-cncr2820700328>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 34.Liu XL, Li FQ, Liu LX, Li B, Zhou ZP. TNF-α, HGF and macrophage in peritumoural liver tissue relate to major risk factors of HCC Recurrence. Hepatogastroenterology. 2013;60:1121–1126. doi: 10.5754/hge12982. [DOI] [PubMed] [Google Scholar]


