Abstract
The Gram-negative bacterium, Vibrio parahaemolyticus, is a major cause of sea-food-derived food poisoning throughout the world. The pathogenicity of V. parahaemolyticus is attributed to several virulence factors, including two type III secretion systems (T3SS), T3SS1 and T3SS2. Herein, we compare the virulence of V. parahaemolyticus POR strains, which harbor a mutation in the T3SS needle apparatus of either system, to V. parahaemolyticus CAB strains, which harbor mutations in positive transcriptional regulators of either system. These strains are derived from the clinical RIMD 2210633 strain. We demonstrate that each mutation affects the virulence of the bacterium in a different manner. POR and CAB strains exhibited similar levels of swarming motility and T3SS effector production and secretion, but the CAB3 and CAB4 strains, which harbor a mutation in the T3SS2 master regulator gene, formed reduced biofilm growth under T3SS2 inducing conditions. Additionally, while the cytotoxicity of the POR and CAB strains was similar, the CAB2 (T3SS1 regulatory mutant) strain was strikingly more invasive than the comparable POR2 (T3SS1 structural mutant) strain. In summary, creating structural or regulatory mutations in either T3SS1 or T3SS2 causes differential downstream effects on other virulence systems. Understanding the biological differences of strains created from a clinical isolate is critical for interpreting and understanding the pathogenic nature of V. parahaemolyticus.
Keywords: T3SS, Vibrio parahaemolyticus, bacterial virulence, invasion, biofilm
Introduction
Virulent strains of the Gram-negative halophilic bacterium, Vibrio parahaemolyticus, are emerging in coastal regions throughout the world (Nair et al., 2007). Vibrio parahaemolyticus thrives in warm brackish waters and is one of the leading causes of seafood-derived food poisoning due to the consumption of raw or undercooked shellfish. Infection of this pathogen can lead to acute gastroenteritis and even septicemia (Broberg et al., 2011). Additionally, strains of V. parahaemolyticus have recently been associated with a lethal infection of shrimp, resulting in the devastation of shrimp farms (Tran et al., 2013). The global dissemination of V. parahaemolyticus underscores the need for a better understanding of the bacterium’s virulence traits.
Principle to the pathogenicity of V. parahaemolyticus is thermostable direct hemolysins (TdhA/S) and two type III secretion systems (T3SS1 and T3SS2) (Broberg et al., 2011). The T3SSs form a needle-like apparatus to translocate effector proteins directly into the host cytoplasm to manipulate host cells (Fig. 1a). T3SS1 contains several effectors that systematically kill host cells by causing autophagy, membrane blebbing, cell-rounding, and finally lysis (Burdette et al., 2008; Broberg et al., 2010). T3SS2 is necessary for the enterotoxicity seen in piglet and infant rabbit models (Pineyro et al., 2010; Ritchie et al., 2012). Effectors from T3SS2 promote bacterial cell invasion, inactivation of the host immune pathway, and disruption of the gut epithelial barrier (Ritchie et al., 2012; Zhang et al., 2012; Zhou et al., 2013).
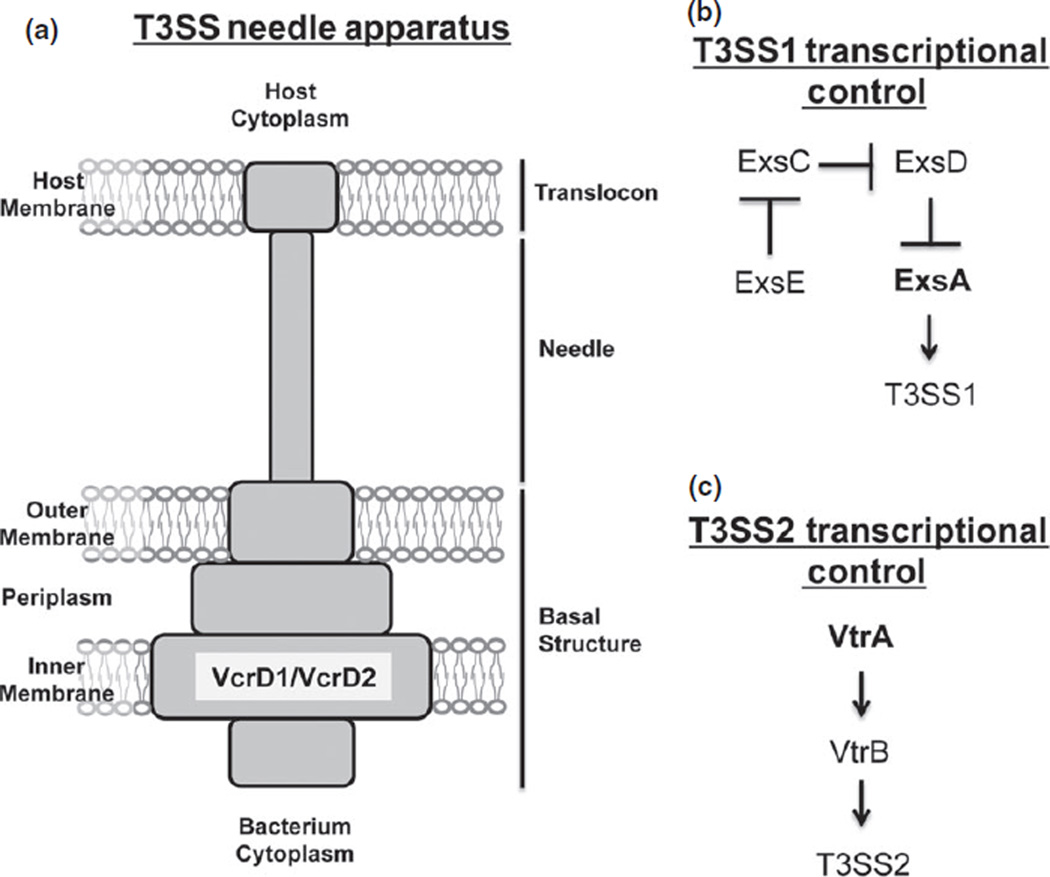
Fig. 1.
Schematic representation of T3SS needle apparatus and the transcriptional networks controlling T3SS1 and T3SS2 gene expression. (a) The T3SS is composed of a needle that spans the inner and outer membranes of the bacterium, and injects effector proteins directly into the host cytoplasm. The POR strains contain mutations in the vcrD1 and vcrD2 genes that are an essential component of the inner membrane ring of the needle’s basal structure in T3SS1 and T3SS2, respectively. (b) T3SS1 gene expression is controlled by the ExsACDE cascade. The exsA gene was deleted in the CAB2 strain to prevent induction of T3SS1 genes. (c) T3SS2 gene expression is controlled by the sequential activation of the transcriptional regulators VtrA and VtrB. The vtrA gene was deleted in the CAB3 strain to prevent induction of T3SS2 genes.
The two T3SSs of V. parahaemolyticus are regulated by separate transcriptional networks. Low calcium levels induce T3SS1 gene expression, which is regulated by the ExsACDE cascade (Fig. 1b) (Kodama et al., 2010a). ExsA is an AraC family transcription factor that binds to the promoter region of T3SS1 genes to promote their expression (Zhou et al., 2008). Dulbecco’s modified Eagle’s medium (DMEM) with low calcium is often used to prime bacteria for T3SS1-mediated infection (Burdette et al., 2008). T3SS2 genes are induced by bile salts through activation of the transcription factors VtrA and VtrB (Fig. 1c) (Gotoh et al., 2010). Both of these transcription factors are positive regulators that work in a sequential fashion to promote expression of T3SS2 genes (Kodama et al., 2010b).
Two sets of strains, PORs and CABs, were created with mutations in either a T3SS needle apparatus component or a transcriptional regulator, respectively (Table 1). These strains were derived from the POR1 strain, which was originally derived from the virulent O3:K6 serotype termed RIMD 2210633 by deleting the tdhA/S toxins (Park et al., 2004a). Therefore, the POR and CAB strains carry mutations in two toxin genes, tdhA and tdhS, to eliminate the toxic effects of these hemolysins on host cells (Hiyoshi et al., 2010). POR2 (POR1ΔvcrD1) and POR3 (POR1ΔvcrD2) contain a deletion in a gene that encodes the inner membrane ring of the needle apparatus belonging to either T3SS1 or T3SS2, respectively (Fig. 1a). POR4 contains mutations in both T3SS1 and T3SS2 needles and therefore lacks the ability to secrete all T3SS effector proteins (Broberg et al., 2011). In contrast, the CAB strains contain mutations that disrupt gene expression. CAB2 (POR1ΔexsA) and CAB3 (POR1ΔvtrA) contain a deletion of a master regulator responsible for controlling expression of either T3SS1 or T3SS2 genes, respectively (Fig. 1b and c). CAB4 contains both deletions and is entirely deficient in T3SS1 and T3SS2 gene expression (Zhang et al., 2012).
Table 1.
Effector production and secretion from POR and CAB strains. The symbol ‘+/−’ indicates a reduced level of effector protein
| Produce T3SS1 effectors |
Secrete T3SS1 effectors |
Produce T3SS2 effectors |
Secrete T3SS2 effectors |
|
|---|---|---|---|---|
| POR1 | + | + | + | + |
| POR2 | +/− | − | + | + |
| POR3 | + | + | + | − |
| POR4 | +/− | − | + | − |
| CAB2 | − | − | + | + |
| CAB3 | + | + | − | − |
| CAB4 | − | − | − | − |
Mutations in the transcriptional networks of each T3SS can have unexpected consequences that must be evaluated for proper interpretation of experimental results. For example, ExsA and VtrA have been shown by microarray and RNAseq analysis to affect gene expression outside the pathogenicity island of T3SS1 and T3SS2 (Gotoh et al., 2010; Kodama et al., 2010a, b; Gode-Potratz et al., 2011; Nydam et al., 2014). This study compares the different virulence-related aspects of strains with a mutation in a T3SS transcriptional regulator gene as opposed to a mutation in a T3SS structural gene. We examined effector production and secretion, swarming, biofilm formation, cytotoxicity, and invasion of host cells. From this study, we found that biofilm growth is inversely regulated with T3SS2 activation in a VtrA-dependent manner and that CAB2, a T3SS1 regulatory mutant, invades HeLa cells with an efficacy over 10-fold higher than that of POR2, a T3SS1 structural mutant. These traits highlight the impact of a structural vs. regulatory manipulation of T3SSs on the overall virulence ability of V. parahaemolyticus and suggest the T3SS transcriptional regulators impact other virulence-related systems.
Materials and methods
Strains and media
The POR2 (POR1ΔvcrD1), POR3 (POR1ΔvcrD2), POR4 (POR1ΔvcrD1/vcrD2) (Park et al., 2004b), CAB2 (POR1ΔexsA), CAB3 (POR1ΔvtrA), CAB4 (POR1ΔexsA/vtrA) (Zhang et al., 2012), POR1ΔlafK, and POR1Δhns (Salomon et al., 2014) strains were derived from POR1 (ΔtdhA/tdhS) (Park et al., 2004a), which was derived from the parental RIMD 2210633 strain (Makino et al., 2003). Vibrio parahaemolyticus strains were routinely grown in marine LB (MLB) media (Luria–Bertani broth containing 3% NaCl) at 30 °C. To induce T3SS1 or T3SS2 genes expression, bacteria were grown overnight and diluted to OD600 = 0.3 in DMEM or MLB with 0.05% bile salts, respectively (Broberg et al., 2010; Gotoh et al., 2010). Bacteria were then transferred to 37 °C for 1 h for infection assays and 3 h for swarming experiments and effector secretion assays. HeLa cells (ATCC) were cultured in DMEM (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (Sigma) at 37 °C in 5% CO2.
Growth curve
POR and CAB strains were diluted to OD600 = 0.1 in 30 mL of MLB at 30 °C. OD600 was measured every 30 min for each strain.
Secretion assay
After T3SS1 and T3SS2 induction, bacterial cells and supernatant were harvested as previously described (Salomon et al., 2013). Samples were examined by Western blot analysis using antibodies: rabbit anti-VopQ (Burdette et al., 2009), rabbit anti-VopS (Yarbrough et al., 2009), rabbit anti-VopA, and rabbit anti-VopL (Liverman et al., 2007). Effector antibodies were generated by Strategic Biosolutions (Newark, DE) using recombinant proteins. The experiment was performed twice with similar results and a representative experiment is shown.
Swarming motility assay
For swarming motility assays, bacteria were grown overnight and normalized to OD600 = 1. After T3SS1/T3SS2 induction or no induction, 2 µL of culture was spotted onto heart infusion (HI) plates containing 2% NaCl and 1.5% agar. Swarming plates were incubated at 30 °C for 24 h and then swarming diameter was measured.
Biofilm growth
Vibrio parahaemolyticus cultures were grown overnight and diluted to OD600 = 0.1 in MLB media, DMEM, or MLB media with 0.05% bile salts. Bacteria were grown aerobically in 96-well microtiter plate at 37 °C for 24 h. Bacteria cultures were then removed, washed, and stained with 0.1% crystal violet for 15 min as described (O’Toole, 2011). Wells were washed and dried overnight. The remaining crystal violet was resuspended in 30% acetic acid, and the absorbance was measured at 595 nm. Each experiment was performed three times with similar results and representative experiments are shown.
LDH release assay
After T3SS1 and T3SS2 induction, V. parahaemolyticus strains were added at a multiplicity of infection (MOI) of 10 to HeLa cells in DMEM containing no serum or phenol red. Plates were centrifuged at 1000 g for 5 min to synchronize infection, and then incubated at 37 °C. Next, samples were collected and LDH activity in the medium was measured using a Cytotoxicity Detection kit (Takara). Percentage cytotoxicity is compared to total lysed cells in 1% Triton X-100. Each experiment was performed three times with similar results and representative experiments are shown.
Confocal microscopy
After infection, HeLa cells were fixed in 3.2% paraformaldehyde and then permeabilized with 0.5% Triton X-100. The nuclei were stained with Hoescht (Sigma), and the actin was stained with Rhodamine-phalloidin (Molecular probes). Images were taken with a Zeiss LSM 510 scanning confocal microscope.
Invasion assay
Invasion of V. parahaemolyticus strains was performed as described (Zhang et al., 2012). In brief, pre-induced V. parahaemolyticus strains were added at a MOI of 10 to HeLa cells, and the plates were centrifuged at 1000 g for 5 min. After a 2-h incubation at 37 °C, 100 µg mL−1 gentamicin was added to kill extracellular bacteria. At 3 h, after the addition of gentamicin, bacterial numbers were determined by serially diluting samples on minimal marine media (77 mM K2HPO4, 35 mM KH2PO4, 20 mM NH4Cl, 5 mM K2SO4, 2% NaCl, 0.4% galactose, and 1.5% agar) plates. Experiment was performed twice with similar results and representative experiment is shown.
Results
POR and CAB strains exhibit similar growth rates
To assess the differences in virulent traits of mutant strains lacking a T3SS needle apparatus component or transcriptional regulator, we compared the virulence phenotypes of the POR and CAB strains. Importantly, the T3SS mutations did not affect the bacteria’s growth rate (Supporting Information, Fig. S1) and, therefore, the differences seen are independent of this factor.
While POR and CAB strains differ in effector production, they appear to secrete similar levels of effector protein
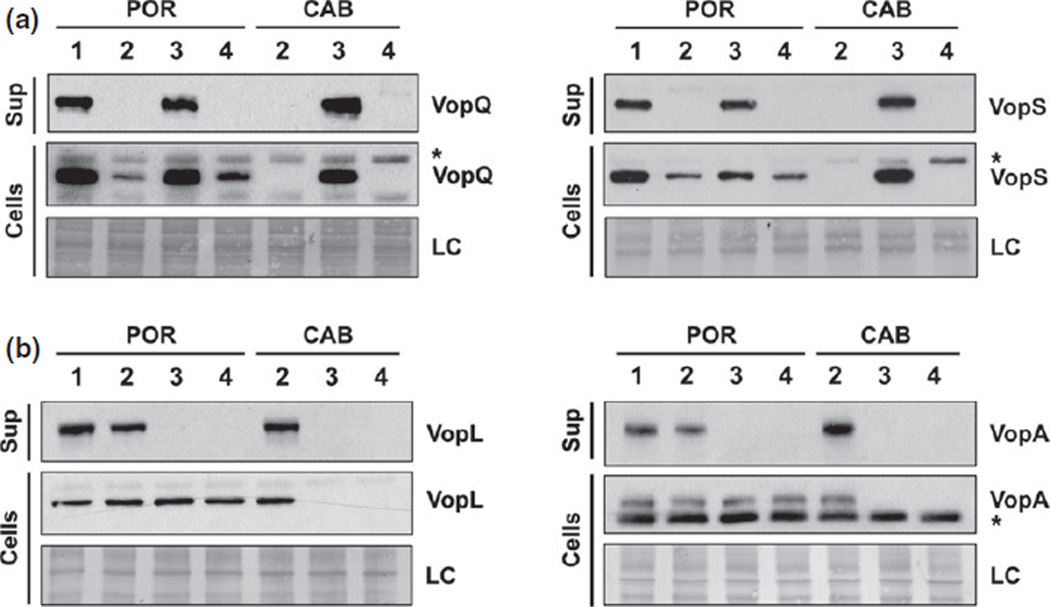
Next, we examined if POR and CAB strains produce and secrete similar levels of effector proteins. To test this, we induced each T3SS for 3 h and detected effector levels in bacterial lysates and culture supernatants. When T3SS1 was induced and functional (POR1, POR3, and CAB3), the T3SS1 effectors VopQ and VopS were produced and secreted at similar levels between POR and CAB strains (Fig. 2a). Interestingly, there was a difference in effector production within POR strains, which supports the hypothesis that T3SS1 needle construction is necessary for optimal effector production. Additionally, when T3SS2 was induced and functional (POR1, POR2, and CAB2), the T3SS2 effectors VopA and VopL were produced and secreted at similar levels in all strains (Fig. 2b). Overall, structural or regulatory mutations of each T3SS affect the secretion of its cognate effectors but do not affect secretion of effectors by the other T3SS.
Fig. 2.
T3SS1 and T3SS2 effectors are produced and secreted at similar levels by POR and CAB strains. (a) Secretion of T3SS1 effectors, VopQ and VopS, from T3SS1-induced strains grown in DMEM for 3 h at 37 °C. (b) Secretion of T3SS2 effectors, VopA and VopL, from T3SS2-induced strains grown in MLB with 0.05% bile salts for 3 h at 37 °C. Proteins from bacterial lysates or TCA-precipitated media detected by immunoblot analysis using anti-VopQ, anti-VopS, anti-VopA, and anti-VopL antibodies. Asterisk represents nonspecific band recognized by antibody and LC is abbreviation for loading control.
Swarming is similar for POR and CAB strains
Lateral flagella are produced by V. parahaemolyticus to transport the bacterium through viscous environments and along surfaces. Swarming is important during infection and is co-regulated with T3SS1 genes by surface contact, quorum sensing, and levels of calcium and iron (Gode-Potratz et al., 2010, 2011; Gode-Potratz & McCarter, 2011). We tested the swarming abilities of POR and CAB strains on HI swarm agar for 24 h and found no differences in migration (Fig. S2a). As a negative control, we mutated the gene lafK, which encodes an essential regulator of swarming gene expression (Stewart & McCarter, 2003). We then induced T3SS1 and T3SS2 prior to spotting the bacteria on the swarm plates. Again, there was no difference in bacterial migration upon T3SS1 (Fig. S2b) or T3SS2 induction (Fig. S2c), supporting the proposal that the mutations within the POR and CAB strains have no effect on the regulation of swarming.
VtrA represses biofilm growth under bile salt induction
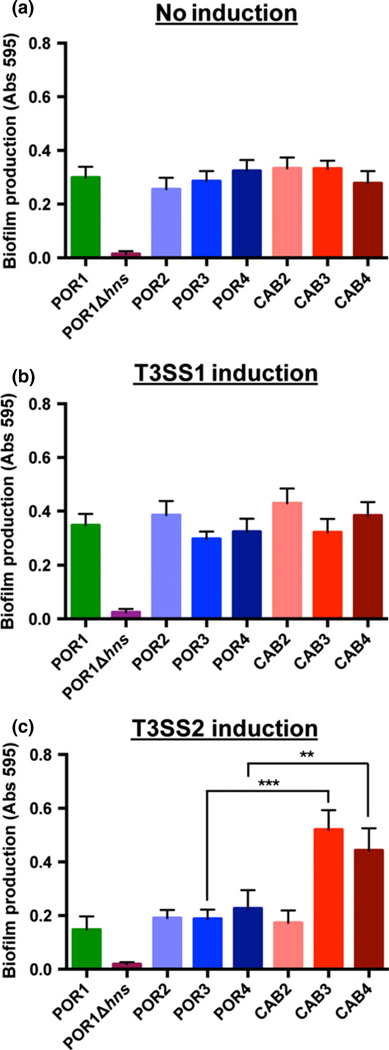
Biofilm production is important for bacteria survival in different environments and shares an inverse relationship with swarming and T3SS1 gene induction in V. parahaemolyticus (Boles & McCarter, 2002; Kim & McCarter, 2007; Gode-Potratz & McCarter, 2011). To assess biofilm production, we grew the POR and CAB strains statically in microtiter plates for 24 h and stained the resulting biomass with crystal violet. A mutant lacking the hns gene served as a negative control, as it does not exhibit biofilm growth (Gode-Potratz & McCarter, 2011). Growing the bacteria in non-inducing or T3SS1-inducing conditions caused no variation in biofilm production between POR and CAB strains (Fig. 3a and b). However, when grown in T3SS2-inducing conditions, CAB3 and CAB4 produced almost twofold higher amounts of biofilm growth when compared to the POR strains or CAB2 strain, which also produced less biofilm growth than non-induced strains (Fig. 3c). These experiments show that biofilm production is optimal in the absence of VtrA activation.
Fig. 3.
Bile salts reduce biofilm formation through vtrA. Levels of biofilm growth from strains grown statically in (a) non-inducing (MLB) media, (b) T3SS1 inducing (DMEM) media, and (c) T3SS2-inducing (MLB + 0.05% bile salts) media. Resulting biomass was stained with 0.1% crystal violet and measured at absorbance 595 nm. Asterisks indicate statistical significant differences using two-tailed t-test (**P < 0.005, ***P < 0.0005), and error bars show standard deviation (n = 8).
Cytotoxicity is similar between POR3 and CAB3 strains under T3SS1-inducing conditions
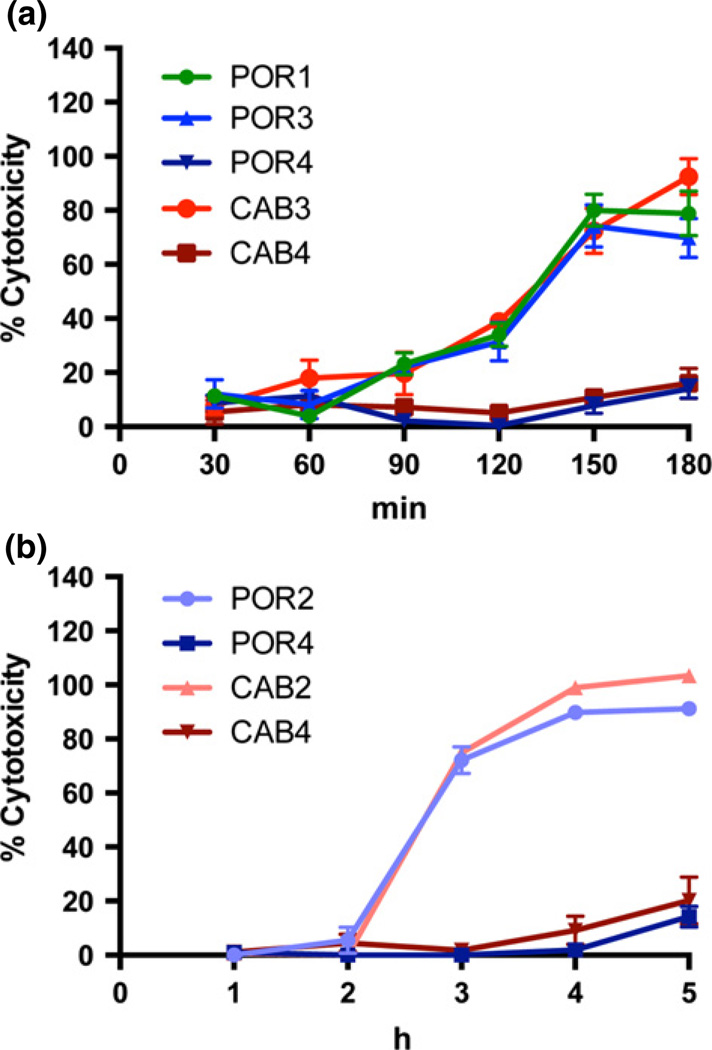
We next assessed whether the POR3 and CAB3 strains exhibit different levels of T3SS1-dependent cytotoxicity toward HeLa cells. After pre-inducing T3SS1, we infected HeLa cells and measured LDH release every 30 min. Both POR3 and CAB3 caused host cell lysis within 2.5 h at a rate similar to the POR1 parental strain. As expected, POR4 and CAB4 were not cytotoxic (Fig. 4a).
Fig. 4.
POR and CAB strains exhibit similar rates of cytotoxicity. Cytotoxicity measured as LDH release from HeLa cells infected with POR and CAB strains pre-induced in (a) T3SS1-inducing (DMEM) media, and (b) T3SS2-inducing (MLB + 0.05% Bile salts) media. Error bars show standard deviation (n = 3).
In addition to cell lysis, we examined the ability of POR3 and CAB3 to cause T3SS1-dependent cell rounding, which is a well-documented phenotype resulting from actin cytoskeleton disruption (Burdette et al., 2008; Yarbrough et al., 2009). In accordance with the LDH release assay, POR3 and CAB3 caused rounding at a rate similar to that of the POR1 parental strain. The negative control strains, POR4 and CAB4, did not cause any noticeable alteration in the actin cytoskeleton (Fig. S3). These results show that POR3 and CAB3 strains exhibit comparable levels of cytotoxicity upon induction of T3SS1.
Cytotoxicity is similar between POR2 and CAB2 strains upon T3SS2 induction
We next examined the cytotoxicity rates of POR2 and CAB2 after T3SS2 induction. POR1 was omitted from these experiments as this strain contains an active T3SS1 which induces faster lysis than T3SS2. After pre-inducing T3SS2, we infected HeLa cells and measured LDH release every hour. We found that POR2 and CAB2 caused c. 100% cytotoxicity at a similar rate within 4 h. POR4 and CAB4 started causing detectable lysis at 5 h (Fig. 4b), but this was likely due to overgrowth of the bacteria (data not shown).
POR and CAB strains differ in T3SS2-mediated invasion efficiency
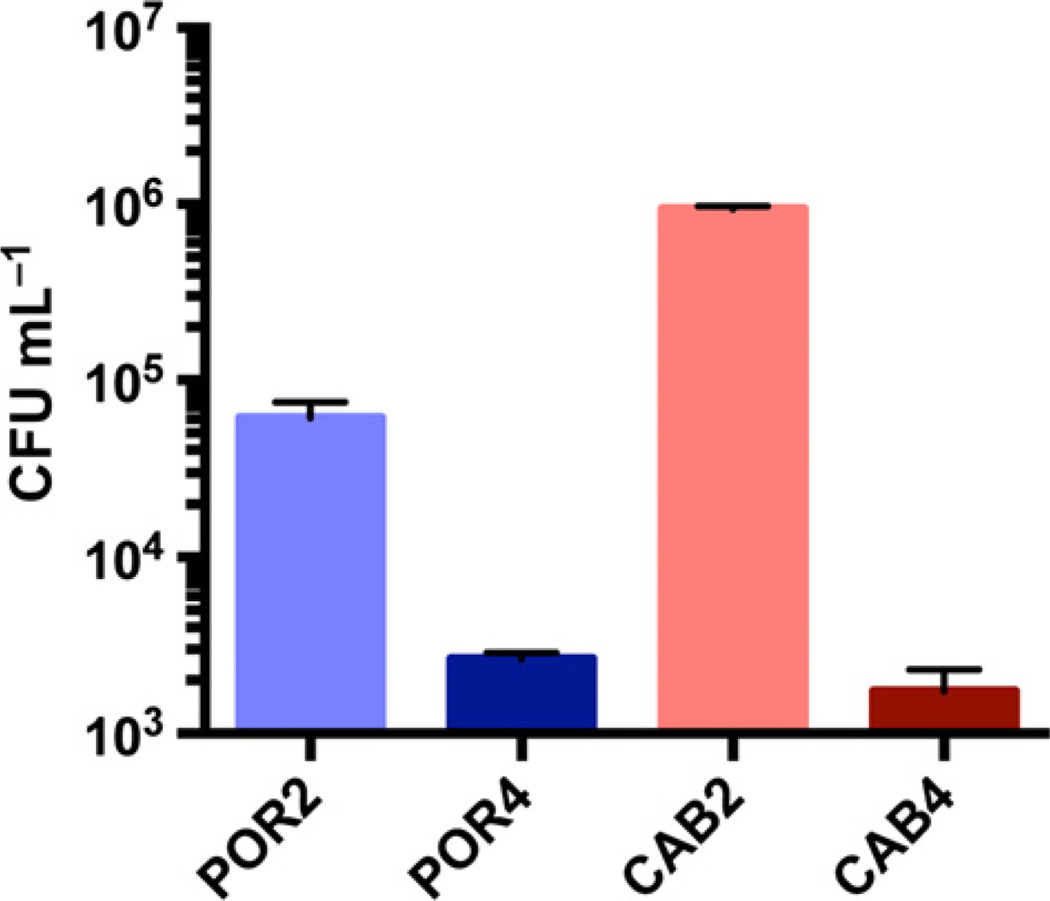
Recently, it was shown that V. parahaemolyticus invades host cells in a T3SS2-dependent manner (Zhang et al., 2012). Therefore, we tested the ability of POR2 and CAB2 strains to invade HeLa cells during an infection. HeLa cells were incubated with bacteria for 2 h and then inoculated with gentamicin. This antibiotic is nonpermeable to membranes and will only kill extracellular bacteria (Zhang et al., 2012). At 3 h after gentamicin addition, CAB2 exhibited over 10-fold higher levels of invasion compared to the POR2 strain, and this difference was maintained throughout the course of the infection. The negative control strains, POR4 and CAB4, exhibited very low levels of invasion (Fig. 5). We also tested bacterial invasion using 293 T cells and found a similar trend in which the CAB2 strain exhibited a twofold higher level of invasion than the POR2 strain (data not shown). This experiment shows that CAB2 invades host cells more efficiently than POR2 by an unknown mechanism.
Fig. 5.
ExsA mutant exhibits better invasion than T3SS structural mutant. HeLa cell invasion with POR and CAB strains. HeLa cells were incubated with bacteria at MOI of 10 for 2 h in 1 mL total volume prior to inoculation with 100 µg mL−1 gentamicin to kill extracellular bacteria. Intracellular bacteria quantified 3 h after gentamicin treatment as colony forming units per milliliter (CFU mL−1). Error bars show standard deviation (n = 3).
Discussion
In this study, we investigated the virulence differences between structural or regulatory mutants of the V. parahaemolyticus T3SSs. Previous studies of V. parahaemolyticus reported that swarming and T3SS1 gene expression are co-regulated, and biofilm production and T3SS1 genes expression are inversely regulated (Gode-Potratz & McCarter, 2011; Ferreira et al., 2012). Yet despite this connection, we found no variation in swarming between the POR and CAB strains under a variety of induction conditions, and no difference in biofilm growth during noninduction or T3SS1 induction. Interestingly, we found that T3SS2 regulatory mutants could produce larger biofilms than other mutants during T3SS2 induction. Furthermore, POR1-4 and CAB2 strains (Fig. 3c) produced less biofilm mass than when these strains were not induced (Fig. 3a). This suggests that biofilm formation is repressed in a VtrA-dependent manner upon induction with bile salts.
One interesting observation from this study was that POR1 and POR3, in which T3SS1 is functional, exhibited higher levels of effector production than POR2 and POR4, in which T3SS1 is nonfunctional, during T3SS1 induction. This result would support the hypothesis that a factor dependent on a functional needle apparatus alters the expression levels of T3SS1 genes. ExsE, a negative regulator of this system, is likely the factor responsible as it is secreted from the bacterium during induction of T3SS1 (Fig. 1b) (Kodama et al., 2010a, b). Therefore, the repressive role of ExsE is likely maintained in POR2 and POR4 strains as it is accumulating inside the cell, and this could explain the low levels of T3SS1 effector production.
While the POR and CAB strains exhibited similar rates of cytotoxicity, a striking result was observed when both strains were tested for invasion efficiency. CAB2-invaded HeLa cells with an efficacy over 10-fold higher than that of POR2 in HeLa cells. The T3SS2 effector, VopC, mediates invasion by constitutively activating Rac and CDC42 through deamidation (Zhang et al., 2012; Okada et al., 2013). However, POR2 and CAB2 appear to produce and secrete similar levels of effector proteins, which suggest the T3SS transcriptional regulators may alter the expression of non-T3SS genes that affect invasion efficiency. We are currently exploring this hypothesis.
In this study, we demonstrated that certain virulence traits differ between T3SS needle mutants and regulatory mutants. The ability of the bacteria to share similar virulence traits while differing in other aspects illustrates the importance of performing multiple assays to illuminate strain variation. This point is demonstrated best by T3SS2-mediated phenotypes. While POR2 and CAB2 did not differ in cytotoxic ability, they differed greatly in invasion efficiency. Therefore, subtle differences do exist between strains harboring structural or regulatory mutations, and these differences are only found by examining all aspects of infection. Future studies on the expression profiles of these various strains should elucidate specific genes involved in the infection process.
Acknowledgements
We thank members of the Orth laboratory for helpful discussions. This work is supported by NIH Allergy and Infectious Disease (R01-AI087808) and the Welch Foundation (I-1561.) K.O. is a Burroughs Wellcome Investigator in Pathogenesis of Infectious Disease and a W.W. Caruth Jr. Biomedical Scholar and an Earl A. Forsythe Chair in Biomedical Science. TC is supported by T32 AI007520.
Footnotes
We have no conflict of interest to declare.
Author contributions
T.C. and K.O. conceived and designed the experiments. T.C., V.A., J.K., J.F., and M.S. performed the experiments. T.C., D.S., A.K., and K.O. analyzed the data. T.C. and K.O. contributed reagents/materials/analysis tools. T.C. and K.O. wrote the paper.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. POR and CAB strains grow at similar rates.
Fig. S2. POR and CAB strains exhibit similar levels of swarming.
Fig. S3. The T3SS1 proficient strains, POR3 and CAB3, cause similar rates of actin rearrangement in HeLa cells.
References
- Boles BR, McCarter LL. Vibrio parahaemolyticus scrABC, a novel operon affecting swarming and capsular polysaccharide regulation. J Bacteriol. 2002;184:5946–5954. doi: 10.1128/JB.184.21.5946-5954.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broberg CA, Zhang L, Gonzalez H, Laskowski-Arce MA, Orth K. A Vibrio effector protein is an inositol phosphatase and disrupts host cell membrane integrity. Science. 2010;329:1660–1662. doi: 10.1126/science.1192850. [DOI] [PubMed] [Google Scholar]
- Broberg CA, Calder TJ, Orth K. Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes Infect. 2011;13:992–1001. doi: 10.1016/j.micinf.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette DL, Yarbrough ML, Orvedahl A, Gilpin CJ, Orth K. Vibrio parahaemolyticus orchestrates a multifaceted host cell infection by induction of autophagy, cell rounding, and then cell lysis. P Natl Acad Sci USA. 2008;105:12497–12502. doi: 10.1073/pnas.0802773105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette DL, Seemann J, Orth K. Vibrio VopQ induces PI3-kinase-independent autophagy and antagonizes phagocytosis. Mol Microbiol. 2009;73:639–649. doi: 10.1111/j.1365-2958.2009.06798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira RB, Chodur DM, Antunes LC, Trimble MJ, McCarter LL. Output targets and transcriptional regulation by a cyclic dimeric GMP-responsive circuit in the Vibrio parahaemolyticus Scr network. J Bacteriol. 2012;194:914–924. doi: 10.1128/JB.05807-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gode-Potratz CJ, McCarter LL. Quorum sensing and silencing in Vibrio parahaemolyticus. J Bacteriol. 2011;193:4224–4237. doi: 10.1128/JB.00432-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gode-Potratz CJ, Chodur DM, McCarter LL. Calcium and iron regulate swarming and type III secretion in Vibrio parahaemolyticus. J Bacteriol. 2010;192:6025–6038. doi: 10.1128/JB.00654-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gode-Potratz CJ, Kustusch RJ, Breheny PJ, Weiss DS, McCarter LL. Surface sensing in Vibrio parahaemolyticus triggers a programme of gene expression that promotes colonization and virulence. Mol Microbiol. 2011;79:240–263. doi: 10.1111/j.1365-2958.2010.07445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh K, Kodama T, Hiyoshi H, et al. Bile acid-induced virulence gene expression of Vibrio parahaemolyticus reveals a novel therapeutic potential for bile acid sequestrants. PLoS ONE. 2010;5:e13365. doi: 10.1371/journal.pone.0013365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyoshi H, Kodama T, Iida T, Honda T. Contribution of Vibrio parahaemolyticus virulence factors to cytotoxicity, enterotoxicity, and lethality in mice. Infect Immun. 2010;78:1772–1780. doi: 10.1128/IAI.01051-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, McCarter LL. ScrG, a GGDEF-EAL protein, participates in regulating swarming and sticking in Vibrio parahaemolyticus. J Bacteriol. 2007;189:4094–4107. doi: 10.1128/JB.01510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama T, Yamazaki C, Park KS, Akeda Y, Iida T, Honda T. Transcription of Vibrio parahaemolyticus T3SS1 genes is regulated by a dual regulation system consisting of the ExsACDE regulatory cascade and H-NS. FEMS Microbiol Lett. 2010a;311:10–17. doi: 10.1111/j.1574-6968.2010.02066.x. [DOI] [PubMed] [Google Scholar]
- Kodama T, Gotoh K, Hiyoshi H, et al. Two regulators of Vibrio parahaemolyticus play important roles in enterotoxicity by controlling the expression of genes in the Vp-PAI region. PLoS ONE. 2010b;5:e8678. doi: 10.1371/journal.pone.0008678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liverman AD, Cheng HC, Trosky JE, et al. Arp2/3-independent assembly of actin by Vibrio type III effector VopL. P Natl Acad Sci USA. 2007;104:17117–17122. doi: 10.1073/pnas.0703196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino K, Oshima K, Kurokawa K, et al. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet. 2003;361:743–749. doi: 10.1016/S0140-6736(03)12659-1. [DOI] [PubMed] [Google Scholar]
- Nair GB, Ramamurthy T, Bhattacharya SK, Dutta B, Takeda Y, Sack DA. Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin Microbiol Rev. 2007;20:39–48. doi: 10.1128/CMR.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nydam SD, Shah DH, Call DR. Transcriptome analysis of Vibrio parahaemolyticus in type III secretion system 1 inducing conditions. Front Cell Infect Microbiol. 2014;4:1. doi: 10.3389/fcimb.2014.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada R, Zhou X, Hiyoshi H, et al. The Vibrio parahaemolyticus effector VopC mediates Cdc42-dependent invasion of cultured cells but is not required for pathogenicity in an animal model of infection. Cell Microbiol. 2013;16:938–947. doi: 10.1111/cmi.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole GA. Microtiter dish biofilm formation assay. J Vis Exp. 2011;47:2437. doi: 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KS, Ono T, Rokuda M, Jang MH, Iida T, Honda T. Cytotoxicity and enterotoxicity of the thermostable direct hemolysin-deletion mutants of Vibrio parahaemolyticus. Microbiol Immunol. 2004a;48:313–318. doi: 10.1111/j.1348-0421.2004.tb03512.x. [DOI] [PubMed] [Google Scholar]
- Park KS, Ono T, Rokuda M, Jang MH, Okada K, Iida T, Honda T. Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect Immun. 2004b;72:6659–6665. doi: 10.1128/IAI.72.11.6659-6665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineyro P, Zhou X, Orfe LH, Friel PJ, Lahmers K, Call DR. Development of two animal models to study the function of Vibrio parahaemolyticus type III secretion systems. Infect Immun. 2010;78:4551–4559. doi: 10.1128/IAI.00461-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie JM, Rui H, Zhou X, et al. Inflammation and disintegration of intestinal villi in an experimental model for Vibrio parahaemolyticus-induced diarrhea. PLoS Pathog. 2012;8:e1002593. doi: 10.1371/journal.ppat.1002593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon D, Guo Y, Kinch LN, Grishin NV, Gardner KH, Orth K. Effectors of animal and plant pathogens use a common domain to bind host phosphoinositides. Nat Commun. 2013;4:2973. doi: 10.1038/ncomms3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon D, Klimko JA, Orth K. H-NS regulates the Vibrio parahaemolyticus type VI secretion system 1. Microbiology. 2014;160:1867–1873. doi: 10.1099/mic.0.080028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart BJ, McCarter LL. Lateral flagellar gene system of Vibrio parahaemolyticus. J Bacteriol. 2003;185:4508–4518. doi: 10.1128/JB.185.15.4508-4518.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L, Nunan L, Redman RM, Mohney LL, Pantoja CR, Fitzsimmons K, Lightner DV. Determination of the infectious nature of the agent of acute hepatopancreatic necrosis syndrome affecting penaeid shrimp. Dis Aquat Organ. 2013;105:45–55. doi: 10.3354/dao02621. [DOI] [PubMed] [Google Scholar]
- Yarbrough ML, Li Y, Kinch LN, Grishin NV, Ball HL, Orth K. AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science. 2009;323:269–272. doi: 10.1126/science.1166382. [DOI] [PubMed] [Google Scholar]
- Zhang L, Krachler AM, Broberg CA, Li Y, Mirzaei H, Gilpin CJ, Orth K. Type III effector VopC mediates invasion for Vibrio species. Cell Rep. 2012;1:453–460. doi: 10.1016/j.celrep.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Shah DH, Konkel ME, Call DR. Type III secretion system 1 genes in Vibrio parahaemolyticus are positively regulated by ExsA and negatively regulated by ExsD. Mol Microbiol. 2008;69:747–764. doi: 10.1111/j.1365-2958.2008.06326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Gewurz BE, Ritchie JM, et al. A Vibrio parahaemolyticus T3SS effector mediates pathogenesis by independently enabling intestinal colonization and inhibiting TAK1 activation. Cell Rep. 2013;3:1690–1702. doi: 10.1016/j.celrep.2013.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]