Abstract
Brown and beige adipose tissues have a significant capacity for energy expenditure that may be exploited as a treatment for obesity and metabolic disease. However, the limited volumes of these tissues in adults hinders realization of this potential. Engineering beige adipose tissue may provide an alternative source of this tissue. In this paper we describe the preparation of poly(ethylene glycol) (PEGDA) hydrogels with mechanical properties similar to native adipose tissue. Adipose derived stem cells (ASC) were cultured in hydrogels without adhesive sequences or degradable monomers. Cells were able to differentiate, independent of scaffold properties and were maintained as a viable and functioning adipose tissue mass. The cells expressed their own basement membrane proteins consistent with the composition of adipose tissue. The ASCs could be induced to express uncoupling protein-1 (UCP-1) and cIDEA, makers of beige adipocytes with expression level varying with hydrogel stiffness. This hydrogel-based culture system serves as a first step in engineering beige adipose tissue.
Graphical Abstract
Introduction
The incidence of obesity is increasing globally, generating a burden greater than $190 billion annually on the American healthcare system alone1. These costs result primarily from morbidity and mortality associated with the increased incidence of type 2 diabetes, hypertension, heart disease, and certain malignancies in obese and overweight individuals. Complications from obesity result not only from weight gain directly, but more importantly, from negative metabolic consequences associated with obesity. Adipose is a key endocrine tissue that regulates systemic metabolic function. Studies in pre-clinical models have shown that improvements in the metabolic function of adipose tissue enhances overall metabolic performance2-4.
There are two main types of adipose tissue, white adipose tissue and brown adipose tissue. White adipose (including subcutaneous white adipose and visceral white adipose) makes up nearly all adipose tissue in adults, with brown adipose present in only small regions of supraclavicular, mediastinal and peri-adrenal depots5,6. White adipose is involved in energy storage, structural support and hormone regulation. A number of researchers have investigated biomaterial-based approaches for engineering white adipose, based on the clinical need for new adipose sources for breast reconstruction, soft tissue augmentation and composite tissue reconstruction7-19. However, engineering metabolically active brown or beige adipose tissue has not previously been pursued.
Brown adipose tissue has a high capacity for energy expenditure and thermogenesis, and thus has been considered a potential target for the treatment of obesity and metabolic disease20-23. Increasing the presence of brown adipose tissue in an individual would be expected to improve overall metabolic function24, 25, but the small volume of brown adipose in adults suggests that its therapeutic potential may be limited when targeted directly. Alternatively, white adipose tissue can be induced to behave in a manner similar to brown adipose tissue. There is currently incredible enthusiasm for therapeutic induction of this so-called “beige” adipose tissue to increase adipose thermogenic capacity as a treatment for metabolic disease22, 26-31. An alternative approach could be to use tissue engineering to generate beige adipose tissue from autologous adipose derived stromal cells (ASCs) that can be transplanted into a patient.
Poly (ethylene glycol) diacrylate (PEGDA) hydrogels have been investigated extensively as scaffolds for tissue engineering. PEGDA hydrogels are resistant to non-specific protein adsorption and cell adhesion, providing an inert starting point for biomaterial design. The hydrogels can be engineered with mechanical properties similar to soft tissues, and fairly straightforward chemistry can be used to incorporate biological signals into the network structure. PEGDA hydrogels are typically prepared such that they are not susceptible to degradation under ambient and/or physiologic conditions.32 Segments susceptible to hydrolysis or enzymatic cleavage can be readily introduced within network crosslinks allowing for controlled hydrogel degradation. Previous studies have utilized PEG-based hydrogels functionalized with extracellular matrix (ECM)-based attachment sites and enzymatically degradable sequences for 3D adipocyte differentiation12, 33. Mesenchymal stem cells encapsulated in PEG hydrogels can be differentiated into adipocytes34, and thin films of PEG have been utilized to investigate the role of sub-micron topography on preadipocyte behavior35. However, PEG hydrogels have not been used as an environment for engineering beige adipose tissue.
In this study, we investigated beige adipocyte formation from ASCs in 3D culture. PEGDA hydrogels were used as a cell culture platform to provide a controlled 3D environment for cell growth and to investigate the role of matrix stiffness on ASC function. The fundamental design goal for the hydrogels was to mimic the stiffness of adipose tissue. This would allow for investigation of the role of mechanical properties on ASC differentiation into beige adipocytes. The hydrogels were prepared without adhesion sequences or degradable crosslinks. This decouples the mechanical effects from the effects of cell adhesion or degradability of the hydrogel. The hydrogels engineered in a range of mechanical properties approximating those found in adipose tissue by varying the crosslinker content, and the viability and differentiation of ASCs was evaluated. The expression of beige adipose makers, adipogenic markers and extracellular matrix proteins were measured as a function of hydrogel elastic modulus. To our knowledge, this is the first study to apply concepts from tissue engineering to the generation of brown or beige adipose tissue.
Materials and Methods
Chemicals
DMEM/F12, penicillin/streptomycin, and BODIPY 493/503 (4,4-Difluoro-1,3,5,7,8-Pentamethyl-4-Bora-3a,4a-Diaza-s-Indacene) were purchased from Fisher Scientific. Collagenase type I, FBS, HEPES sodium salt, NaCl, HCl, NaOH, eosin Y, N-vinylpyrrolidone (NVP), triethanolamine (TEA), carboxymethylcellulose, indomethacin, dexamethasone (DEX), 3-Isobutyl-1-methylxanthine (IBMX), rosiglitazone, 3,3',5-Triiodo-L-thyronine (T3), and insulin were purchased from Sigma. PEG-diacrylate (PEGDA) macromers were synthesized according to previously published method36. The PEGDA purity and structure were verified by 1H NMR (Advance 300 Hz; Bruker). The product was dissolved in CDCl3 with 0.05% tetramethylsilane as an internal standard, to perform 1H NMR.
Cell Isolation and Culture
Subcutaneous adipose depots were harvested from euthanized C57BL/6 male 12-15 week old mice and digested in 500 units activity/ml collagenase type I. All animal procedures were approved by the Institutional Animal Care and Use Committee at the University of Chicago. The adipose derived stem cells (ASC) were enzymatically isolated following a published protocol37. Briefly, digestion was performed in an orbital shaker at 37 °C for 60 minutes. The digest was then centrifuged with the floating adipocytes layer discarded. The pelleted stromal vascular fraction was washed twice with media and plated in tissue culture flask. The media used was DMEM/F12 with 10% FBS and 1% penicillin/streptomycin. Cells were incubated at 37 °C and 5% CO2. Media was changed every 2-3 days.
Spheroid Formation
Cells were trypsinized, counted, and resuspended in DMEM/F12 supplemented with 10% FBS, 1% penicillin/streptomycin, and 0.24% (w/v) carboxymethylcellulose (Sigma) at a density of 20,000 cells/150ul. The solution was pipetted into a non-adherent 96-well U-bottom plate (150 μL/well) (Sigma) and cultured at 37°C and 5% CO2. Cell spheroids were formed after 24 hours.
PEGDA Hydrogel Formation and Spheroid Encapsulation
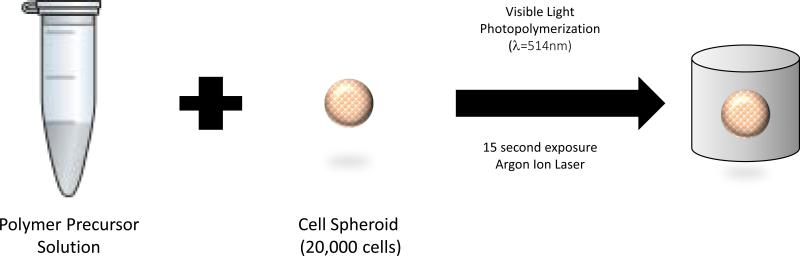
Hydrogel precursor solutions were prepared with 8kDa PEGDA at concentrations of 3, 4, 5, 6, and 7% (w/v) in 10 mM HEPES (500 mL ddH2O, 4 g NaCl, and 1.302 mg HEPES sodium salt; pH balanced to 7.4 with HCl or NaOH) with 0.05 mM eosin Y as the initiator, 225 mM triethanolamine (TEA) as a co-initiator and 37 mM N-vinylpyrrolidone (NVP). Precursor solutions (100 μL/hydrogel) were photopolymerized in the presence of cell spheroids by exposure to visible light (λ = 514 nm) for 15 seconds using an Argon Ion Laser (Coherent, Inc., Santa Clara, CA) at a laser flux of 100 mW/cm2 (Figure 1). The hydrogels with cell spheroids were removed from the wells, transferred to a 24-well plate and washed 3 times with HEPES buffer solution, immersed in media and incubated at 37 °C and 5% CO2.
Figure 1.
Cartoon of the procedure for preparing hydrogels containing the cell spheroids.
Cell Culture and Differentiation
The hydrogels were cultured for 24 hours in DMEM/F12 with 10% FBS and 1% penicillin/streptomycin. Induction media consisted of DMEM/F12 with 10% FBS, 1% penicillin/streptomycin, 125 μM Indomethacin, 2 μg/mL DEX, 0.5 mM IBMX, 1 nM 3,3',5-Triiodo-L-thyronine (T3), and 5 μg/mL insulin38. Induction media was given to the spheroids for 48 hours to induce adipocyte differentiation. Thiazolidinedione (TZD, 0.5 μM rosiglitazone) was added to the media to promote differentiation into beige adipocytes. After 2 days of induction, cells were given maintenance media consisting of DMEM/F12 with 10% FBS, 1% penicillin/streptomycin, 1 nM T3 and 5 μg/mL insulin. Based on published protocols on beige adipocyte stimulation25, 38, 39, the first two media changes after induction included rosiglitazone. On the first change (day 3), the spheroids were given 0.5 μM rosiglitazone in the maintenance media. On the second change (day 5), the spheroids were supplemented with 1.0 μM rosiglitazone. Maintenance media changes past day 7 did not include rosiglitazone. The hydrogels were cultured for a total of 20 days.
Staining
A LIVE/DEAD® assay kit (Molecular Probes, Inc. Eugene, OR) was used to evaluate cell viability at day 20. Briefly, hydrogels were rinsed with PBS and incubated with LIVE/DEAD staining reagents calcein AM (green fluorescent dye stains live cells) and ethidium homodimer-1 (red fluorescent dye stains dead cells) for 1 h following the manufacturer's protocol. Samples were imaged using confocal laser scanning microscopy Axiovert 200 5x objective (Carl Zeiss MicroImaging, Inc., Thornwood, NY). The percentage of viable cells was quantified by estimating the area of cells in the spheroid stained green (arealive) and those stained red (areadead) using confocal microscopy:
Lipid loading was evaluated with BODIPY staining. Hydrogels containing adipocyte spheroids were incubated in 4% paraformaldehyde overnight. After paraformaldehyde was removed hydrogels were washed two times for thirty minutes each in phosphate buffered saline (PBS). The hydrogels were stained with BODIPY 493/503 (4,4-Difluoro-1,3,5,7,8-Pentamethyl-4-Bora-3a,4a-Diaza-s-Indacene) at 0.01 mg/ml in PBS at 37°C for 1 hour. Hydrogels were washed two times with PBS and imaged using an Axiovert 200 inverted confocal microscope 10x objective (Carl Zeiss MicroImaging, Inc., Thornwood, NY) at excitation wavelength 488 nm and long pass filter 505 nm.
Swelling Ratio and Mesh Size
Hydrogel swelling was calculated by measuring the ratio of the mass of the swollen gel (MS) 24 h postphotopolymerization to the mass of the dried gel (MD). The mass of the swollen hydrogel was measured by removing excess fluid from the hydrogel surface. The mass of the dried hydrogel was measured after drying the hydrogels under vacuum for 24 hours at 50°C. The hydrogel mass swelling ratio (Qm) was calculated as:
Where MS is the mass of the swollen gel and MD is the mass of the dried gel. The mesh size (ξ) of the swollen hydrogel was quantified based on the theory of rubber elasticity as previously described40:
In the above V2,S represents the polymer volume fraction of the hydrogel in the swollen state which is also equal to the reciprocal of the volumetric swelling ratio (Q):
in which VP refers to the hydrogel volume in the dried state and VS refers to the hydrogel volume in swollen state. The root mean square unperturbed end to end distance of the polymer chain between crosslinks, is given by the following equation:
The average bond length between the C-C and C-O bonds in the PEG repeat unit is l (l = 1.46 Å), the characteristic ratio of PEG is Cn (Cn = 4) and the molecular weight of the PEG repeat unit is Mr (Mr = 44 g/mol). The average molecular weight between cross-links is , this value was calculated according to the following equation41:
in which v is the specific volume of the polymer (0.84 cm3/g), V1 is the molar volume of the solvent (18 cm3/mol) and V2,r is the polymer volume fraction in the relaxed state immediately after polymerization.
Evaluation of Hydrogel Network Structure on Solute Diffusivity
A theoretical model developed by Lustig and Peppas based on free-volume theory was used to describe the relationship between solute diffusivity and network structure The influence of the hydrogel on solute diffusion was estimated as42:
Where Dg is the diffusion coefficient of the solute in the hydrogel, ξ is the mesh size, and V2 is the polymer volume fraction in the swollen state. Y is the ratio of the required critical volume for successful translational movement of the encapsulated solute and the average free volume per solvent molecule. A reasonable approximation for Y is unity. The diffusion coefficient of the solute in the pure solvent, Do, can be estimated using the Stokes-Einstein equation:
where k is Boltzmann's constant (1.38 × 10-23 m2kg/s2K); T is temperature (T = 37 °C = 310 K ); η is the viscosity of the solvent (6.92 × 10−4 kg/ms for water at 37 °C) and Rs is the Stokes radius of the solute 42. Insulin is a solute in the media that is critical for the cell spheroids within the hydrogel network to receive. Insulin has a hydrodynamic radius of 1.47 nm.
Quantitative Real-time PCR
The hydrogels were flash frozen in liquid nitrogen on days 1 and 20, minced using a razor blade, and processed using Qiagen RNeasy Mini kit for extraction following the manufacturer instructions. The purity and concentration of the isolated RNA was assessed using a Nanodrop 2000. The cDNA was synthesized with Quanta Biosciences Qscript. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using SYBR green on a Bio-Rad MyiQ RT-PCR detection system (Bio-Rad, Hercules, California). Primers (Table 1) from Integrated DNA Technologies (Coralville, Iowa) were selected from literature or designed using Mouse Primer Depot (http://mouseprimerdepot.nci.nih.gov/) and Primer-Blast (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). Melt curve analysis was used to assess primer specificity.18s rRNA was used as the reference gene to control for total mRNA recovery. Gene expression levels were evaluated by the ΔΔ threshold cycle (Ct) method43.
Table 1.
qRT-PCR Primer sequences
| gene | Marker Type | Forward Primer | Reverse Primer |
|---|---|---|---|
| 18S | reference | AACCCGTTGAACCCCATT | CCATCCAATCGGTAGTAGCG |
| Adiponectin | adipogenic | GAATCATTATGACGGCAGCA | TCATGTACACCGTGATGTGGTA |
| PPARG | adipogenic | TCTTCCATCACGGAGAGGTC | GATGCACTGCCTATGAGCAC |
| Pref-1 | preadipocyte | TGTGCAGGAGCATTCGTACT | CGGGAAATTCTGCGAAATAG |
| Collagen IV | basement membrane | CACATTTTCCACAGCCAGAG | GTCTGGCTTCTGCTGCTCTT |
| Fibronectin-1 | basement membrane | ACTGGATGGGGTGGGAAT | GGAGTGGCACTGTCAACCTC |
| Lama2 | basement membrane | TGCATTCGAAGCAAGATTCA | CTCGAAGGCTCCCAGACT |
| Lama4 | basement membrane | AGGGTGCACATTCTCCTGAC | GAGACTAGCGACTCAGGCGT |
| cIDEA | beige | TGCTCTTCTGTATCGCCCAGT | GCCGTGTTAAGGAATCTGCTG |
| PGC1α | beige | TGAGAACCGCTAGCAAGTTT | TGTAGCGACCAATCGGAAAT |
| PRDM16 | beige | ACGCAGAACTTCTCGCTACC | ATGGGAGATGCTGACGGATA |
| UCP-1 | beige | ACTGCCACACCTCCAGTCATT | CTTTGCCTCACTCAGGATTGG |
Statistics
The significant differences between the groups of data were determined using analysis of variance (ANOVA) followed with Tukey post test. In all cases, p<0.05 was considered significant.
Results
Physical and Mechanical Characterization of PEGDA Hydrogels
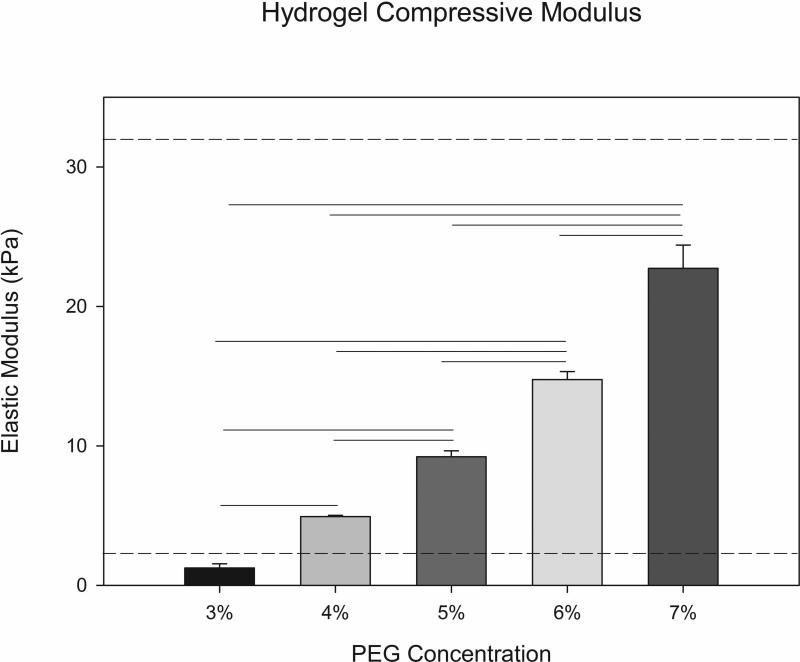
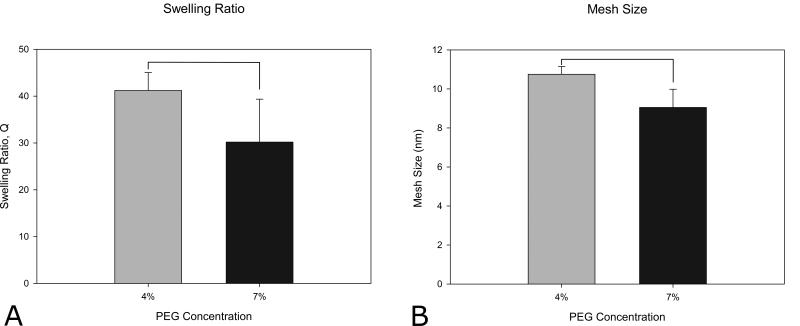
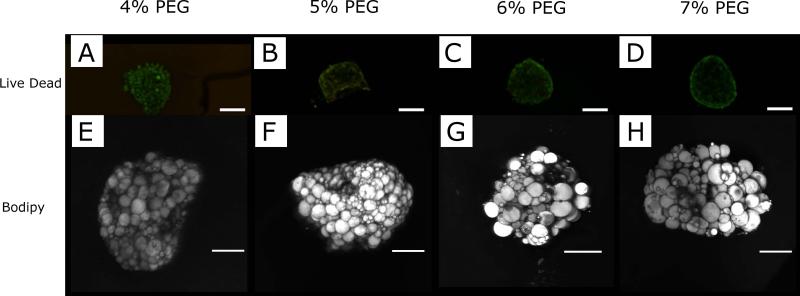
PEGDA hydrogels were synthesized using free-radical photopolymerization in the presence of visible light. Hydrogel mechanical properties were varied by altering the PEGDA macromer crosslinker concentration (3%-7% w/vol). The compressive modulus of the PEG hydrogels was found to increase with increasing crosslinker content (Figure 2). The elastic modulus values achieved are within the published range of moduli previously reported for adipose tissue (2-32 kPa) 44, 45. Increasing the concentration of PEGDA from 4% to 7% also resulted in a statistically significant decrease in both the swelling ratio and mesh size (Figure 3) of the hydrogel network. The 4% and 5% PEGDA hydrogels had a less rigid disc morphology macroscopically compared to the higher compressive modulus hydrogels (6% and 7%) that retained a more defined disc shape after swelling. The ASC cell spheroids on day 1 (Figure 4A-D) the cells appear smooth and elongated within the spheroid after differentiating the cells within the spheroid have rounded and lipid loaded at day 20 (Figure 4E-H and Figure 5A-D). The hydrogel swelling ratio ranged from 41.19 ± 3.83 (4%) to 30.17 ± 9.17 (7%) and the mesh size (ξ) ranged between 10.75 ± 0.399 (4%) and 9.05 ± 0.933nm (7%).
Figure 2.
Compressive modulus of the PEGDA hydrogels as a function of macromer crosslinker concentration. The range of values for adipose tissue are indicated by dotted lines (n=4, p<0.05). Standard error shown as error bars.
Figure 3.
Swelling ratio (A) and mesh size (B) for 4% and 7% PEGDA macromer concentrations. (n=5, p<0.05) Standard deviation shown as error bars. Bars indicate statistically significant differences (p < 0.05).
Figure 4.
Brightfield images of cell spheroids day 1 (A,B,C,D) and day 20 (E,F,G,H) in 4%, 5%, 6%, 7% PEGDA hydrogels, respectively. Scale bar is 200 μm.
Figure 5.
Viable cells are present throughout the spheroid at day 20 in PEGDA hydrogels as indicated by (A,B,C,D) confocal images of a LIVE/DEAD stain. Live cells stain green and dead cells in red. Scale bar is 200 μm. 98-99% viability was observed in all in all PEGDA concentrations. Bodipy stained spheroids (E,F,G,H) at day 20 in PEGDA hydrogels show the formation of lipid loaded cells with both unilocular and multilocular appearance throughout the spheroid volume. Scale bar is 200 μm.
The influence of the network structure on diffusion within the hydrogels was also estimated from theoretical models of free-volume theory. The diffusion coefficient (Do) of insulin, an important mediator of adipocyte function, in media is 2.23 × 10−6 cm2/s. Based on the polymer volume fraction and mesh size the diffusion coefficient for insulin within the PEGDA hydrogel (Dg) would range from 1.88 × 10−6 cm2/s in the 4% PEGDA (Dg/Do=0.84) to 1.81 × 10−6 cm2/s (Dg/Do=0.81) in the 7% PEGDA hydrogel. These results suggest there is little difference in diffusive resistance between the hydrogel conditions. TZDs have been shown to be helpful for promoting beige adipocyte formation. TZD, rosiglitazone, (C18H19N3O3S, ~357 Da molecular weight) is much smaller in size than insulin (C256H381N65O76S6, ~5800 Da molecular weight). Less diffusive resistance would be expected for the smaller rosiglitazone in the hydrogels.
Maintenance of a Differentiated Cells in 3D Culture
The spheroids in the hydrogels had a smooth and relatively consistent structure at day 1 independent of crosslinker concentration (Figure 4A-D). After differentiation the spheroid surfaces were rougher with evidence of lipid loading in the cells (Figure 4E-H). The viability of cell spheroids encapsulated in the hydrogels was high (98-99%) under all PEGDA concentrations out to the final time point used in this study (20 days, Figure 5A-D) ). Lipid loading was observed throughout the spheroid volume at all crosslinker concentrations (Figure 5E-H). Both unilocular and multilocular adipocytes were observed within the spheroids in the hydrogels. The cell spheroids did not exhibit expression of the adipocyte marker adiponectin on day 1. However, after 20 days of culture in differentiation media, all groups express adiponectin and peroxidase proliferator-activated receptor gamma (PPARG), markers of adipocyte differentiation, without significant differences in expression level with hydrogels stiffness (Figure 6). Pref-1 a marker of preadipocytes was not expressed at day 20. The cells exhibited high viability and differentiation rates despite using PEGDA hydrogels that did not contain degradable linkages or cell adhesion motifs. PEGDA hydrogels were able to maintain viable, differentiated adipocytes in absence of any exogenous extracellular matrix proteins or peptides, over a range of mechanical properties and physical network structure.
Figure 6.
Adiponectin (A) and PPARG (B) are expressed by in cell spheroids in the PEGDA hydrogels. Data are shown for Day 20 in TZD treated spheroids as a function of PEGDA Concentration. (n=3)
Beige Markers
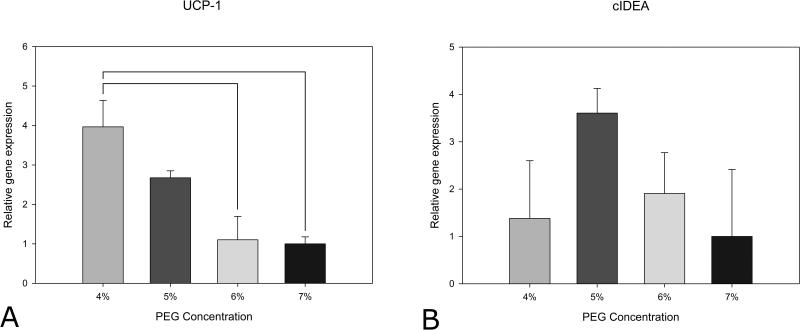
Mitochondrial uncoupling protein 1 (UCP-1) is utilized in the thermogenesis process in brown and beige adipocytes to uncouple respiration and generate heat. UCP-1 mRNA expression is found in brown and beige adipocytes but is not typically expressed at significant levels in white adipocytes. Cell death-inducing DNA fragmentation factor-alpha-like effector A (cIDEA) is a gene that plays a role in adipocyte thermogenesis and is expressed in beige and brown adipocytes. Under differentiation conditions without the TZD rosiglitazone, the cells did not express markers of beige adipocytes (data not shown). When exposed to rosiglitazone as well as the standard differentiation cocktail, mRNA for the beige markers, UCP-1 and cIDEA, were both upregulated (Figure 7). This agrees with previous studies that show that TZD administration is needed to optimize the development of beige adipocytes46, 47. Neither PGC1α nor the brown transcription factor PRDM16 were expressed by the adipocytes treated with TZD.
Figure 7.
UCP-1 (A) and cIDEA (B) expression in cell spheroids in the PEGDA hydrogels. Data are shown for day 20 in TZD treated spheroids as a function of PEGDA Concentration. UCP-1 expression decreased with PEGDA concentration. (n=3, statistical significance indicated by lines p<0.05)
Interestingly, UCP-1 expression (Figure 7A) showed an inverse relationship with PEGDA concentration. UCP-1 expression decreased with increasing hydrogel stiffness. cIDEA levels (Figure 7B) shared a similar trend of expression decreasing with increasing hydrogel stiffness, however the differences were not significant. Mechanical stiffness has not previously been investigated as playing a role in beige adipocyte formation.
Basement Membrane Proteins
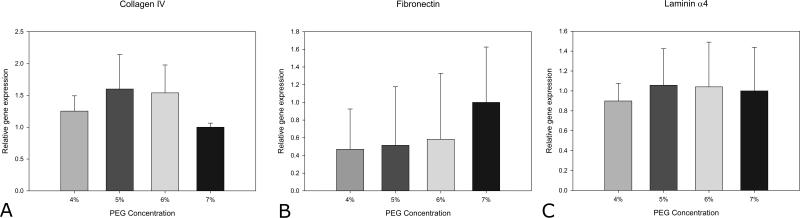
Adipocytes in native tissue are surrounded by a thin extracellular matrix (ECM) structure known as the basement membrane. Collagen IV, laminin α4, laminin α2, and fibronectin-1 are all components of adipose tissue basement membrane. The adipocytes in 3D culture expressed the basement membrane proteins collagen IV, laminin α4, and fibronectin-1 (Figure 8). Laminin α2 was not expressed. The level of the basement membrane components did not vary with hydrogel composition.
Figure 8.
Collagen iv (A), fibronectin-1 (B), laminin α4 (C) in the PEGDA hydrogels. Data are shown for day 20 in TZD treated spheroids as a function of PEGDA Concentration (n=3).
Discussion
Biomaterials have been investigated for engineering many different tissues and organs. However, to our knowledge, there have not been previous studies using biomaterials to generate brown or beige adipose tissue. There is significant interest in the ability to generate beige adipose as a potential treatment for metabolic disease and obesity22, 26-31. The transplantation of adipose tissue with “good” metabolic function has been shown to benefit overall metabolic parameters48, 49 We propose that appropriately engineered beige adipose tissue could be transplanted into diseased patients to improve overall metabolic function.
The starting point for this research was to generate a hydrogel environment that mimics native adipose tissue. PEGDA hydrogels were synthesized with compressive moduli within the range of values previously published for adipose tissue44, 45. There are many options for incorporating biological signals into PEGDA hydrogels. However, adipocytes are surrounded by a complex basement membrane that is difficult to recreate in synthetic systems, and the composition of this basement membrane is important in regulating adipose tissue function50. The ASCs were cultured in the gels as a spheroid enabling them to produce their own matrix. The adipocytes expressed genes for collagen IV, laminin α4, and fibronectin-1 proteins known to be present in adipose basement membrane. Previous studies have shown that many cells exhibit poor viability in PEG in the absence of ECM-based peptides or proteins42, 51-53. However, the cell-to-cell contact and production of their own basement membrane in our system likely allows long term viability and function in the hydrogels.
Under standard adipogenic culture conditions the ASCs differentiated into adipocytes but did not express any markers indicating a beige adipocyte phenotype. However, the addition of the TZD rosiglitazone to the media resulted in both UCP-1 and cIDEA expression. PRDM16, a brown and beige adipocyte marker, was not expressed at significant levels in this study. PRDM16 has been shown to be expressed by beige adipocytes differentiated from subcutanous depot stromal vascular fraction cells cultured in 2D for short term culture (4-8 days) while continuously supplied TZD25, 29, 39. However, previous work has shown PRDM16 expression decreased when subcutaneous depot preadipcytes are dosed with TZD for 4 days and then not supplied with TZD for 4 additional in vitro culture days39. The mitochondrogenesis-promoting gene PGC1α was also not expressed at day 20 in TZD-treated cells either. Other primary cell studies have shown that PGC1α is expressed by adipocytes differentiated from epididymal depot preadipocytes cultured in 2D for short term culture (4-8 days) while continuously supplied TZD29, 39, 46. Possibly, PRDM16 and PGC1α expression was not observed due to the transient nature of the TZD exposure. In this study, TZD was supplemented in the culture media for the initial 7 day culture period; for the following 13 days the cells did not receive any TZD.
Interestingly, UCP-1 expression varied with PEGDA stiffness. cIDEA also showed a trend of decreasing levels with PEGDA stiffness. These data suggest that the mechanical properties of the substrate environment influence expression of beige markers. It is not clear why beige markers would vary with hydrogel stiffness. To our knowledge there are no data on the influence of mechanical properties on beige adipose formation. White adipocytes are able to sense physical attributes of 3D environment, including mechanical forces54. Adipose tissue ECM in collagen VI knockout mice is characterized by decreased stiffness resulting in enlarged adipocytes without the typically associated necrosis and inflammation55. It is also possible that properties other than stiffness that vary with crosslinker concentration could drive cell behavior, such as resistance to nutrient transport. However, there was little differences in the estimated diffusion coefficients of insulin between the hydrogel conditions. Regardless, these in vitro findings support that the properties of the substrate influences adipocyte beiging. This information will be used to further refine our approach for engineering beige adipose tissue.
In addition to the beige adipose markers, the cells expressed adiponectin and PPARG in 3D culture. Adiponectin upregulation and absence of Pref-1 a marker associated with preadipocytes, indicates that the ASCs differentiated into adipocytes. Lipid staining showed both multilocular and unilocular lipid storage in the differentiated cells. Unlike beige adipose markers, expression of these more general adipocyte markers did not vary with hydrogel stiffness. Adiponectin has been suggested to exhibit insulin-sensitizing properties. When combined with the expression of beige adipocyte markers, this suggests the formation of adipose tissue with appropriate metabolic function.
Conclusion
ASCs were differentiated into beige adipocytes in 3D PEGDA hydrogels with mechanical properties approximating the native adipose environment. The cells expressed ECM protein genes and were viable for nearly three weeks in the absence of hydrogel modifications. Beige adipocytes could be generated in the hydrogels and the expression of beige adipocyte markers varied with hydrogel composition. Overall these results are a first step towards engineering beige adipose tissue.
Acknowledgements
This work was funded, in part, by the University of Chicago Diabetes Research and Training Center (NIH, P30 DK020595), the University of Chicago Institute for Translational Medicine (NIH, CTSA UL1 TR000430), The Pritzker Institute of Biomedical Science and Engineering, and the Veterans Administration.
References
- 1.Cawley J, Meyerhoefer C. J Health Econ. 2012;31:219–230. doi: 10.1016/j.jhealeco.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Gunawardana SC, Piston DW. Am J Physiol Endocrinol Metab, American Journal of Physiology - Endocrinology and Metabolism. 2015:ajpendo 00570 02014. [Google Scholar]
- 3.Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, Lambert PD, Mataki C, Elliott PJ, Auwerx J. Cell Metab. Vol. 8. United States: 2008. pp. 347–358. [DOI] [PubMed] [Google Scholar]
- 4.Gunawardana SC. World J Diabetes. 2014;5:420–430. doi: 10.4239/wjd.v5.i4.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sacks H, Symonds ME. Diabetes. Vol. 62. United States: 2013. pp. 1783–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richard D, Monge-Roffarello B, Chechi K, Labbe SM, Turcotte EE. Front Endocrinol (Lausanne) 2012;3:36. doi: 10.3389/fendo.2012.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patrick CW. Seminars in Surgical Oncology. 2000;19:302–311. doi: 10.1002/1098-2388(200010/11)19:3<302::aid-ssu12>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 8.Toriyama K, Kawaguchi N, Kitoh J, Tajima R, Inou K, Kitagawa Y, Torii S. Tissue Engineering. 2002;8:157–165. doi: 10.1089/107632702753503144. [DOI] [PubMed] [Google Scholar]
- 9.Masuda T, Furue M, Matsuda T. Tissue Engineering. 2004;10:1672–1683. doi: 10.1089/ten.2004.10.1672. [DOI] [PubMed] [Google Scholar]
- 10.Alhadlaq A, Tang M, Mao JJ. Tissue Engineering. 2005:11. doi: 10.1089/ten.2005.11.556. [DOI] [PubMed] [Google Scholar]
- 11.Hemmrich K, von Heimburg D, Rendchen R, Di Bartolo C, Milella E, Pallua N. Biomaterials. 2005;26:7025–7037. doi: 10.1016/j.biomaterials.2005.04.065. [DOI] [PubMed] [Google Scholar]
- 12.Brandl FP, Seitz AK, Tessmar JKV, Blunk T, Gopferich AM. Biomaterials. 2010;31:3957–3966. doi: 10.1016/j.biomaterials.2010.01.128. [DOI] [PubMed] [Google Scholar]
- 13.Choi JH, Gimble JM, Lee K, Marra KG, Rubin JP, Yoo JJ, Vunjak-Novakovic G, Kaplan DL. Tissue Engineering Part B-Reviews. 2010:16. doi: 10.1089/ten.teb.2009.0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang XL, Reagan MR, Kaplan DL. Journal of Mammary Gland Biology and Neoplasia. 2010;15:365–376. doi: 10.1007/s10911-010-9192-y. [DOI] [PubMed] [Google Scholar]
- 15.Sorrell JM, Baber MA, Traktuev DO, March KL, Caplan AI. Biomaterials. 2011;32:9667–9676. doi: 10.1016/j.biomaterials.2011.08.090. [DOI] [PubMed] [Google Scholar]
- 16.Ladewig K, Abberton K, O'Connor AJ. Journal of Biomaterials and Tissue Engineering. 2012;2:1–13. [Google Scholar]
- 17.Wu I, Nahas Z, Kimmerling KA, Rosson GD, Elisseeff JH. Plastic and Reconstructive Surgery. 2012:129. doi: 10.1097/PRS.0b013e31824ec3dc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lilja HE, Morrison WA, Han XL, Palmer J, Taylor C, Tee R, Moller A, Thompson EW, Abberton KM. Stem Cells and Development. 2013;22:1602–1613. doi: 10.1089/scd.2012.0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uriel S, Huang JJ, Moya ML, Francis ME, Wang R, Chang SY, Cheng MH, Brey EM. Biomaterials. 2008;29:3712–3719. doi: 10.1016/j.biomaterials.2008.05.028. [DOI] [PubMed] [Google Scholar]
- 20.Seale P, Lazar MA. Diabetes. Vol. 58. United States: 2009. pp. 1482–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cesari F. Nature Reviews Molecular Cell Biology. 2008;9:742–742. [Google Scholar]
- 22.Harms M, Seale P. Nat Med. 2013;19:1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- 23.Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng Y-H, Doria A, Kolodny GM, Kahn CR. New England Journal of Medicine. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Himms-Hagen J, Cui J, Danforth E, Jr., Taatjes DJ, Lang SS, Waters BL, Claus TH. Am J Physiol. 1994;266:R1371–1382. doi: 10.1152/ajpregu.1994.266.4.R1371. [DOI] [PubMed] [Google Scholar]
- 25.Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, Cohen P, Cinti S, Spiegelman BM. J Clin Invest. 2011;121:96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spiegelman BM. Diabetes. 62:1774–1782. doi: 10.2337/db12-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu J, Cohen P, Spiegelman BM. Genes Dev. 27:234–250. doi: 10.1101/gad.211649.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bi P, Shan T, Liu W, Yue F, Yang X, Liang XR, Wang J, Li J, Carlesso N, Liu X, Kuang S. Nat Med. 2014;20:911–918. doi: 10.1038/nm.3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, Khandekar M, Virtanen KA, Nuutila P, Schaart G, Huang K, Tu H, van Marken Lichtenbelt WD, Hoeks J, Enerback S, Schrauwen P, Spiegelman BM. Cell. Vol. 150. 2012 Elsevier Inc; United States: 2012. pp. 366–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lidell ME, Betz MJ, Dahlqvist Leinhard O, Heglind M, Elander L, Slawik M, Mussack T, Nilsson D, Romu T, Nuutila P, Virtanen KA, Beuschlein F, Persson A, Borga M, Enerback S. Nat Med. Vol. 19. United States: 2013. pp. 631–634. [DOI] [PubMed] [Google Scholar]
- 31.Keipert S, Jastroch M. Biochim Biophys Acta. 2014;1837:1075–1082. doi: 10.1016/j.bbabio.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 32.Lee CY, Teymour F, Camastral H, Tirelli N, Hubbell JA, Elbert DL, Papavasiliou G. Macromolecular Reaction Engineering. 2014;8:314–328. [Google Scholar]
- 33.Patel PN, Gobin AS, West JL, Patrick CW. Tissue Engineering. 2005:11. doi: 10.1089/ten.2005.11.1498. [DOI] [PubMed] [Google Scholar]
- 34.Stosich MS, Bastian B, Marion NW, Clark PA, Reilly G, Mao JJ. Tissue Engineering. 2007;13:2881–2890. doi: 10.1089/ten.2007.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fozdar DY, Wu X, Patrick CW, Jr., Chen S. Biomedical Microdevices. 2008;10:839–849. doi: 10.1007/s10544-008-9198-z. [DOI] [PubMed] [Google Scholar]
- 36.Chiu YC, Cheng MH, Uriel S, Brey EM. J Tissue Viability. 2009 doi: 10.1016/j.jtv.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 37.Yu G, Wu X, Kilroy G, Halvorsen YD, Gimble JM, Floyd ZE. Methods Mol Biol. 2011;702:29–36. doi: 10.1007/978-1-61737-960-4_3. [DOI] [PubMed] [Google Scholar]
- 38.Aune UL, Ruiz L, Kajimura S. J Vis Exp. 2013 doi: 10.3791/50191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohno H, Shinoda K, Spiegelman BM, Kajimura S. Cell Metab, A. Vol. 15. 2012 Elsevier Inc; United States: 2012. pp. 395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raeber GP, Lutolf MP, Hubbell JA. Acta Biomaterialia. 2007:3. doi: 10.1016/j.actbio.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 41.Canal T, Peppas NA. J Biomed Mater Res. 1989;23:1183–1193. doi: 10.1002/jbm.820231007. [DOI] [PubMed] [Google Scholar]
- 42.Weber LM, Lopez CG, Anseth KS. Journal of Biomedical Materials Research Part A. 2009;90A:720–729. doi: 10.1002/jbm.a.32134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Livak KJ, Schmittgen TD. Methods. Vol. 25. 2001 Elsevier Science (USA); United States: 2001. pp. 402–408. [DOI] [PubMed] [Google Scholar]
- 44.Samani A, Bishop J, Luginbuhl C, Plewes DB. Physics in Medicine and Biology. 2003;48:2183–2198. doi: 10.1088/0031-9155/48/14/310. [DOI] [PubMed] [Google Scholar]
- 45.Alkhouli N, Mansfield J, Green E, Bell J, Knight B, Liversedge N, Tham JC, Welbourn R, Shore AC, Kos K, Winlove CP. American Journal of Physiology-Endocrinology and Metabolism. 2013;305:E1427–E1435. doi: 10.1152/ajpendo.00111.2013. [DOI] [PubMed] [Google Scholar]
- 46.Petrovic N, Walden TB, Shabalina IG, Timmons JA, Cannon B, Nedergaard J. J Biol Chem. Vol. 285. United States: 2010. pp. 7153–7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson-Fritch L, Nicoloro S, Chouinard M, Lazar MA, Chui PC, Leszyk J, Straubhaar J, Czech MP, Corvera S. J Clin Invest. 2004;114:1281–1289. doi: 10.1172/JCI21752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tran TT, Kahn CR. Nat Rev Endocrinol. 2010;6:195–213. doi: 10.1038/nrendo.2010.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tran TT, Yamamoto Y, Gesta S, Kahn CR. Cell Metab. 2008;7:410–420. doi: 10.1016/j.cmet.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vaicik MK, Thyboll Kortesmaa J, Moverare-Skrtic S, Kortesmaa J, Soininen R, Bergstrom G, Ohlsson C, Chong LY, Rozell B, Emont M, Cohen RN, Brey EM, Tryggvason K. PLoS One. Vol. 9. United States: 2014. p. e109854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burdick JA, Anseth KS. Biomaterials. 2002;23:4315–4323. doi: 10.1016/s0142-9612(02)00176-x. [DOI] [PubMed] [Google Scholar]
- 52.Peyton SR, Raub CB, Keschrumrus VP, Putnam AJ. Biomaterials. 2006;27:4881–4893. doi: 10.1016/j.biomaterials.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 53.Salinas CN, Anseth KS. Journal of Tissue Engineering and Regenerative Medicine. 2008;2:296–304. doi: 10.1002/term.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shoham N, Gefen A. Journal of Biomechanics. 2012:45. doi: 10.1016/j.jbiomech.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 55.Khan T, Muise ES, Iyengar P, Wang ZV, Chandalia M, Abate N, Zhang BB, Bonaldo P, Chua S, Scherer PE. Mol Cell Biol. 2009;29:1575–1591. doi: 10.1128/MCB.01300-08. [DOI] [PMC free article] [PubMed] [Google Scholar]