Abstract
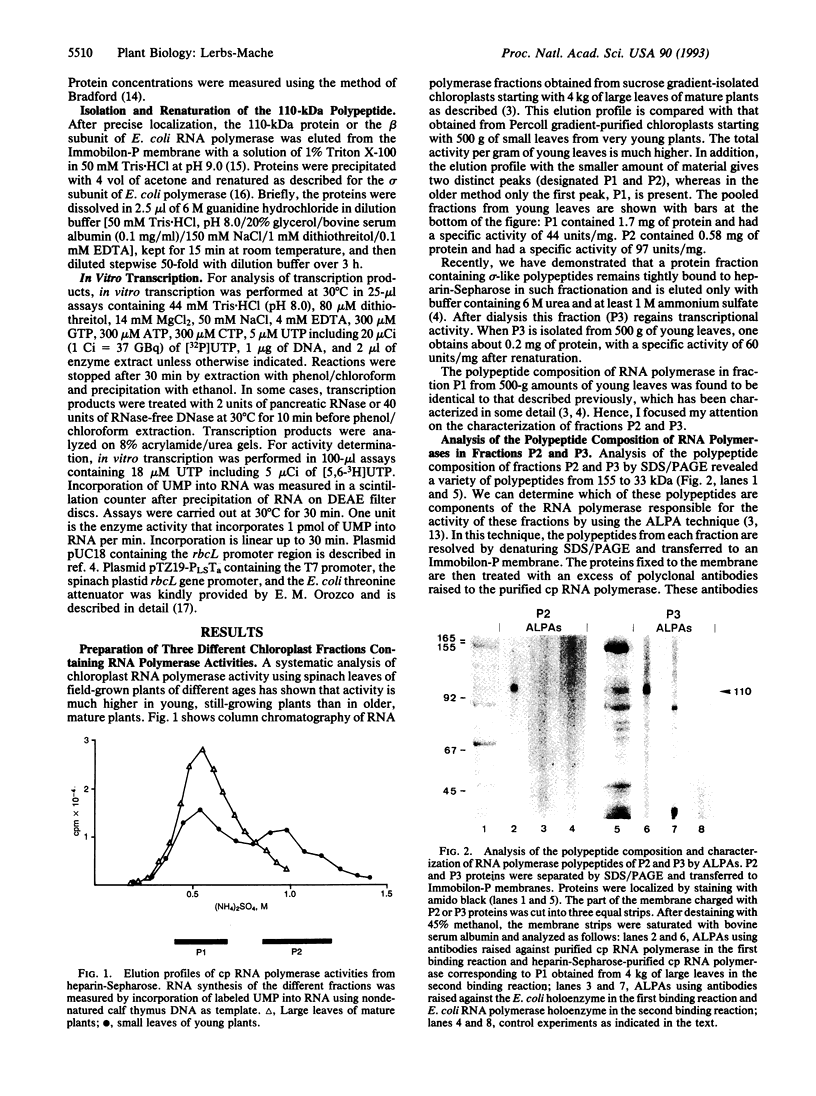
Highly purified RNA polymerase preparations from spinach chloroplasts contain seven major polypeptides of 150, 145, 110, 102, 80, 75, and 38 kDa. I find that RNA polymerase activity can be separated under defined conditions into three different fractions by heparin-Sepharose chromatography. Immunological analysis has shown that the first fraction contains RNA polymerase activity associated with all seven major polypeptides, and other studies have shown that some of these polypeptides (150, 145, 80, and 38 kDa) are associated with an RNA polymerase similar to the Escherichia coli enzyme. However, similar analyses of the remaining fractions show activity associated only with the 110-kDa polypeptide, suggesting the existence of a second kind of chloroplast RNA polymerase. Samples of this 110-kDa polypeptide purified by SDS/PAGE actively synthesize RNA in a reaction dependent on a supercoiled DNA template and the four ribonucleoside triphosphates. Hence, this polypeptide has all of the properties expected of a single-subunit RNA polymerase of the T7 bacteriophage type.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baeza L., Bertrand A., Mache R., Lerbs-Mache S. Characterization of a protein binding sequence in the promoter region of the 16S rRNA gene of the spinach chloroplast genome. Nucleic Acids Res. 1991 Jul 11;19(13):3577–3581. doi: 10.1093/nar/19.13.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldacci G., Chérif-Zahar B., Bernardi G. The initiation of DNA replication in the mitochondrial genome of yeast. EMBO J. 1984 Sep;3(9):2115–2120. doi: 10.1002/j.1460-2075.1984.tb02099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Briat J. F., Mache R. Properties and characterization of a spinach chloroplast RNA polymerase isolated from a tanscriptionally active DNA-protein complex. Eur J Biochem. 1980 Oct;111(2):503–509. doi: 10.1111/j.1432-1033.1980.tb04966.x. [DOI] [PubMed] [Google Scholar]
- Chang D. D., Clayton D. A. Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc Natl Acad Sci U S A. 1985 Jan;82(2):351–355. doi: 10.1073/pnas.82.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. D., Hauswirth W. W., Clayton D. A. Replication priming and transcription initiate from precisely the same site in mouse mitochondrial DNA. EMBO J. 1985 Jun;4(6):1559–1567. doi: 10.1002/j.1460-2075.1985.tb03817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. J., Orozco E. M., Jr Recognition of prokaryotic transcription terminators by spinach chloroplast RNA polymerase. Nucleic Acids Res. 1988 Sep 12;16(17):8411–8431. doi: 10.1093/nar/16.17.8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D. A. Transcription of the mammalian mitochondrial genome. Annu Rev Biochem. 1984;53:573–594. doi: 10.1146/annurev.bi.53.070184.003041. [DOI] [PubMed] [Google Scholar]
- Fuller C. W., Beauchamp B. B., Engler M. J., Lechner R. L., Matson S. W., Tabor S., White J. H., Richardson C. C. Mechanisms for the initiation of bacteriophage T7 DNA replication. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):669–679. doi: 10.1101/sqb.1983.047.01.078. [DOI] [PubMed] [Google Scholar]
- Greenberg B. M., Narita J. O., DeLuca-Flaherty C., Gruissem W., Rushlow K. A., Hallick R. B. Evidence for two RNA polymerase activities in Euglena gracilis chloroplasts. J Biol Chem. 1984 Dec 10;259(23):14880–14887. [PubMed] [Google Scholar]
- Gruissem W., Elsner-Menzel C., Latshaw S., Narita J. O., Schaffer M. A., Zurawski G. A subpopulation of spinach chloroplast tRNA genes does not require upstream promoter elements for transcription. Nucleic Acids Res. 1986 Oct 10;14(19):7541–7556. doi: 10.1093/nar/14.19.7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager D. A., Burgess R. R. Elution of proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: results with sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase, and other enzymes. Anal Biochem. 1980 Nov 15;109(1):76–86. doi: 10.1016/0003-2697(80)90013-5. [DOI] [PubMed] [Google Scholar]
- Hess W. R., Prombona A., Fieder B., Subramanian A. R., Börner T. Chloroplast rps15 and the rpoB/C1/C2 gene cluster are strongly transcribed in ribosome-deficient plastids: evidence for a functioning non-chloroplast-encoded RNA polymerase. EMBO J. 1993 Feb;12(2):563–571. doi: 10.1002/j.1460-2075.1993.tb05688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Bogorad L. Maize chloroplast RNA polymerase: the 180-, 120-, and 38-kilodalton polypeptides are encoded in chloroplast genes. Proc Natl Acad Sci U S A. 1990 Feb;87(4):1531–1535. doi: 10.1073/pnas.87.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson G. S., Holton T. A., Whitfield P. R., Bottomley W. Spinach chloroplast rpoBC genes encode three subunits of the chloroplast RNA polymerase. J Mol Biol. 1988 Apr 20;200(4):639–654. doi: 10.1016/0022-2836(88)90477-9. [DOI] [PubMed] [Google Scholar]
- Khanna N. C., Lakhani S., Tewari K. K. Identification of the template binding polypeptide in the pea chloroplast transcriptional complex. Nucleic Acids Res. 1992 Jan 11;20(1):69–74. doi: 10.1093/nar/20.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna N. C., Lakhani S., Tewari K. K. Photoaffinity labelling of the pea chloroplast transcriptional complex by nascent RNA in vitro. Nucleic Acids Res. 1991 Sep 25;19(18):4849–4855. doi: 10.1093/nar/19.18.4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U., De Camp J. D., Bogorad L. Two types of chloroplast gene promoters in Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3453–3457. doi: 10.1073/pnas.89.8.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klement J. F., Moorefield M. B., Jorgensen E., Brown J. E., Risman S., McAllister W. T. Discrimination between bacteriophage T3 and T7 promoters by the T3 and T7 RNA polymerases depends primarily upon a three base-pair region located 10 to 12 base-pairs upstream from the start site. J Mol Biol. 1990 Sep 5;215(1):21–29. doi: 10.1016/s0022-2836(05)80091-9. [DOI] [PubMed] [Google Scholar]
- Lerbs-Mache S. Quantification of DNA-dependent RNA polymerase subunits and initiation factor(s) by antibody-linked polymerase assays. FEBS Lett. 1988 Jul 18;234(2):392–394. doi: 10.1016/0014-5793(88)80123-6. [DOI] [PubMed] [Google Scholar]
- Lerbs S., Bräutigam E., Parthier B. Polypeptides of DNA-dependent RNA polymerase of spinach chloroplasts: characterization by antibody-linked polymerase assay and determination of sites of synthesis. EMBO J. 1985 Jul;4(7):1661–1666. doi: 10.1002/j.1460-2075.1985.tb03834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little M. C., Hallick R. B. Chloroplast rpoA, rpoB, and rpoC genes specify at least three components of a chloroplast DNA-dependent RNA polymerase active in tRNA and mRNA transcription. J Biol Chem. 1988 Oct 5;263(28):14302–14307. [PubMed] [Google Scholar]
- Masters B. S., Stohl L. L., Clayton D. A. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987 Oct 9;51(1):89–99. doi: 10.1016/0092-8674(87)90013-4. [DOI] [PubMed] [Google Scholar]
- Morden C. W., Wolfe K. H., dePamphilis C. W., Palmer J. D. Plastid translation and transcription genes in a non-photosynthetic plant: intact, missing and pseudo genes. EMBO J. 1991 Nov;10(11):3281–3288. doi: 10.1002/j.1460-2075.1991.tb04892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourioux G., Douce R. Slow Passive Diffusion of Orthophosphate between Intact Isolated Chloroplasts and Suspending Medium. Plant Physiol. 1981 Mar;67(3):470–473. doi: 10.1104/pp.67.3.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita J. O., Rushlow K. E., Hallick R. B. Characterization of a Euglena gracilis chloroplast RNA polymerase specific for ribosomal RNA genes. J Biol Chem. 1985 Sep 15;260(20):11194–11199. [PubMed] [Google Scholar]
- Nielsen B. L., Rajasekhar V. K., Tewari K. K. Pea chloroplast DNA primase: characterization and role in initiation of replication. Plant Mol Biol. 1991 Jun;16(6):1019–1034. doi: 10.1007/BF00016074. [DOI] [PubMed] [Google Scholar]
- Orozco E. M., Jr, Mullet J. E., Chua N. H. An in vitro system for accurate transcription initiation of chloroplast protein genes. Nucleic Acids Res. 1985 Feb 25;13(4):1283–1302. doi: 10.1093/nar/13.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekhar V. K., Sun E., Meeker R., Wu B. W., Tewari K. K. Highly purified pea chloroplast RNA polymerase transcribes both rRNA and mRNA genes. Eur J Biochem. 1991 Jan 1;195(1):215–228. doi: 10.1111/j.1432-1033.1991.tb15697.x. [DOI] [PubMed] [Google Scholar]
- Reiss T., Link G. Characterization of transcriptionally active DNA-protein complexes from chloroplasts and etioplasts of mustard (Sinapis alba L.). Eur J Biochem. 1985 Apr 15;148(2):207–212. doi: 10.1111/j.1432-1033.1985.tb08826.x. [DOI] [PubMed] [Google Scholar]
- Sexton T. B., Christopher D. A., Mullet J. E. Light-induced switch in barley psbD-psbC promoter utilization: a novel mechanism regulating chloroplast gene expression. EMBO J. 1990 Dec;9(13):4485–4494. doi: 10.1002/j.1460-2075.1990.tb07899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. B., Gruissem W. Control of plastid gene expression: 3' inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell. 1987 Dec 24;51(6):1145–1157. doi: 10.1016/0092-8674(87)90600-3. [DOI] [PubMed] [Google Scholar]
- Szewczyk B., Laver W. G., Summers D. F. Purification, thioredoxin renaturation, and reconstituted activity of the three subunits of the influenza A virus RNA polymerase. Proc Natl Acad Sci U S A. 1988 Nov;85(21):7907–7911. doi: 10.1073/pnas.85.21.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitlin D., Hu J., Bogorad L. Binding and transcription of relaxed DNA templates by fractions of maize chloroplast extracts. Proc Natl Acad Sci U S A. 1989 Feb;86(3):876–880. doi: 10.1073/pnas.86.3.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dePamphilis C. W., Palmer J. D. Loss of photosynthetic and chlororespiratory genes from the plastid genome of a parasitic flowering plant. Nature. 1990 Nov 22;348(6299):337–339. doi: 10.1038/348337a0. [DOI] [PubMed] [Google Scholar]
- van der Meer J., Dorssers L., Zabel P. Antibody-linked polymerase assay on protein blots: a novel method for identifying polymerases following SDS-polyacrylamide gel electrophoresis. EMBO J. 1983;2(2):233–237. doi: 10.1002/j.1460-2075.1983.tb01411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]