Abstract
Cross-reactive immunological material (CRIM) status is an important prognostic factor in patients with infantile Pompe disease (IPD) being treated with enzyme replacement therapy. Western blot analysis of cultured skin fibroblast lysates has been the gold standard for determining CRIM status. Here, we evaluated CRIM status using peripheral blood mononuclear cell (PBMC) protein. For 6 of 33 patients (18%) CRIM status determination using PBMC was either indeterminate or discordant with GAA genotype or fibroblast CRIM analysis results. While the use of PBMCs for CRIM determination has the advantage of a faster turnaround time, further evaluation is needed to ensure the accuracy of CRIM results.
Abbreviations: CRIM, cross-reactive immunological material; GAA, acid alpha-glucosidase; IPD, infantile Pompe disease; ITI, immune tolerance induction; PBMCs, peripheral blood mononuclear cells
Keywords: Acid alpha-glucosidase, Cross reactive immunological material, Western blot analysis, Pompe disease, Enzyme replacement therapy
1. Introduction
Pompe disease is caused by a deficiency of the lysosomal enzyme, acid alpha-glucosidase (GAA; EC 3.2.1.20) [1]. Patients with a severe deficiency of GAA activity present in infancy with cardiomyopathy and skeletal myopathy [1], [2], [3]. Cross-reactive immunological material (CRIM) status is an important prognostic factor for patients with infantile Pompe disease (IPD) being treated with enzyme replacement therapy (ERT) [4], [5]. About 30% of patients with IPD make no detectable GAA protein based on Western blot analysis using skin fibroblasts, and are classified as CRIM-negative [5], [6]. These patients usually fare poorly due to the development of high, sustained anti-rhGAA IgG antibodies that significantly reduce the efficacy of ERT. In contrast, CRIM-positive patients with IPD make some residual GAA protein, although with severely reduced or deficient GAA activity. They usually have low antibody titers and a better clinical outcome, however a subset of CRIM-positive cases also develop high sustained antibody titers [4]. Immune tolerance induction (ITI) for CRIM-negative patients, which prevents the production of anti-rhGAA IgG antibodies, is most effective when initiated in the ERT-naïve setting with the first dose of ERT [7]. This observation, coupled with the knowledge that early treatment in IPD results in the best prognosis, has necessitated development of a rapid method for determining CRIM status. Historically, CRIM status for IPD patients has been determined using Western blot analysis of skin fibroblast lysates; different bands representing inactive precursor (110 kDa), intermediate form (95 kDa), and active forms (76 and 70 kDa) of GAA can be distinguished in normal skin fibroblast protein [8], [9], [10], [11]. However, this process takes several weeks due to the time needed for culturing skin fibroblasts from skin biopsy tissue. In addition, some patients may make small amounts of GAA protein that are below the limits of detection by Western blot [6]. Recently, we have published that CRIM status can be predicted based on GAA gene mutations for over 90% of patients [12]. For example, premature stop codons caused by nonsense or frameshift mutations usually result in CRIM-negative alleles, and missense mutations typically result in CRIM-positive alleles. However, these predictions depend on the specific mutation, and prediction of CRIM status may not be possible or accurate for novel mutations in all patients. Therefore, a fast, accurate method of CRIM determination for all IPD patients would be beneficial. A new method for determining CRIM status, using peripheral blood mononuclear cells (PBMCs) has been reported recently [13], with promising results in a small group of patients. Here, we present results obtained using the same method to determine CRIM status in 33 IPD patients and comparison of those results with CRIM status predicted by GAA mutations, and/or CRIM status determined using skin fibroblasts, where available.
2. Patients and methods
All subjects were diagnosed with Pompe disease and were enrolled in a Duke Medicine Institutional Review Board-approved protocol (IRB#Pro00001562; LDN6709 Site 206; Clinicaltrials.gov Identifier: NCT01665326). Samples for analysis were sent from across the USA to the Duke Biochemical Genetics Laboratory. CRIM status was determined by Western blot analysis of PBMCs and cultured skin fibroblast protein, as previously published [12], [13]. Specifically, blood samples for PBMC analysis were collected in BD Vacutainer® CPT™ (Cell Preparation Tube) with Sodium Citrate (Becton Dickinson, REF 362760, Franklin Lakes, New Jersey). Upon arrival in the laboratory, PBMCs were isolated within 24–48 h of sample collection when possible. Cell lysates were prepared and Western blot analysis performed as previously described [13]. None of the patients in this study were on ERT at the time of sample collection. Protein samples from normal human fibroblasts, normal human PBMCs and known affected Pompe patients were used as the assay control for comparison (Corielle Institute for Medical Resarch, Camden, NJ). β-actin antibody was used as a loading control and to assess the integrity of the patient protein samples [13].
Sequence analysis of the GAA gene (NM_000152) was performed in a CLIA and CAP certified laboratory. To predict CRIM status from GAA mutations, we used our database which correlates GAA mutations and CRIM status of about 140 patients with IPD [12]. If a mutation had been previously found in a CRIM-negative patient, it was designated as CRIM-negative. If a mutation had previously been found in homozygous state in a CRIM-positive patient, it was designated as CRIM-positive. If a mutation was not in our database, or was in a compound heterozygote CRIM-positive patient, we made the prediction of CRIM status as previously described [12]. Mutations resulting in a premature stop codon (nonsense, frameshift) are expected to be CRIM-negative, unless the stop codon occurs in the last exon of the gene or up to about 50 nucleotides from the 3'end of the penultimate exon, where it can be missed by the nonsense-mediated decay machinery [14]. Missense mutations are usually CRIM-positive unless the nucleotide affects a splice junction, or the change is in the first ATG codon of the gene, which can result in CRIM-negative status. The effect of splice site mutations can be difficult to predict unless the mutation has previously been associated with CRIM status, or in vitro studies of protein production have been done.
3. Results
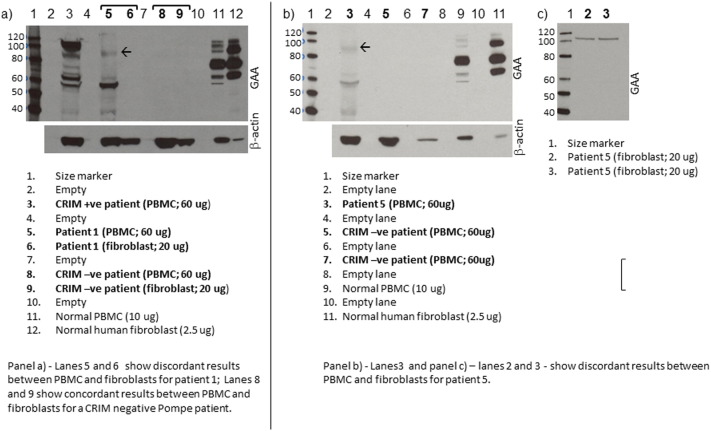
Thirty-three patients were included in this study. Results of Western blot analysis of normal PBMC protein were comparable to those previously reported [13] (see Supplementary Fig. S1). In 27 of the 33 patients (82%), the CRIM status in PBMCs was concordant with the CRIM status predicted by GAA gene mutations (20 CRIM-positive, 7 CRIM-negative). For 8 of these 27 patients, skin fibroblast CRIM status was also available and was concordant with CRIM status in PBMCs and CRIM status predicted by GAA gene mutations (7 CRIM-positive, 1 CRIM-negative). For the remaining 6 patients (18%), CRIM status determination using PBMCs was either indeterminate or discordant with that determined by skin fibroblasts analysis and/or as predicted by GAA gene mutations (Table 1; see Supplementary Fig. S1). For five of these cases (Patients 1–5), a ~ 90 kDa band was observed on Western blots of PBMC protein; four of these patients (Patients 1–4) were found to be CRIM-negative on skin fibroblast Western blot analysis and/or by prediction based on GAA gene mutations. Patient 5 was predicted to be CRIM-positive based on GAA gene mutations, and was confirmed to be CRIM positive by Western blot analysis of fibroblast protein. For the remaining one patient with discordant results (Patient 6), there were no bands in the size range of GAA protein on Western blot analysis of PBMC protein, but the patient was predicted to be CRIM-positive based on GAA genotype. Western blot using an anti β-actin antibody indicated that the protein was intact. A skin fibroblast sample from Patient 6 was not available for Western blot analysis.
Supplementary Fig. S1.
Representative Western blot showing GAA binding patterns in PBMCs and fibroblasts. The arrow marks the 90 kDa band seen in Western blots of PBMC protein from Patients 1–5. Bands migrating above 110 kDa and below 70 kDa – outside the expected size range for GAA processing forms – were seen in some samples. We do not know the identity of these bands.
Table 1.
CRIM status in PCMCs and fibroblasts, and GAA mutations, in 6 patients with inconclusive PBMC CRIM status results.
| Patient | CRIM status in PBMC | CRIM status in skin fibroblasts | Predicted CRIM status based on GAA mutations |
GAA mutations |
|
|---|---|---|---|---|---|
| Allele 1 | Allele 2 | ||||
| 1 | Indeterminate (90 kDa band) | Negative | Negative | c.437delT (p.Met146ArgfsX7)a | c.2237G > A (p.Trp746X)b |
| 2 | Indeterminate (90 kDa band) | Negative | Negative | c.1754 + 2T > Ac | c.1822C > T (p.Arg608X)d |
| 3 | Indeterminate (90 kDa band) | Negative | Negative | c.2560C > T (p.Arg854X)e | c.2560C > T (p.Arg854X)e |
| 4 | Indeterminate (90 kDa band) | NA | Negative | c.2560C > T (p.Arg854X)e | c.2560C > T (p.Arg854X)e |
| 5 | Indeterminate (90 kDa band) | Positive (~ 110 kDa band) | Positive | c.1827delC (p.Tyr609X)f | c.2481 + 102_2646 + 31del (p.Gly828_Asn882del)g |
| 6 | Negative (60 kDa band) | NA | Positive | c.2297A > C (p.Tyr766Ser)h | c.2297A > C (p.Tyr766Ser)h |
Further references and information about previously published mutations are available at www.pompecenter.nl/(Pompe Center at Erasmus Medical Center).
p.Met146ArgfsX7 is predicted to create a CRIM-negative allele due to introduction of a premature stop codon. To our knowledge, this mutation has not been found in other patients.
c.2237G > A (p.Trp746X) was first reported by Beesley et al [15], and is predicted to create a CRIM-negative allele due to introduction of a premature stop codon. It was previously found in patients who were CRIM-negative on fibroblast analysis [7], [12].
To our knowledge, c.1754 + 2T > A has not been found in any other patients. However, we have previously identified c.1754 + 1G > A in a patient who was CRIM-negative on fibroblast analysis, suggesting that abolishment of this splice site could result in a CRIM-negative allele [12]
c.1822C > T (p.Arg608X) was previously reported as a “severity class A” mutation with no predicted expression of the protein [16].
c.2560C > T (p.Arg854X) is common among patients with Pompe disease of African descent [17], [18], [19], [20]. In cDNA studies, the allele carrying this mutation was found not to be expressed [21]. Patients who are homozygous for this mutation have been reported to be CRIM negative [7], [22], [23].
1827delC (p.Tyr609X) [24] is predicted to create a CRIM-negative allele due to introduction of a premature stop codon.
p.Gly828_Asn882del is common among patients with Pompe disease of Dutch ancestry and is also found in other populations [25], [26], [27], [28], [29], [30]. Previous studies show that this allele is transcribed and produces protein [12], [29].
To our knowledge, p.Tyr766Ser has not been previously reported. We have found p.Tyr766Ser in homozygosity in three patients who are CRIM-positive, but not in any CRIM-negative patients (unpublished data).
4. Discussion
Development of a rapid method for determining CRIM status in patients with IPD is important so that appropriate treatment can be initiated as soon as possible. A blood-based method using PBMC for determination of CRIM-status would have the advantage of faster turnaround time as compared to use of cultured skin fibroblasts, the current gold standard [13]. However, our results indicate that the results of CRIM status determined by Western blot analysis of PBMC may not always be clear or concordant with those obtained by analysis of skin fibroblast protein and CRIM status predicted from GAA gene mutations. The reason for this discrepancy in a subset of patients (18% in this cohort) is unclear. All PBMC samples in this study were prepared using the same standard protocol, and processed within 24–48 h of collection. Western blot analysis with an anti β-actin antibody indicated that the protein was intact for all the discrepant samples.
Several PBMC samples were not included in this study because the protein sample was degraded and the results could not be considered reliable. This emphasizes the challenges of handling and shipment of these samples from distant places. The need for good internal controls to assess protein integrity and quality, and methods to stabilize protein during transportation cannot be emphasized enough. Additional challenges for the PBMC assay include the need for specialized cell preparation tubes (CPT), and limitations to obtaining sufficient blood sample volumes from patients who may be very young or fragile.
It is possible that the 90 kDa band observed on Western blot analysis of PBMCs in Patients 1–5 may represent non-specific binding of the anti-GAA antibody. However, why this should occur in only some and not all samples is unknown. This band is particularly troubling because it is very close to the size range of GAA protein band (95 kDa). In Patient 6, a band of about 60 kDa, smaller than the expected size for GAA protein, was observed. There was no evidence of degradation of the sample on β-actin analysis. Of interest, this 60 kDa band has been observed in a previous study as well [13].
In conclusion, while a blood-based assay to determine CRIM status has the advantage of rapid results for faster initiation of treatment, further method development is needed to ensure the accuracy of the results. Important future work should include analysis of the 90 kDa and 60KDa bands that are present in some samples, but which do not correspond to any known GAA processing forms. Isolation and sequencing of this protein will be essential to determine whether its presence on Western blot is due to non-specific antibody binding, or whether it represents a breakdown product of GAA in PBMCs. Additionally, before initiating ERT, CRIM results obtained by analysis of blood PBMC should be confirmed by correlation with CRIM status as predicted by GAA mutations. Now that newborn screening for Pompe disease is a reality, more presymptomatic and early symptomatic cases are being identified. GAA mutation analysis is important not only for confirmation of the diagnosis in these cases, but also helpful in determining CRIM status in conjunction with Western blot results in PBMCs and/or fibroblasts if possible.
Acknowledgments
We are grateful to the patients with Pompe disease and their families for participating in this research. This research was supported by a grant from the Genzyme Corporation, a Sanofi Company (Cambridge, MA) and in part by the Lysosomal Disease Network. The Lysosomal Disease Network (U54NS065768) is a part of the National Institutes of Health (NIH) Rare Diseases Clinical Research Network (RDCRN), supported through collaboration between the NIH Office of Rare Diseases Research (ORDR) at the National Center for Advancing Translational Science (NCATS), the National Institute of Neurological Disorders and Stroke (NINDS) and National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The following is the supplementary data related to this article.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ymgmr.2015.10.012.
Contributor Information
Deeksha S. Bali, Email: deeksha.bali@dm.duke.edu.
Jennifer L. Goldstein, Email: jennifer.goldstein@dm.duke.edu.
Catherine Rehder, Email: catherine.rehder@dm.duke.edu.
Zoheb B. Kazi, Email: zoheb.kazi@mc.duke.edu.
Kathryn L. Berrier, Email: katie.berrier@dm.duke.edu.
Jian Dai, Email: jian.dai@mc.duke.edu.
Priya S. Kishnani, Email: priya.kishnani@dm.duke.edu.
References
- 1.Hirschhorn R. The Metabolic and Molecular Bases of INherited Disease. 8th ed. McGraw-Hill; New York: 2000. Glycogen Storage Disease type II: acid alpha-glucosidae (acid maltase) deficiency; pp. 3389–3420. [Google Scholar]
- 2.Kishnani P.S., Steiner R.D., Bali D., Berger K., Byrne B.J., Case L.E., Crowley J.F., Downs S., Howell R.R., Kravitz R.M., Mackey J., Marsden D., Martins A.M., Millington D.S., Nicolino M., O'Grady G., Patterson M.C., Rapoport D.M., Slonim A., Spencer C.T., Tifft C.J., Watson M.S. Pompe disease diagnosis and management guideline. Genet Med. 2006;8:267–288. doi: 10.1097/01.gim.0000218152.87434.f3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gungor D., Reuser A.J. How to describe the clinical spectrum in Pompe disease? Am. J. Med. Genet. A. 2013;161A:399–400. doi: 10.1002/ajmg.a.35662. [DOI] [PubMed] [Google Scholar]
- 4.Banugaria S.G., Prater S.N., Ng Y.K., Kobori J.A., Finkel R.S., Ladda R.L., Chen Y.T., Rosenberg A.S., Kishnani P.S. The impact of antibodies on clinical outcomes in diseases treated with therapeutic protein: lessons learned from infantile Pompe disease. Genet. Med. 2011;13:729–736. doi: 10.1097/GIM.0b013e3182174703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kishnani P.S., Goldenberg P.C., DeArmey S.L., Heller J., Benjamin D., Young S., Bali D., Smith S.A., Li J.S., Mandel H., Koeberl D., Rosenberg A., Chen Y.T. Cross-reactive immunologic material status affects treatment outcomes in Pompe disease infants. Mol. Genet. Metab. 2010;99:26–33. doi: 10.1016/j.ymgme.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berrier K.L., Kazi Z.B., Prater S.N., Bali D.S., Goldstein J., Stefanescu M.C., Rehder C.W., Botha E.G., Ellaway C., Bhattacharya K., Tylki-Szymanska A., Karabul N., Rosenburg A.S., Kishnani P.S. CRIM-negative infantile Pompe disease: characterization of immune responses in patients treated with ERT monotherapy. Genet. Med. 2015 doi: 10.1038/gim.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Messinger Y.H., Mendelsohn N.J., Rhead W., Dimmock D., Hershkovitz E., Champion M., Jones S.A., Olson R., White A., Wells C., Bali D., Case L.E., Young S.P., Rosenberg A.S., Kishnani P.S. Successful immune tolerance induction to enzyme replacement therapy in CRIM-negative infantile Pompe disease. Genet. Med. 2012;14:135–142. doi: 10.1038/gim.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moreland R.J., Jin X., Zhang X.K., Decker R.W., Albee K.L., Lee K.L., Cauthron R.D., Brewer K., Edmunds T., Canfield W.M. Lysosomal acid alpha-glucosidase consists of four different peptides processed from a single chain precursor. J. Biol. Chem. 2005;280:6780–6791. doi: 10.1074/jbc.M404008200. [DOI] [PubMed] [Google Scholar]
- 9.Hasilik A., Neufeld E.F. Biosynthesis of lysosomal enzymes in fibroblasts. Synthesis as precursors of higher molecular weight. J. Biol. Chem. 1980;255:4937–4945. [PubMed] [Google Scholar]
- 10.Reuser A.J., Kroos M., Oude Elferink R.P., Tager J.M. Defects in synthesis, phosphorylation, and maturation of acid alpha-glucosidase in glycogenosis type II. J. Biol. Chem. 1985;260:8336–8341. [PubMed] [Google Scholar]
- 11.Wisselaar H.A., Kroos M.A., Hermans M.M., van Beeumen J., Reuser A.J. Structural and functional changes of lysosomal acid alpha-glucosidase during intracellular transport and maturation. J. Biol. Chem. 1993;268:2223–2231. [PubMed] [Google Scholar]
- 12.Bali D.S., Goldstein J.L., Banugaria S., Dai J., Mackey J., Rehder C., Kishnani P.S. Predicting cross-reactive immunological material (CRIM) status in Pompe disease using GAA mutations: lessons learned from 10 years of clinical laboratory testing experience. Am. J. Med. Genet. C Semin. Med. Genet. 2012;160C:40–49. doi: 10.1002/ajmg.c.31319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z., Okamoto P., Keutzer J. A new assay for fast, reliable CRIM status determination in infantile-onset Pompe disease. Mol. Genet. Metab. 2014;111:92–100. doi: 10.1016/j.ymgme.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 14.Miller J.N., Pearce D.A. Nonsense-mediated decay in genetic disease: friend or foe? Mutat. Res. Rev. Mutat. Res. 2014;762:52–64. doi: 10.1016/j.mrrev.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beesley C.E., Child A.H., Yacoub M.H. The identification of five novel mutations in the lysosomal acid a-(1-4) glucosidase gene from patients with glycogen storage disease type II. Mutations in brief no. 134. Hum. Mutat. 1998;11:413. doi: 10.1002/(SICI)1098-1004(1998)11:5<413::AID-HUMU16>3.0.CO;2-I. Online. [DOI] [PubMed] [Google Scholar]
- 16.Kroos M., Pomponio R.J., van Vliet L., Palmer R.E., Phipps M., Van der Helm R., Halley D., Reuser A. Update of the Pompe disease mutation database with 107 sequence variants and a format for severity rating. Hum. Mutat. 2008;29:E13–E26. doi: 10.1002/humu.20745. [DOI] [PubMed] [Google Scholar]
- 17.Adams E.M., Becker J.A., Griffith L., Segal A., Plotz P.H., Raben N. Glycogenosis type II: a juvenile-specific mutation with an unusual splicing pattern and a shared mutation in African Americans. Hum. Mutat. 1997;10:128–134. doi: 10.1002/(SICI)1098-1004(1997)10:2<128::AID-HUMU5>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 18.Becker J.A., Vlach J., Raben N., Nagaraju K., Adams E.M., Hermans M.M., Reuser A.J., Brooks S.S., Tifft C.J., Hirschhorn R., Huie M.L., Nicolino M., Plotz P.H. The African origin of the common mutation in African American patients with glycogen-storage disease type II. Am. J. Hum. Genet. 1998;62:991–994. doi: 10.1086/301788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oba-Shinjo S.M., da Silva R., Andrade F.G., Palmer R.E., Pomponio R.J., Ciociola K.M., Gutierrez S.C.M.P.S., Porta G., Marrone C.D., Munoz V., Grzesiuk A.K., Llerena J.C., Jr., Berditchevsky C.R., Sobreira C., Horovitz D., Hatem T.P., Frota E.R., Pecchini R., Kouyoumdjian J.A., Werneck L., Amado V.M., Camelo J.S., Jr., Mattaliano R.J., Marie S.K. Pompe disease in a Brazilian series: clinical and molecular analyses with identification of nine new mutations. J. Neurol. 2009;256:1881–1890. doi: 10.1007/s00415-009-5219-y. [DOI] [PubMed] [Google Scholar]
- 20.Nino M.Y., Mateus H.E., Fonseca D.J., Kroos M.A., Ospina S.Y., Mejia J.F., Uribe J.A., Reuser A.J., Laissue P. Identification and functional characterization of GAA mutations in Colombian patients affected by pompe disease. JIMD Rep. 2013;7:39–48. doi: 10.1007/8904_2012_138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hermans M.M., de Graaff E., Kroos M.A., Wisselaar H.A., Willemsen R., Oostra B.A., Reuser A.J. The conservative substitution Asp-645–>Glu in lysosomal alpha-glucosidase affects transport and phosphorylation of the enzyme in an adult patient with glycogen-storage disease type II. Biochem. J. 1993;289(Pt 3):687–693. doi: 10.1042/bj2890687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kishnani P.S., Nicolino M., Voit T., Rogers R.C., Tsai A.C., Waterson J., Herman G.E., Amalfitano A., Thurberg B.L., Richards S., Davison M., Corzo D., Chen Y.T. Chinese hamster ovary cell-derived recombinant human acid alpha-glucosidase in infantile-onset Pompe disease. J. Pediatr. 2006;149:89–97. doi: 10.1016/j.jpeds.2006.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elder M.E., Nayak S., Collins S.W., Lawson L.A., Kelley J.S., Herzog R.W., Modica R.F., Lew J., Lawrence R.M., Byrne B.J. B-cell depletion and immunomodulation before initiation of enzyme replacement therapy blocks the immune response to acid alpha-glucosidase in infantile-onset Pompe disease. J. Pediatr. 2013;163:847–854. doi: 10.1016/j.jpeds.2013.03.002. e841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hermans M.M., van Leenen D., Kroos M.A., Beesley C.E., Van Der Ploeg A.T., Sakuraba H., Wevers R., Kleijer W., Michelakakis H., Kirk E.P., Fletcher J., Bosshard N., Basel-Vanagaite L., Besley G., Reuser A.J. Twenty-two novel mutations in the lysosomal alpha glucosidase gene (GAA) underscore the genotype-phenotype correlation in glycogen storage disease type II. Hum. Mutat. 2004;23:47–56. doi: 10.1002/humu.10286. [DOI] [PubMed] [Google Scholar]
- 25.Dagnino F., Stroppiano M., Regis S., Bonuccelli G., Filocamo M. Evidence for a founder effect in Sicilian patients with glycogen storage disease type II. Hum. Hered. 2000;50:331–333. doi: 10.1159/000022938. [DOI] [PubMed] [Google Scholar]
- 26.Montalvo A.L., Bembi B., Donnarumma M., Filocamo M., Parenti G., Rossi M., Merlini L., Buratti E., De Filippi P., Dardis A., Stroppiano M., Ciana G., Pittis M.G. Mutation profile of the GAA gene in 40 Italian patients with late onset glycogen storage disease type II. Hum. Mutat. 2006;27:999–1006. doi: 10.1002/humu.20374. [DOI] [PubMed] [Google Scholar]
- 27.Huie M.L., Chen A.S., Brooks S.S., Grix A., Hirschhorn R. A de novo 13 nt deletion, a newly identified C647W missense mutation and a deletion of exon 18 in infantile onset glycogen storage disease type II (GSDII) Hum. Mol. Genet. 1994;3:1081–1087. doi: 10.1093/hmg/3.7.1081. [DOI] [PubMed] [Google Scholar]
- 28.Van der Kraan M., Kroos M.A., Joosse M., Bijvoet A.G., Verbeet M.P., Kleijer W.J., Reuser A.J. Deletion of exon 18 is a frequent mutation in glycogen storage disease type II. Biochem. Biophys. Res. Commun. 1994;203:1535–1541. doi: 10.1006/bbrc.1994.2360. [DOI] [PubMed] [Google Scholar]
- 29.Boerkoel C.F., Exelbert R., Nicastri C., Nichols R.C., Miller F.W., Plotz P.H., Raben N. Leaky splicing mutation in the acid maltase gene is associated with delayed onset of glycogenosis type II. Am. J. Hum. Genet. 1995;56:887–897. [PMC free article] [PubMed] [Google Scholar]
- 30.Hirschhorn R., Huie M.L. Frequency of mutations for glycogen storage disease type II in different populations: the delta525T and delta exon 18 mutations are not generally “common” in white populations. J. Med. Genet. 1999;36:85–86. [PMC free article] [PubMed] [Google Scholar]