Abstract
The Genotype-Tissue Expression (GTEx) project, sponsored by the NIH Common Fund, was established to study the correlation between human genetic variation and tissue-specific gene expression in non-diseased individuals. A significant challenge was the collection of high-quality biospecimens for extensive genomic analyses. Here we describe how a successful infrastructure for biospecimen procurement was developed and implemented by multiple research partners to support the prospective collection, annotation, and distribution of blood, tissues, and cell lines for the GTEx project. Other research projects can follow this model and form beneficial partnerships with rapid autopsy and organ procurement organizations to collect high quality biospecimens and associated clinical data for genomic studies. Biospecimens, clinical and genomic data, and Standard Operating Procedures guiding biospecimen collection for the GTEx project are available to the research community.
Introduction
The aim of the Genotype-Tissue Expression (GTEx) Project of the U.S. National Institutes of Health (NIH) Common Fund (https://commonfund.nih.gov/GTEx) is to determine how genetic variation affects normal gene expression in human tissues, and ultimately to assess how this relationship correlates with the development of disease. To achieve this goal, the project planned to collect multiple different human tissues from each of hundreds of donors, isolate nucleic acids from the tissues and perform genotyping, gene expression profiling, whole genome sequencing, and RNA sequencing, and analyze the data to identify expression quantitative trait loci (eQTL).1–3 The scientific goals of the project required that the donors and their biospecimens present with no evidence of disease (henceforth termed “normal tissues” or “normal biospecimens”).
The project began as a 2.5-year pilot study to assess the feasibility of collecting tissue from up to 40 different tissue types from women and 34 different tissue types from men, from hundreds of individual donors.1,4 The resulting biospecimens needed to yield RNA with an RNA Integrity Number (RIN)5 of at least 6 for optimal RNA sequencing results. The National Cancer Institute (NCI)'s Biorepositories and Biospecimen Research Branch (BBRB) worked with the National Human Genome Research Institute (NHGRI), the National Institute of Mental Health (NIMH), and other GTEx project partners to develop a plan for collecting the normal human biospecimens for GTEx.
A group of experts working in the ethical, scientific, and operational aspects of biobanking was assembled to identify key challenges of collecting normal biospecimens and to help develop a framework for the project. This planning group recognized that donations of normal biospecimens from living donors were rare, typically only occurring as a secondary research donation when surgery is performed for cancer treatment, limb amputation, or other surgical disease treatments. Thus it was envisioned that the GTEx biospecimens would be collected from deceased donors, due to the project's need for multiple tissues per individual donor.
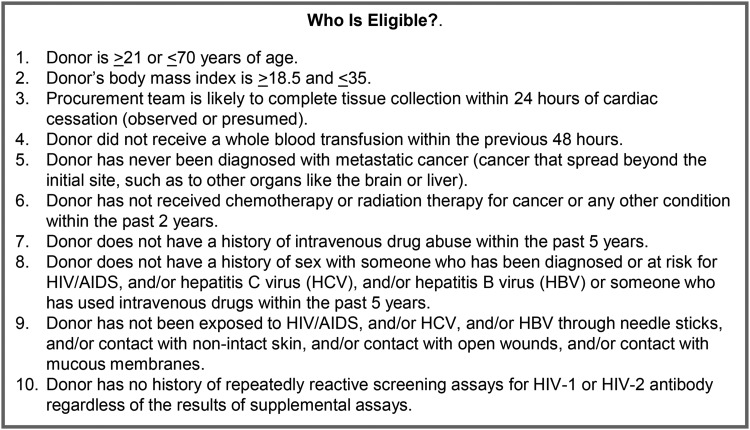
The GTEx project set out to partner with organ procurement organizations (OPOs) and institutions with rapid autopsy programs to obtain the high number of normal biospecimens needed for this study.4 The expert planning group established a comprehensive set of recommendations for acquiring high-quality, normal biospecimens. A summary of the recommendations has been published4 and the full recommendations are posted for public use (http://biospecimens.cancer.gov/global/pdfs/caHUB_ANTWG_Postmortem_BPs.pdf). GTEx management used these recommendations to develop the eligibility criteria for GTEx donors (Fig. 1).
FIG. 1.
GTEx Eligibility Requirements. A set of eligibility requirements were developed to align with the scientific needs of the project and to reduce the risk of collecting tissue that was diseased, autolyzed, or otherwise unsuitable for molecular analysis. The eligibility criteria were developed within a framework that considered the limited data immediately available in the timeframe surrounding a potential donor's death and the feasibility of the Biospecimen Source Sites to obtain the tissues and associated data in a timely manner.
Herein we provide an overview of how we developed a tissue collection platform focused on meeting the ethical, scientific, informatics, and operational challenges of biospecimen procurement for the GTEx project. The success of this project depended on tissue donations from families who have lost a loved one. With sincere respect and gratitude, we thank the GTEx donors and their families for their generous contributions.
Materials and Methods
Consenting donors and addressing ethical, legal and social issues
The medical institutions and OPOs that collected the biospecimens for the GTEx pilot study, Biospecimen Source Sites (BSS), chose to either submit a GTEx research protocol and undergo full or expedited IRB review, or upon consultation with their Office of Research Subject Protection determined that the research does not constitute human subjects research and did not require further review because the donors were deceased. The GTEx pilot study required explicit next-of-kin or legally authorized representative authorization for participation in the project.
Under the law, deceased individuals are not considered to be human subjects and do not require consent for research; however, GTEx management decided to require authorization due to the large amount of sequencing data to be produced and made publically available. With respect to consent, the BSSs sometimes had different approaches as to the specific manner in which authorization was obtained, but project staff worked closely with the BSSs to ensure that the basic requirements and essential consent elements (Table 1) for the GTEx project were met.
Table 1.
Essential GTEx Consent Elements
| A description that genetic and genomic research may be conducted on the donated biospecimens |
| The donated biospecimens may be shared with researchers who are approved by an access committee, including international researchers |
| The donated biospecimens may be used for broad future research |
| Commercial products may be developed using the donated biospecimens however the donor families will not financially profit from these products |
| There may be a risk of loss of privacy and confidentiality |
| The biospecimens may be withdrawn; however, molecular data may not be retrieved once it is generated |
| No individual genetic information will be returned to the next-of-kin or legal representative; however, results from the collective GTEx biospecimen set will be available on the GTEx portal ((http://www.gtexportal.org/home) and the NIH's National Center for Biotechnology Information's database of Genotypes and Phenotypes (dbGAP) http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000424.v1.p1 |
Consent was obtained in person as well as over the phone. The GTEx project included a sub-study that evaluated the attitudes and concerns of family decision-makers regarding the consent process and other ethical issues concerning the donation process.6 This sub-study provides information to help ensure that the project effectively addresses the concerns and expectations of the study participants. This sub-study also created and provided novel training materials for consenting personnel, in order to improve interactions with and understanding of donor families. The training can be found at http://gtextraining.org/.
Privacy and confidentiality of donor information also received careful consideration. Although the project collected a limited data set from the BSSs, only de-identified data (http://www.hhs.gov/ocr/privacy/hipaa/administrative/privacyrule/index.html) will be distributed to collaborators in the GTEx project or to downstream secondary researchers. An electronic data capture system, the Comprehensive Data Resource described later in this article, controlled the display and access to data based on user roles and entitlements. BSSs could only view data from their own sites, and only a limited number of approved staff members within the GTEx project could view protected health information. In addition, the project established a combined material transfer agreement/data use agreement that covered all parties receiving either biospecimens or data.
The template agreement is posted publicly at http://biospecimens.cancer.gov/global/pdfs/caHUB_Material_Transfer_and_Data_Use_Agreement_072512-508.pdf. While the development and negotiation of such an agreement required an initial time investment, the agreement clearly laid out responsibilities and requirements for privacy protection for all parties and was written broadly enough to cover the collection process throughout the course of the project.
Developing an infrastructure to support the collection of human biospecimens
To meet the challenging biospecimen requirements of the GTEx pilot study, NCI's BBRB and its partners developed a novel infrastructure for collecting normal human biospecimens for research purposes. BBRB, together with the Frederick National Laboratory for Cancer Research, developed an operational plan to contract medical centers and OPOs to screen potential donors, consent next-of-kin, and collect and ship biospecimens. Each BSS complied with their respective institution's biospecimen handling policies to procure GTEx biospecimens that were sent to a central, separately contracted Comprehensive Biospecimen Resource (CBR).
The CBR inventoried and divided biospecimens for three purposes: (1) shipment to the separately contracted GTEx Laboratory, Data Analysis, and Coordinating Center (LDACC) for molecular analysis, (2) histology for pathology review, and (3) local storage for future analysis. Whole brains were sent directly from the BSSs to a separate Brain Bank for proper sectioning and were then subsequently sent to the LDACC for analysis. Also, blood and skin samples were sent directly to the LDACC to generate EBV-transformed lymphoblastoid cell lines and fibroblasts, respectively.
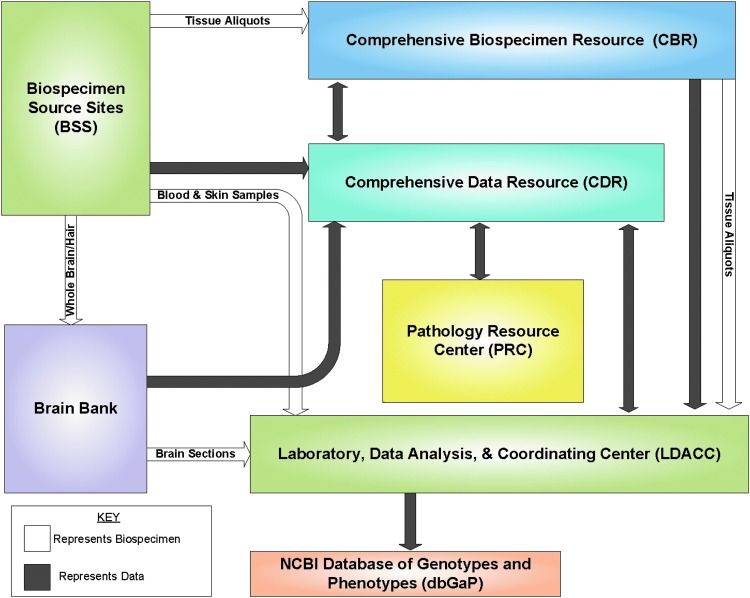
The entire biospecimen collection, processing, storage, and transfer operation was coordinated through a central quality control program and uses a custom web portal for data entry. A Pathology Resource Center (PRC), comprised of board-certified pathologists, evaluated the quality of the biospecimens. Figure 2 outlines the biospecimen platform and the flow of biospecimens and data to and from each site within the infrastructure. Detailed SOPs can be found at http://biospecimens.cancer.gov/resources/sops/library.asp that describe tissue procurement, shipping, kit utilization, data entry, pathology review, and other aspects of the GTEx project.
FIG. 2.
The GTEx Biospecimen Collection Infrastructure. The GTEx Biospecimen Source Sites were responsible for donor recruitment, tissue procurement and processing, and data collection. Brain and hair samples were sent to the Miami Brian Bank for quality control purposes, coronal sectioning of brain tissue, and storage of brain tissue. The Comprehensive Biospecimen Resource handled biospecimen receipt, processing, distribution and storage, histology and imaging, and kit development and production. The Comprehensive Data Resource is a data repository that served as an honest broker to keep limited data set information confidential and distribute de-identified data. The Pathology Resource Center performed case review through tissue identification and quality assessments. The Laboratory, Data Analysis, and Coordinating Center conducted molecular and data analysis as well as served as a project management and data-coordinating center. Clinical, demographic, handling, genetic and molecular data from GTEx biospecimens can be accessed through the National Center for Biotechnology Information's database of Genotypes and Phenotypes. Color images available online at www.liebertpub.com/bio
Pathology review
The PRC was created and implemented in the GTEx project to validate tissue origin, content, and integrity, and to ensure that the collected tissues meet prescribed quality standards. The PRC reviewed the disease state of the tissue for evidence of cancer, infectious disease, or inflammatory disease to confirm that collected biospecimens were “normal” or non-diseased, and subsequently annotate tissue dimensions and composition as well as determine acceptability of the final biospecimen for inventory.
After tissues were sectioned and stained at the CBR, tissue sections were scanned using a digital whole slide imaging system (Aperio).7 The PRC pathologists were able to review these images remotely via an Internet web portal. The PRC assessed and generated a report on multiple parameters, including verifying that the correct target tissue has been obtained and is of the correct size; the degree of autolysis; the presence of clinically unsuspected malignancy or infection; and the presence of significant “contaminant” but normal adjacent tissue, such as excessive adherent fat around an aliquot.
When all quality control measures were completed, the reports were made available to the LDACC to assess whether to go forward with genomic processing and to provide critical, real-time feedback in process improvement for the BSSs. Table 2 lists the criteria required for a biospecimen to be included in the molecular analysis pipeline.
Table 2.
Criteria Required for a Biospecimen to Be Included in The Molecular Analysis Pipeline
| Pathology review confirms that the correct target tissue has been obtained and comprises at least 50% of the sample (with few exceptions) |
| Pathology review confirms that the correct size of the target tissue has been obtained |
| Pathology review confirms that there is no presence of malignancy |
| RNA extracted from the biospecimen has at least 500 ng of total RNA |
| RNA extracted from the biospecimen has a RIN of 5.7 or higher |
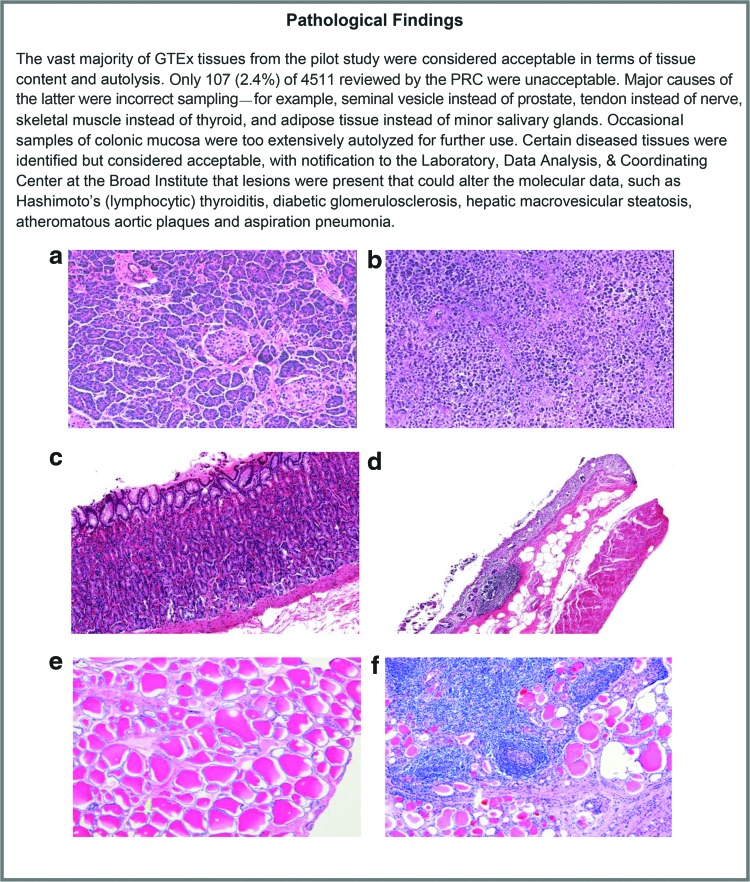
The pathology review added another layer of confidence when interpreting data from GTEx samples. Instances occurred when a biospecimen's gene expression profile did not correlate with the gene expression profiles of other samples from the same biospecimen type or same donor, for example, when a GTEx donor was the recipient of a donated organ. The pathology report helped to identify tissue types not suitable for the GTEx project due to tissue sampling inconsistencies, problems with poor preservation, and tissue heterogeneity issues (Fig. 3). Pathology review via rapid digital pathologic assessment of biospecimens streamlined the targeted collection of appropriate tissue types with consistent quality.
FIG. 3.
GTEx Pathology Review. Certain tissues were not considered acceptable for GTEx analysis purposes. (a, b) Autolysis. (a) Well-preserved pancreas with distinguishable exocrine and endocrine elements (RNA integrity number [RIN] 6.3). (b) Severely autolyzed pancreas (RIN 2.4). (c, d) Heterogeneous tissue sampling. (c) Well-preserved gastric mucosa with a RIN of 7.3. The higher RIN reflects multiple cell types: the abundant well-fixed gastric glands in the mucosal layer combined with muscularis mucosa. (d) Poorly preserved colon mucosa with a RIN of 7.3. Although the mucosa was also the intended target in this biospecimen, it was badly autolyzed and the RIN reflects the residual colon muscularis propria. (e, f) Acceptable diseased tissues. (e) Normal thyroid. (f) Thyroid with Hashimoto's germinal center formation was identified but was still considered to be eligible for the GTEx study. Color images available online at www.liebertpub.com/bio
Developing a total quality program
The establishment and implementation of a robust quality management program was integral for obtaining GTEx biospecimens that were suitable for genomic analysis. Hallmarks of established quality management methodology including data management, Standard Operating Procedure (SOP) development, and auditing have been adopted by the project.
A major challenge for GTEx was in the management of data associated with the biospecimens; each case could include more than 500 data elements. To help manage the data, a system for data queries was implemented and this process proved to be an effective method for producing well-organized, easily accessible, and reliable data. Implementation of data management techniques can be time consuming and burdensome. However, such techniques provided real time feedback to the BSSs on their protocol conduct, which resulted in site improvements and improved adherence to protocol.
Queries were also used to guide data collection best practices, resulting in an increase in data fidelity and a decrease in the issuance of certain query types. The development of these standards allowed for harmonization across all BSSs and was an important contributor to the project's successful acquisition of high quality biospecimens.
One lesson learned from the GTEx pilot study is that there is not a “one size fits all approach” to SOP development for biospecimen procurement; this lesson was largely based on institutional differences, many of which were unavoidable. The BSSs were consulted when new SOPs were developed to ensure that the SOPs were operationally feasible. Document control software was also utilized to ensure that current versions of SOPs were being used and training was conducted to ensure comprehension of new procedures. Approximately 100 supporting quality documents were developed to provide consistency and clarity to this complex project. Many of these documents are available to the public, including SOPs, workflows and project-related tools (http://biospecimens.cancer.gov/resources/sops/library.asp).
Results
Biospecimen annotation
The utility of a biospecimen for research purposes depends a great deal on the degree of annotation associated with the biospecimen. The GTEx pilot study used the Comprehensive Data Resource (CDR) to facilitate the input and analysis of multiple data types related to informed consent, the donor's medical history, biospecimen collection and handling, and pathology annotations. The CDR is a distributed, multi-tenant informatics platform that controls display and access to data based on user roles and entitlements. Personally identifiable information and protected health information were restricted to a limited data set and to individuals with authorized access through dynamic content redaction.
Web service application program interfaces (APIs) connected remote Laboratory Information Management Systems, whole-slide imaging systems and molecular analysis APIs for real-time, two-way data transfers. The CDR proved to be user-friendly, durable, and scalable. Its common data model was easy to query and easy to integrate with outside information technology systems for the benefit of sharing and exchanging data. The CDR was accessed by multiple users around the clock for entering and accessing data necessary for the collection, shipment, processing, and analysis of biospecimens.
The challenging biospecimen collection setting and differences among partner organizations complicated the process of uniform data collection for donor characteristics. For example, data related to the health of the donor could have been derived from multiple sources, including the donor's medical record (if available) and information from the donor's next-of-kin. Accordingly, additional CDR data fields were included to annotate the source of various information reported. Terminology related to data elements, such as medication name and medical condition or cause of death, varied across the BSSs, most often due to the use of similar but different terms or synonyms.
The CDR interfaced with a common vocabulary system, derived in part from the International Statistical Classification of Diseases (ICD-10-CM), Current Procedural Terminology (CPT4) and the National Drug Code, to standardize the data collected about GTEx donors and to improve comparability across information sets from different BSSs. These vocabulary elements were presented dynamically and in context to data entry personnel during data entry.
Data from the CDR was exported to the LDACC-developed GTEx web portal (http://www.gtexportal.org/home), where members of the public can request access to GTEx samples, and to dbGAP (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000424.v1.p1), where the research community can request access to genomic data and corresponding biospecimen data, such as clinical, demographic, and biospecimen handling data.
Number of biospecimens collected
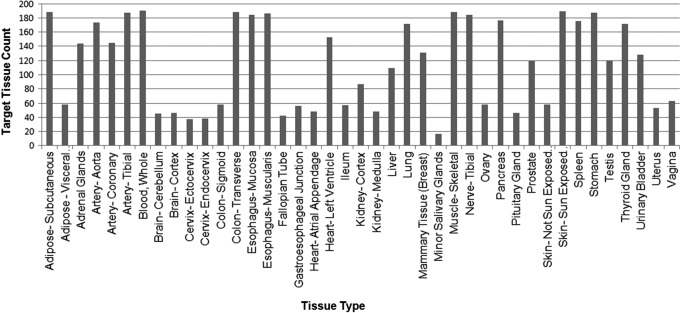
The GTEx pilot biospecimen collection included up to 41 different PAXgene preserved postmortem target tissue types (Fig. 4). The PAXgene Tissue preservation system9 developed by Qiagen was selected for GTEx because it facilitates good histological analysis as well as extraction of high-quality RNA and DNA. Hair, whole brain (frozen), and sections from 11 brain sub-regions (frozen) were also collected. The tissue types collected for GTEx were chosen based on their relevance to the scientific goals of the project, their clinical significance, logistical feasibility, and their relevance to the research community. Recovering target tissues from donors in rapid autopsy programs, where the donors were not donating tissue for transplant purposes, was frequently achievable. The OPO setting proved to be more challenging due to several factors, including the need to prioritize tissue and organ donations to living recipients and delays due to cause-of-death investigations; however this obstacle did not hinder GTEx from reaching its collection goal, ultimately procuring 10,152 PAXgene preserved tissue aliquots from190 donors.
FIG. 4.
The number and type of GTEx tissues collected. 41 different PAXgene preserved tissues types were collected for the GTEx project. Six tissues were female specific and two tissues were male specific. Only five tissues were mandatory for each case collected, which partially explains the significant differences among the number of tissues collected per tissue type.
RNA quality
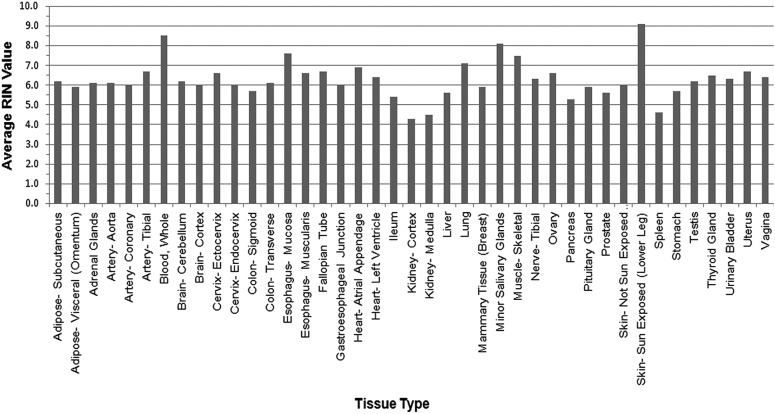
High quality RNA was extracted from the majority of the biospecimens collected for the GTEx project. Fig. 5 shows the average RIN values5 derived by the LDACC, on a per tissue basis for tissues collected from the first 190 GTEx cases (see Supplementary Text S1 for the percentages of individual tissue types that have RIN values greater than 6; Supplementary material is available online at www.liebertpub.com/bio). Eleven tissue types had at least 70% of the samples with RIN greater than 6, and more than 60% of all RNA from GTEx tissues had RIN values greater than 6, making them suitable for high dimensional genomic analyses. A RIN value of 6 was set as a goal because, in general, tissues with a RIN of 6 and above yield high quality RNA sequencing data, whereas tissues with a RIN value below 6 are more likely to have samples with failed sequencing reactions.8 At the outset of the project, we also tested RNAs of different quality (RIN 2.0–9.0) across multiple different library construction protocols for RNA sequencing. The most scalable and robust library construction protocol was the Illumina TruSeq protocol with Poly-A selection, which performed best with more intact RNA with RIN values of 6.0 or higher.
FIG. 5.
GTEx RIN values. Here we show the average RNA integrity number (RIN) values by tissue type for 190 cases. Twenty-four tissue types had average RNA integrity values greater than 6 during the pilot study.
Because autolysis sets in immediately after death and may adversely affect the quality of RNA from postmortem tissues, well-planned measures were taken to reduce the postmortem interval (PMI) for GTEx biospecimens. PMI is defined as the interval between the time of death or the cessation of blood flow and the time that the tissue is placed in preservative. The average RIN value was 8.6 for tissues with a PMI less than 4 hours, and 6.7 for tissues with a PMI between 4 and 8 hours, while tissues with a PMI of >8 hours resulted in average RIN values below 6 (Fig. 6). An hourly correlation between PMI and RIN is presented in Supplementary Text S2. Based on these findings, the GTEx project made an effort to collect all tissues from each GTEx case within 8 hours from time of death or cessation of blood flow to maximize the number of tissues that will present with RIN values ≥6.
FIG. 6.
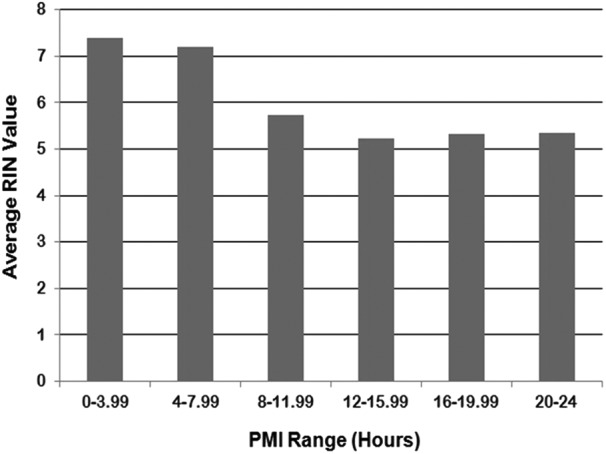
The effect of ischemic time on RIN values. RIN values greatly decreased when the postmortem interval (PMI) was ≥8 hours. PMI is based on interval between the time of death or the cessation of blood flow and the time that the last tissue is placed in preservative.
Discussion
A novel biospecimen collection platform was needed to address the many unique aspects of the GTEx project. Most established tissue networks do not contain a large number of normal, non-disease biospecimens, but instead have a collection of retrospective disease or condition-specific biospecimens, frequently with limited annotation available. BBRB met the GTEx pilot requirements by creating the infrastructure described here for the acquisition of normal biospecimens through contractual relationships with OPOs and rapid autopsy programs.
The biospecimen collection project of the GTEx pilot faced many challenges. Collecting biospecimens for research purposes using a novel preservation method (PAXgene Tissue) presented a new challenge for OPOs due to the fact that the staff needed to be trained in proper use of the preservative including the required step of switching from fix to stabilizer solutions. Ongoing feedback from project partners was essential to synergize project needs and local operations. Throughout the initiation, start-up and steady state phases of the pilot GTEx project, collaboration with all partners was paramount to the success of GTEx analysis, and continues to be an essential factor in the project.
The pilot phase GTEx biospecimens and data are already valuable resources for the scientific community. RNA sequencing data and expression array data generated by the LDACC, as well as clinical annotation for the GTEx biospecimen collection, are publicly available on dbGAP10 (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000424.v1.p1) and are being analyzed by an international team of bioinformatics experts.1
Multiple aliquots of GTEx tissues have been made available to the scientific community through a Request for Application funding mechanism (http://grants.nih.gov/grants/guide/rfa-files/RFA-RM-12-009.html) and a biospecimen access policy has been developed for already-funded investigators (http://www.gtexportal.org/static/form/GTEx_sample_access_policy_to_the_public_v20131024.doc). In addition, 40 SOPs from the GTEx project are available for public use and are a major contribution to the research biobanking community (http://biospecimens.cancer.gov/resources/sops/default.asp).
A meeting entitled “The GTEx Symposium: All Things Considered- Biospecimens, Omics and Data” was held on the NIH campus on May 20 and 21, 2015, that included presentations on each of the topics discussed in this report and some molecular analysis presentations. The meeting videocast was archived and can be accessed at http://videocast.nih.gov/Summary.asp?File=19024&bhcp=1 (May 20) and http://videocast.nih.gov/Summary.asp?File=19026&bhcp=1 (May 21). Project policy mandates that all data generated by researchers from the GTEx biospecimens must be made publicly available. Such availability will greatly enhance the value of GTEx resources by complementing other initiatives aimed at identifying functional elements in the human genome, such as the ENCyclopedia Of DNA Elements (ENCODE) project (http://www.genome.gov/10005107) and the Library of Integrated Network-based Cellular Signatures (LINCS) program (http://commonfund.nih.gov/lincs), in translating genome-wide association study findings to help prioritize the advancement of therapeutic targets.
One goal of the GTEx pilot study was to assess the feasibility of collecting a large number of normal, high-quality biospecimens. This goal was met in 2.5 years with the collection of samples from 190 donors. The first set of molecular analysis data from the GTEx pilot, including RNA sequencing data was published in May 2015.11–17 Due to the overwhelming proof of feasibility during the pilot, the NIH Common Fund committed support to continue the GTEx project. Therefore the GTEx study is ongoing and will continue through the end of 2015 and aims to collect a total of 900 cases.
To meet the demands of such a large biospecimen collection beyond the pilot phase, the GTEx project developed relationships with several additional BSSs and worked with all other partners to ensure that proper staffing and resources are in place to accommodate the influx of biospecimens collected.
The project has added the collection of frozen tissue samples for a limited number of tissue types based on feedback from a request for information (RFI) regarding the potential uses of stored GTEx biospecimens (http://grants.nih.gov/grants/guide/notice-files/NOT-RM-12-028.html). Adding frozen biospecimens will increase the utility of the biospecimen set for the research community in terms of being able to compare the GTEx tissue directly to past and future studies with frozen tissue since PAXgene preserved tissue is not yet widely utilized. The novel infrastructure put in place by BBRB and its partners is an essential part of the groundbreaking GTEx scientific project, which has the potential to change our understanding of gene regulation and how gene interactions contribute to disease development.
Supplementary Material
Acknowledgments
Members of the GTEx consortium include Laura Barker, Margaret Basile, Alexis Battle, Joy Boyer, Debra Bradbury, Jason P. Bridge, Amanda Brown, Robin Burges, Christopher Choi, Deborah Colantuoni, Nancy Cox, Emmanouil T. Dermitzakis, Leslie K. Derr, Michael J. Dinsmore, Kenyon Erickson, Johnelle Fleming, Timothée Flutre, Barbara A. Foster, Eric R. Gamazon, Gad Getz, Bryan M. Gillard, Roderic Guigó, Kenneth W. Hambright, Pushpa Hariharan, Rick Hasz, Hae K. Im, Scott Jewell, Ellen Karasik, Manolis Kellis, Pouya Kheradpour, Susan Koester, Daphne Koller, Anuar Konkashbaev, Tuuli Lappalainen, Roger Little, Jun Liu, Edmund Lo, John T. Lonsdale, Chunrong Lu, Daniel G. MacArthur, Harold Magazine, Julian B. Maller, Yvonne Marcus, Deborah C. Mash, Mark I. McCarthy, Jeffrey McLean, Bernadette Mestichelli, Mark Miklos, Jean Monlong, Magboeba Mosavel, Michael T. Moser, Sara Mostafavi, Dan L. Nicolae, Jonathan Pritchard, Liqun Qi, Kimberly Ramsey, Manuel A. Rivas, Barnaby E. Robles, Daniel C. Rohrer, Mike Salvatore, Michael Sammeth, John Seleski, Saboor Shad, Laura A. Siminoff, Matthew Stephens, Jeff Struewing, Timothy Sullivan, Susan Sullivan, John Syron, David Tabor, Mehran Taherian, Jorge Tejada, Gary F. Temple, Jeffrey A. Thomas, Alexander W. Thomson, Denee Tidwell, Heather M. Traino, Zhidong Tu, Dana R. Valley, Simona Volpi, Gary D. Walters, Lucas D. Ward, Xiaoquan Wen, Wendy Winckler, Shenpei Wu, and Jun Zhu. The authors would like to thank the donors and their families for their generous tissue donation to the GTEx study.
The authors would also like to thank the following individuals for their previous and ongoing efforts in the GTEx study: Assya Abdallah, Anjene Addington, James M. Anderson, Patrick K. Bender, Mark Cosentino, Norma Diaz-Mayoral, Theresa Engel, Fernando Garci, Allen Green, Tiffanie Hammond, Katherine Jaffe, Judy Keen, Mary Kennedy, Peter Kigonya, Brent Lander, Sreenath Nampally, Cathy Ny, James Robb, Vikram Santhanum, Nataliya Sharopova, Shilpi Singh, Conrado Soria, Anne Sturcke, Surendra Sukari, Elizabeth J. Thomson, Magda Tomaszewski, Casandra Trowbridge, Ferdinand Udoye, David Vanscoy, Negin Vatanian, Elizabeth L. Wilder, and Penelope Williams.
This work was supported by the National Institutes of Health (HHSN261200800001E (Leidos Prime contract with NCI); 10XS170 (NDRI), 10XS171 (Roswell Park Cancer Institute), 10X172 (Science Care Inc.), 12ST1039 (IDOX); 10ST1035 (Van Andel Institute); HHSN268201000029C (Broad Institute); and R01 DA006227-17 (U Miami Brain Bank).
Author Disclosure Statement
The authors disclosed no conflicting financial interests. J.V is a member of the Board of Directors of NDRI.
References
- 1.The GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nature Genetics 2013;45:580–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun W, Hu Y. eQTL mapping using RNA-seq data. Stat Biosci 2013;5:198–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The GTEx Consortium. The Genotype-Tissue Expression (GTEx) Pilot Analysis: Multi-tissue gene regulation in humans. Science 2015;348:648–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mucci NR, Moore H, Brigham LE, et al. Meeting research needs with postmortem biospecimen donation: Summary of recommendations for postmortem recovery of normal human biospecimens for research. Biopreserv Biobank 2013;11:77–82 [DOI] [PubMed] [Google Scholar]
- 5.Schroeder A, Mueller O, Stocker S, et al. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol 2006;7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siminoff LA, Traino HM, Mosavel M, Barker L, Gudger G, Undale A. Family decision maker perspectives on the return of genetic results in biobanking research. Genet Med 2015. Epub ahead of print 9 April 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Staniszewski W. Virtual microscopy, data management and image analysis in Aperio ScanScope system. Folia Histochem Cytobiol 2009;47:699–701 [DOI] [PubMed] [Google Scholar]
- 8.Gallego Romero I, Pai AA, Tung J, Gilad Y. RNA-seq: Impact of RNA degradation on transcript quantification. BMC Biol 2014;12:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groelz D, Sobin L, Branton P, Compton C, Wyrich R, Rainen L. Non-formalin fixative versus formalin-fixed tissue: A comparison of histology and RNA quality. Exper Mol Pathol 2013;94:188–194 [DOI] [PubMed] [Google Scholar]
- 10.Mailman MD, Feolo M, Jin Y, et al. The NCBI dbGaP database of genotypes and phenotypes. Nature Genetics 2007;39:1181–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015;348:648–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baran Y, Subramaniam M, Biton A, et al. The landscape of genomic imprinting across diverse adult human tissues. Genome Res 2015;25:927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibson G. GTEx detects genetic effects. Science 2015;348:640–641 [DOI] [PubMed] [Google Scholar]
- 14.Mele M, Ferreira PG, Reverter F, et al. The human transcriptome across tissues and individuals. Science 2015;348:660–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pierson E, Consortium GT, Koller D, Battle A, Mostafavi S. Sharing and specificity of co-expression networks across 35 human tissues. PLoS Comput Biol 2015;11:e1004220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rivas MA, Pirinen M, Conrad DF, et al. Effect of predicted protein-truncating genetic variants on the human transcriptome. Science 2015;348:666–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roosing S, Hofree M, Kim S, et al. Functional genome-wide siRNA screen identifies as mutated in Joubert syndrome. eLife. 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.