Abstract
Arbuscular mycorrhiza fungi (AMF) can colonize the roots of Amorpha fruticosa, a perennial leguminous woody shrub, and form arbuscular mycorrhiza (AM). AMF have significant promoting effects on A. fruticosa growth as the intensity of fungal colonization increases. Taking AMF-A. fruticosa symbionts as the experimental material, gel-free isobaric tags for relative and absolute quantification (iTRAQ) coupled with two-dimensional liquid chromatography-tandem mass spectrometry (LC-MS/MS) were used to investigate the expression of A. fruticosa mycorrhizal proteins at the maturation stage. A total of 3,473 proteins were identified, of which 77 showed dramatic changes in their root expression levels; 33 increased, and 44 decreased. We also found nine AMF proteins that were expressed with AMF treatment. The 77 proteins were classified according to function. Plant proteins were assigned into 11 categories: metabolism-related (32%), protein folding and degradation-related (22%), energy-related (10%), protein synthesis-related (8%), stress and defense-related (24%), transcription-related (6%), membrane and transport-related (4%), cellular structure-related (2.5%), signaling transduction-related (11%) and unknown proteins (5%). The results of the study provide a foundation for further investigation of the metabolic characteristics and molecular mechanisms of AM.
Arbuscular mycorrhiza (AM), representing the most widely distributed mutualistic root symbiosis in nature, are the result of long-term evolution between plant and soil fungi. AM fungi (AMF) are obligate symbionts and are recalcitrant to pure culture on synthetic media; they grow only in living plants. Their hereditary variability and heterogeneous characteristics are essential for colonizing a large number of potential host plants. However, different host plants may simultaneously induce the expression of different symbiosis-related genes. AMF are some of the most widespread microorganisms, and they can form symbionts with more than two-thirds of the vascular plants in natural or artificial ecosystems. These plants include important agricultural species, such as wheat, rice, and the model plant Populus trichocarpa1,2. The foundation of mycorrhizal symbiosis is the ability of AMF, using their multicore hyphae, to provide nutrients (especially phosphorous) to host plants that have long-distance illiquidity3. New physiological and molecular evidence has shown that, for phosphorus, the mycorrhizal pathway (MP) is operational regardless of plant growth responses. Meanwhile, the contribution of the direct pathway (DP) is decreased, which results in a greater dependence of host plants on the nutrients that AMF provide4. AMF can utilize only simple carbon and nitrogen sources from their hosts to complete their life cycles. This may be due to the loss of some enzyme-encoding genes and to macromolecular synthesis defects that have arisen during the long-term evolution of symbiosis with plants5,6.
AM play a significant role in promoting the growth of host plants, and researchers have increased their efforts to study the interactions between AMF and host plants. Recorbet and colleagues have compared the root proteome responses of Medicago truncatula upon colonization with two AM fungi, i.e., Glomus mosseae (GM) and G. intraradices, using two-dimensional electrophoresis (2-DE)7. They found 42 symbiosis proteins; of these, 32 could be confidently identified and retrieved following MS/MS and matching with a database encompassing 21 fungal proteins. To test the mechanisms by which shoots of Cd-treated mycorrhizal plants avoid metal toxicity, Aloui has performed a 2-DE/MALDI-TOF-based comparative proteomic analysis of the M. truncatula shoot responses upon mycorrhization and Cd exposure8. finding that Cd triggers an opposite response than mycorrhization, which is coupled with an increase in molecular chaperones in the shoots of mycorrhizal plants relative to those that are metal-free. Wang has studied the dynamic changes in maize leaf protein expression profiles under AMF colonization9. In that study, the differentially expressed proteins in maize leaves were separated by 2-DE, and the results reveal 21 differentially expressed gel spots in maize leaves. Among them, 8 proteins were successfully identified. With the development of molecular biology techniques, quantitative analysis of the differences in protein expression profiles during the colonization process of pathogenic or symbiotic microorganisms has become possible; these techniques have played a critical role in analyzing pathogenic mechanisms. Isobaric tags for relative and absolute quantification (iTRAQ) represents one of the new and powerful techniques for simultaneous analysis of multiple samples and provide relative quantification of hundreds of proteins. iTRAQ reagents produce high-quality, reproducible results from complex samples, and iTRAQ has thus become widely used. However, iTRAQ-based studies on the symbiotic mechanisms of AMF and host plants have rarely been reported. Our group has studied the symbiotic relationship between plants and fungi at the mRNA level, Zhang xingxing identified 30 symbiosis-related genes expressed in Amorpha fruticosa roots colonized by GM at different stages by using mRNA differential-display PCR (DDRT-PCR). The expressed genes were confirmed by reverse Northern blotting. Eleven fragments were sequenced and putatively identified by homologous alignment, and these genes were found to relate to defense and signal transduction10. Kong xiangshi also has found 47 symbiosis-related unigenes during AMF treatment by using suppression subtractive hybridization (SSH) and subsequent Gene Ontology (GO) database, BLAST annotation and literature searches to categorize each of the identified genes. Among the expressed genes, those related to plant metabolism and stress and defense show important roles during the symbiotic process of AMF-A. fruticosa11. Based on our previous work, we have continued to use A. fruticosa, a perennial leguminous woody shrub plant, as a host. Using AMF-A. fruticosa symbionts as the experimental materials, we used iTRAQ combined with 2-D LC-MS/MS to investigate the expression of A. fruticosa mycorrhizal proteins at the maturation stage. The results of the study provide a theoretical basis for the further analysis of the metabolic characteristics and molecular mechanisms of symbiosis between AMF and A. fruticosa.
Results
AMF Colonization
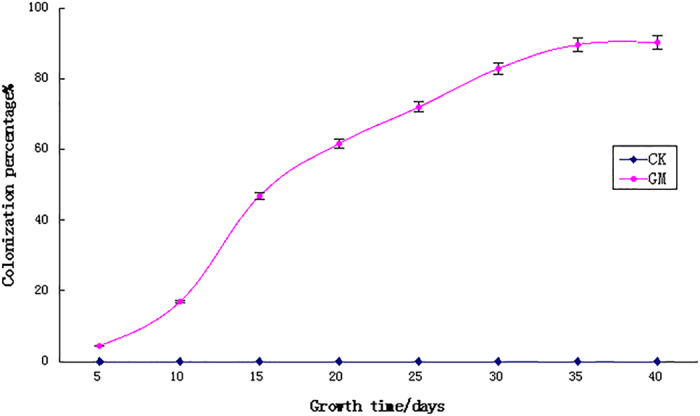
The colonization percentage of A. fruticosa roots is shown in Fig. 1. At day 5, the roots were relatively small, and only a few hyphae were detected. Most of the spores were not in contact with the host roots and were mainly in their vegetative growth stage12,13. The infection rate began to increase at day 10, and the growth status of GM-inoculated seedlings was significantly better than that of non-inoculated plants. The colonization percentage increased rapidly at day 15. At that time, there was a large quantity of mycelium-infected roots that increased over time. The colonization percentage reached its peak at the vigorous phase (i.e., 30 days), and a large number of vesicles and arbuscules were observed within the roots. At 40 days, we observed that the hyphae had extended from the plant root surface and had infected neighboring roots. No colonization by AM was observed in the non-inoculated plants, because the mixed soil used for culture had been autoclaved thoroughly.
Figure 1. The colonization percentage of Amorpha fruticosa roots by AMF changes with time.
Identification of Symbiosis-Related Proteins using iTRAQ LC-MS/MS
The roots of A. fruticosa changed their protein levels when colonized by GM, and mutualistic symbionts formed. Using an iTRAQ approach, 86 differentially expressed symbiosis-related proteins were successfully identified. Among them, 77 were plant proteins, with 33 proteins showing increases and 44 showing decreases (Table 1), and 9 were fungal proteins (Table 2). More detailed information in supplementary information. AMF proteins play an import role in symbiotic systems, but they show high expression in only the AMF themselves. The low overall concentration of AMF proteins, and the limitations of the technology, resulted in few AMF proteins being detected14.
Table 1. Identified symbiosis related proteins in A. fruticosa roots colonized by G. mosseae.
| Accession | Protein name | Plant species | Unused score | FM/control |
|---|---|---|---|---|
| Metabolism | ||||
| 356523620 | 3-oxoacyl-[acyl-carrier-protein] synthase I | Glycine max | 6 | 6.88 |
| 502118474 | chorismate synthase, chloroplastic-like isoform X1 | Cicer arietinum | 2.2 | 1.96 |
| 17026394 | UDP-glucose pyrophosphorylase | Amorpha fruticosa | 52 | 0.43 |
| 502122125 | pyruvate kinase, cytosolic isozyme | Cicer arietinum | 26 | 0.51 |
| 502134148 | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | Cicer arietinum | 24 | 0.52 |
| 356505594 | sucrose synthase 2 | Glycine max | 22 | 0.43 |
| 356551144 | alpha-1,4 glucan phosphorylase L isozyme | Glycine max | 22 | 0.33 |
| 357453895 | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase | Medicago truncatula | 20 | 0.51 |
| 356557483 | carbamoyl-phosphate synthase large chain | Glycine max | 20 | 0.53 |
| 356576733 | zeta-carotene desaturase, chloroplastic/chromoplastic | Glycine max | 20 | 0.51 |
| 356542858 | beta-amylase | Glycine max | 19 | 0.27 |
| 502081957 | UDP-glucuronic acid decarboxylase 6-like isoform X3 | Cicer arietinum | 14 | 0.67 |
| 17063848 | 4-coumarate:CoA ligase | Amorpha fruticosa | 14 | 0.47 |
| 356566195 | alpha-glucan phosphorylase, Hisozyme | Glycine max | 13 | 0.32 |
| 356552274 | glucose-1-phosphate adenylyltransferase small subunit | Glycine max | 12 | 0.49 |
| 502151454 | probable sucrose-phosphate synthase | Cicer arietinum | 12 | 0.54 |
| 355481047 | Neutral invertase | Medicago truncatula | 11 | 0.29 |
| 356536154 | methylthioribose kinase | Glycine max | 8.6 | 0.46 |
| 357445053 | Leukotriene-A4 hydrolase | Medicago truncatula | 7.6 | 0.17 |
| 84514155 | p-coumaroyl-shikimate 3'-hydroxylase | Trifolium pratense | 6.7 | 0.19 |
| 502131944 | carotenoid 9,10(9',10')-cleavage dioxygenase 1-like isoform X1 | Cicer arietinum | 6.2 | 0.08 |
| 68264915 | beta-conglycinin alpha subunit | Glycine max | 5 | 0.20 |
| 356509826 | thromboxane-A synthase | Glycine max | 5.2 | 0.78 |
| 356549592 | caffeic acid 3-O-methyltransferase | Glycine max | 2.8 | 3.91 |
| 502134043 | neutral ceramidase | Cicer arietinum | 6.3 | 12.65 |
| Signaling | ||||
| 357446813 | Somatic embryogenesis receptor -like kinase | Medicago truncatula | 12 | 2.21 |
| 502110137 | rho GDP-dissociation inhibitor 1 | Cicer arietinum | 2.4 | 2.86 |
| Stress and defense | ||||
| 356523918 | agglutinin-2 | Glycine max | 5.8 | 11.47 |
| 356527464 | transaldolase | Glycine max | 17 | 0.52 |
| 310704426 | stearoyl-acyl carrier protein desaturase | Phaseolus lunatus | 5.6 | 0.54 |
| 356518971 | pantothenate kinase 2 | Glycine max | 5.9 | 0.61 |
| 502180482 | abscisic acid 8'-hydroxylase 3 | Cicer arietinum | 2.4 | 13.80 |
| 357438243 | Germin-like protein subfamily 1 member | Medicago truncatula | 2.4 | 16.44 |
| Membrance and transport | ||||
| 356526157 | coatomer subunit beta'-2 | Glycine max | 16 | 1.58 |
| 356532026 | K(+) efflux antiporter 2, chloroplastic | Glycine max | 1.5 | 24.89 |
| 75219328 | Protein TIC110, chloroplastic | Pisum sativum (pea) | 10 | 0.42 |
| Energy | ||||
| 356521795 | dihydrolipoyl dehydrogenase | Glycine max | 22 | 3.44 |
| 356552735 | isocitrate dehydrogenase [NAD] regulatory subunit 1 | Glycine max | 2.2 | 1.67 |
| 502163841 | aldehyde dehydrogenase family 2 member C4 | Cicer arietinum | 2.2 | 3.98 |
| 48927683 | putative inorganic pyrophosphatase | Arachis hypogaea | 5.6 | 0.26 |
| 372450305 | ATPase subunit 1 (mitochondrion) | Lotus japonicus | 41 | 0.30 |
| 356520768 | stellacyanin | Glycine max | 2.1 | 0.43 |
| 9280616 | NADH dehydrogenase subunit 9 | Lupinus angustifolius | 16 | 0.38 |
| Protein folding and degradation | ||||
| 502132065 | proteasome subunit beta type-6 | Cicer arietinum | 21 | 2.34 |
| 355518872 | Bi-ubiquitin | Medicago truncatula | 18 | 2.51 |
| 356547865 | serine carboxypeptidase II-3 | Glycine max | 8.3 | 42.54 |
| 356500665 | serine carboxypeptidase 24-like isoform 2 | Glycine max | 5.7 | 3.20 |
| 356540970 | serine carboxypeptidase-like 34 | Glycine max | 5 | 2.78 |
| 163914235 | subtilase | Lotus japonicus | 3 | 2.72 |
| 356514109 | subtilisin-like protease SDD1 | Glycine max | 2.1 | 1.31 |
| 502162590 | oligopeptidase A | Cicer arietinum | 17 | 0.59 |
| 49257109 | protein disulfide isomerase | Glycine max | 18 | 1.28 |
| 356548123 | 26S proteasome regulatory subunit 4 homolog A | Glycine max | 15 | 0.55 |
| 502142068 | 26S protease regulatory subunit 8 homolog A | Cicer arietinum | 28 | 0.48 |
| 356553349 | probable 26S proteasome non-ATPase regulatory subunit 3 | Glycine max | 16 | 0.61 |
| 357474441 | 26S proteasome non-ATPase regulatory subunit | Medicago truncatula | 26 | 0.61 |
| 502090101 | T-complex protein 1 subunit zeta | Cicer arietinum | 24 | 0.50 |
| 356513012 | T-complex protein 1 subunit delta-like isoform 2 | Glycine max | 22 | 0.44 |
| 357493557 | T-complex protein 1 subunit eta | Medicago truncatula | 19 | 0.58 |
| 355515447 | Peptidyl-prolyl cis-trans isomerase E | Medicago truncatula | 6 | 0.10 |
| Protein synthesis | ||||
| 356508574 | 40S ribosomal protein S9–2 | Glycine max | 24 | 3.34 |
| 356521522 | 40S ribosomal protein S16 | Lupinus polyphyllus | 10 | 1.87 |
| 356571876 | Nascent polypeptide-associated complex subunit alpha | Medicago truncatula | 2.1 | 1.43 |
| 2500521 | Eukaryotic initiation factor 4A-15 | Nicotiana tabacum | 34 | 0.62 |
| 356548401 | lysyl-tRNA synthetase | Glycine max | 15 | 0.48 |
| 502168167 | methionine aminopeptidase 1A | Cicer arietinum | 4.1 | 0.77 |
| Translation related | ||||
| 194466266 | perchloric acid soluble translation inhibitor protein | Arachis hypogaea | 9.2 | 3.20 |
| Transcription related | ||||
| 357479669 | Histone H4 | Medicago truncatula | 14 | 6.36 |
| 357485127 | Histone H3 | Medicago truncatula | 4.7 | 2.60 |
| 502098976 | small nuclear ribonucleoprotein-associated protein B'-like isoform X2 | Cicer arietinum | 5.5 | 1.92 |
| 356547438 | pre-mRNA-processing-splicing factor 8 | Glycine max | 31 | 0.66 |
| Cellular structure | ||||
| 502138074 | tubulin beta-1 chain | Cicer arietinum | 8.1 | 6.32 |
| 356545743 | myosin-Vb | Glycine max | 3.6 | 0.77 |
| Unkonw | ||||
| 291047846 | unknown | Glycine max | 22 | 0.69 |
| 359807666 | unknown | Medicago truncatula | 6 | 3.37 |
| 257688087 | unknown | Glycine max | 5.6 | 7.47 |
| 351725945 | unknown | Glycine max | 4 | 2.03 |
Note: Unused score represent this data was significant; FM/control represent the changes of more than 1.2 or less than 0.8 fold were considered as significant.
Table 2. Fungal proteins expressed in AM identified by iTRAQ approach.
| Accession | Protein name | Fungus species | Unused score | FM/control |
|---|---|---|---|---|
| Protein folding | ||||
| 90970323 | heat shock protein 60 | Rhizophagus intraradices | 19.71 | 14.5199 |
| 76780890 | binding protein | Rhizophagus intraradices | 2.21 | 16.0671 |
| Cellular structure | ||||
| 52626570 | alpha-tubulin | Glomus diaphanum | 8.82 | 18.8814 |
| 219553143 | beta-tubulin | Rhizophagus clarus | 13.04 | 15.417 |
| Metabolism | ||||
| 378404947 | fumarate reductase | Rhizophagus intraradices | 11.97 | 7.7834 |
| 8134607 | Phosphoglycerate kinase | Funneliformis mosseae | 3.37 | 25.881 |
| 38146200 | glutamine synthetase | Funneliformis mosseae | 3.21 | 11.6025 |
| Protein synthesis | ||||
| 82792162 | elongation factor 1-alpha, partial | Scutellospora heterogama | 8.03 | 6.8289 |
| Energy | ||||
| 254212205 | F-ATPase beta subunit, partial (mitochondrion) | Glomus custos | 8.03 | 15.8362 |
Note: Unused score represent this data was significant; FM/control represent the changes of more than 1.2 or less than 0.8 fold were considered as significant.
Classification of Symbiosis-Related Proteins
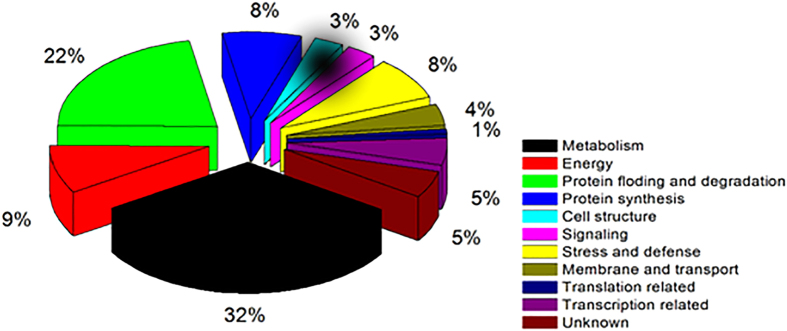
The GO database, BLAST annotations and information reported in the literature were used to categorize each of the identified proteins11. The 77 differentially expressed proteins in A. fruticosa were categorized into different functional classes and assigned to 11 categories. The functional categories are shown in Fig. 2; they include metabolism-related (32%), protein folding and degradation-related (22%), energy-related (10%), protein synthesis-related (8%), stress and defense-related (24%), transcription-related (6%), membrane and transport-related (4%), cellular structure-related (2.5%), signaling transduction-related (11%) and unknown (5%). Among these classes, proteins related to plant metabolism, protein folding and degradation, and energy (totaling 64% of the identified proteins) play important roles during the symbiotic process of AMF-A. fruticosa.
Figure 2. Functional classification of the AM symbiotic-related proteins.
Discussion
Previous studies have demonstrated that colonization is a multi-step, genetically regulated process under the control of specific loci15,16. AMF interact with host plants as cell walls, cell membranes and cellular components undergo dramatic changes17. During the colonization process, functional proteins are induced to express and regulate this process, ultimately forming stable mutualistic symbionts.
Signaling-related proteins
Mutualistic symbionts are the result of a mutual recognition and interaction process between AMF and plant signaling molecules. During the colonization process, signal transduction occurs so that the symbiotic partners recognize each other and the host plants decrease their defense responses. At the same time, AMF are prepared to colonize and to form appressoria and, subsequently, to form arbuscules, vesicles and spores. Because of their mutual nutritional relationship, a real-time dynamic signal dialogue between fungi and host plants is continually present. In this study, we found that the protein levels of Rho GDP-dissociation inhibitor 1 and somatic embryogenesis receptor-like kinase were significantly increased in the symbiotic roots.
Rho GDP-dissociation inhibitor 1, a regulator of Rho GTPase, regulates the balance of Rho GTPase bound to GTP or GDP. There are 2 conformational states of Rho GTPase: the GTP-bound ‘active’ state, and the GDP-bound ‘inactive’ state, in which GTP has been hydrolyzed to GDP18. As a member of the subfamily of small G proteins, Rho GTPase regulates a number of important signal-transduction pathways in eukaryotic cells. Rho GTPase, called Rop (Rho-related GTPase) in plants, has different isomers in animals and fungi19. Rho GTPases are widely distributed in plants, and the corresponding genes in Arabidopsis, maize, barley, rice, peas and alfalfa have been cloned20,21,22. Rho GTPases participate in the regulation of a variety of cellular processes, e.g., gene expression, cell wall synthesis, H2O2 production, actin rearrangement processes, signal transduction pathways of MAP kinase23,24, and cytoskeletal assembly and reassembly, to produce a variety of cellular responses. As a regulatory factor, RhoGDI1 was significantly increased in A. fruticosa AM. Clearly, this protein is closely related to signal transduction between A. fruticosa and GM.
Multiple somatic embryogenesis receptor-like kinases (SERKs) have been defined, including the leucine-rich repeat receptor-like kinase (LRR-RLK) subfamily members and a family of transmembrane signal-transduction proteins25. They are characterized by a predicted signal sequence, a single transmembrane region, and a cytoplasmic kinase domain. These features suggest that some SERK family protein kinases may play pivotal roles in communication between cells and the environment or in cell-cell interactions. Currently, SERK genes have been cloned from various plant species. The AtSERK3 gene participates in the brassinolide (brassinosteroid, BR) signal-transduction pathway. BR is an important hormone that regulates plant growth and development. Functional analysis has shown that the Arabidopsis thaliana mutant became a dwarf when the AtSERK3 gene is knocked out26. The overexpression of the OsSERK1 gene in rice cultivars leads to an increase in host resistance to blast fungus27; in contrast, transcripts of the lettuce LsSERK gene not only are decreased in in vitro somatic embryonic structures but also easily infect Sclerotinia28. Studies have also shown that the SERK gene is closely related to antibiotic stress. Plant root colonization by AMF results in increased levels of somatic embryogenesis receptor-like kinase, which plays a major role in promoting plant growth and enhancing plant disease resistance.
Stress and defense-related proteins
Inoculation with AMF has strong growth-promoting effects on A. fruticosa, especially at the mature stage of symbiont formation. These effects are mediated by increased action of SERK in BR signal-transduction pathways, which have a key role in the regulation of autoimmune responses and of plant root cell elongation and division. However, such regulation is not determined by a single factor. At an early stage of symbiosis, a weak defense response emerges when roots are stimulated by AMF colonization. Lectin plays a crucial role in this defense response by recognizing and binding to the sugar molecules of intruders and interfering with their function on plants. Many plant lectins can bind to glucose, mannitol, galactose or other monosaccharides, and they exhibit high affinity to the oligosaccharides of alien plants. Studies have shown that lectins on leguminous tree surfaces can gather rhizobia around the roots29. As AMF infect the roots of A. fruticosa, plant defense responses are initiated, resulting in agglutinin-2 accumulation. Agglutinin-2 is an important factor for the identification of AMF, similarly to rhizobia.
When A. fruticosa is colonized by AMF, the abscisic acid (ABA) content increases rapidly, leading to the closing of plant stomata and decreased transpiration; this response also activates the genes encoding soluble osmolytes, thus decreasing stress injuries and the impact of stress-induced reactive oxygen and ethylene30. Therefore, ABA accumulation may stimulate metabolic enzymes to produce a feedback effect31. The major ABA catabolic route is decomposition via ABA 8′-hydroxylase to form phaseic acid. Therefore, ABA 8′-hydroxylase accumulation in A. fruticosa may represent a mechanism for regulating ABA levels.
In multiple rice mapping populations, germin-like protein (GLP) markers have been associated with quantitative trait loci (QTL) for resistance to rice blast pathogens. At the early stage of rice blast fungus infection or mechanical damage, some OsGLPs are transiently induced and expressed. Varying 5′ regulatory regions and the differential expression of some protein family members between resistant and susceptible cultivars correspond with differential hydrogen peroxide (H2O2) accumulation levels after fungal infection32. Wang discovered a new wheat germin-like protein33 that is up-regulated in both resistant and susceptible plants. It has been speculated to be involved in wheat defense responses. GLP is significantly increased at the early stage of AMF infection in roots of A. fruticosa, and it may participate in biotic stress responses.
Protein folding and degradation-related proteins
During the symbiosis process, the modification and degradation of peptides and proteins are critical for maintaining cell function. Protein disulfide isomerase, bi-ubiquitin, serine carboxypeptidase, proteasome subunit beta type-6 and subtilisin-like protease SDD1 accumulate in plant roots to ensure proper cell function.
Plants use the proteasome pathway for selective protein degradation, and the proteasome plays pivotal roles in removing abnormally modified proteins and non-targeted proteins. Interactions between bi-ubiquitin and proteasome subunit beta type-6 provide an effective way to degrade proteins. Bi-ubiquitin is highly conserved in eukaryotes, and it is covalently bound to target proteins through post-translational modification to mediate degradation.
Serine carboxypeptidase (SCP), an enzyme that catalyzes the hydrolysis of proteins in eukaryotes, has been found in rice, Arabidopsis and peas. It has been shown that SCP has broad functions in plants, including protein turnover and secondary metabolism synthesis, and it plays an important role in improving plant stress resistance. Liu showed that the expression of OsBISCPL1 was induced by rice blast fungi and antiviral signaling molecules (salicylic acid and jasmonic acid)34 and that overexpression of OsBISCPL1 could enhance disease resistance, oxidative stress tolerance and ABA sensitivity in transgenic Arabidopsis plants. OsBISCPL1 is expressed ubiquitously and differentially in rice, and it is induced by antiviral signaling molecules (BTH, JA, SA and ACC) and is up-regulated by incompatible interactions between rice and the blast fungus. Liu has shown that the expression of the ZmSCP gene in corn is up-regulated under induction by Rhizoctonia solani and that the ZmSCP protein are associated with various abiotic stresses35.
The subtilisin-like protease SDD1 is a member of the processing-type proteases in eukaryotes. As a preproprotein, it can direct peptides for transport to the cytoplasm. SDD1 is a crucial gene that regulates stomatal development and encodes a subtilisin-like serine protease. As a processive enzyme, it may activate a protein molecule or a signal that directs receptors into contact with epidermal cells during stomatal development processes. Liang has shown that the serine protease-encoding gene SDD1 is widely expressed acts on the development of stomata and is also necessary for normal root development36.
Protein disulfide isomerase (PDI), a multifunctional protein, is distributed widely in eukaryotic organisms and is involved in modifying/folding newly synthesized proteins. The catalytic thiol-disulfide exchange reaction to form disulfide is involved in many physiological processes, such as auxiliary protein folding in the endoplasmic reticulum, reconstruction of misfolded proteins, and the repair and refolding of damaged proteins under stress37. Additionally, as a chaperone, PDI can assemble heterogeneous protein peptides and regulate disulfide bonds in an ATP-dependent manner, and it may also be closely related to sugar transport, protein synthesis and other metabolic processes in eukaryotic organisms.
Energy-related Proteins
During the symbiosis process, dihydrolipoyl dehydrogenase, aldehyde dehydrogenase and isocitrate dehydrogenase [NAD] regulatory subunit 1 accumulated in plant roots. AMF colonization significantly enhances the energy metabolism of plants. The Krebs cycle provides more energy than glycolysis, and it is an important pathway not only an important for sugar metabolism but also for the metabolism of lipids, proteins and nucleic acids, which are eventually oxidized to carbon dioxide and water. Isocitrate dehydrogenase (IDH) is considered to be the rate-limiting enzyme of the Krebs cycle; it catalyzes decarboxylation to ketoglutarate while reducing NAD+ to NADH38. Therefore, the activity of NAD-IDH has a significant impact on cellular metabolism. Isocitrate dehydrogenase [NAD] regulatory subunit 1, a regulatory factor, controls the activity of NAD-IDH and thus affects metabolic activity.
Kuhlemeier has explored the energy metabolism of tobacco pollen and has found that, in vegetative tissues39, pyruvate enters the Krebs cycle by pyruvate dehydrogenase (PDH); however, in reproductive organs, it is converted to acetaldehyde by pyruvate decarboxylase (PDC) and then enters the Krebs cycle via aldehyde dehydrogenase (ALDH) and acetyl coenzyme A synthetase (ACS). Thus, ALDH plays an important role in the pyruvate metabolism pathway of PDC/ALDH/ACS. Under stress conditions, plant cells quickly accumulate excessive reactive oxygen species (ROS), which cause oxidative stress and result in the accumulation of large amounts of toxic substance and eventually in plant death40. Aldehydes are an important component of peroxidation reaction products, and they play a crucial role in the oxidation of carboxylic aldehydes, the removal of toxic aldehydes, and the reduction of lipid peroxidation, thereby improving plant tolerance41. As an important member of the pyruvate dehydrogenase family, dihydrolipoyl dehydrogenases ensure the production of oxidatively decarboxylated pyruvate CoA, and CoA then enters the Krebs cycle to produce large amounts of energy for plant growth.
Cellular structure-related proteins
Dramatic changes in plant morphology and in the penetrating mycelium, dynamic reorganization of cytoskeletal elements and organelle transformation occur when arbuscular vesicles develop42. Tubulin is an important component of the cytoskeleton, and it plays an important role in maintaining intracellular structural order and cell morphology. Meanwhile, tubulin is closely related to cellular transport, cell differentiation, cell motility, signal recognition, cell division and other developmental activities. Mills has revealed dramatic changes in both microtubules and actin arrangement in the host cell, and further studies have found that microtubules and actin rearrangement in the host cell are necessary for expression in non-host plants43. Studies on plant tubulin have primarily been focused on annual plants, such as Arabidopsis, tobacco, and rice, but study of the tubulin gene in perennial trees has been rare44.
Membrane and transport-related proteins
A K+ efflux antiporter and coatomer, which are membrane and transport-related proteins, respectively, were found in the A. fruticosa mycorrhizea. The K+ efflux antiporter is mainly responsible for maintaining the intracellular ion balance and regulating the cells’ osmotic pressure. During AMF colonization, AMF invasion affects the ion balance of plant root cells, and plants maintain the intracellular ion balance to stimulate K+ increases. Coatomer, a coat protein, transports vesicles, and vesicle-mediated non-selective transport ensures the accurate transport of proteins and lipids.
Metabolism-related proteins
During mycorrhizal symbiosis, increased levels of 3-oxoacyl-[acyl-carrier-protein] synthase, neutral ceramidase and caffeic acid 3-O-methyltransferase (COMT), which are metabolism-related proteins, were observed.
Lipid metabolism is one of the basic metabolic pathways in plants. The β-ketoacyl-acyl carrier protein synthase (KASI)-mediated acyl chain extension is important in the de novo synthesis of fatty acids. Ceramides, which are central molecules in the sphingolipid signaling pathway, play important roles as second messengers in plants and participate in many significant plant signaling pathways, such as cell growth, proliferation, differentiation, senescence and apoptosis45. Neuraminidase is a key enzyme that regulates ceramide. Neutral neuraminidase hydrolyzes ceramide to form sphingosine (ref). Liu has found that AtCER is involved in H2O2-induced oxidative stress46.
During cell morphogenesis, lignin plays an important role in the growth and development of vascular tissues and is involved in cell wall lignification, which increases the hardness or compressive strength of the cell wall. It also promotes the formation of mechanical tissues while also having a major impact on plant lodging, disease and stress resistance47. There are 3 types of monomeric lignin biosynthesis pathways,: the shikimate pathway, the phenylketonuria pathway and the lignin biosynthesis-specific pathway48. COMT is a key enzyme in the specific lignin pathway and is involved in the synthesis of S-lignin49. AMF colonization enhanced the synthesis of woody amorpha lignin, thus affecting the growth and development of plants.
Transcription and protein synthesis-related proteins
Ribosomal proteins are important components of the ribosome, and they have important roles in translation efficiency and ribosome stability. They also participate in important cellular processes, such as DNA repair, apoptosis and regulation of gene expression; e.g., 40S ribosomal proteins showed significant accumulation in plant roots after AMF invasion.
Nascent polypeptide-associated complex subunit alpha (NAC), which is located at the top of the newly synthesized polypeptide, can reversibly bind to eukaryotic ribosomes and guide the correct distribution and translocation of newly synthesized polypeptides in the cell. The observed increases in 40S ribosomal protein and NAC levels, combined with folding- and degradation-associated proteins, ensure the fast and accurate synthesis and distribution of AM symbiosis-related proteins.
Transcriptional regulation is an important aspect of the regulation of gene expression. The results show significant accumulation of histone H3 and histone H4 in the host plant roots. Nucleosomes constitute the basic unit of chromatin in eukaryotes. Histones, which are structural proteins of chromosomes, play important roles in DNA folding and packing, protecting DNA from digestive enzymes, and gene regulation, tumor formation, and apoptosis. The N-terminal amino acids of histones participate in acetylation, methylation, phosphorylation, ubiquitination and other covalent modifications. Studies have shown that histones may change the structure of chromatin via post-translational modifications, thus modulating gene expression50.
Unknown proteins
During AMF symbiosis, the expression levels of proteins within A. fruticosa roots were changed; some proteins disappeared, and new symbiosis proteins arose. The functional analysis of symbiotic proteins in A. fruticosa, a non-model plant, is not difficult. Because these proteins were differentially expressed in the symbiotic system, they are targets for future studies.
AM-A. fruticosa molecular regulation model
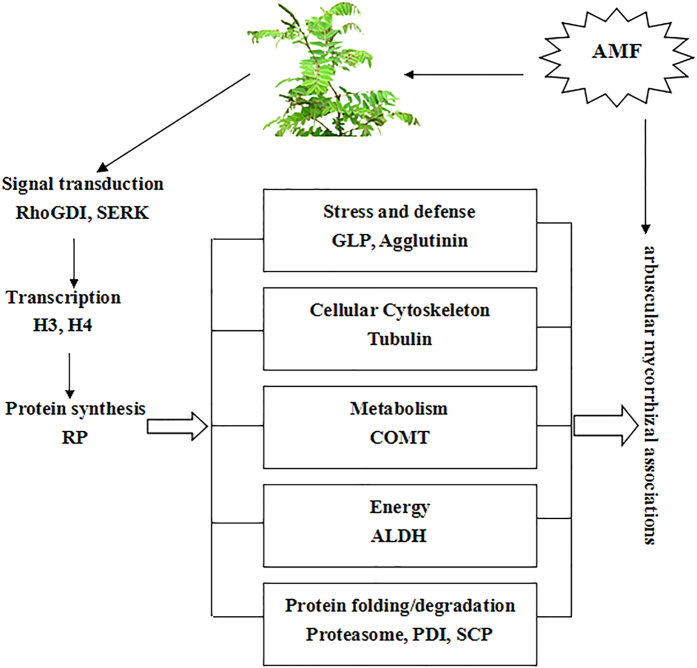
By using bioinformatics analysis, we found that mycorrhizal proteins were involved in several biological processes and cellular activities (Fig. 3), and we verified that the symbiosis formed between AMF and A. fruticosa is a uniform and harmonious result of symbiotic interactions.
Figure 3. Several biological processes of cellular activities, and it was drawn by dandan Qi.
Methods
G. mosseae (GM) was harvested from sorghum, which was supplied by the Ecology Laboratory of Heilongjiang University, by co-culturing for longer than 40 days. Inocula contained a mixture of the rhizosphere that consisted of AM fungal spores, hyphae and mycorrhizal fragments. The inocula contained approximately 500 spores per 20 g.
Seedling culture
Seeds of A. fruticosa were purchased from the Academy of Agricultural Sciences of Heilongjiang Province. A. fruticosa seeds were sterilized with 0.4% K2MnO4 for 20 min, rinsed, and then covered with a layer of white gauze to keep them moist. Germination was conducted in an incubator at 30 °C for 60 h after soaking for 24 h. The growing medium was 50% peat soil, 30% vermiculite and 20% sand. It was sterilized in an autoclave at 121°C for 2 h and then air dried for 1 week before the start of the experiments10,11.
Mycorrhizal colonization percentage determination
The germinated seeds were then planted in a pre-sterilized mixed matrix and grown under a 16-h photoperiod at temperatures of 25/18 °C (day/night) with 60% relative humidity. One group was inoculated with GM inoculum, and the other was inoculated with sterilized inoculum as a control (CK). Each treatment was repeated 10 times. A total of 20 pots were arranged randomly and watered every 2 days. The mycorrhizal colonization percentage of the seedlings was determined using the Phillip and Hayman staining method (KOH bleaching-acid fuchsin stain) with some modifications (Phillips J M, 1970)51.
Protein extraction, protein quantification and SDS-PAGE
At the maturation stage, A. fruticosa roots were harvested, and total root protein was precipitated with 10% (w/v) trichloroacetic acid (TCA) in acetone at −20 °C overnight. After centrifugation at 40,000 × g at 4 °C for 1 h, the pellets were washed 3 times with cold 80% acetone. A 2-D Quant kit (GE Healthcare, USA) was used to determine the protein concentrations. SDS-polyacrylamide gel (12%) electrophoresis was performed with 30-μg samples at 120-V constant voltage for 2 h. The gel was stained with Coomassie blue and visualized52,53.
iTRAQ labeling
The CK group and the GM group each included 3 biological replicates. After digesting with trypsin, the proteins from the non-infected and infected samples were labeled with iTRAQ reagents 115 (CK1), 116 (CK2), 117 (CK3), 118 (GM1), 119 (GM2), and 121 (GM3) and were then combined following the manufacturer’s protocol at a ratio of 1:1:1:1:1:1 for LC-MS/MS analysis54,55.
LC-MS/MS measurements
The labeled samples were pooled and purified using a strong cation-exchange chromatography (SCX) column and were then separated on an analytical column (1.7 μm, 100 μm × 100 mm) at a flow rate of 300 nL/min using a linear gradient of 5–35% acetonitrile (ACN) over 40 min. The ion spray voltage was 4.5 kV, and nitrogen was used as a nebulizing gas (30 psi) and a curtain gas (15 psi). From each MS scan, the 30 most intense precursor ions were selected for MS/MS fragmentation and were detected at a mass resolution of 30,000 at m/z 40056. Data analysis was performed with a Triple TOF 5600 System, and then the iTRAQ data were compared with the protein sequences of homologous species after genome annotation.
Protein identification
Protein Pilot 4.0 (AB Sciex Inc., USA) was used to simultaneously identify and quantify proteins57,58. Differentially expressed proteins were required to satisfy 3 conditions for identification: (1) each confident protein identification involved at least 1 unique peptide; (2) the P-value was less than 0.05; and (3) changes of greater than 1.2-fold or less than 0.8 fold were considered significant. All of the identified proteins were classified according to the annotations acquired by using the UniProt knowledge base and the GO database.
Additional Information
How to cite this article: Song, F. et al. Proteomic analysis of symbiotic proteins of Glomus mosseae and Amorpha fruticosa. Sci. Rep. 5, 18031; doi: 10.1038/srep18031 (2015).
Supplementary Material
Acknowledgments
We sincerely thank Prof. sixue Chen for greatly improved the manuscript. This study was supported by National Natural Science Foundation of China (31070576 and 31270535), Natural Science Foundation of Heilongjiang Province of China (No. ZD201206), Excellent Youth Foundation of Heilongjiang Province of China (No. JC201306) and High-level Talents Support Program of Heilongjiang University (Ecological Restoration Team).
Footnotes
Author Contributions F.S. designed the research, D.Q. wrote the main manuscript text of chinese and interpreted the proteomics data, X.K. and X.L. wrote the main manuscript text of english and edited language, Y.G., Z.Z. and Q.W. extract proteins. All authors reviewed the manuscript.
References
- Fitter A., Heinemeyer A. & Staddon P. The impact of elevated CO2 and global climate change on arbuscular mycorrhizas: a mycocentric approach. New Phytologist. 147, 179–187 (2000). [Google Scholar]
- Rosendahl S. Populations and individuals of arbuscular mycorrhizal fungi. New Phytologist. 178, 253–266 (2008). [DOI] [PubMed] [Google Scholar]
- Smith S. & Read D. Mycorrhizal symbiosis from cellular to ecosystem scales. Annual review of plant biology. 62, 227–250 (2008). [DOI] [PubMed] [Google Scholar]
- Smith S. E. & Smith F. A. Roles of arbuscular mycorrhizas in plant nutrition and growth: new paradigms from cellular to ecosystem scales. Annual review of plant biology. 62, 227–250 (2011). [DOI] [PubMed] [Google Scholar]
- Kiers E. T. et al. Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science. 333, 880–882 (2011). [DOI] [PubMed] [Google Scholar]
- Trépanier M. et al. Dependence of arbuscular-mycorrhizal fungi on their plant host for palmitic acid synthesis. Applied and environmental microbiology. 71, 5341–5347 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recorbet G. et al. Identification of in plant-expressed arbuscular mycorrhizal fungal proteins upon comparison of the root proteomes of Medicago truncatula colonised with two Glomus species. Fungal Genetics and Biology. 47, 608–618 (2010). [DOI] [PubMed] [Google Scholar]
- Aloui A. et al. Arbuscular mycorrhizal symbiosis elicits shoot proteome changes that are modified during cadmium stress alleviation in Medicago truncatula. BMC plant biology. 11, 75 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. H., Yuan K. & Yang L. F. Identification and Functional Analysis of Maize Leaf Proteins Responding to the Abuscular Mycorrhizal Fungi (AMF). Chinese Journal of Tropical Agriculture. 33, 40–44 (2013). [Google Scholar]
- Song F. Q., Li J. Z. & Zhang X. X. Characterization of expressed genes in the establishment of arbuscular mycorrhiza between Amorpha fruticosa and Glomus mosseae. Journal of Forestry Research. 25, 541–548 (2014). [Google Scholar]
- Song F. Q., Kong X. S., Li J. Z. & Chang W. Screening the related genes in the AM Fungi and Amorpha fruticosa symbiosis with the subtractive hybridization technique. Scienta Slivae Sinicae. 50, 64–74 (2014). [Google Scholar]
- Heikham E., Rupam K. & Bhoopander G. Arbuscular mycorrhizal fungi in alleviation of salt stress: a review. Annals of Botany. 104, 1263–1280 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song F. Q., Kong X. S., Dong A. R. & Liu X. F. Impact of arbuscular mycorrhizal fungi on the growth and related physiological indexes of Amorpha fruticosa. Journal of Medicinal Plants Research. 6, 3648–3655 (2012). [Google Scholar]
- Claudia H. & Helge K. A roadmap of cell-type specific gene expression during sequential stages of the arbuscular mycorrhiza symbiosis. BMC Genomics. 14, 306–324 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfante P. et al. The Lotus japonicus LjSym4 gene is required for the successful symbiotic infection of root epidermal cells. Molecular plant-microbe interactions: MPMI. 13, 1109–1120 (2000). [DOI] [PubMed] [Google Scholar]
- Novero M. et al. Dual requirement of the LjSym4 gene for mycorrhizal development in epidermal and cortical cells of Lotus japonicus roots. New phytologist. 154, 741–749 (2002). [DOI] [PubMed] [Google Scholar]
- Bonfante P. et al. At the interface between mycorrhizal fungi and plants: the structural organization of cell wall, plasma membrane and cytoskeleton. Fungal Associations. 9, 45–61 (2001). [Google Scholar]
- Etienne Manneville S. & Hall A. Rho GTPases in cell biology. Nature. 420, 629–635 (2002). [DOI] [PubMed] [Google Scholar]
- Li H. et al. Arabidopsis Rho-related GTPases: differential gene expression in pollen and polar localization in fission yeast. Plant Physiology. 118, 407–417 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winge P., Brembu T., Kristensen R. & Bones A. M. Genetic structure and evolution of RAC-GTPases in Arabidopsis thaliana. Genetics. 156, 1959–1971 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen T. M. et al. Conserved subgroups and developmental regulation in the monocot rop gene family. Plant physiology. 133, 1791–1808 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z. L. & Yang Z. The Rop GTPase: an emerging signaling switch in plants. Plant molecular biology. 44, 1–9 (2000). [DOI] [PubMed] [Google Scholar]
- Takai Y., Sasaki T. & Matozaki T. Small GTP-binding proteins. Physiological reviews. 81, 153–208 (2001). [DOI] [PubMed] [Google Scholar]
- Yang Z. et al. Small GTPases versatile signaling switches in plants. The Plant Cell. 14, 375–388 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S. H. & Bleecker A. B. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proceedings of the National Academy of Sciences. 98, 10763–10768 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T. et al. Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiology. 121, 743–752 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Xiong L. & Yang Y. Rice SERK1 gene positively regulates somatic embryogenesis of cultured cell and host defense response against fungal infection. Planta. 222, 107–117 (2005). [DOI] [PubMed] [Google Scholar]
- Santos M. et al. Suppression of SERK gene expression affects fungus tolerance and somatic embryogenesis in transgenic lettuce. Plant Biology. 11, 83–89 (2009). [DOI] [PubMed] [Google Scholar]
- Yin A. H. & Han S. F. Relationship of Nodulation with Reactions of Letins of Leguminous Trees with EPS of Rhizobium. Journal of Nanjing Forestry University. 29, 88–90 (2005). [Google Scholar]
- Verslues P. & Zhu J. Plant signaling from genes to biochemistry. Biochemical Society Transactions. 33 (2005). [DOI] [PubMed] [Google Scholar]
- Cutler A. J. & Krochko J. E. Formation and breakdown of ABA. Trends in plant science. 4, 472–478 (1999). [DOI] [PubMed] [Google Scholar]
- Davidson R. M. et al. Rice germin-like proteins: Allelic diversity and relationships to early stress responses. Rice. 3, 43–55 (2010). [Google Scholar]
- Wang J. M. et al. Cloning and Chromosome Mapping of a Germin-Like Protein Gene in Wheat and Its Expression in Response to Infection with Wheat Powdery Mildew. Scientia Agricultura Sinica. 42, 3104–3111 (2009). [Google Scholar]
- Liu H. et al. A rice serine carboxypeptidase-like gene OsBISCPL1 is involved in regulation of defense responses against biotic and oxidative stress. Gene. 420, 57–65 (2008). [DOI] [PubMed] [Google Scholar]
- Liu L. et al. Cloning and Expression Analysis of Serine Carboxypeptidases in Maize (Zea mays L.). Acta Agronomica Sinica. 39, 164–171 (2013). [Google Scholar]
- Liang K. et al. Identification and Genetic Analysis of Two Putative SDD1 Alleles in Arabidopsis. Chin Bull Bot. 3, 318–326 (2010). [Google Scholar]
- Kim Y. J. et al. Protein disulfide isomerase-like protein 1-1 controls endosperm development through regulation of the amount and composition of seed proteins in rice. PlOS one. 7, 44493 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R. D. & Gadal P. Structure, functions and regulation of NAD and NADP dependent isocitrate dehydrogenases in higher plants and in other organisms. Plant Physiology and Biochemistry. 28, 411–427 (1990). [Google Scholar]
- Kuhlemeier C. Aldehyde dehydrnase in tobacco pollen. Plant Mol Bio. 1, 355–365 (1997). [DOI] [PubMed] [Google Scholar]
- Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends in plant science. 7, 405–410 (2002). [DOI] [PubMed] [Google Scholar]
- Li X. L., Yang C. P. & Xu X. L. Molecular Cloning and Expression Analysis of Aldehyde Dehydrogenase (ALDH) Gene Segment in Leymus Chinensis. Chinese Agricultural Science Bulletin. 23, 115–120 (2007). [Google Scholar]
- Genre A. & Bonfante P. Actin versus tubulin configuration in arbuscule-containing cells from mycorrhizal tobacco roots. New phytologist. 140, 745–752 (1998). [DOI] [PubMed] [Google Scholar]
- Mills D., Kunoh H., Keen N. & Mayama S. Molecular aspects of pathogenicity and resistance: requirement for signal transduction. American Phytopathological Society, 257–267 (1996). [Google Scholar]
- Rao G. D. & Zhang J. G. Advance of Studies on Plant Tubulin Gene. World Forestry Research. 26, 11–16 (2013). [Google Scholar]
- Liang H. et al. Ceramides modulate programmed cell death in plants. Genes & Development. 17, 2636–2641 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R. H., Jiang W. B. & Yu D. Q. The Response of AtCER to Oxidative Stress in Arabidopsis thaliana. Acta Botanica Yunnaica. 31, 326–334 (2009). [Google Scholar]
- Li B. et al. Advance on Key Enzyme Gene (COMT) Involved in Lignin Biosynthesis. Molecular Plant Breeding. 1, 117–124 (2010). [Google Scholar]
- Hamada K. et al. 4-Coumarate: coenzyme A ligase in black locust (Robinia pseudoacacia) catalyses the conversion of sinapate to sinapoyl-CoA. Journal of plant research. 117, 303–310 (2004). [DOI] [PubMed] [Google Scholar]
- Kang C. et al. Carbon Emission Flow in Networks. Sci. Rep. 2, 479, 10.1038/srep00479 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley D. M. et al. The role of laccase in lignification. The Plant Journa. 4, 751–757 (1993). [Google Scholar]
- Wei Q. D. et al. Advancement of Histone and Its Relationship with Regulation of Gene Expression. Journal of Tropical Medicine. 6, 596–598 (2006). [Google Scholar]
- Phillips J. & Hayman D. Improved procedures for clearing roots and staining parasitic and vesicular-arbuscular mycorrhizal fungi for rapid assessment of infection. Transactions of the British mycological Society. 55, 158–161 (1970). [Google Scholar]
- Sophie A., Leslie M. H. & Sona P. ABA-Dependent and Independent G-Protein Signaling in Arabidopsis Roots Revealed through an iTRAQ Proteomics Approach. Journal of proteome research. 10, 3107–3122 (2011). [DOI] [PubMed] [Google Scholar]
- Zhang K. R., Carolyn M., Charles H. H. & Michael A. D. The Medicago truncatula Small Protein Proteome and Peptidome. Journal of Proteome Research. 5, 3355–3367 (2006). [DOI] [PubMed] [Google Scholar]
- Liu J. Y. et al. Comparative analysis of proteomic profile at different development stages of Volvariella volvacea by iTRAQ-coupled 2D LC-MSMS. Microbiology China. 39, 853–864 (2012). [Google Scholar]
- Pan X. Q. et al. iTRAQ Protein Profile Analysis of Tomato Green-ripe Mutant Reveals New Aspects Critical for Fruit Ripening. Journal of Proteome Research. 13, 1979–1993 (2014). [DOI] [PubMed] [Google Scholar]
- Alexey C., Consuelo M. V., Neus V. & Roman A. Z. Functional Identification of Target by Expression Proteomics (FITExP) reveals protein targets and highlights mechanisms of action of small molecule drugs. Scientific Reports. 5, 11176, 10.1038/srep11176 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose A. M. et al. Quantitative proteomic analysis of host—pathogen interactions: a study of Acinetobacter baumannii responses to host airways. BMC Genomics. 16, 422–443 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.