Abstract
Background:
Inflammation has been hypothesized to increase the risk of cancer development as an initiator or promoter, yet no large-scale study of inherited variation across cancer sites has been conducted.
Methods:
We conducted a cross-cancer genomic analysis for the inflammation pathway based on 48 genome-wide association studies within the National Cancer Institute GAME-ON Network across five common cancer sites, with a total of 64 591 cancer patients and 74 467 control patients. Subset-based meta-analysis was used to account for possible disease heterogeneity, and hierarchical modeling was employed to estimate the effect of the subcomponents within the inflammation pathway. The network was visualized by enrichment map. All statistical tests were two-sided.
Results:
We identified three pleiotropic loci within the inflammation pathway, including one novel locus in Ch12q24 encoding SH2B3 (rs3184504), which reached GWAS significance with a P value of 1.78 x 10–8, and it showed an association with lung cancer (P = 2.01 x 10–6), colorectal cancer (GECCO P = 6.72x10-6; CORECT P = 3.32x10-5), and breast cancer (P = .009). We also identified five key subpathway components with genetic variants that are relevant for the risk of these five cancer sites: inflammatory response for colorectal cancer (P = .006), inflammation related cell cycle gene for lung cancer (P = 1.35x10-6), and activation of immune response for ovarian cancer (P = .009). In addition, sequence variations in immune system development played a role in breast cancer etiology (P = .001) and innate immune response was involved in the risk of both colorectal (P = .022) and ovarian cancer (P = .003).
Conclusions:
Genetic variations in inflammation and its related subpathway components are keys to the development of lung, colorectal, ovary, and breast cancer, including SH2B3, which is associated with lung, colorectal, and breast cancer.
Inflammation has been hypothesized to increase the risk of cancer development as an initiator or promoter through three primary processes: increased genetic mutations, anti-apoptotic signaling, and increased angiogenesis—all pivotal processes in tumor development and relevant for most of the cancer sites (1–3). Many studies have demonstrated that genomic variants in the inflammation pathways are relevant to cancer susceptibility (4–10), and The Pan-Cancer project coordinated by The Cancer Genome Atlas (TCGA) has demonstrated the importance and value of analyzing somatic data across tumor types (11–13). However, no large-scale study of inherited variation across cancer sites has been conducted. Therefore we investigated the potential pleiotropic impact of sequence variants in the inflammation-related pathways across five cancer sites within the Genetic Associations and Mechanisms in Oncology (GAME-ON) Network established by the National Cancer Institute (NCI) and the Genetic and Epidemiology of Colorectal Cancer Consortium (GECCO) (14).
The GAME-ON Network was launched by NCI to capitalize on the extensive investment in cancer genome-wide association studies (GWAS), with the overarching goal to integrate post-GWAS research and to facilitate analyses that address research questions that are common across multiple cancer sites. The GAME-ON Network is focused on tumors that currently represent major public health burden, including cancer of the lung, ovary, breast, prostate, and colorectum, and has assembled extensive genomic data from these five cancer consortia, which constitute the basis of our cross-cancer analysis of inflammation pathway.
The goal of this investigation is not only to estimate the effect of single genetic variants in the inflammation-related pathways, but also to estimate the contribution of the genetic variations in subcomponents within the inflammation pathway, such as immune response, cytokines, and inflammatory response, among others. Standard pathway analysis approaches, such as gene-set enrichment analysis, have the limitations of potential gene size biases and typically rely on the most significant single variants in a specific gene or pathway (15–17). On the other hand, hierarchical modeling (HM) based on Bayesian framework represents an alternative for addressing some of the shortcomings of standard pathway analysis by incorporating pathway information in the second-stage model, which accounts for the information from the full dataset, instead of the most significant variants (4,18,19). It also has the advantage of providing effect size estimation in addition to the significance level, which is lacking in most of the other pathway-based approaches. Therefore, we employed hierarchical modeling to estimate the effect of inflammation-related pathways across five common cancer sites based on the genomic data available in the GAME-ON network.
Methods
Study Population
Within the GAME-ON Network (http://epi.grants.cancer.gov/gameon/) and GECCO, forty-eight studies from North America and Europe participated in this investigation. All studies frequency-matched case patients and control patients on at least age and sex, and all subjects were of European descent. The study characteristics are summarized in Supplementary Table 1 (available online) (20–30). In total, 64 591 cancer patients and 74 467 control patients were included in the current analysis. Table 1 summarizes the characteristics of the studies participating in this analysis, with the majority of the studies using Illumina genotyping platforms. All studies included have obtained approval from the institutional ethics review board, and informed consents were obtained from each study participant by the individual study coordinating center.
Table 1.
Characteristics of the participating studies from lung, colorectal, breast, prostate and ovary cancer consortium
| Cancer | Study, No. | Case patients, No. | Control patients, No. | Genotyping platform | Reference panel | Covariates | Refs |
|---|---|---|---|---|---|---|---|
| Lung - overall | 9 | 14 900 | 29 485 | Illumina 317K/550K/610K | HapMap2 | Age, sex, PCs | (20) |
| Adenocarcinoma | 9 | 4813 | 28 489 | Illumina 317K/550K/610K | HapMap2 | Age, sex, PCs | |
| Squamous cell | 9 | 4110 | 28 643 | Illumina 317K/550K/610K | HapMap2 | Age, sex, PCs | |
| Colorectal - GECCO | 13 | 10 314 | 12 857 | Illumina 550K/610K/CytoSNP/Omni Affymetrix 100K/500K |
1000 Genome | Age, sex (when applicable), PCs | (26) |
| Colorectal - CORECT | 6 | 5100 | 4831 | Affymetrix Axiom | 1000 Genome | Age, sex, PCs | (28) |
| Ovary overall | 3 | 4369 | 9123 | Illumina 317K/370K/550K/610K/670K/2.5M | 1000 Genome | Site, PCs | (27) |
| Serous | 3 | 2556 | 9123 | Illumina 317K/370K/550K/610K/670K/2.5M | 1000 Genome | Site, PCs | |
| Endometroid | 3 | 715 | 9123 | Illumina 317K/370K/550K/610K/670K/2.5M | 1000 Genome | Site, PCs | |
| Breast overall | 11 | 15 748 | 18 084 | Illumina 317K//370K/550K/660K/1.2M | 1000 Genome | Age, PCs (vary by studies) | (23,30) |
| ER-negative | 8 | 4939 | 13 128 | Illumina 317K/370K/550K/660K/1.2M | 1000 Genome | Age, PCs (vary by studies) | (29) |
| Prostate overall | 6 | 14 160 | 12 724 | Illumina and Affymetrix arrays | 1000 Genome | Age, study | |
| Aggressive | 6 | 4939 | 12 724 | Illumina and Affymetrix arrays | 1000 Genome | Age, study, PCs | |
| Total* | 48 | 64 591 | 74 467 |
* Number of unique individuals after accounting for tumor subtypes and overlapping control patients. CORECT = ColoRectal Transdisciplinary Study; ER = estrogen receptor; GECCO = Genetics and Epidemiology of Colorectal Cancer Consortium; PCs = principal components representing residual European ancestry.
Gene and Variants Selection, Pathway Assignment
To identify relevant genes of interest we conducted keyword searches in pathway databases such as Gene Ontology (GO, including biological process, molecular function, and cellular component), Kyoto Encyclopedia of Genes and Genomes (KEGG), and The Pharmacogenomics Knowledge Base (PharmaGKB), as well as literature searches using keywords related to inflammation, immune response, and cytokine. In addition, investigators from the participating studies could nominate genetic variants in the inflammation pathways based on preliminary results shown in their own cancer-specific study with P value of less than .01 and a minimum sample size of 500 case-control pairs. A total of 921 genes were identified through the keyword searches and nomination. This list was then merged with the Illumina 550K BeadChip annotation database, resulting in a list of 12 370 genomic variants that are within 10kb of a gene coding region and present on the Illumina 550K BeadChip for the subsequent statistical analysis. These 12 370 variants were categorized into 53 subcomponents related to the inflammation pathway based on the Gene Ontology headings and KEGG keywords (Supplementary Table 2, available online). Seventy-eight of the 921 genes that could not be assigned automatically through Gene Ontology and KEGG were then assigned to the most suitable category based on their biological function and literature curation. In principle, there are three levels of the pathways for illustrative purposes. Note that genes can belong to multiple subpathway components depending on their biological functions. In addition, a pathway component in a higher level can contain more genes than those included in its substructure, and the sublevel pathway components are not exclusively restricted to the genes in their Level 1 pathway. For example, IL6 is part of humoral immune response, immune system development, and regulation of immune response. These relationships are specified through the second stage covariate matrix in the hierarchical modeling. The distribution of the variants and genes of 12 370 markers in subpathway components are shown in Supplementary Table 2 (available online).
Quality Control Criteria of the Genomic Data
The main quality control (QC) criteria for each cancer consortium are summarized in Supplementary Table 3 (available online). There are small variations across the cancer-specific consortium regarding the QC criteria, but in general, all cancer-specific analysis has excluded subjects with sex discrepancy, high missing rate, non-European ancestry, unexpected duplicates or relatedness, and excessive global heterozygosity. The variants with low call rates, low minor allele frequencies, and extreme departure from Hardy-Weinberg Equilibrium were excluded. The cutoffs used in each cancer-specific analysis are summarized in Supplementary Table 3 (available online).
Imputation
All imputation was conducted based on the 1000 Genome March 2012 reference panel using either MACH or IMPUTE (31–33), with the exception of the lung cancer studies that were imputed to the HapMap2 reference panel. The difference in the imputation reference panel was not expected to have any meaningful effect on the results, as the variants included in our analysis are present on Illumina 550K BeadChips. Therefore, they were either available as the directly genotyped data or can be reasonably captured by either 1000Genome or HapMap2 reference panels.
Statistical Analysis
The association between genetic variants and cancer risk was estimated with odds ratios (ORs) and 95% confidence intervals (CIs) based on unconditional logistic regression. All effect estimates from each study and pooled estimates were based on log-additive models and represent per-allele odds ratios adjusted for age, principal components, and sex, if applicable. The study-specific results were first combined within each cancer site by a fixed effects model. The methodology and the results of the cancer-specific results have been described previously (20,23,26–30). The cancer-specific results were then combined using the association analysis based on subsets (ASSET) meta-analytic approach, which allows for disease heterogeneity and potential opposite directions of the same genetic variant on different cancer sites (34). It searches for the most parsimonious grouping based on the test statistics and the outcome variable can be any of the five cancers, not a single specific tumor type. In addition to overall cancer risk, we have also included major subtypes of each tumor site, by lung cancer histology (adenocarcinoma and squamous cell carcinoma) and ovary cancer histology (serous and endometrioid cancers), aggressiveness for prostate cancer, and estrogen receptor (ER) status for breast cancer. The overlapping subjects amongst cancer subtypes (eg, overlapping controls for lung adenocarcinoma and squamous cell carcinoma) and across cancer types (eg, UK ovary and UK breast GWAS both used control patients from Wellcome Trust Case Control Consortium, WTCCC) were accounted for in the covariance matrix when estimating the standard errors. All statistical tests were two-sided.
LD Pruning
To avoid over-representation of pathways with high linkage disequilibrium (LD), we pruned out those variants that were in LD (R2 > 0.7) before conducting the hierarchical modeling analysis. The variants with stronger statistical significances in the LD pairs were retained. The LD pruning was done based on SNAP (Broad Institute) (35). After LD pruning at R-square threshold of 0.7, 5066 markers remained for the pathway analysis.
Hierarchical Modeling (HM)
One of the major strengths of the HM approach is that prior knowledge of biological function and genomic properties can be incorporated into effect estimation for the genetic variants of interest. This information is incorporated via a second-stage covariate matrix, which was developed with gene-specific columns to represent the pathway membership of specific genes.
Gene-specific columns were created based on the function of the genes using the biological process subontologies within the Gene Ontology (Supplementary Table 2, available online). Variables were created for each subcomponent related to the inflammation pathway. The HM model was based on the methodology described by Chen and Witte and others (4,18). It provides for a single distribution of effects of the variants and uses the second-stage covariate matrix to further emphasize those believed more strongly a priori to be causal. Complete statistical descriptions of the model have been published previously (18).
For variants with effects in opposite directions for different cancer types (such as TERT [36,37]), the magnitude of the associations was estimated using the average of the absolute values of all regression coefficients. The standard errors were estimated based on folded normal distribution (38); the overlapping subjects were accounted for in the covariance using the equation described by Lin et al. (39), and the hierarchical modeling was conducted using R software.
Network Map
A Network Map was produced to visualize how each subpathway component is related to every other based on the size of the subcomponent (nodes), the overlapping coefficient (edges), and statistical significance of the subcomponents (40). The overlapping coefficient is estimated based on the number of overlapping genes between subpathways divided by the minimum of the respective sizes of each subpathway. The subpathway components that are more closely related would then be plotted in vicinity with edges connected. The Network map is produced by Cytoscape network visualization software (40).
Results
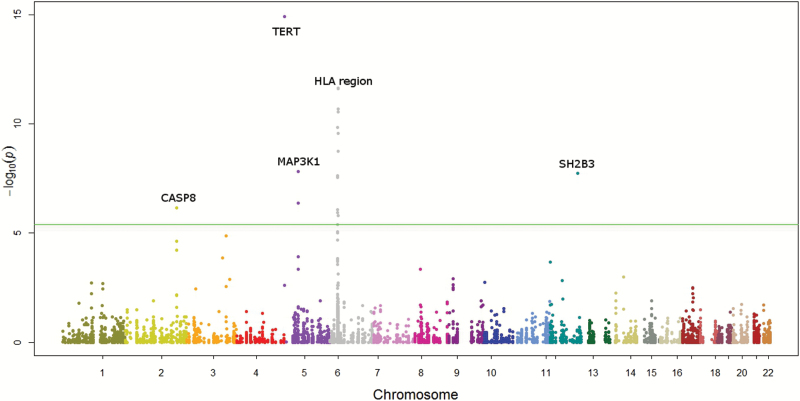
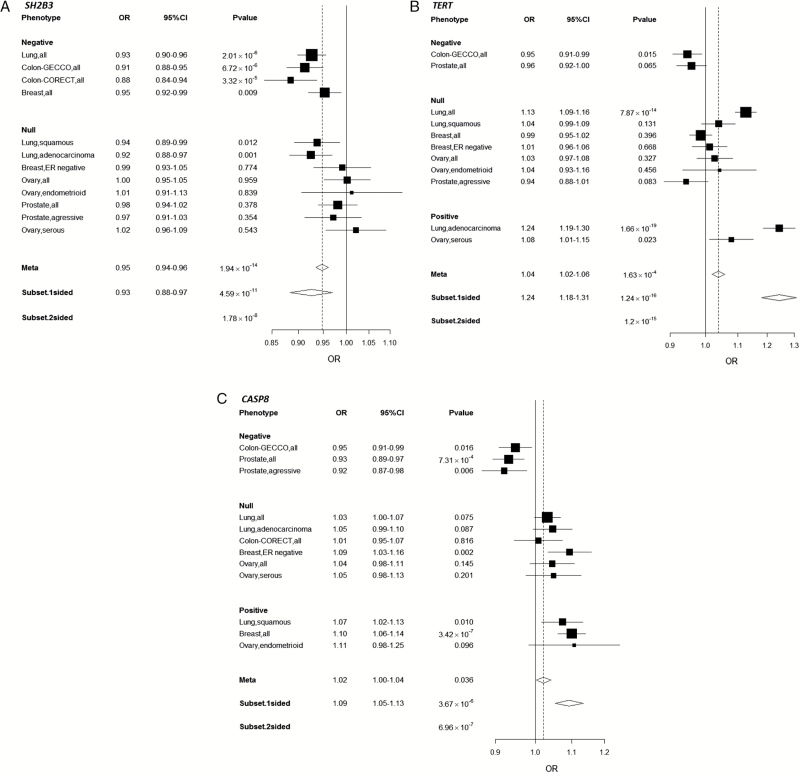
Figure 1 shows the associations between the genetic variants and cancer risk based on the ASSET single marker analysis in Manhattan plot. Sixteen variants representing five independent regions reached pathway-level significance (P < 4x10-6): Four of them were accounted for by previously known cancer loci: CASP8, MAP3K1, TERT, HLA-BAT3 region (29,36,41–53). The results for the top 16 variants are shown in Supplementary Table 4 (available online). Notably, one variant (rs3184504 in SH2B3) that was not previously known to be associated with cancer risk at chromosome 12q24 reached a GWAS significance level P value of 1.78 x 10–8, which accounted for the subset searches by ASSET. This variant was associated with risk of lung cancer (OR = 0.93, 95% CI = 0.90 to 0.96, P = 2.01 x 10–6), colorectal cancer (GECCO OR = 0.91, 95% CI = 0.88 to 0.95, P = 6.72x10-6; CORECT OR = 0.88, 95% CI = 0.84 to 0.94, P = 3.32x10-5), and breast cancer (OR = 0.95, 95% CI = 0.92 to 0.99, P = .009) (Figure 2A). It is not associated with prostate or ovarian cancer with odds ratios of 0.98 (95% CI = 0.94 to 1.02) and 1.00 (95% CI = 0.95 to 1.05), respectively.
Figure 1.
Manhattan plot for the associations between 12 370 variants and cancer risk based on two-sided association analysis based on subsets (ASSET) analysis. The green line denotes the pathway-level significance threshold at a P value of 4x10-6.
Figure 2.
Odds ratio and 95% confidence interval for the following variants: (A) rs3184504 located in gene SH2B3 at Ch12q24 (the reference allele C; the effect allele T); (B) rs2736100 located in gene TERT at 5p15 (the reference allele A; the effect allele C); (C) rs10931936 located in gene CASP8 at 2q33 (the reference allele C, the effect allele T). CI = confidence interval; CORECT = ColoRectal Transdisciplinary Study; GECCO = Genetics and Epidemiology of Colorectal Cancer Consortium; ER = estrogen receptor; OR = odds ratio;
In addition to SH2B3, the previously known cancer region TERT (as represented by rs2736100) demonstrated pleiotropic effects on lung adenocarcinoma (P = 1.66x 10–19), colorectal cancer (P = .015), and ovarian cancer serous subtype (P = .023), and CASP8 (represented by rs10931936) also had pleiotropic effect on breast cancer (P = 3.42x10-7), prostate cancer (P = 7.31x10-4), and lung squamous cell carcinoma (P = .01) (Figure 2, B and C).
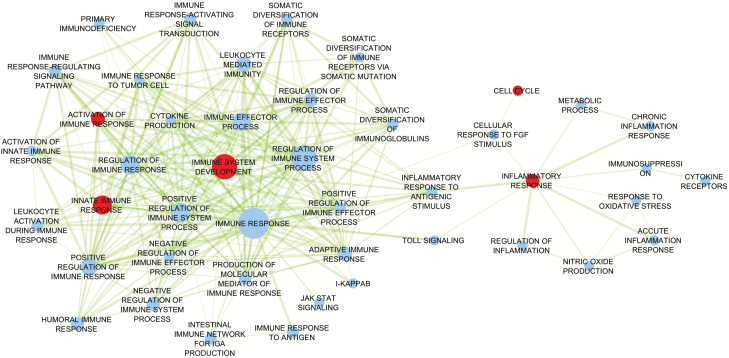
The hierarchical model suggested five subpathway components related to inflammation having a more prominent role in carcinogenesis: immune system development (P = .025), activation of immune response (P = .028), innate immune response (P = .030), inflammatory response (P = .008), and cell cycle genes that are relevant for inflammation (P = .0001) (Table 2). To assess whether these associations were driven by a single cancer site or were common across cancer sites, we also estimated the pathway effect by cancer sites, and the results are reported in Table 2. The genes related to innate immune response were associated with colorectal (P = .022) and ovarian cancer (P = .003), while the rest of the statistically significant pathways were mainly associated with one cancer site: The variants in the genes related to immune system development were shown to be associated with breast cancer (P = .001); the variants in cell cycle genes were only associated with lung cancer (P = 1.35x10-6); the genetic variants related to activation of immune response were only associated with ovarian cancer (P = .009), and the variants in inflammatory response were only associated with colorectal cancer (P = .006). In general, the subcomponents of the inflammation pathway are intricately related. The relation between the subcomponents is presented in the Network Map (Figure 3).
Table 2.
Number of genes and variants and significance level of the five significant inflammation-related subpathways*
| Cancer type | Inflammation- related cell cycle | Inflammatory response | Immune system development | Activation of immune response | Innate immune response |
|---|---|---|---|---|---|
| No. genes (no. variants) | 14 (37) | 83 (975) | 324 (4910) | 90 (1306) | 197 (2411) |
| No. genes (No. variants) - After LD pruning at R2 of 0.7 | 11 (22) | 66 (410) | 302 (1979) | 85 (528) | 173 (925) |
| Overall P | 1.23x10-4 | .008 | .025 | .028 | .030 |
| Lung cancer P | 1.35x10-6 | .952 | .239 | .068 | .054 |
| Colorectal cancer P | .333 | .006 | .518 | .973 | .022 |
| Ovary cancer P | .121 | .594 | .265 | .009 | .003 |
| Breast cancer P | .902 | .058 | .001 | .114 | .302 |
| Prostate cancer P | .810 | .941 | .566 | .587 | .194 |
* The pathway significance levels were estimated based on the hierarchical modeling based on the method previously described in Chen et al. (18). LD = linkage disequilibrium.
Figure 3.
Network map for inflammation pathways. Each node is a subcomponent, and the size of the node represents the number of the genes in that subcomponent. The edges are drawn if the overlapping coefficient between the two nodes is greater than 0.3. The thickness of the edges represents the degree of similarity between the two subpathway components. The red node represents the subpathway component with statistical significance level of less than .05 based on the hierarchical modeling.
Discussion
Based on extensive genomic data from five common cancer sites, the current analysis identified three loci in the inflammatory pathways associated with multiple cancers, including one novel pleiotropic locus at chromosome 12q24 associated with cancer of the lung, colorectum, and breast. The identification of three pleiotropic loci in the inflammation pathway is more than expected by chance under null hypothesis with pathway-level significance level, and it provides encouraging evidence that common genetic mechanisms may underlie multiple cancers. The pathway analysis after accounting for correlated variants indicated that genes related to inflammatory response, immune system development, activation of immune response, innate immune response, and cell cycle genes related to inflammation have effects across different cancers.
The locus at chromosome 12q24 represented by rs3184504 is mapped to a gene encoding SH2B adapter protein 3 (SH2B3), a key negative regulator of cytokine signaling (54,55). This missense variant results in an amino acid change from tryptophan to arginine at codon 262. This locus has been previously shown to be associated with several immunological characteristics (such as platelet counts [56,57), eosinophil counts [58], red blood cell counts [59]) and risk of chronic diseases such as rheumatoid arthritis (60), Type 1 diabetes (61), and coronary heart diseases (62). It was recently implicated in colorectal cancer risk (63). Our analysis showed a strong association between this locus and risk of lung, colorectal, and breast cancers, clearly demonstrating the pleiotropic effect of this new locus.
Pathway effects estimated by hierarchical modeling helped to identify the inflammation subcomponents most relevant to cancer, specifically inflammatory response, immune system development, activation of immune response, innate immune response, and cell cycle genes related to inflammation. Our results suggest that although the genetic variations of inflammation pathway are important for most of the tumor types, the driving subcomponents within inflammation are different by cancer type with some degree of commonality across tumors. Each cancer site investigated is associated with multiple subcomponents of the inflammation pathway, except for prostate cancer. Overall, genetic variation in innate immune response contributes to both colorectal and ovarian cancer, while genetic variants related to inflammatory response are more relevant to colorectal cancer and those related to activation of immune response are particularly important for ovarian cancer. Genetic variants related to immune system development are mainly associated with breast cancer, and those related to cell cycle control genes are specifically related to lung cancer. Nevertheless, genetic variants related to inflammation pathways did not seem to have an association with prostate cancer risk overall.
The role of inflammatory response and colorectal cancer is an active research area, particularly because of its strong association with inflammatory bowel disease and the use of nonsteroidal anti-inflammatory drugs (64). In addition, the intestinal microflora are also important in maintaining the homeostatic immune function and regulation of inflammatory response (8,65). Previous studies have shown that genetic variants and biomarkers (such as C-reactive protein, Interleukins, Serum Amyloid A) related to inflammatory response are associated with colorectal cancer risk (2,64). Our results are consistent with these previous observations and suggest that the subcomponent related to inflammatory response is particularly important for colorectal cancer development (3).
Innate immune response is a cell defense system (eg, neutrophils and macrophages) that does not involve recognition of a specific antigen, as opposed to the adaptive immune response (eg, B-cells and T-cells), which is a specific response to an antigen (66,67). The innate immune cells are involved in tissue remodeling and repair, and the genes that are involved in this process include complement components (Cs), collectins, clusterins, killer cell lectins, mitogen-activated protein kinase (eg, MAP3K1), macrophage receptors, and toll-like receptors (TLRs), among others. Both innate and adaptive immune systems are crucial for the immune response to tumor cells, but it has been suggested that an environment with abundant innate immune cells as a result of chronic inflammation can in turn promote angiogenesis and cell proliferation and lead to cancer progression (66–68). In addition, previous animal studies have shown that innate immune response to intestinal bacteria is sufficient to promote colorectal carcinoma in mice (65,69–71). This is consistent with our finding that genetic variants in the innate immune response appear to be associated with increased cancer risk, in particular for colorectal and ovarian cancer, while the genetic variants in adaptive immune response did not show an association as a whole (P = .67).
In addition to those related to innate immune response, our results indicated that genes related to the activation of the immune response also contribute to development of ovarian cancer. There is compelling evidence that factors related to immune response can alter the pathogenesis of ovarian cancers as well as the initiation of ovarian cancer through genetic and protein analysis (72–74). Our results are compatible with the previous reports that demonstrated the role of immune response in the initiation of ovarian cancer through genetic and protein analysis.
The immune system plays an instrumental role in maintaining tissue homeostasis, cell regeneration, and prevention of infection and cell transformation. The development of the immune system is a fundamental process that occurs early in life but has profound effects on the efficiency of an individual’s immune response later on. The majority of the immune cells are derived from hematopoietic stem cells and then differentiate into different cell lineages based on cell interactions and cytokines. The main genes that are involved in this developmental process are interleukins (ILs), colony-stimulating factors (CSFs), genes related to Cluster of Differentiation (CDs), and SH2B3, among others. Our results indicated that those events that occur early in life are particularly important for breast cancer. Although there is a wealth of literature on immune response and the breast cancer prognosis (75,76), to our knowledge this is the first study to indicate a role of immune system development in the initiation of breast cancer.
Our results based on hierarchical modeling suggest that cell cycle genes that are related to inflammation response form a biologically important subcomponent for lung cancer. Specifically, TERT and BAG6/BAT3, both known cancer susceptibility genes, are in this category: BAG6 (BAG Family Molecular Chaperone Regulator 6, previously known as BAT3) was first characterized as part of the human major histocompatibility complex class III region and was also shown to be involved in DNA damage–induced apoptosis (77,78); telomeres, regulated in part by TERT, are a center piece for anti-apoptosis, and cellular clock and telomere dysfunction was shown to be involved in chronic inflammation in various different health conditions, including chronic obstructive pulmonary diseases (79–88). The results of our analysis are in line with the evidence established in previous studies and highlight the power of large sample sizes.
We applied hierarchical modeling to detect subpathway effects, as it has the advantages of not solely relying on the most significant variant in the pathway; instead, it models the effect of all variants that belong to the same subcomponent through the second-stage prior matrix. One limitation of this approach is the possible violation of the exchangeability assumption. For example, some variants in the same subcomponent can have a larger effect and some can have a very modest effect. When the exchangeability assumption is violated, the effect estimates of truly causal variants may be shrunk toward the wrong prior mean. In most cases, this would be brought toward the null, underestimating the effect estimates. Nevertheless, previous simulation studies have demonstrated that hierarchical modeling is relatively robust to the alteration of the priors, provided that the priors specified are reasonable (4,89,90). In our analysis, we used the absolute value of the regression coefficients from the first stage because the main research emphasis here is the size of the effect rather than its direction, which could vary from one variant to another and from one cancer site to another. Using the absolute value allows us to model the magnitude of the effect without concerns of heterogeneity of the directions across cancer sites and variants.
This study has notable strengths, including the large sample size and information derived from 48 genome-wide association studies across five cancer sites with a total of 139 058 individuals. It is the first large-scale genomic analysis for inflammation pathways across major cancer types. We were not able to conduct another independent study with five cancer sites and an equal sample size for replication, given the uniqueness of this dataset. However, our results provided robust estimation based on ASSET and hierarchical modeling; both approaches aim to reduce the potential of false-positive results through multiple-testing penalty or estimation shrinkage.
In summary, we have identified novel regions with pleiotropic effects in the inflammation pathways and identified several key subcomponents within the inflammation pathway that are important for lung, colorectal, breast, and ovarian cancers. These results provide further insight into the etiology of these cancers and identify the differences and commonality related to the etiological role of inflammation across tumor types.
Funding
TRICL (Transdisciplinary Research for Cancer of Lung) and International Lung Cancer Consortium (ILCCO): National Institute of Health U19 CA148127-01 (PI: Amos), Canadian Cancer Society Research Institute (no. 020214, PI: Hung).
DRIVE (Discovery, Biology, and Risk of Inherited Variants in Breast Cancer): National Institute of Health U19 CA148065.
CORECT (ColoRectal Transdisciplinary Study): National Institute of Health U19 CA148107; R01 CA81488, P30 CA014089.
ELLIPSE (ELLIPSE, Elucidating Loci in Prostate Cancer Susceptibility): This work was support by the GAME-ON U19 initiative for prostate cancer (ELLIPSE), U19 CA148537.
FOCI (Transdisciplinary Cancer Genetic Association and Interacting Studies): National Institutes of Health U19 CA148112-01 (PI: Sellers), R01-CA122443, P50-CA136393, P30-CA15083 (PI: Goode), Cancer Research UK (C490/A8339, C490/A16561, C490/A10119, C490/A10124 [PI: Pharoah]).
GECCO (Genetics and Epidemiology of Colorectal Cancer Consortium): National Cancer Institute, National Institutes of Health, US Department of Health and Human Services (U01 CA137088; R01 CA059045). ASTERISK: a Hospital Clinical Research Program (PHRC) and supported by the Regional Council of Pays de la Loire, the Groupement des Entreprises Françaises dans la Lutte contre le Cancer (GEFLUC), the Association Anne de Bretagne Génétique and the Ligue Régionale Contre le Cancer (LRCC). DACHS: German Research Council (Deutsche Forschungsgemeinschaft, BR 1704/6-1, BR 1704/6-3, BR 1704/6-4, and CH 117/1-1), and the German Federal Ministry of Education and Research (01KH0404 and 01ER0814). DALS: National Institutes of Health (R01 CA48998 to MLS); HPFS is supported by the National Institutes of Health (P01 CA 055075, UM1 CA167552, R01 137178, R01 CA 151993, and P50 CA 127003), NHS by the National Institutes of Health (R01 CA137178, P01 CA 087969, R01 CA151993, and P50 CA 127003), and PHS by the National Institutes of Health (R01 CA042182). OFCCR: National Institutes of Health, through funding allocated to the Ontario Registry for Studies of Familial Colorectal Cancer (U01 CA074783); see CFR section. Additional funding toward genetic analyses of OFCCR includes the Ontario Research Fund, the Canadian Institutes of Health Research, and the Ontario Institute for Cancer Research, through generous support from the Ontario Ministry of Research and Innovation. PLCO: Intramural Research Program of the Division of Cancer Epidemiology and Genetics and supported by contracts from the Division of Cancer Prevention, National Cancer Institute, NIH, DHHS. Additionally, a subset of control samples were genotyped as part of the Cancer Genetic Markers of Susceptibility (CGEMS) Prostate Cancer GWAS (Yeager M, et al. Nat Genet. 2007;39(5):645–649), Colon CGEMS pancreatic cancer scan (PanScan) (Amundadottir L, et al. Nat Genet. 2009;41(9):986–990 and Petersen GM, et al. Nat Genet. 2010;42(3):224–228), and the Lung Cancer and Smoking study. The prostate and PanScan study datasets were accessed with appropriate approval through the dbGaP online resource (http://cgems.cancer.gov/data/) accession numbers phs000207.v1.p1 and phs000206.v3.p2, respectively, and the lung datasets were accessed from the dbGaP website (http://www.ncbi.nlm.nih.gov/gap) through accession number phs000093.v2.p2. Funding for the Lung Cancer and Smoking study was provided by National Institutes of Health (NIH), Genes, Environment, and Health Initiative (GEI) Z01 CP 010200, NIH U01 HG004446, and NIH GEI U01 HG 004438. For the lung study, the GENEVA Coordinating Center provided assistance with genotype cleaning and general study coordination, and the Johns Hopkins University Center for Inherited Disease Research conducted genotyping. PMH: National Institutes of Health (R01 CA076366 to PA Newcomb). VITAL: National Institutes of Health (K05 CA154337). WHI: The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, US Department of Health and Human Services through contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C, and HHSN271201100004C.
Supplementary Material
GECCO: The authors would like to thank all those at the GECCO Coordinating Center for helping bring together the data and people that made this project possible. The authors acknowledge Dave Duggan and team members at TGEN (Translational Genomics Research Institute), the Broad Institute, and the Génome Québec Innovation Center for genotyping DNA samples of case patients and control patients, and for scientific input for GECCO. ASTERISK: We are very grateful to Dr. Bruno Buecher, without whom this project would not have existed. We also thank all those who agreed to participate in this study, including the patients and the healthy control persons, as well as all the physicians, technicians, and students. DACHS: We thank all participants and cooperating clinicians and Ute Handte-Daub, Renate Hettler-Jensen, Utz Benscheid, Muhabbet Celik, and Ursula Eilber for excellent technical assistance. HPFS, NHS, and PHS: We would like to acknowledge Patrice Soule and Hardeep Ranu of the Dana Farber Harvard Cancer Center High-Throughput Polymorphism Core who assisted in the genotyping for NHS, HPFS, and PHS under the supervision of Dr. Immaculata Devivo and Dr. David Hunter, Qin (Carolyn) Guo and Lixue Zhu who assisted in programming for NHS and HPFS, and Haiyan Zhang who assisted in programming for the PHS. We would like to thank the participants and staff of the Nurses’ Health Study and the Health Professionals Follow-Up Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. In addition, this study was approved by the Connecticut Department of Public Health (DPH) Human Investigations Committee. Certain data used in this publication were obtained from the DPH. The authors assume full responsibility for analyses and interpretation of these data. PLCO: The authors thank Drs. Christine Berg and Philip Prorok, Division of Cancer Prevention, National Cancer Institute, the Screening Center investigators and staff or the Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial, Mr. Tom Riley and staff, Information Management Services, Inc., Ms. Barbara O’Brien and staff, Westat, Inc., and Drs. Bill Kopp, Wen Shao, and staff, SAIC-Frederick. Most importantly, we acknowledge the study participants for their contributions to making this study possible. The statements contained herein are solely those of the authors and do not represent or imply concurrence or endorsement by NCI. PMH: The authors would like to thank the study participants and staff of the Hormones and Colon Cancer study. WHI: The authors thank the WHI investigators and staff for their dedication, and the study participants for making the program possible. A full listing of WHI investigators can be found at: http://www.whi.org/researchers/Documents%20%20Write%20a%20Paper/WHI%20Investigator%20Short%20List.pdf.
DRIVE: The DRIVE GAME-ON consortium (http://epi.grants.cancer.gov/gameon/) would like to thank the following key investigators (institution and location): Muriel Adank (VU University Medical Center, Amsterdam, the Netherlands), Habibul Ahsan (University of Chicago, Chicago, IL), Irene Andrulis (Mount Sinai Hospital, Toronto, Canada), Kristiina Aittomäki (University of Helsinki and Helsinki University Central Hospital, Helsinki, Finland), Lars Beckman (Institute for Quality and Efficieny in Health Care, Cologne, Germany), Carl Blomquist (University of Helsinki and Helsinki University Central Hospital, Helsinki, Finland), Federico Canzian (German Cancer Research Center, Heidelberg, Germany), Jenny Chang-Claude (Deutsches Krebsforschungszentrum [DKFZ], Heidelberg, Germany), Laura Crisponi (Istituto di Ricerca Genetica e Biomedica, Consiglio Nazionale delle Ricerche, Cagliari, Italy), Kamila Czene (Karolinska Institut, Stockholm, Sweden), Norbert Dahmen (University of Mainz, Mainz, Germany), Isabel dos Santos Silva (London School of Hygiene and Tropical Medicine, London, UK), Olivia Fletcher (The Institute of Cancer Research, London, UK), Lorna Gibson (London School of Hygiene and Tropical Medicine, London, UK), Per Hall (Karolinska Institut, Stockholm, Sweden), HEBON (Hereditary Breast and Ovarian Cancer Research Group Netherlands), Rebecca Hein (Deutsches Krebsforschungszentrum, Heidelberg, Germany, and University of Cologne, Cologne, Germany), Albert Hofman (Erasmus Medical Center, Rotterdam, the Netherlands), John L. Hopper (Melbourne School of Population Health, University of Melbourne, Melbourne, Victoria, Australia), Astrid Irwanto (Genome Institute of Singapore, Singapore), Rudolf Kaaks (German Cancer Research Center, Heidelberg, Germany), Muhammad G. Kibriya (University of Chicago, Chicago, IL), Peter Lichtner (German Research Center for Environmental Health, Neuherberg, Germany), Jianjun Liu (Genome Institute of Singapore, Singapore), Enes Makalic (Melbourne School of Population Health, University of Melbourne, Melbourne, Victoria, Australia), Alfons Meindl (Technische Universität München, Munich, Germany), Hanne Meijers-Heijboer (VU University Medical Center, Amsterdam, the Netherlands), Bertram Müller-Myhsok (Max Planck Institute of Psychiatry, Munich, Germany), Taru A. Muranen (University of Helsinki and Helsinki University Central Hospital, Helsinki, Finland), Heli Nevanlinna (Univesity of Helsinki and Helsinki University Central Hospital, Helsinki, Finland), Julian Peto (London School of Hygiene and Tropical Medicine, London, UK), Ross L. Prentice (Fred Hutchinson Cancer Research Center, Seattle, WA), Nazneen Rahman (Institute of Cancer Research, Sutton, UK), Daniel F. Schmidt (Melbourne School of Population Health, University of Melbourne, Melbourne, Victoria, Australia), Rita K. Schmutzler (University of Cologne, Cologne, Germany), Melissa C. Southey (The University of Melbourne, Melbourne, Victoria, Australia), Clare Turnbull (Institute of Cancer Research, Sutton, UK), Andre G. Uitterlinden (Erasmus Medical Center, Rotterdam, the Netherlands), Rob B. van der Luijt (University Medical Center Utrecht, Utrecht, the Netherlands), Quinten Waisfisz (VU University Medical Center, Amsterdam, the Netherlands), Alice S. Whittemore (Stanford University, Stanford, CA), and Wei Zheng (Vanderbilt University, Nashville, TN).
FOCI: FOCI consortium would like to thank the following investigators for their contribution: Mike Birrer, Ann Chen, Julie Cunningham, Ed Iversen, John McLaughlin, Steven Narod, Harvey Risch, Jenny Permuth-Wey, Paul Pharoah, Simon Gayther, and Susan Ramus. UK ovarian cancer GWAS: We thank all the individuals who took part in this study. We thank all the researchers, clinicians, and administrative staffs who have enabled the many studies contributing to this work. This study made use of data generated by the Wellcome Trust Case Control consortium with its project funding provided by the Wellcome Trust under award 076113. We thank the support of the UK National Institute for Health Research Biomedical Research Centres at the University of Cambridge and University College Hospital.
References
- 1. Elinav E, Nowarski R, Thaiss CA, et al. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer. 2013;13 (11):759–771. [DOI] [PubMed] [Google Scholar]
- 2. Brenner DR, Scherer D, Muir K, et al. A review of the application of inflammatory biomarkers in epidemiologic cancer research. Cancer Epidemiol Biomarkers Prev. 2014;23 (9):1729–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mantovani A, Allavena P, Sica A, et al. Cancer-related inflammation. Nature. 2008;454 (7203):436–444. [DOI] [PubMed] [Google Scholar]
- 4. Brenner DR, Brennan P, Boffetta P, et al. Hierarchical modeling identifies novel lung cancer susceptibility variants in inflammation pathways among 10,140 cases and 11,012 controls. Hum Genet. 2013;132 (5):579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pu X, Hildebrandt MA, Lu C, et al. Inflammation-related Genetic Variations and Survival for Advanced Non-Small Cell Lung Cancer Receiving First-line Chemotherapy. Clin Pharmacol Ther. 2014;96 (3):360–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Spitz MR, Gorlov IP, Amos CI, et al. Variants in inflammation genes are implicated in risk of lung cancer in never smokers exposed to second-hand smoke. Cancer Discov. 2011;1 (5):420–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Spitz MR, Gorlov IP, Dong Q, et al. Multistage analysis of variants in the inflammation pathway and lung cancer risk in smokers. Cancer Epidemiol Biomarkers Prev. 2012;21 (7):1213–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang H, Taverna D, Stram DO, et al. Genetic variation in the inflammation and innate immunity pathways and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2013;22 (11):2094–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. White KL, Schildkraut JM, Palmieri RT, et al. Ovarian cancer risk associated with inherited inflammation-related variants. Cancer Res. 2012;72 (5):1064–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Amirian ES, Scheurer ME, Liu Y, et al. A novel approach to exploring potential interactions among single-nucleotide polymorphisms of inflammation genes in gliomagenesis: an exploratory case-only study. Cancer Epidemiol Biomarkers Prev. 2011;20 (8):1683–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weinstein JN, Collisson EA, Mills GB, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45 (10):1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505 (7484):495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502 (7471):333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheng I, Kocarnik JM, Dumitrescu L, et al. Pleiotropic effects of genetic risk variants for other cancers on colorectal cancer risk: PAGE, GECCO and CCFR consortia. Gut. 2014;63 (5):800–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fehringer G, Liu G, Briollais L, et al. Comparison of pathway analysis approaches using lung cancer GWAS data sets. PLoS One. 2012;7 (2):e31816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Holden M, Deng S, Wojnowski L, et al. GSEA-SNP: applying gene set enrichment analysis to SNP data from genome-wide association studies. Bioinformatics. 2008;24 (23):2784–2785. [DOI] [PubMed] [Google Scholar]
- 17. Damian D, Gorfine M. Statistical concerns about the GSEA procedure. Nat Genet. 2004;36 (7):663; author reply 663. [DOI] [PubMed] [Google Scholar]
- 18. Chen GK, Witte JS. Enriching the analysis of genomewide association studies with hierarchical modeling. Am J Hum Genet. 2007;81 (2):397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lewinger JP, Conti DV, Baurley JW, et al. Hierarchical Bayes prioritization of marker associations from a genome-wide association scan for further investigation. Genet Epidemiol 2007;31 (8):871–882. [DOI] [PubMed] [Google Scholar]
- 20. Timofeeva MN, Hung RJ, Rafnar T, et al. Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Hum Mol Genet. 2012;21 (22):4980–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goode EL, Chenevix-Trench G, Song H, et al. A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat Genet. 2010;42 (10):874–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Song H, Ramus SJ, Tyrer J, et al. A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat Genet. 2009;41 (9):996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Siddiq A, Couch FJ, Chen GK, et al. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum Mol Genet. 2012;21 (24):5373–5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Amin Al Olama A, Kote-Jarai Z, Schumacher FR, et al. A meta-analysis of genome-wide association studies to identify prostate cancer susceptibility loci associated with aggressive and non-aggressive disease. Hum Mol Genet. 2013;22 (2):408–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peters U, Hutter CM, Hsu L, et al. Meta-analysis of new genome-wide association studies of colorectal cancer risk. Hum Genet. 2012;131 (2):217–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peters U, Jiao S, Schumacher FR, et al. Identification of Genetic Susceptibility Loci for Colorectal Tumors in a Genome-Wide Meta-analysis. Gastroenterology. 2013;144 (4):799–807 e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pharoah PD, Tsai YY, Ramus SJ, et al. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat Genet. 2013;45 (4):362–370, 370e1–370e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang H, Burnett T, Kono S, et al. Trans-ethnic genome-wide association study of colorectal cancer identifies a new susceptibility locus in VTI1A. Nat Comm. 2014;5:4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Garcia-Closas M, Couch FJ, Lindstrom S, et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat Genet. 2013;45 (4):392–398, 398e1–398e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Michailidou K, Hall P, Gonzalez-Neira A, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45 (4):353–361, 361e1–361e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Howie B, Fuchsberger C, Stephens M, et al. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44 (8):955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marchini J, Howie B, Myers S, et al. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39 (7):906–913. [DOI] [PubMed] [Google Scholar]
- 33. Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 Bethesda. 2011;1 (6):457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bhattacharjee S, Rajaraman P, Jacobs KB, et al. A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits. Am J Hum Genet. 2012;90 (5):821–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Johnson AD, Handsaker RE, Pulit SL, et al. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24 (24):2938–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rafnar T, Sulem P, Stacey SN, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet. 2009;41 (2):221–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mocellin S, Verdi D, Pooley KA, et al. Telomerase reverse transcriptase locus polymorphisms and cancer risk: a field synopsis and meta-analysis. J Natl Cancer Inst. 2012;104 (11):840–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Elandt RC. The folded normal distribution. Technometrics. 1961;3 (4):11. [Google Scholar]
- 39. Lin DY, Sullivan PF. Meta-analysis of genome-wide association studies with overlapping subjects. Am J Hum Genet. 22009;85 (6):862–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Merico D, Isserlin R, Stueker O, et al. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5 (11):e13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McKay JD, Hung RJ, Gaborieau V, et al. Lung cancer susceptibility locus at 5p15.33. Nat Genet. 2008;40 (12):1404–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Truong T, Hung RJ, Amos CI, et al. Replication of lung cancer susceptibility loci at chromosomes 15q25, 5p15, and 6p21: a pooled analysis from the International Lung Cancer Consortium. J Natl Cancer Inst. 2010;102 (13):959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Haiman CA, Chen GK, Vachon CM, et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor-negative breast cancer. Nat Genet. 2011;43 (12):1210–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kote-Jarai Z, Olama AA, Giles GG, et al. Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat Genet. 2011;43 (8):785–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rothman N, Garcia-Closas M, Chatterjee N, et al. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat Genet. 2010;42 (11):978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Petersen GM, Amundadottir L, Fuchs CS, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet. 2010;42 (3):224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cox A, Dunning AM, Garcia-Closas M, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39 (3):352–358. [DOI] [PubMed] [Google Scholar]
- 48. Shephard ND, Abo R, Rigas SH, et al. A breast cancer risk haplotype in the caspase-8 gene. Cancer Res. 2009;69 (7):2724–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447 (7148):1087–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thomas G, Jacobs KB, Kraft P, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1). Nat Genet. 2009;41 (5):579–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Turnbull C, Ahmed S, Morrison J, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42 (6):504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang Y, Broderick P, Webb E, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet. 2008;40 (12):1407–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Landi MT, Chatterjee N, Yu K, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 22009;85 (5):679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Devalliere J, Charreau B. The adaptor Lnk (SH2B3): an emerging regulator in vascular cells and a link between immune and inflammatory signaling. Biochem Pharmacol. 2011;82 (10):1391–1402. [DOI] [PubMed] [Google Scholar]
- 55. Roberts KG, Morin RD, Zhang J, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22 (2):153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shameer K, Denny JC, Ding K, et al. A genome- and phenome-wide association study to identify genetic variants influencing platelet count and volume and their pleiotropic effects. Hum Genet. 2014;133 (1):95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gieger C, Radhakrishnan A, Cvejic A, et al. New gene functions in megakaryopoiesis and platelet formation. Nature. 2011;480 (7376):201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gudbjartsson DF, Bjornsdottir US, Halapi E, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41 (3):342–347. [DOI] [PubMed] [Google Scholar]
- 59. van der Harst P, Zhang W, Mateo Leach I, et al. Seventy-five genetic loci influencing the human red blood cell. Nature. 2012;492 (7429):369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stahl EA, Raychaudhuri S, Remmers EF, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42 (6):508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Barrett JC, Clayton DG, Concannon P, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41 (6):703–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schunkert H, Konig IR, Kathiresan S, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43 (4):333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schumacher FR, Stenzel SL, Jiao S, et al. Genome-wide association study of colorectal cancer identifies six new susceptibility loci. Nat Comm. 2015; In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Terzic J, Grivennikov S, Karin E, et al. Inflammation and colon cancer. Gastroenterology. 2010;138 (6):2101–2114 e5. [DOI] [PubMed] [Google Scholar]
- 65. Rao VP, Poutahidis T, Ge Z, et al. Innate immune inflammatory response against enteric bacteria Helicobacter hepaticus induces mammary adenocarcinoma in mice. Cancer Res. 2006;66 (15):7395–7400. [DOI] [PubMed] [Google Scholar]
- 66. Disis ML. Immune regulation of cancer. J Clin Oncol. 2010;28 (29):4531–4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14 (10):1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Degen JL, Palumbo JS. Hemostatic factors, innate immunity and malignancy. Thrombosis Res. 2012;129(Suppl 1):S1–S5. [DOI] [PubMed] [Google Scholar]
- 69. Palumbo JS, Barney KA, Blevins EA, et al. Factor XIII transglutaminase supports hematogenous tumor cell metastasis through a mechanism dependent on natural killer cell function. J Thrombosis Haemostasis. 2008;6 (5):812–819. [DOI] [PubMed] [Google Scholar]
- 70. Camerer E, Qazi AA, Duong DN, et al. Platelets, protease-activated receptors, and fibrinogen in hematogenous metastasis. Blood. 2004;104 (2):397–401. [DOI] [PubMed] [Google Scholar]
- 71. Santaolalla R, Sussman DA, Ruiz JR, et al. TLR4 activates the beta-catenin pathway to cause intestinal neoplasia. PLoS One. 2013;8 (5):e63298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Charbonneau B, Moysich KB, Kalli KR, et al. Large-scale evaluation of common variation in regulatory T cell-related genes and ovarian cancer outcome. Cancer Immunol Res. 2014;2 (4):332–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Block MS, Charbonneau B, Vierkant RA, et al. Variation in NF-kappaB Signaling Pathways and Survival in Invasive Epithelial Ovarian Cancer. Cancer Epidemiol Biomarkers Prev. 2014;23 (7):1421–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Charbonneau B, Goode EL, Kalli KR, et al. The immune system in the pathogenesis of ovarian cancer. Crit Rev Immunol. 2013;33 (2):137–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kees T, Egeblad M. Innate immune cells in breast cancer--from villains to heroes? J Mammary Gland Biol Neoplasia. 2011;16 (3):189–203. [DOI] [PubMed] [Google Scholar]
- 76. Curigliano G. Immunity and autoimmunity: revising the concepts of response to breast cancer. Breast. 2011;20(Suppl 3):S71–S74. [DOI] [PubMed] [Google Scholar]
- 77. Grover A, Izzo AA. BAT3 regulates Mycobacterium tuberculosis protein ESAT-6-mediated apoptosis of macrophages. PLoS One. 2012;7 (7):e40836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kawahara H, Minami R, Yokota N. BAG6/BAT3: emerging roles in quality control for nascent polypeptides. J Biochem. 2013;153 (2):147–160. [DOI] [PubMed] [Google Scholar]
- 79. Amsellem V, Gary-Bobo G, Marcos E, et al. Telomere dysfunction causes sustained inflammation in chronic obstructive pulmonary disease. Am J Respiratory Crit Care Med. 2011;184 (12):1358–1366. [DOI] [PubMed] [Google Scholar]
- 80. Stewart JA, Chaiken MF, Wang F, et al. Maintaining the end: roles of telomere proteins in end-protection, telomere replication and length regulation. Mutation Res. 2012;730(1–2):12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Prescott J, Wentzensen IM, Savage SA, et al. Epidemiologic evidence for a role of telomere dysfunction in cancer etiology. Mutation Res. 2012;730(1–2):75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Murnane JP. Telomere dysfunction and chromosome instability. Mutation Res. 2012;730(1–2):28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Podlevsky JD, Chen JJ. It all comes together at the ends: telomerase structure, function, and biogenesis. Mutation Res. 2012;730(1–2):3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Armanios M. Telomerase and idiopathic pulmonary fibrosis. Mutation Res. 2012;730(1–2):52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Cifuentes-Rojas C, Shippen DE. Telomerase regulation. Mutation Res. 2012;730(1–2):20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wolkowitz OM, Mellon SH, Epel ES, et al. Leukocyte telomere length in major depression: correlations with chronicity, inflammation and oxidative stress--preliminary findings. PLoS One. 2011;6 (3):e17837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bhattacharjee RN, Banerjee B, Akira S, et al. Telomere-mediated chromosomal instability triggers TLR4 induced inflammation and death in mice. PLoS One. 2010;5 (7):e11873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Aikata H, Takaishi H, Kawakami Y, et al. Telomere reduction in human liver tissues with age and chronic inflammation. Exp Cell Res. 2000;256 (2):578–582. [DOI] [PubMed] [Google Scholar]
- 89. Greenland S. Principles of multilevel modelling. Int J Epidemiol. 2000;29 (1):158–167. [DOI] [PubMed] [Google Scholar]
- 90. Greenland S, Poole C. Empirical-Bayes and semi-Bayes approaches to occupational and environmental hazard surveillance. Arch Environ Health. 1994;49 (1):9–16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.