Abstract
Memories are thought to be formed in response to transient experiences in part through changes in local protein synthesis at synapses. In Drosophila, the amyloidogenic (prion-like) state of the RNA binding protein Orb2 has been implicated in long-term memory, but how conformational conversion of Orb2 promotes memory formation is unclear. Combining in vitro and in vivo studies, we find that the monomeric form of Orb2 represses translation and removes mRNA polyA tails, while the oligomeric form enhances translation and elongates the polyA tails, and imparts its translational state to the monomer. The CG13928 protein, which binds only to monomeric Orb2, promotes deadenylation, whereas the putative polyA binding protein CG4612 promotes oligomeric Orb2-dependent translation. Our data support a model in which monomeric Orb2 keeps target mRNA in a translationally dormant state, and experience-dependent conversion to the amyloidogenic state activates translation, resulting in persistent alteration of synaptic activity and stabilization of memory.
Introduction
Synthesis of new proteins at the synapse has emerged as one of the potential biochemical mechanisms for producing experience-dependent persistent change in synaptic physiology and animal behavior(Richter and Klann, 2009). The cytoplasmic polyadenylation element binding protein (CPEB) CPEB, a family of sequence-specific RNA binding proteins, is one of the regulators of synaptic protein synthesis (Richter, 2007). Unexpectedly, one form of neuronal CPEB carries amino acid sequences characteristic of self-sustaining amyloid or prion- like proteins(Fiumara et al., 2010; Heinrich and Lindquist, 2011; Majumdar et al., 2012; Raveendra et al., 2013; Si et al., 2010; Si et al., 2003b; Stephan et al., 2015). The CPEB homolog in Aplysia (ApCPEB), Drosophila (Orb2), and in mouse (CPEB3) form amyloidogenic oligomers, which are critical for the persistence of long-term synaptic facilitation and behavioral memory(Fioriti et al., 2015; Kacsoh et al., 2015; Keleman et al., 2007; Kruttner et al., 2012; Kruttner et al., 2015; Majumdar et al., 2012; Si et al., 2010). These observations led to a model for long-term memory in which external stimuli recruit a self-sustaining amyloidogenic form of neuronal CPEB in the activated synapse, where CPEB then maintains memory through the sustained and regulated synthesis of a specific set of synaptic proteins. However, the prevailing view is that amyloid formation in general leads to inactivation of proteins. Therefore a central tenet of the hypothesis remains untested; how does neuronal CPEB regulate synaptic protein synthesis and what is the consequence of conversion to the self-sustaining amyloidogenic state?
The Drosophila CPEB family is comprised of two genes, Orb1 and Orb2 (Hafer, 2011 #33). All family members share an RNA binding domain (RRM) and a zinc finger domain in the C-terminal end but differ significantly in the N-terminal end, suggesting functional differences among family members. Orb2 has two protein isoforms, Orb2A and Orb2B (Majumdar et al., 2012; Mastushita-Sakai et al., 2010). Both isoforms contain an N-terminal prion-like domain, but the short Orb2A isoform has 8 and the longer isoform Orb2B has 162 amino acids ahead of the prion-like domain. The two isoforms also differ in their biophysical properties; Orb2A forms amyloids more efficiently than Orb2B both in vitro and in vivo (Kruttner et al., 2012; Majumdar et al., 2012). Although present in very low levels, deletion of Orb2A or a point mutation in Orb2A, Orb2AF5>Y5, reduces the oligomerization of the abundant Orb2B and impairs persistence of memory (Kruttner et al., 2012; Kruttner et al., 2015; Majumdar et al., 2012). Previous studies indicated Orb2 has translation repressive functions (Mastushita-Sakai et al., 2010; Xu et al., 2012; Xu et al., 2014), and regulates translation of reporter mRNA in the mushroom body (Kruttner et al., 2015). However, the biochemical functions of the two isoforms of Orb2, and more importantly, the function of amyloidogenic states in translation remain unknown. In this study, using an in vitro reconstituted translational assay, as well as in vivo behavior-dependent translation of Orb2-target genes, we set out address the following questions: 1) Does Orb2 act as a translation activator or repressor or both? 2) What is the role of monomeric and amyloidogenic-oligomeric Orb2 in protein synthesis? 3) How does Orb2 regulate translation?
Results
Two distinct sequence elements in the Tequila 3′UTR are important for Orb2 recruitment
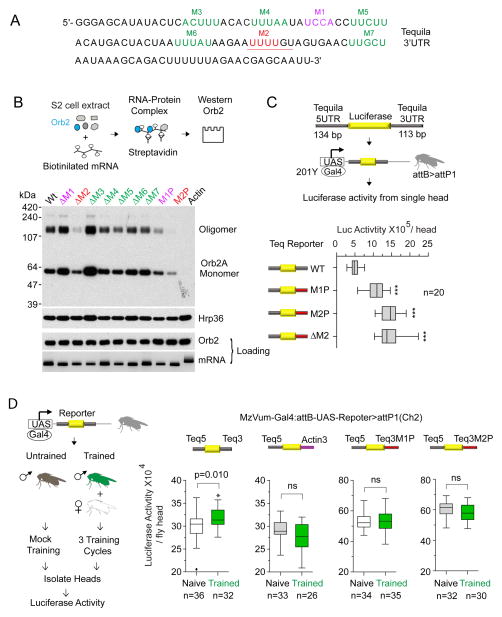
To assess the biochemical consequence of Orb2 conformational switch, we set out to design an Orb2-dependent translation assay. To this end we first sought to determine the sequences that are important for the recruitment of Orb2 to one of its target mRNA, Tequila, a human neurotrypsin homologue. We choose Tequila because, 1) it is required for long-term memory formation(Didelot et al., 2006), 2) it has a very short 3′UTR of 113 nucleotides (Fig 1A), 3) both isoforms and both physical states of Orb2 bind to the 3′UTR of Tequila (Fig S1A), and 4) Tequila expression in the adult brain is regulated by Orb2 (Mastushita-Sakai et al., 2010). Using an in vitro RNA pull down assay and a series of small deletions and nucleotide substitutions of the 3′UTR we identified two sequence elements, UCCACCUU/G (Mutation1 or M1) and UUUUGU (Mutation2 or M2), that are involved in Orb2 recruitment to Tequila 3′UTR (Fig 1B & Fig S1B). The M2 sequence is analogous to canonical CPEB-binding element UUUUAU (Richter, 2007), and mutation of UUUUGU to GCUUGU (M2P) or deletion of four U residues (ΔM2) significantly reduced both monomeric and oligomeric Orb2-recruitment (Fig 1B & Fig S1B). A U/CUUU/GU sequence motif, similar to M2 sequence, is also detected in the 3′UTR of Tequila gene of other Drosophila species (Fig S1C) as well as in other Orb2 targets (Mastushita-Sakai et al., 2010). While mutation or deletion of the M1 UCCAAXU sequence motif (Fig S1D) resulted in variable effects, irrespective of the nature of mutation, M2 consistently reduced Orb2 recruitment. Therefore we concluded the M2 sequence encompasses the primary Orb2 recruitment site and used this sequence element in subsequent studies to probe Orb2-dependency.
Figure 1. A CPE-like elements recruits Orb2 to the Tequila 3′UTR and mutation of this sequence increases protein expression.
(A) Sequence of 113 nucleotides long 3′UTR of Tequila gene. M1-M7 indicates sequences that were mutated. The conserved M1 and M2 sequences important for Orb2 recruitment are indicated in pink and red respectively. (B) Deletion (ΔM2) or mutation (M2P) of M2 elements reduced recruitment of both Orb2 monomer and oligomer to the mRNA. Top panel: Schematic of the RNA-protein binding assay using S2 cell lysate. Lower panel: Western blot of RNA bound Orb2A protein. The Actin88F (Actin) 3′UTR controls for non-specific Orb2 binding and Hrp36 serves as a control for general RNA binding. The amount of RNA and Orb2 protein used in the assay are shown below as loading. (C) Decrease in Orb2 recruitment increases protein expression in adult mushroom body neurons. The statistical significance was measured by one way ANOVA. (D) Male courtship conditioning enhances wild type reporter expression. Left panel: Schematic of the experimental design. Right panel: the training-dependent increase was observed when the reporter was expressed in all mushroom body lobes using MzVum-Gal4. The statistical significance was measured by unpaired two-tailed t-test. Data is expressed as mean ± SEM. ns, not significant and * ≤0.05, ** ≤0.01 and *** ≤0.001 indicates p values. Please also see supplementary figure 1.
Orb2-recruitment is important for training dependent increase in Tequila expression
Is Orb2-recruitment important for in vivo protein expression? To this end luciferase reporter containing the wild type Tequila 5′UTR and either the wild type 3UTR or M2P, ΔM2 or M1P mutant 3′UTR were integrated into the same genomic location using the attP-attB system to control for any chromosomal position effect on transcript expression (Fig S1E). Expression of the reporters in the adult mushroom body neurons (Fig S1F) using the UAS-Gal4 system revealed that compared to wild type, mutations in the 3′UTR that reduce Orb2 recruitment increase reporter expression (Fig 1C). Male courtship suppression training, a training paradigm that produces Orb2-dependent long-term memory(Keleman et al., 2007), increased expression from only the wild type reporter, but not the mutant reporter or a reporter bearing actin 3′UTR (Fig 1D & S1G). The training-dependent increase was observed when the reporter was either expressed in all lobes of the mushroom body (MzVUm-Gal4) or in the γ-lobe only (201Y-Gal4) but not in the α-β lobe (17D-Gal4) (Fig 1D & S1G, H). These several results suggest Orb2-recruitment is important for training-dependent expression of Tequila in the γ-lobe neurons of the mushroom body. These results are consistent with the observations that Orb2 activity is important in γ-lobe neurons for long-term memory(Keleman et al., 2007), that behavioral training increases Tequila expression in the mushroom body (Didelot et al., 2006) and that Orb2 acts as a translation repressor (Mastushita-Sakai et al., 2010; Xu et al., 2012; Xu et al., 2014).
Monomeric Orb2B acts as a translation repressor
The cell based studies can’t distinguish a direct effect on translation from an indirect effect on mRNA/protein stability. Therefore we sought to establish an in vitro reconstituted Orb2-dependent translation system (Fig S2A). The Orb2B protein and components of the translation regulatory complexes that interact with Orb2 in the adult brain are also present in the early Drosophila embryo (White-Grindley et al., 2014; Xu et al., 2012; Xu et al., 2014). The embryo extract has also been used to study translational regulation (Lie and Macdonald, 2000). Therefore using embryo extract we developed an in vitro dual reporter system in which firefly luciferase reports Orb2-dependent translation, while renilla luciferase serves as an internal control (Please see Fig S2A–C for further details of the assay).
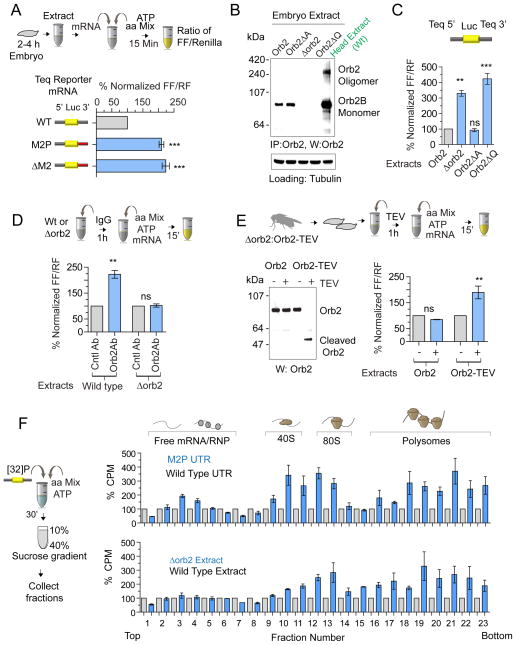
As in the adult fly head, M2P or ΔM2 that reduced Orb2 recruitment to the 3′UTR also enhanced protein expression from reporter mRNA in Drosophila embryo translation extract (Fig 2A) but not in the control mammalian reticulocyte lysate (Fig S2D). In a reciprocal experiment we prepared translation extracts from various orb2 mutant embryos (Fig 2B). The expression from the wild type reporter mRNA was significantly higher in extracts prepared from orb2 null (Δorb2), or the strong hypomorphic mutant (Orb2ΔQ) flies but not from flies lacking the Orb2A isoform (Orb2ΔA), which is not detectable in the embryo extract (Fig 2B&C). Unlike the wild type reporter, the expression from mutant reporters (Fig S2E, F) or control Actin88F 3′UTR was unaffected by Orb2 levels (Fig S2G). Moreover, reporter mRNA containing the 5′ and 3′ UTR of other Orb2 target genes such as neuroligin and atypical protein kinase C were also increased in the Orb2 deficient extracts (Fig S2H & I) (Mastushita-Sakai et al., 2010; Xu et al., 2014).
Figure 2. Orb2 acts as a translational repressor.
(A) Loss of Orb2 recruitment increases protein expression in vitro. Top panel: Schematic of the in vitro translation assay using Drosophila embryo extract. Lower panel: normalized ratio of firefly and Renilla enzyme activity of 3 independent experiments. Each experiment was comprised of three technical repeats. (B) Embryonic translation extract contains only monomeric Orb2B unlike adult head extract, which contains both Orb2 monomer and oligomer. The Δorb2 flies lack the entire Orb2 locus, the Orb2ΔQ lacks the Q-rich 80 amino acids common to Orb2A/B and Orb2ΔA lacks only the Orb2A-specific exon. Tubulin serves as loading control. (C) The absence of Orb2 enhances wild type Tequila reporter expression. (D) Treatment of embryo extract with anti-Orb2 antibody enhances reporter expression. Addition of Orb2 antibody in extract lacking Orb2 (Δorb2) has no effect. (E) Acute inactivation of Orb2 enhances translation. Top panel: A genomic rescue fragment in orb2 null background that contains a TEV-protease site within the Orb2 gene. Left panel: Western blot analysis of TEV enzyme treated wild type and Orb2-TEV embryo extract. Right panel: addition of TEV in Orb2-TEV, but not wild type (Orb2) embryo extract enhances reporter expression. (F) Polysome profile analysis of in vitro translation reaction shows the relative abundance of translating mRNA. Left panel: schematic of polysome assay using [P32]-label mRNA. Right upper panel: mutant reporter (M2P) mRNA is more abundant in polysome fractions compared to wild type. Right lower panel: wild type reporter is more abundant in polysome when translated in Δorb2 embryo extract. Data is expressed as mean ± SEM and * <0.01, ** <0.001 and *** <0.0001 indicates p value. Please also see supplementary figure 2.
The enhanced expression of Orb2 target genes in the orb2 mutant extracts was not due to a secondary consequence of chronic Orb2 protein depletion: acute inactivation of Orb2 by adding anti-Orb2 antibodies to the wild type extract also enhanced expression (Fig 2D). Moreover, we generated a fly strain in which both copies of the Orb2 gene carried a TEV protease recognition site (Kapust and Waugh, 2000) sequence (Δorb2:Orb2-TEV) at amino acid position 369. The Orb2-TEV fully substituted the wild type Orb2 function. In translation extract prepared from Orb2-TEV flies, addition of TEV protease cleaved most of the available Orb2 protein within an hour (Fig 2E, left panel) and enhanced reporter expression (Fig 2E, right panel). Polysome analysis of [32P] labelled mRNA, which measures the 80S ribosome bound translating mRNA, revealed that both the reporter that does not recruit Orb2 (Fig 2F, Top panel) or translation extracts lacking Orb2 (Fig 2F, bottom panel) contain relatively more translating mRNA compared to their wild type counterparts. From these results we conclude that the in vitro translation system indeed reports Orb2-dependent translation of specific mRNA. Since the embryonic translation extract did not have any discernable amount of oligomeric Orb2 (Fig 2B) these results also suggested that the translation inhibition is primarily mediated by monomeric Orb2.
Isolation of monomeric and amyloidogenic oligomeric Orb2 protein
In the mRNA binding assay we did not observe any qualitative difference between Orb2A and Orb2B protein isoforms or the monomeric and a SDS-resistant oligomeric state (Fig 1B, Fig S1A&B). Therefore determination of function of various form of Orb2 requires obtaining both Orb2A and Orb2B in monomeric and amyloid-like states and adding them in a translation extract lacking any endogenous Orb2. Recombinant Orb2 is extremely aggregate prone and Orb2A protein rapidly assembles into oligomers even in 6M urea (Majumdar et al., 2012). However, in insect cells, both monomeric and oligomeric Orb2 can be observed suggesting proper cellular milieu is important for the protein states. Therefore, to obtain monomeric and amyloidogenic Orb2 separated from each other, full length untagged Orb2 protein was expressed in insect Sf9 cells, which lack endogenous Orb2, using Baculovirus (Fig S3A) and whole cell extracts were fractionated in the presence of high salt (500mM NaCl) and detergent using a superose 6 size exclusion column (Fig S3B, left panel). The Orb2 protein was recovered in fractions consistent with two distinct physical states; a monomeric and an oligomeric Orb2 (Fig S3B, right panel). We observed a range of oligomeric species including those that are similar in size to that of the endogenous Orb2-oligomer (Majumdar et al., 2012). We concluded that the oligomeric Orb2 is amyloid in nature based on resistance to heat and detergents, higher binding to Thioflavin T, interaction with the amyloidogenic anti-oligomeric antibody A11, production of proteinase K resistant fractions, and high amyloid-B structure as measured by Fourier Transform Infrared spectroscopy (FTIR) (Fig S3).
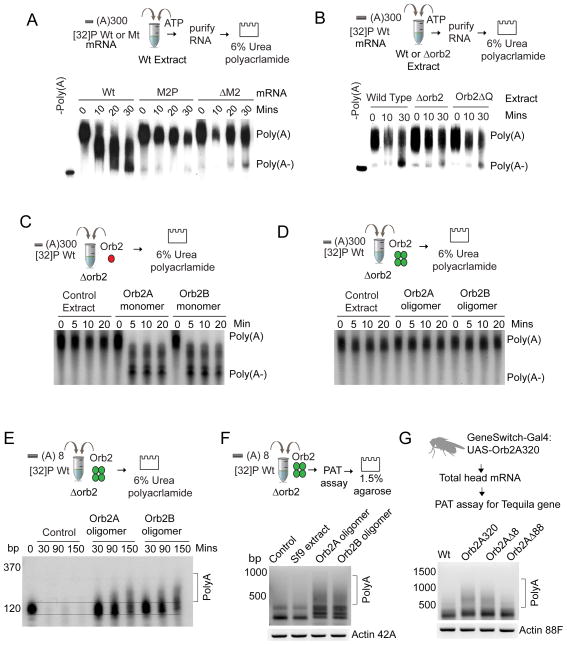
Amyloidogenic oligomeric Orb2 enhances translation
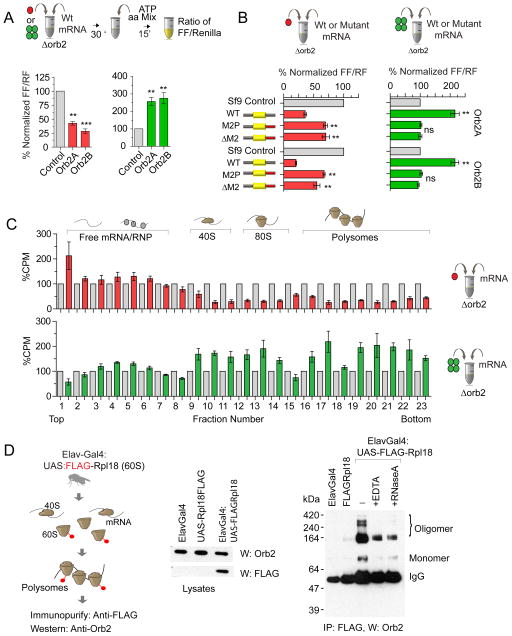
We added the fraction containing monomeric or amyloidogenic oligomeric Orb2 in orb2 null (Fig 3A) or Orb2 hypomorphic (Orb2ΔQ) translation extracts (Fig S4A) with the Tequila reporter mRNA. Equivalent amounts of similarly fractioned untransfected Sf9 cell extracts were used as a control. Addition of monomeric Orb2A or Orb2B protein fractions resulted in significant repression of translation (Fig 3A & Fig S4A) while addition of oligomeric Orb2 resulted in an increase in translation compared to control fractions (Fig 3A & Fig S4A). Mutation of the M2 sequence reduced the monomer-dependent decrease and abolished the oligomer-dependent increase in translation (Fig 3B).
Figure 3. Orb2 monomer acts a translation repressor while amyloidogenic oligomer as an activator.
(A) Orb2 monomer represses translation and Orb2 oligomer increases translation in Δorb2 embryo extract. (B) Mutations of the M2 sequence that reduce Orb2 recruitment also attenuate the translational effect of monomeric and oligomeric Orb2. (C) Polysome profile analysis of in vitro translation reactions shows that the Orb2 monomer reduces, while the oligomer increases the relative abundance of translating mRNA in polysome fractions. (D) Primarily oligomeric Orb2 associates with FLAG-tagged 60S ribosomal subunits that are part of polyribosomes. Treatment with 30mM EDTA (pH8.0) or 20 unit/ml RNaseA that disrupts polysomes also reduces oligomeric Orb2 association. The extracts in the left are immunoprecipitated with anti-FLAG antibodies and probed for Orb2. Data is expressed as mean ± SEM and * <0.01, ** <0.001, *** <0.0001 indicates p value and ns indicates not significant. Please also see supplementary figure 3 and supplementary figure 4.
To determine if the change in translation is indeed due to Orb2 and not due to other co-fractionating proteins, we used different monomeric or oligomeric protein fractions and observed that the translational effect is proportional to the amount of Orb2 protein in a particular fraction (Fig S4B&C). Next we took advantage of the Orb2 protein carrying a TEV protease site (Orb2-TEV) and fractionated Sf9 cells expressing Orb2-TEV protein (Fig S4D). Similar to wild type protein, addition of monomeric Orb2-TEV reduced, whereas the oligomeric form enhanced translation; however, incubation of Orb2-TEV fractions with TEV-protease (Fig S4E&F) significantly attenuated the repressive function of the monomer and the activating function of oligomeric Orb2 (Fig S4E&F). Addition of TEV-protease to the wild type Orb2 had no effect. Taken together these observations suggest the Orb2 protein in the extracts is largely responsible for the changes in translation activity. The change in protein expression is at least partly due to change in translation because addition of monomeric Orb2 decreased the amount of mRNA in polysomal fractions with a corresponding increase of the labelled mRNA in the top of the gradient (Fig 3C, Top panel). Conversely addition of oligomeric Orb2 resulted in an increase in the amount of monosome and polysome bound mRNA and a decrease in full length free mRNA in the top of the gradient compared to control extract (Fig 3C, Bottom panel & Fig S4G).
Next we sought to determine which forms of Orb2 associate with translating ribosomes in the adult nervous system. To this end we used the ribosome-tagging approach, which entails tagging a protein of either the 40S or 60S ribosome, expression of the tagged protein in a specific cell type and immunoprecipitation using the tag (Doyle et al., 2008; Mustroph et al., 2009; Sanz et al., 2009). We found that the Rpl18 protein of the 60S subunit (please see methods for detail), when tagged at the N-terminal end with FLAG, is efficiently incorporated into polysomes in S2 cells and in adult neurons (Fig 3D & S4G). Immunoprecipitation of FLAGRpl18 (Elav-Gal4:UAS-FLAGRpl18), from adult neurons revealed that the Orb2 proteins immunoprecipitated with ribosomes are primarily heat and SDS-resistant oligomers (Fig 3D & S4G). Treatment with 30mM EDTA or ribonuclease RNaseA that are known to disrupt polysomes also reduced immunoprecipitated oligomeric Orb2. Taken together, these results suggest that while the monomeric forms of both Orb2A and Orb2B act as repressors, the amyloidogenic oligomeric Orb2 acts as a translation activator.
The prion-like domain of Orb2A induces oligomerization of Orb2B and enhances translation
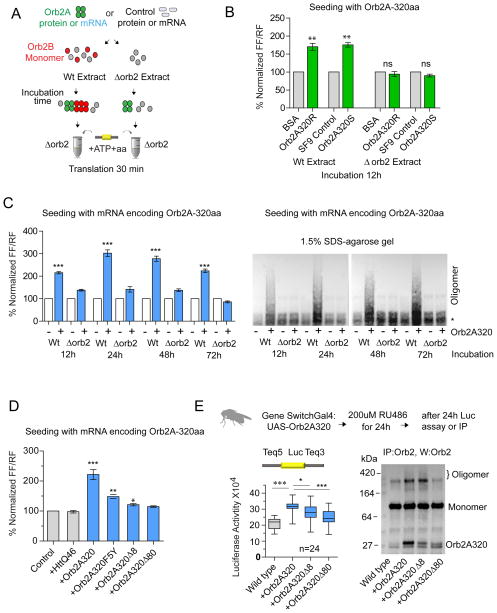
One of the features of self-sustaining amyloidogenic oligomers is that they induce conformational conversion of the monomeric form(Prusiner, 1998). Previous studies indicated that rare but highly amyloidogenic Orb2A induces oligomerization of constitutively expressed Orb2B and only the N-terminal of Orb2A that contains the prion-like domain (PLD), but lacks the RRM domain, is sufficient for long-term memory(Kruttner et al., 2012; Majumdar et al., 2012). The distinct functional states of the monomeric and amyloidogenic oligomeric forms of Orb2B now led us to interrogate the functional consequence of interaction of Orb2B with the Orb2A prion-like domain (Fig 4A). We used just the N-terminal 320 amino acids of Orb2A that contains the prion-like domain but lacks the RRM domain and similar to full length Orb2A, OrbA-320 has both a monomeric and amyloidogenic oligomeric state (Fig S5A). Addition of oligomeric Orb2A-320 isolated from insect cells or from bacteria to translation extract containing monomeric Orb2B enhanced translation (Fig 4B). Addition of oligomeric Orb2A-320 to orb2 null extract had no effect, indicating the enhanced translation was not due to Orb2A-320 and was dependent on Orb2B (Fig 4B& S5B).
Figure 4. Orb2A N-terminal prion-like domain converts monomeric Orb2B to a translational activator.
(A) Schematic representation of the experimental design. The wild type embryo extract was used as source for monomeric Orb2B. The Δorb2 extract, same amount of purified BSA or similar amount of Sf9 cell extracts serve as a controls. (B) The N-terminal 320aa of Orb2A isolated from bacteria (Orb2A320R) or Sf9 cells (Orb2A320S) enhance translation only in wild type extract. The seeding reaction was carried out for 12 hours at 4°C and translation was measured after 15 min. (C) The mRNA expressing Orb2A320 enhances translation and induces Orb2B oligomerization. The seeding reaction was performed for the indicated time and subsequently translation (left panel) and the oligomeric state of Orb2 (right panel) were assessed in 1.5% SDS-agarose gel. The * indicates a non-specific immunoreactive band visible upon long exposure. (D) Mutation that affects the oligomerization of Orb2A also attenuates its ability to enhance translation. (E) Transient expression of the Orb2A320 using Gene switch Gal4 enhances translation in the adult fly brain (left panel) and Orb2-oligomerization (right panel). Removal of the N-terminal 80 amino acids significantly reduces both activities. Data is expressed as mean ± SEM and * <0.05, ** <0.01, *** <0.001 indicates p value and ns indicates not significant. Please also see supplementary figure 5.
The Orb2A-320 proteins can influence Orb2B-dependent translation because of some associated factors. To rule out such a possibility, mRNA encoding Orb2A-320, instead of protein, was added to extract with (wild type), or without Orb2B (Δorb2). The Orb2A320 mRNA produced an equivalent amount of Orb2A320 protein in both extracts (Fig S5B), however, it resulted in a time-dependent increase in Orb2B oligomerization and translation in wild type but not in Δorb2 extract (Fig 4C). Mutation of the M2 sequence abrogated the increase in translation (Fig S5C). The translational enhancement is rapid since a seeding reaction for 30 minutes resulted in a small but significant increase in translation (Fig S5D). The deletion or mutation in the first 8 N-terminal amino acids (Orb2AΔ8) or subsequent 80 amino acids (Orb2AΔ80), or the mutation of the F5 to Y (Orb2A F5Y) reduces Orb2A’s ability to form amyloidogenic oligomers and impairs memory. We observed that compared to wild type Orb2A320, mutant Orb2A mRNA is significantly less effective in enhancing translation (Fig 4D), although the expression of mutant Orb2A fragments was similar to that of wild type Orb2A (Fig S5B). Therefore, mutations that affect Orb2A’s ability to form amyloidogenic oligomers also affect its ability to change Orb2B-dependent translation.
Next we sought to determine whether these in vitro results reflect in vivo translation and transiently overexpressed either full length Orb2A or Orb2A-320 with the Tequila reporter in the adult fly brain. Transient expression of Orb2A (Fig S5E) or Orb2A-320 resulted in a significant increase in reporter expression and in SDS-resistant oligomer (Fig 4E). Unlike wild type Orb2A-320, Orb2A-320Δ80 was less efficient in inducing translation and oligomerization (Fig 4E). Interestingly, unlike in embryo extract expression of Orb2A320Δ8 in the brain resulted in an increase in translation albeit less efficiently than full length Orb2A320 (Fig 4E). Consistent with the translation increase by Orb2A320Δ8 there was also an increase in oligomer, suggesting that the seeding may be more efficient in vivo compared to in vitro. This indicates seeding is influenced by factors that are either limiting or missing in the embryo extract or modification of Orb2 protein (such as phosphorylation) may influence either the seeding capacity of Orb2A or Orb2B to be a substrate of Orb2A-mediated oligomerization. Nonetheless taken together these results suggest that the Orb2A prion-like domain can induce Orb2B oligomerization and enhance translation.
Monomeric Orb2 removes the polyA tail and destabilizes mRNA whereas oligomeric Orb2 stabilizes mRNA and elongates the polyA tail
How does Orb2 affect target mRNA translation? Previously we observed that Orb2 interacts with Transducer of Erb2 (Tob) (White-Grindley et al., 2014), a protein that has been implicated in mRNA deadenylation (Hosoda et al., 2011). To determine whether Orb2-regulates deadenylation, [32P] labelled Tequila 3′UTR with a ~300 nucleotide long polyA tail was added into the in vitro extract. In the wild type 3′UTR, the polyA tail was shortened over time (Fig 5A) and mutations of the M2 sequence reduced polyA tail shortening (Fig 5A). Likewise addition of wild type reporter into extracts lacking Orb2 (Δorb2 or Orb2ΔQ) had low levels of deadenylation compared to the wild type extract suggesting Orb2 is involved in deadenylation of the target mRNA (Fig 5B).
Figure 5. Orb2 monomer reduces whereas Orb2 oligomer stabilizes polyA tail.
(A) Orb2 promotes deadenylation of wild type Teq3′UTR. Mutation of the M2 sequence reduces deadenylation. (B) Deadenylation is reduced in extracts lacking Orb2 (Δorb2 or Orb2ΔQ) compared to wild type extract. (C) Only Orb2 monomer but not (D) Orb2 oligomer efficiently deadenylates the 3′UTR. (E) Orb2 oligomer stabilizes and polyadenylates capped and short polyA tailed mRNA in vitro. (F) PCR based PolyA tail (PAT) assay shows addition of Orb2 oligomer increases polyA tail length in vitro. Actin serves as a control for total mRNA used. (G) Expression of Orb2A320 and Orb3A320Δ8 but not Orb2A 320Δ80 increases Tequila mRNA polyA tail length. Actin serves as amount of mRNA used in the reactions.
Next we sought to determine whether the functional difference between monomeric and oligomeric Orb2 arises from their differential mRNA deadenylation activity and carried out the deadenylation assay in orb2 null extracts in the presence of exogenously added monomeric or oligomeric Orb2. Addition of monomeric Orb2 rapidly deadenylated (Fig 5C) while the addition of oligomeric Orb2 did not reduce the polyA tail (Fig 5D). We wondered whether the oligomer is simply an inactive version of monomer or rather it is actively participating in polyA tail maintenance either by protecting the existing polyA tail from decay and/or elongation of the polyA tail. Therefore instead of ~300 nucleotides, we used an 8-nucleotide long polyA tailed mRNA, which is sufficient to trigger polyadenylation (Benoit et al., 1999). Unlike long-polyadenylated mRNA, the short polyA-tailed mRNA was degraded in control extracts or in extract containing Orb2 monomer within 30 minutes (Fig S5F). However, in extract containing Orb2 oligomer there was significantly less degradation of the mRNA and there was also a low but significant increase in size of the mRNA (Fig 5E & S5F). PCR-based polyA tail length assay or PAT assay (please see methods for details of PAT assay) revealed that the incubation with Orb2 oligomer indeed results in longer polyA tail length (Fig 5F). To determine whether Orb2 affects the polyA tail length of Tequila mRNA in the adult brain we transiently expressed Orb2A320 and measured the polyA tail length using the PAT assay. Consistent with reporter translation and Orb2 oligomerization (Fig 4E) expression of Orb2A320 or Orb2A320Δ8 but not Orb2A320Δ80 resulted in an increase in polyA tail length (Fig 5G). Taken together these results indicate that while monomeric Orb2 reduces polyA tail length and eventually destabilizes mRNA, oligomeric Orb2 protects/elongates the polyA tail and stabilizes mRNA.
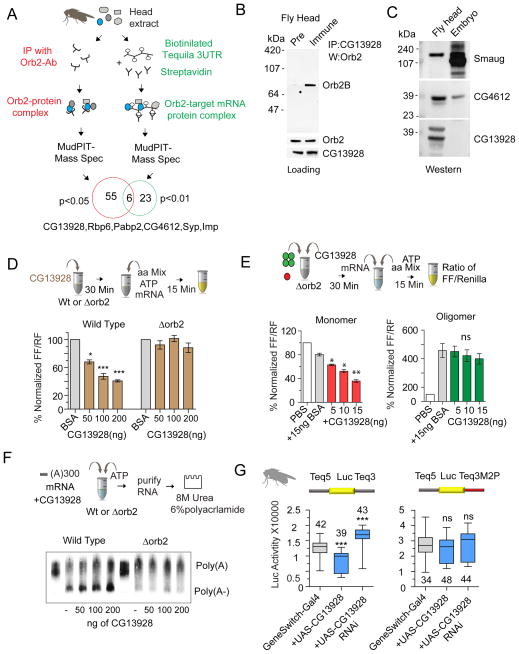
Identification of Orb2 and Orb2-target mRNA associated proteins in the adult head
What is the molecular basis of the differential activity of the monomeric and oligomeric Orb2? Since both states require the same M2 sequence we reasoned that the two forms may form distinct protein complexes. Previously we identified Orb2-interacting proteins in the adult fly head by immunopurifying the Orb2-protein complex in presence of RNAse (White-Grindley et al., 2014). These complexes include proteins that are involved in Orb2-dependent translation as well as stability, localization or modification of Orb2. To determine the mechanism of Orb2-dependent translation we sought to identify protein complexes from adult fly head that are specifically assembled in the Orb2 target mRNA Tequila compared to an Actin88F control using MudPIT-mass spectrometry (Fig 6A & S6A). Comparing 6 independent control and experimental groups we identified 29 proteins that were significantly (p<0.01) enriched in the Tequila 3′UTR affinity purification (Table S1). Six out of 29 proteins -CG13928, Rbp6, Pabp2, CG4612, Syncrip, and Imp- were also identified as part of an Orb2-protein complex previously and three of these proteins, Pabp2, Rbp6 and CG13928 strongly interacted with monomeric Orb2 (Fig S6B &C). We also identified Smaug, a known translational regulator that represses translation by promoting deadenylation (Smibert et al., 1996), as an Orb2 interacting protein (Fig S6D). Addition of recombinant Smaug protein reduced wild type reporter translation in the presence of Orb2 (Fig S6E), suggesting Smaug is one of the accessory factors for Orb2-mediated translation repression.
Figure 6. CG13928 promotes Orb2 monomer-dependent translation repression and deadenylation.
(A) Schematic of the complementary proteomics analyses from adult fly heads. Orb2-interacting proteins from fly head lysate were either immunoprecipitated with Orb2 antibody or RNA-affinity purified using an Orb2 target 3′UTR. (B) CG13928 interacts with monomeric Orb2 in the adult fly head. Co-immunoprecipitation of endogenous Orb2 with preimmune or CG13928 serum. Lowers panels show the loading controls. (C) CG13928 is enriched in the nervous system. Western blot analysis of Smaug, CG13928 and CG4612 protein in embryo extract and in adult fly head. (D) CG13928 represses translation in an Orb2-dependent manner. The effect of increasing the amount of purified CG13928 protein was compared with respect to equivalent amounts of BSA. (E) CG13928 imparts translation repression through monomeric Orb2. Fixed amounts of Orb2 (100 ng of monomer and oligomer fractions) were incubated with increasing amounts of CG13928 recombinant protein. CG13928 enhanced repression but had no significant effect on oligomer-mediated translation increase. (F) CG13928 enhances deadenylation in an Orb2-dependent manner. Addition of increasing amounts of CG13928 protein proportionally enhances deadenylation only in wild type extract but not in Orb2 depleted (Δorb2) extract. (G) CG13928 negatively regulates Orb2-dependent reporter translation in the adult fly head. Translation of wild type and M2 mutant reporters were measured from single fly heads expressing FLAG-tagged CG13928 or CG13928 RNAi under RU484 inducible Gene-switch Gal4. Data is expressed as mean ± SEM and * <0.05, ** <0.01 and *** <0.001 indicates p value. Please also see supplementary figure 6.
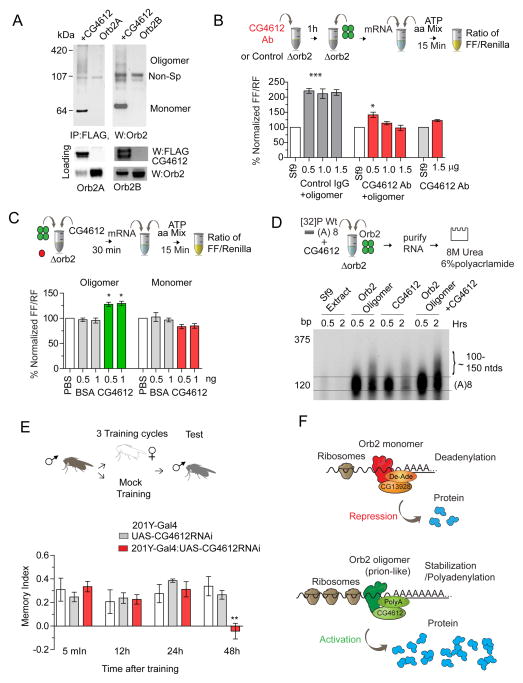
CG13928 contributes to the repressive function of monomeric Orb2, while CG4612 contributes to the translation activating function of oligomeric Orb2
What protein factors contribute to Orb2 mediated translation repression? To this end we specifically focused on one of the Orb2-interacting proteins, CG13928 because, i) it interacts with Orb2 (Fig 6B) and Smaug (Fig S6F), ii) it is enriched in the nervous system (Fig 6C) and iii) both in adult Drosophila brain (Fig 6B) and in vitro (Fig S6G), it interacts only with monomeric but not oligomeric Orb2. Addition of recombinant CG13928 protein reduced translation in a concentration dependent manner in wild type extract but not in extracts lacking Orb2 (Fig 6D). When incubated with a limiting amount of monomeric or oligomeric Orb2, CG13928 enhanced the translation repressive function of monomeric Orb2 (Fig 6E, left panel) but had no significant effect on the oligomeric Orb2 activity (Fig 6E, right panel). Consistent with translation repression, CG13928 also enhanced Orb2 monomer-mediated deadenylation (Fig 6F). Similar to the in vitro results, in the adult brain overexpression of CG13928 (ElavGeneSwitch: UAS-Teq-Reporter; UAS-FLAG-CG13928) reduced translation, whereas decrease in CG13928 (ElavGeneSwitch: UAS-Teq-Reporter; UAS- CG13928-dsRNA) increased reporter expression (Fig 6G). Neither overexpression nor reduction of CG13928 had any significant effect on the M2 sequence mutated reporter (Fig 6G & S6H). Taken together these results suggest CG13928 contributes to the repressive function of monomeric Orb2.
What protein factors contribute to Orb2-mediated translation activation? To this end we focused on two Orb2-interacting proteins identified from the proteomic studies, Pabp2 and CG4612 for the following reasons: i), they are polyA binding proteins (PABP) and PABP proteins are known to promote polyadenylation and enhance translation(Benoit et al., 1999; Smith et al., 2014) and ii), unlike CG13928, both of these proteins also bind to oligomeric Orb2 (Fig 7A & S6G). Addition of anti-CG4612 antibody reduced oligomeric-Orb2 mediated translation (Fig 7B) and addition of recombinant purified CG4612 resulted in a small but significant increase in the oligomeric Orb2-dependent translation (Fig 7C). One plausible explanation for the small increase in translation by exogenously added CG4612 protein is that the translation extract already contained CG4612 (Fig 6C). Nonetheless, the effect of CG4612 in Orb2 oligomer-mediated translation is specific since addition of CG4612 protein to extract lacking Orb2 or to wild type extract that contains only monomeric Orb2 had no significant effect on translation (Fig 7B, C & Fig S7B), and the addition of a similar amount of purified Pabp2 had no effect on Orb2-oligomer-mediated translation (Fig S7C). To determine whether CG4612 affects polyA tail length, Tequila 3UTR with a short polyA tail was incubated with oligomeric Orb2 in the presence or absence of CG4612. Consistent with a small increase in translation there was also a similar increase in the amount of long polyA tailed mRNA (Fig 7D). Taken together these results suggest that the two proteins, CG13928 and CG4612, contribute to the translation repressive and activating function of Orb2 respectively (Fig 7F).
Figure 7. CG4612 contributes to Orb2-oligomer mediated translation activation.
(A) CG4612 interacts with Orb2 monomer and oligomers. Untagged Orb2 was co-transfected with Flag-tagged CG4612 and Flag immunoprecipitate was western blotted for Orb2. (B) CG4612 activity is required for Orb2-oligomer mediated translation increase. Incubation of the translation extract with anti-CG4612 antibody, but not control IgG, significantly reduced the translation enhancing activity of oligomeric Orb2. Addition of CG4612 antibody did not result in a general translation inhibition. (C) Recombinant CG4612 enhances translation only when combined with Orb2B oligomer but not monomer. (D) CG4612 enhances Orb2 oligomer-mediated polyadenylation of short polyA tailed mRNA. A limiting amount of Orb2 oligomers was used to score the effect of CG4612. For ease of visualization the position of the starting mRNA has been indicated with two lines. Addition of CG4612 itself has some mRNA stabilizing activity, mostly apparent at the 30 min time point. (E) CG4612 is required for long-term male courtship suppression memory. Flies expressing CG4612 RNAi had similar short-term and intermediate-term memory but had significantly reduced long-term memory measured at 48h. (F) Model for Orb2-monomer mediated translation repression and oligomer-mediated translation activation. Data is expressed as mean ± SEM and * <0.05, ** <0.01 and *** <0.001 indicates p value. Please also see supplementary figure 7.
CG4612 is required for long-term memory
Since Orb2 oligomerization is important for long-term memory we sought to determine whether CG4612, which is involved in Orb2-oligomer dependent translation, is also important for long-term memory. To this end we expressed dsRNA against CG4612 in the mushroom body γ-lobe neurons using the 201Y-Gal4 driver (201Y-Gal4:UAS-CG4612RNAi) and tested these flies in a male courtship suppression paradigm (Fig 7E & Fig S7D). In this paradigm male flies learn to suppress their courtship behavior upon repeated exposure to an unreceptive female. The 201Y-Gal4: UAS-CG4612RNAi flies developed normally and when tested they did not display a deficit in short-term memory or memory up to a day. However, when tested 48 hours after training there was significant impairment in learned suppression of male courtship (Fig 7E). The memory phenotype due to impairment of CG4612 activity is reminiscent of the phenotype observed with selective perturbation of Orb2 oligomerization (Majumdar et al., 2012) and inhibition of the polyA polymerase Gld2 activity in the adult brain (Kwak et al., 2008).
Discussion
Previous studies have suggested that the self-sustaining amyloidogenic state of specific isoforms of neuronal CPEB produces an enduring mark in the activated synapse (Si et al., 2003a). However the biochemical consequence of this enduring marking of the synapse remains largely unknown. Here we report that when Drosophila Orb2 adopts the amyloid-like state it gains a new function and the translational repressive function of the monomeric form of the protein changes to a translation activating function. Therefore Drosophila Orb2 provides an example of an amyloid-based protein switch that turns a repressor into an activator. Recently, regulated aggregation–dependent transcriptional or translational control have reported by others (Berchowitz et al., 2015; Cai et al., 2014; Kwon et al., 2013). Therefore, it is conceivable that in multicellular eukaryotes, amyloid-like protein aggregations are used to create altered activity states and in some cases to create persistent change in cellular physiology from transient stimuli.
Translational regulation by monomeric and oligomeric Orb2
Similar to mammalian CPEB1, Orb2 acts as a repressor as well as an activator of translation via regulating polyA tail length (Udagawa et al., 2012). A U-rich sequence in the 3′UTR is important for the recruitment of Orb2 monomer and oligomer. The recruitment of the monomer results in shortening of the polyA tail and reduction in translation, while the oligomer protects/increases polyA tail length and translation. The following in vitro observations suggest that the oligomerization results in not just loss of monomeric function but gain of a new function: one, addition of Orb2 oligomer in extract lacking any monomeric protein (orb2−/−) increases translation (Fig 3A, B) suggesting oligomeric Orb2 has activities of its own in addition to inhibiting the monomeric Orb2 function. Two, the oligomeric Orb2 binds mRNA and it not only lacks the deadenylating activity of monomeric Orb2 but it also can add to the polyA tail and stabilize mRNA. Finally, oligomeric Orb2 is the primary form of Orb2 that associates with polyribosomes in the adult neuron. Therefore the net outcome of Orb2 oligomerization is reduction in deadenylation, increase in polyA tail length, stabilization of mRNA and enhanced translation. However, the relative contributions of inhibition of deadenylation and elongation of the polyA tail in Orb2-oligomer mediated translation is unclear.
Mechanistically, the zinc-finger domain containing protein CG13928 that only interacts with monomeric Orb2 enhances the repressive function. Since embryo extract lacking CG13928 can still support Orb2-dependent translation repression, CG13928 likely facilitates the recruitment of deadenylation complex to the mRNA. The mechanistic basis of Orb2 oligomerization-mediated translation enhancement is less clear. Unlike monomeric Orb2, no oligomer-specific interacting protein has been identified and although CG4612 is required for oligomerization-mediated translation, it binds to both forms of the Orb2 protein. However, oligomer may enhance translation not only by recruitment of a specific protein but also by increasing the affinity for existing interactors. For example, an mRNA bound by oligomeric Orb2 will have more CG4612 compared to a monomer bound mRNA and therefore more of the CG4612-associated proteins. Such a mechanism would be reminiscent of CPEB1-mediated polyadenylation (Mendez et al., 2000): unphosphorylated CPEB1 has weak affinity for the CPSF160 complex and phosphorylation increases the affinity of CPEB1 for CPSF160 leading to polyadenylation and translation activation. In mammals, PABPs are multifunctional proteins, and in some cases are known to activate translation by interacting with the translation initiation complex(Smith et al., 2014). Curiously, the other PABP, Pabp2, interacts with both forms of Orb2 yet does not have a significant effect on Orb2 function, except for translation inhibition at a high concentration (Fig S7C). Further studies would be required to dissect the detailed mechanism of translational activation and the role of PABPs and other proteins in Orb2-mediated translation activation.
A plausible model for seeded prion-like conversion of Orb2 and persistent translation
Among the two isoforms of Orb2, Orb2A and Orb2B, we did not observe any significant difference in mRNA binding, translation activity or the kind of protein complexes they form. However, unlike Orb2B, which is widely distributed Orb2A is extremely low in abundance, expressed only in a subset of neurons (Kruttner et al., 2012; Majumdar et al., 2012) and is primarily synaptic (Kruttner et al., 2015). Therefore, based on their abundance and distribution we postulate that Orb2B regulates Orb2-target mRNA distribution, stability and translation, while Orb2A controls where and when amyloid-like oligomerization will occur and translational activation will ensue. Indeed, just the N-terminal prion-like domain of Orb2A is sufficient to support long-term memory and we find that the N-terminal domain of Orb2A is sufficient to induce Orb2-oligomerization and change the translational state of Orb2B.
Although direct evidence that the amyloidgenic oligomer regulates translation only in the activated synapse is still lacking, based on these observations we proposed a plausible model for synapse-specific persistent translation (Fig 7F). We posit that Orb2B binds to the target mRNA and the bound mRNA is transported to the synapses and kept in a repressed state via association with the deadenylation complex. Synaptic activation increases the local concentration of the low abundant Orb2A protein via phosphorylation (White-Grindley et al., 2014) and/or other yet unknown mechanisms. Increase in the Orb2A protein level triggers self-sustaining amyloidogenic-oligomerization of Orb2A-Orb2B. Binding of the oligomer to the 3′UTR prevents deadenylation and recruits polyadenylation complex and both of these events result in enhanced translation. Because of the self-sustaining and stable nature of the amyloid state, this creates a local and self-sustaining translation activation of Orb2-target mRNA, maintaining the changed state of synaptic activity over time.
CPEB-mediated translation regulation and long-term memory in mammals
In mammals there are four CPEB proteins, CPEB1-4, and all of them are expressed in the adult nervous system. Among these four isoforms recent studies suggest a functional role of aggregated mammalian CPEB3 similar to Aplysia and Drosophila (Fioriti et al., 2015; Stephan et al., 2015). CPEB3 forms amyloid-like oligomers in the adult hippocampus and removal of CPEB3 from the hippocampus affects both the consolidation and expression of long-term memory (Fioriti et al., 2015). Functionally, CPEB3 regulates neuronal protein synthesis and ubiquitination and SUMOylation regulates translation inhibitory function and aggregation of CPEB3 respectively (Drisaldi et al., 2015; Pavlopoulos et al., 2011). Intriguingly, constitutive removal of CPEB3 improved some forms of memory (Huang et al., 2014), although the mechanistic basis of the consequences of constitutive removal of CPEB3 remains unclear. Among the other CPEB family members, variants of CPEB2 have putative prion-like domains, although it remains to be seen whether they confer prion-like properties and whether CPEB2 is required for long-term memory.
Does amyloidogenic CPEB (Orb2) provide a biochemical basis of long-lasting memory?
Models for the biochemical basis of long-lasting memory anticipated an experience -dependent molecular switch and included autolytic kinases (Crick, 1984; Lisman, 1994) and even prions (Tompa and Friedrich, 1998). It has been proposed that such a biochemical switch of long-lasting memory would possess certain properties: i) be engaged by a temporally defined physiological stimulus; ii) form in response to some but not all experiences; iii) produce a change in the neuronal properties that elicit appropriate behavioral responses; and iv) deal with the natural turnover of individual proteins to enable a persistent change in behavioral output(Crick, 1984; Dudai, 2002; Lynch and Baudry, 1984).
The emerging evidence from Aplysia, Drosophila and Mouse suggests that the self-sustaining aggregates of CPEB may be a biochemical switch involved in at least some forms of long-term memory for the following reasons: first, activity-dependent conversion to the amyloidogenic state suggests it can be engaged by behavioral training; second, phosphorylation, SUMOylation and other mechanisms can confer the specificity and selectivity to amyloidogenic conversion; third, Orb2-dependent translation of mRNAs can alter the protein composition of the synapse, thereby altering synaptic properties and neuronal output. Finally, once triggered, the stable and self-sustaining capacity of the amyloidogenic state would outlast turnover of individual molecules to sustain memory over the long term. However some key questions remain unanswered: how and what ensures selective engagement of the self-sustaining state of neuronal CPEB only in response to long-term memory inducing stimuli? Once engaged how long does it persist and is the continued presence of the amyloidogenic state necessary for the persistence of memory? Is amyloid formation correlated with or predictive of long-lasting memory? Can a transient memory be stabilized by artificial recruitment of the amyloidogenic state?
Experimental Procedure
(Please see the supplementary section for the details of the methods).
RNA binding assay
The RNA binding assay was performed as described before in Sakai et. al (Mastushita-Sakai et al., 2010).
In vitro translation assay
Embryo extract was prepared according to the protocol described by Jeske et al (Jeske and Wahle, 2008). A 25 μL reaction consisting of 50ng (~0.75pM) translation reporter, 40% (v/v) embryo extract, and translation mix were incubated at 26ºC for 15 minutes unless indicated otherwise. Luciferase activity was measured using the dual-glo luciferase assay system (Promega) and normalized using renila luciferase reporter.
In vitro seeding assay
The seeding reactions were first assembled in WT or Δorb2 embryo extract with ~100 ng Orb2A320 oligomer or a corresponding control fractions for 12h at 4°C or 30 min at 26ºC. For purified recombinant proteins 10 ng of recombinant Orb2 or 10ng BSA was used. For seeding using mRNA 5ng of Orb2A320 mRNAs were translated in WT or Δorb2 embryo extract for an hour at 26ºC and incubated at 4ºC for additional 12–72h. For each reaction a corresponding mRNA blank control was carried out. The translation reporters were pre-incubated for 30 mins with new oligomers, followed by translation in Δorb2 embryo extract for 30 mins.
In vitro deadenylation and polyadenylation assay
The deadenylation assay was performed according to the protocol of Jeske et.al (Jeske and Wahle, 2008) and the polyadenylation assay as described by Olga et al (Coll et al., 2014). In deadenylation assay 5 to 20 nM [32p]-labeled uncapped-polyA tailed mRNA and in the polyadenylation assay m7GpppG-capped RNA with 8 adenine residues at the 3′end were used. Total RNA was extracted using phenol-chloroform and separated in 8M urea-6% polyacrylamide gel according to standard protocol.
In vivo single fly head translation assay
For GeneSwitch-Gal4 inducible expression, starved 3–4 day old adult flies were exposed to 200μM of RU486 (Mifepristone) in 2% sucrose. After 24 hrs individual fly heads were collected in 96-well plate, crushed in 50 μl of PBS+0.1% NP-40+0.1% Triton-X 100 buffer and luciferase activity was measured.
Supplementary Material
Acknowledgments
We are grateful to Dr. Tomoko Sakai for initiating the RNA-binding protein proteomics study. We also thank Blake Ebner for critical comments on the manuscript and Mark Miller for graphics. We are grateful to Prof. Elmar Wahle (Martin Luther University, Germany) and Dr. Craig Simbert (University of Toronto, Canada) for sharing reagents.
Footnotes
Author Contributions:
KS and RMK conceived the project and designed the experiments. RMK contributed to all experiments except Fig 7E. LL and PC performed the behavioral experiments and AS and LF performed the proteomics experiments. BS and JU helped in the biophysical analysis of the Orb2 amyloids. KS and BS wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Benoit B, Nemeth A, Aulner N, Kuhn U, Simonelig M, Wahle E, Bourbon HM. The Drosophila poly(A)-binding protein II is ubiquitous throughout Drosophila development and has the same function in mRNA polyadenylation as its bovine homolog in vitro. Nucleic acids research. 1999;27:3771–3778. doi: 10.1093/nar/27.19.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz LE, Kabachinski G, Walker MR, Carlile TM, Gilbert WV, Schwartz TU, Amon A. Regulated Formation of an Amyloid-like Translational Repressor Governs Gametogenesis. Cell. 2015;163:406–418. doi: 10.1016/j.cell.2015.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Chen J, Xu H, Liu S, Jiang QX, Halfmann R, Chen ZJ. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156:1207–1222. doi: 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll O, Villalba A, Gebauer F. Cytoplasmic polyadenylation assays. Methods in molecular biology. 2014;1125:53–63. doi: 10.1007/978-1-62703-971-0_5. [DOI] [PubMed] [Google Scholar]
- Crick F. Memory and molecular turnover. Nature. 1984;312:101. doi: 10.1038/312101a0. [DOI] [PubMed] [Google Scholar]
- Didelot G, Molinari F, Tchenio P, Comas D, Milhiet E, Munnich A, Colleaux L, Preat T. Tequila, a neurotrypsin ortholog, regulates long-term memory formation in Drosophila. Science. 2006;313:851–853. doi: 10.1126/science.1127215. [DOI] [PubMed] [Google Scholar]
- Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drisaldi B, Colnaghi L, Fioriti L, Rao N, Myers C, Snyder AM, Metzger DJ, Tarasoff J, Konstantinov E, Fraser PE, et al. SUMOylation Is an Inhibitory Constraint that Regulates the Prion-like Aggregation and Activity of CPEB3. Cell reports. 2015;11:1694–1702. doi: 10.1016/j.celrep.2015.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudai Y. Molecular bases of long-term memories: a question of persistence. Current opinion in neurobiology. 2002;12:211–216. doi: 10.1016/s0959-4388(02)00305-7. [DOI] [PubMed] [Google Scholar]
- Fioriti L, Myers C, Huang YY, Li X, Stephan JS, Trifilieff P, Colnaghi L, Kosmidis S, Drisaldi B, Pavlopoulos E, et al. The Persistence of Hippocampal-Based Memory Requires Protein Synthesis Mediated by the Prion-like Protein CPEB3. Neuron. 2015;86:1433–1448. doi: 10.1016/j.neuron.2015.05.021. [DOI] [PubMed] [Google Scholar]
- Fiumara F, Fioriti L, Kandel ER, Hendrickson WA. Essential role of coiled coils for aggregation and activity of Q/N-rich prions and PolyQ proteins. Cell. 2010;143:1121–1135. doi: 10.1016/j.cell.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich SU, Lindquist S. Protein-only mechanism induces self-perpetuating changes in the activity of neuronal Aplysia cytoplasmic polyadenylation element binding protein (CPEB) Proc Natl Acad Sci U S A. 2011;108:2999–3004. doi: 10.1073/pnas.1019368108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda N, Funakoshi Y, Hirasawa M, Yamagishi R, Asano Y, Miyagawa R, Ogami K, Tsujimoto M, Hoshino S. Anti-proliferative protein Tob negatively regulates CPEB3 target by recruiting Caf1 deadenylase. The EMBO journal. 2011;30:1311–1323. doi: 10.1038/emboj.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WH, Chao HW, Tsai LY, Chung MH, Huang YS. Elevated activation of CaMKIIalpha in the CPEB3-knockout hippocampus impairs a specific form of NMDAR-dependent synaptic depotentiation. Frontiers in cellular neuroscience. 2014;8:367. doi: 10.3389/fncel.2014.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeske M, Wahle E. Cell-free deadenylation assays with Drosophila embryo extracts. Methods in enzymology. 2008;448:107–118. doi: 10.1016/S0076-6879(08)02606-2. [DOI] [PubMed] [Google Scholar]
- Kacsoh BZ, Bozler J, Hodge S, Ramaswami M, Bosco G. A novel paradigm for nonassociative long-term memory in Drosophila: predator-induced changes in oviposition behavior. Genetics. 2015;199:1143–1157. doi: 10.1534/genetics.114.172221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapust RB, Waugh DS. Controlled intracellular processing of fusion proteins by TEV protease. Protein expression and purification. 2000;19:312–318. doi: 10.1006/prep.2000.1251. [DOI] [PubMed] [Google Scholar]
- Keleman K, Kruttner S, Alenius M, Dickson BJ. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nature neuroscience. 2007;10:1587–1593. doi: 10.1038/nn1996. [DOI] [PubMed] [Google Scholar]
- Kruttner S, Stepien B, Noordermeer JN, Mommaas MA, Mechtler K, Dickson BJ, Keleman K. Drosophila CPEB Orb2A mediates memory independent of Its RNA-binding domain. Neuron. 2012;76:383–395. doi: 10.1016/j.neuron.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruttner S, Traunmuller L, Dag U, Jandrasits K, Stepien B, Iyer N, Fradkin LG, Noordermeer JN, Mensh BD, Keleman K. Synaptic Orb2A Bridges Memory Acquisition and Late Memory Consolidation in Drosophila. Cell reports. 2015;11:1953–1965. doi: 10.1016/j.celrep.2015.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JE, Drier E, Barbee SA, Ramaswami M, Yin JC, Wickens M. GLD2 poly(A) polymerase is required for long-term memory. Proc Natl Acad Sci U S A. 2008;105:14644–14649. doi: 10.1073/pnas.0803185105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie S, Corden JL, McKnight SL. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 2013;155:1049–1060. doi: 10.1016/j.cell.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie YS, Macdonald PM. In vitro translation extracts prepared from Drosophila ovaries and embryos. Biochemical and biophysical research communications. 2000;270:473–481. doi: 10.1006/bbrc.2000.2453. [DOI] [PubMed] [Google Scholar]
- Lisman J. The CaM kinase II hypothesis for the storage of synaptic memory. Trends in neurosciences. 1994;17:406–412. doi: 10.1016/0166-2236(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Lynch G, Baudry M. The biochemistry of memory: a new and specific hypothesis. Science. 1984;224:1057–1063. doi: 10.1126/science.6144182. [DOI] [PubMed] [Google Scholar]
- Majumdar A, Cesario WC, White-Grindley E, Jiang H, Ren F, Khan MR, Li L, Choi EM, Kannan K, Guo F, et al. Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory. Cell. 2012;148:515–529. doi: 10.1016/j.cell.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Mastushita-Sakai T, White-Grindley E, Samuelson J, Seidel C, Si K. Drosophila Orb2 targets genes involved in neuronal growth, synapse formation, and protein turnover. Proc Natl Acad Sci U S A. 2010;107:11987–11992. doi: 10.1073/pnas.1004433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez R, Murthy KG, Ryan K, Manley JL, Richter JD. Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Molecular cell. 2000;6:1253–1259. doi: 10.1016/s1097-2765(00)00121-0. [DOI] [PubMed] [Google Scholar]
- Mustroph A, Juntawong P, Bailey-Serres J. Isolation of plant polysomal mRNA by differential centrifugation and ribosome immunopurification methods. Methods in molecular biology. 2009;553:109–126. doi: 10.1007/978-1-60327-563-7_6. [DOI] [PubMed] [Google Scholar]
- Pavlopoulos E, Trifilieff P, Chevaleyre V, Fioriti L, Zairis S, Pagano A, Malleret G, Kandel ER. Neuralized1 activates CPEB3: a function for nonproteolytic ubiquitin in synaptic plasticity and memory storage. Cell. 2011;147:1369–1383. doi: 10.1016/j.cell.2011.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveendra BL, Siemer AB, Puthanveettil SV, Hendrickson WA, Kandel ER, McDermott AE. Characterization of prion-like conformational changes of the neuronal isoform of Aplysia CPEB. Nature structural & molecular biology. 2013;20:495–501. doi: 10.1038/nsmb.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD. CPEB: a life in translation. Trends in biochemical sciences. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Richter JD, Klann E. Making synaptic plasticity and memory last: mechanisms of translational regulation. Genes & development. 2009;23:1–11. doi: 10.1101/gad.1735809. [DOI] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci U S A. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si K, Choi YB, White-Grindley E, Majumdar A, Kandel ER. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell. 2010;140:421–435. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Si K, Giustetto M, Etkin A, Hsu R, Janisiewicz AM, Miniaci MC, Kim JH, Zhu H, Kandel ER. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell. 2003a;115:893–904. doi: 10.1016/s0092-8674(03)01021-3. [DOI] [PubMed] [Google Scholar]
- Si K, Lindquist S, Kandel ER. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 2003b;115:879–891. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- Smibert CA, Wilson JE, Kerr K, Macdonald PM. smaug protein represses translation of unlocalized nanos mRNA in the Drosophila embryo. Genes & development. 1996;10:2600–2609. doi: 10.1101/gad.10.20.2600. [DOI] [PubMed] [Google Scholar]
- Smith RW, Blee TK, Gray NK. Poly(A)-binding proteins are required for diverse biological processes in metazoans. Biochemical Society transactions. 2014;42:1229–1237. doi: 10.1042/BST20140111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan JS, Fioriti L, Lamba N, Colnaghi L, Karl K, Derkatch IL, Kandel ER. The CPEB3 Protein Is a Functional Prion that Interacts with the Actin Cytoskeleton. Cell reports. 2015;11:1772–1785. doi: 10.1016/j.celrep.2015.04.060. [DOI] [PubMed] [Google Scholar]
- Tompa P, Friedrich P. Prion proteins as memory molecules: an hypothesis. Neuroscience. 1998;86:1037–1043. doi: 10.1016/s0306-4522(98)00148-1. [DOI] [PubMed] [Google Scholar]
- Udagawa T, Swanger SA, Takeuchi K, Kim JH, Nalavadi V, Shin J, Lorenz LJ, Zukin RS, Bassell GJ, Richter JD. Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex. Molecular cell. 2012;47:253–266. doi: 10.1016/j.molcel.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White-Grindley E, Li L, Mohammad Khan R, Ren F, Saraf A, Florens L, Si K. Contribution of Orb2A stability in regulated amyloid-like oligomerization of Drosophila Orb2. PLoS biology. 2014;12:e1001786. doi: 10.1371/journal.pbio.1001786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Hafer N, Agunwamba B, Schedl P. The CPEB protein Orb2 has multiple functions during spermatogenesis in Drosophila melanogaster. PLoS genetics. 2012;8:e1003079. doi: 10.1371/journal.pgen.1003079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Tyagi S, Schedl P. Spermatid cyst polarization in Drosophila depends upon apkc and the CPEB family translational regulator orb2. PLoS genetics. 2014;10:e1004380. doi: 10.1371/journal.pgen.1004380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.